Significance
Saccharomyces cerevisiae undergoes a programmed DNA rearrangement to switch between mating types a and alpha. The origins of this complex and multifaceted process, which requires three copies of the mating-type (MAT) locus (with two silenced), have remained unknown. In this study we present a mechanism for mating-type switching in methylotrophic yeasts that shares a common origin with the well-characterized system in S. cerevisiae but has simpler components. This system requires only two copies of the MAT locus, with one copy transcriptionally repressed by proximity to centromeric or telomeric chromatin. Switching between the mating types occurs by recombination between inverted-repeat sequences flanking the MAT loci. This system suggests an ancestral mechanism for mating-type switching in yeasts.
Keywords: Hansenula polymorpha, Pichia pastoris, yeast genetics, comparative genomics
Abstract
Saccharomyces cerevisiae has a complex system for switching the mating type of haploid cells, requiring the genome to have three mating-type (MAT)–like loci and a mechanism for silencing two of them. How this system originated is unknown, because the three-locus system is present throughout the family Saccharomycetaceae, whereas species in the sister Candida clade have only one locus and do not switch. Here we show that yeasts in a third clade, the methylotrophs, have a simpler two-locus switching system based on reversible inversion of a section of chromosome with MATa genes at one end and MATalpha genes at the other end. In Hansenula polymorpha the 19-kb invertible region lies beside a centromere so that, depending on the orientation, either MATa or MATalpha is silenced by centromeric chromatin. In Pichia pastoris, the orientation of a 138-kb invertible region puts either MATa or MATalpha beside a telomere and represses transcription of MATa2 or MATalpha2. Both species are homothallic, and inversion of their MAT regions can be induced by crossing two strains of the same mating type. The three-locus system of S. cerevisiae, which uses a nonconservative mechanism to replace DNA at MAT, likely evolved from a conservative two-locus system that swapped genes between expression and nonexpression sites by inversion. The increasing complexity of the switching apparatus, with three loci, donor bias, and cell lineage tracking, can be explained by continuous selection to increase sporulation ability in young colonies. Our results provide an evolutionary context for the diversity of switching and silencing mechanisms.
Mating-type switching in yeasts is a highly regulated process that converts a haploid cell of one mating type into a haploid of the opposite type (1–5). Switching involves the complete deactivation of one set of regulatory genes and activation of an alternative set, but, unlike most regulatory changes, the switch is achieved by physically replacing the DNA at an expression site that is shared by both types of cell. In Saccharomyces cerevisiae the switching system uses a menagerie of molecular components (3) including three mating-type (MAT)–like loci (the expressed MAT locus and the silent loci HML and HMR); an endonuclease (HO) that creates a double-strand break at the MAT locus, which then is repaired using HMLalpha or HMRa as a donor; a mechanism (Sir1 and Sir2/3/4 proteins) for repressing transcription and HO cleavage at the silent loci; two triplicated sequences (the Z and X regions) that guide repair of the dsDNA break; a donor-bias mechanism (the recombination enhancer, RE) to ensure that switching happens in the correct direction; and a cell lineage-tracking mechanism (Ash1 mRNA localization) to ensure that switching occurs only in particular cells. Most of these components have no function other than facilitating switching.
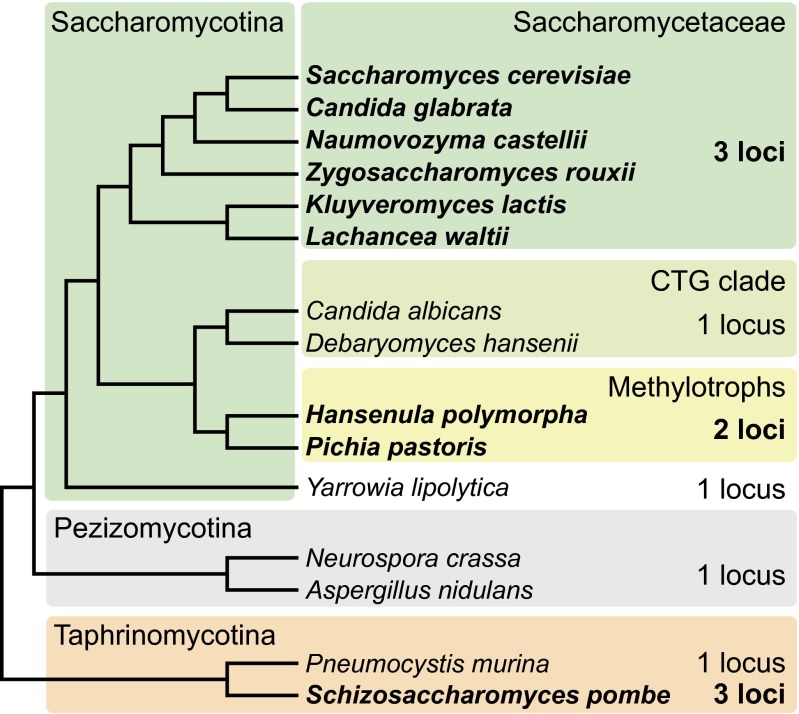
Given its complexity, it is surprising that switching seems to have evolved at least twice (5–10). Only unicellular fungi (yeasts) switch mating type, and these fungi have evolved from multicellular filamentous fungal ancestors on at least five separate lineages (11). Switching has arisen in two of these lineages represented by S. cerevisiae (subphylum Saccharomycotina) and Schizosaccharomyces pombe (Taphrinomycotina). A large clade of nonswitching filamentous ascomycetes, the Pezizomycotina, is related more closely to S. cerevisiae than to S. pombe (Fig. 1). Despite their apparently independent origins, the switching systems of S. cerevisiae (3) and S. pombe (4, 5) have many parallels, including the presence of three MAT-like loci (two of which are silenced) with triplicated guide sequences, a mechanism for specifically inducing a DNA break at MAT, donor bias (27), and cell-lineage tracking. However, the details of switching and silencing are quite different in the two species (3, 4).
Fig. 1.
Cladogram showing numbers of MAT-like loci. Species named in bold have been shown experimentally to switch mating types (3, 4, 12–18). Saccharomycotina, Pezizomycotina, and Taphrinomycotina are subphyla within phylum Ascomycota (19). Saccharomycetaceae, the CTG clade, and the methylotrophs are clades within the Saccharomycotina. The tree topology is based on refs. 20–26.
Switching is an error-prone process (28, 29), so it must confer a benefit to yeast species or it would not have been maintained by natural selection. However, the nature of this evolutionary benefit is debated (2, 6, 8, 30). We and others have proposed that it is related to the control of spore germination in uncertain environments (29, 30). The goal of switching is not to restore diploidy, because some species that switch grow primarily as haploids, but rather is to maximize the ability of a young colony to make new spores if nutrient levels fall. Yeasts are dispersed to new habitats when they are eaten and excreted by insects (31, 32), and ascospores are structures that assist yeast cells to survive passage through the insect digestive tract (33). Although many tetrads (sets of four haploid spores formed by meiosis of a diploid) remain intact, digestion by the insect removes the ascus wall and causes some spores to become isolated (33). If an isolated spore germinates, it has no way of making new spores unless it can find a mating partner of the opposite mating type [the “lonely spore scenario” (2, 7)]. Switching provides a partner, allowing the cells to become diploid and able to make new spores. In S. cerevisiae, new spores can be made just two cell divisions after spore germination (1). Switching enables isolated spores to germinate in very poor environments, replicate for a few cell cycles as permitted by the environment, and then resporulate (29). In contrast, species that cannot switch would need to be more cautious about germinating. Over time, species that can switch are predicted to have a growth advantage over species that cannot switch, because they can risk germinating in poorer environments and so germinate earlier in improving environments. The formation of spores by asexual means does not appear to be an option for most ascomycete yeasts because ascosporulation is linked to meiosis, unlike molds, which can make asexual spores in conidia (19).
In the family Saccharomycetaceae, switching has been reported experimentally in five other genera as well as in Saccharomyces (Fig. 1). Genome sequences show that almost all species in this family have three MAT-like loci and are likely to switch mating types by a mechanism similar to that of S. cerevisiae (8, 29, 34, 35), although some species use an alternative mechanism instead of HO endonuclease to cut the MAT locus (15). Family Saccharomycetaceae is one clade within the subphylum Saccharomycotina, which also includes a large clade of Candida and related species (called the “CTG clade” because of its unusual genetic code). Species in the CTG clade have only one MAT-like locus and do not switch (36, 37). More recently, genomes have been sequenced from a third clade within the Saccharomycotina which includes the methanol-assimilating species Hansenula polymorpha (20) and Pichia pastoris (21, 38) and relatives such as Dekkera bruxellensis (22, 23) and Kuraishia capsulata (24). For convenience we refer to this clade as the “methylotrophs.” Phylogenomic analyses consistently have shown that the methylotrophs are monophyletic and are a sister of the CTG clade, with the Saccharomycetaceae outside the grouping (Fig. 1). H. polymorpha and P. pastoris are regarded as homothallic both on classical grounds [observation of sporulation in colonies grown from single spores or observation of mating between a mother cell and a bud (19)] and genetic grounds [any strain can mate with any other strain (17, 18, 39–41)]. Genetic evidence for mating-type switching has been reported in a few methylotrophs, including H. polymorpha (17, 40) and P. pastoris (18), but the molecular details have not been investigated.
Results
An Inversion Polymorphism Correlates with MAT Gene Expression in H. polymorpha.
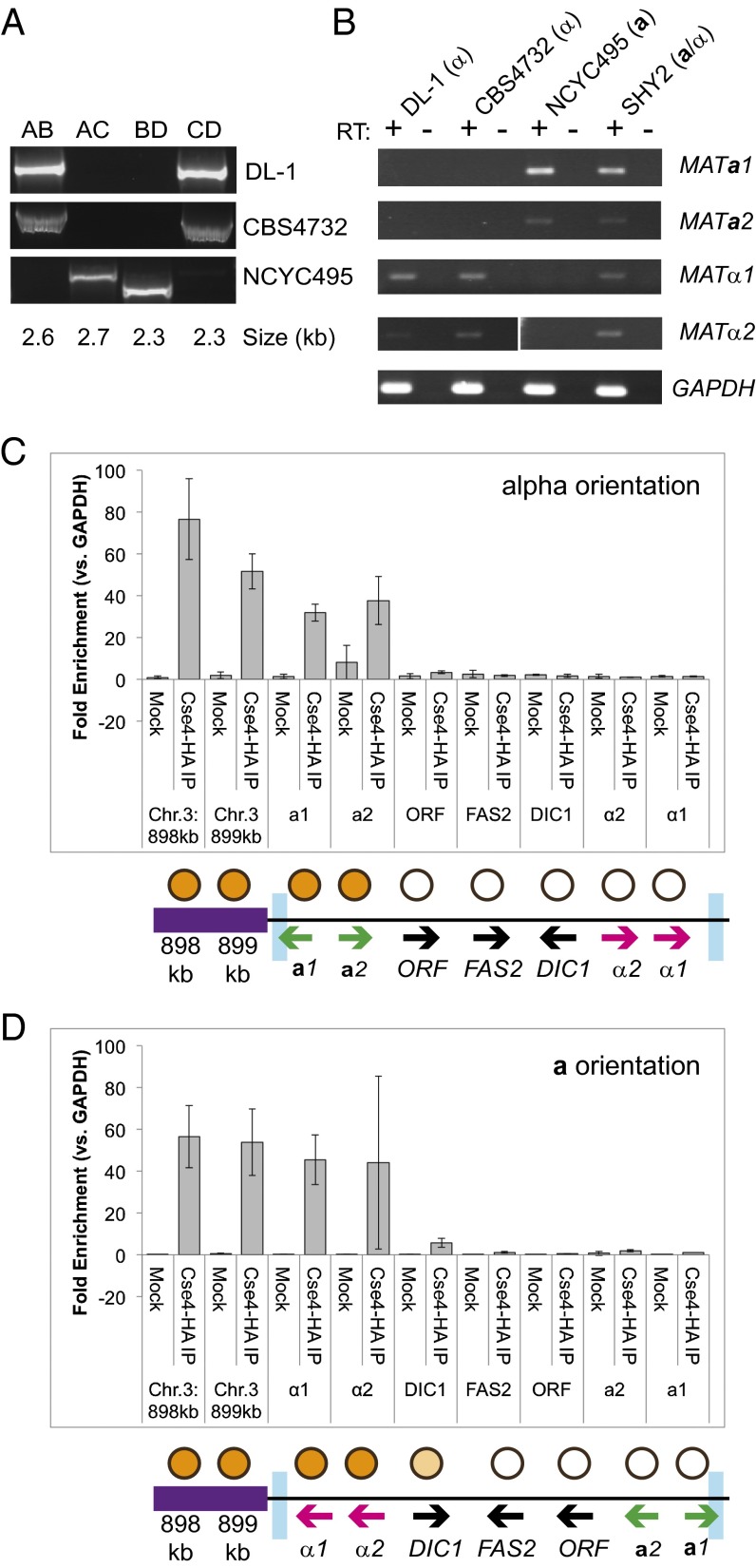
H. polymorpha grows primarily as a haploid and is homothallic (17, 39, 40). The genomes of two strains, NCYC495 and DL-1, have been sequenced. In a taxonomic revision (42) these strains were reclassified as different species, Ogataea polymorpha (NCYC495) and Ogataea parapolymorpha (DL-1); the genomes are about 10% divergent in sequence. We found that they are completely collinear in gene order (no inversions or translocations) throughout the genome except for one 19-kb inversion. The region inverted between the strains has homologs of MATa1 and MATa2 genes at one end and homologs of MATalpha1 and MATalpha2 at the other end, separated by seven other genes whose known functions are unrelated to mating (Fig. 2). These are the only MAT-like loci in the genome. The inverted region is located between two copies of a 2-kb inverted repeat (IR) sequence, derived from the 3′ end of the gene SLA2 and part of MATa1. PCR amplification across the possible inversion endpoints in genomic DNAs from the two strains confirmed that they differ in orientation and that the orientation of a third strain, CBS4732, is the same as that of DL-1 (Fig. 3A). Transcription of the MATalpha or MATa genes depends on the orientation of the 19-kb region (Fig. 3B). In NCYC495, MATa1 and a2 are expressed, and MATalpha1 and alpha2 are repressed, so we designated this orientation as the “a orientation.” The opposite occurs in strains DL-1 and CBS4732, which have the alpha orientation. All four MAT genes are expressed in diploids. The seven other genes in the 19-kb inverted region show expression in both orientations (SI Appendix, Fig. S1).
Fig. 2.
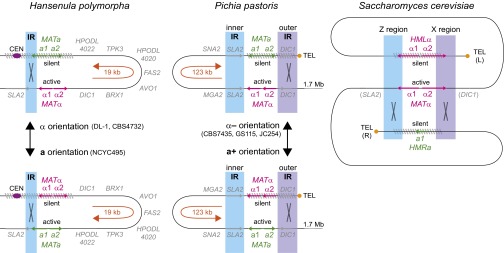
MAT locus organization and mating-type switching in H. polymorpha, P. pastoris, and S. cerevisiae. Blue and purple shading show the IRs. Orange arrows show the orientation of the invertible regions. Two other arrangements of the P. pastoris region (alpha+ and a−), resulting from exchanges in the inner IR that do not change the mating type, are not shown (Fig. 5). In the S. cerevisiae diagram SLA2 and DIC1 indicate the genes that flanked the MAT locus in the ancestor of family Saccharomycetaceae, although they no longer are at this position in S. cerevisiae because of chromosome erosion (29).
Fig. 3.
Structure and expression of the MAT locus in H. polymorpha strains. (A) Orientation of the 19-kb region differs among H. polymorpha haploid strains. PCR products were amplified with primer pairs flanking IRs as shown schematically in Fig. 4A. Products from the AB and CD primer pairs indicate the alpha orientation, and products from the AC and BD pairs indicate the a orientation. (B) Expression of H. polymorpha MAT genes depends on orientation of the 19-kb region. Amplified cDNA was generated from haploid strains in both orientations and from a diploid strain (SHY2, produced by crossing two NCYC495-derived haploid strains with opposite orientations). “+ RT” indicates the addition of reverse transcriptase to cDNA synthesis reaction; “− RT” indicates no reverse transcriptase was added. (C and D) ChIP indicates the presence of Cse4 nucleosomes at the silenced MAT loci in haploids. The graphs present the fold enrichment of the target sequence relative to a control sequence (GAPDH) outside the 19-kb invertible region. The schematics below represent the relative positions of loci examined by ChIP. Positions highlighted in purple indicate putative centromeric regions. Blue boxes represent the IRs. Orange circles indicate the binding of tagged Cse4 to the tested regions. The ORF is the gene HPODL4022. C shows strain SH4330 (alpha orientation), and D shows strain NCYC495 (a orientation).
Gene Expression at the Silent Locus in H. polymorpha Is Repressed by a Centromere.
The region immediately to the left of the silenced MAT genes and the leftmost copy of the 2-kb IR in H. polymorpha (as drawn in Fig. 2) has several characteristics that indicate it could be a regional centromere (20, 43). It coincides with a trough of G+C content (SI Appendix, Fig. S2), and in both NCYC495 and DL-1 the region contains clusters of retrotransposons and LTRs from the Ty5 family, which tends to target centromeres (43). (The sequences and locations of the retroelements differ in the two strains).
We confirmed that the region beside the MAT is a centromere by two methods. First, we overexpressed a tagged version of the centromeric histone Cse4 (CenH3) and used ChIP to measure binding to the region in two haploid strains of opposite orientations derived from an NCYC495 background. The Cse4 signal is high in the putative centromeric region to the left of the IR in both strains. It extends onto the MATa1 and a2 genes only in the alpha-orientation strain (Fig. 3C) and onto the MATalpha1 and alpha2 genes only in the a-orientation strain (Fig. 3D). Second, we reanalyzed Hi-C (chromatin conformation capture) data from a synthetic metagenomics experiment that included H. polymorpha NCYC495 (44) to extract signals resulting from the 3D clustering of its centromeres in the nucleus (45). This analysis revealed centromeric signals from each H. polymorpha chromosome (SI Appendix, Fig. S2) and placed the peak signal from chromosome 3 at position 897 kb, which is 3 kb from the leftmost IR and 5 kb from the MATalpha genes. The centromeric region is transcriptionally silent (SI Appendix, Fig. S2).
Homothallic Mating in H. polymorpha Occurs by Inverting the MAT Region.
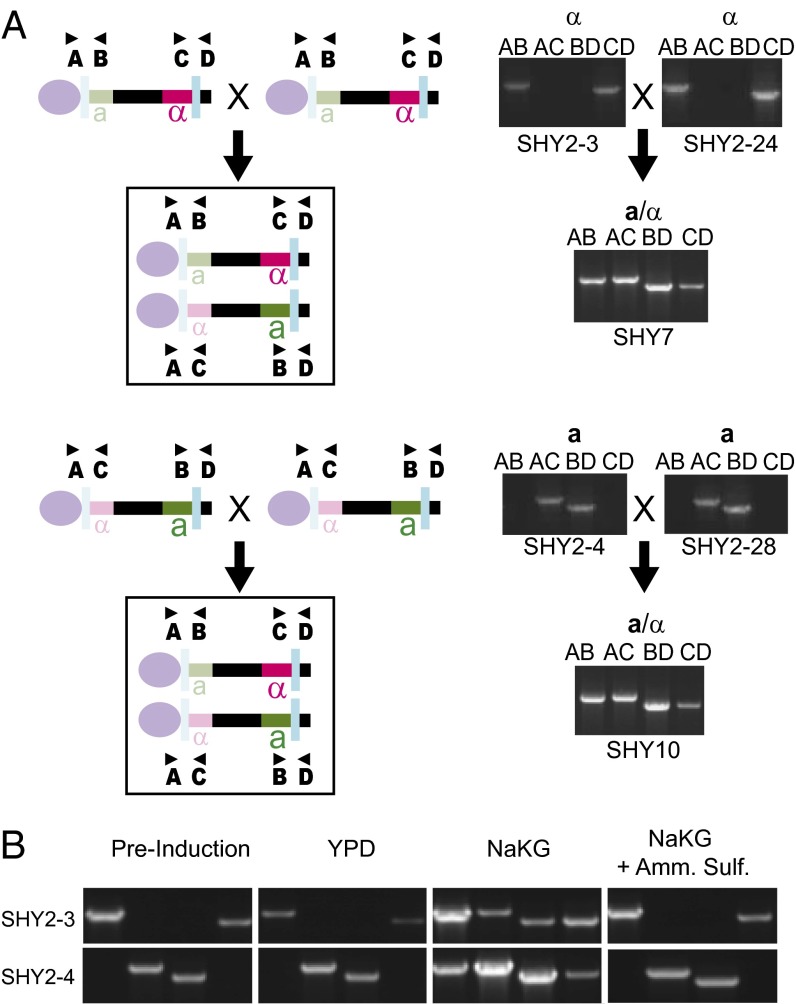
H. polymorpha is homothallic, so any strain can mate with any other strain. If the orientation of the MAT region determines mating type in this species, then mating between two strains with the same orientation should be possible only if one of them first switches mating type by inverting the region. We crossed pairs of NCYC495-derived strains (O. polymorpha) in the alpha orientation or the a orientation by growing them together in mating (nitrogen starvation) medium and using genetic markers to select for diploids (Fig. 4A). In each case, the diploids contained chromosomes with both orientations. This result indicates that one parental chromosome underwent inversion before, during, or after mating. Growth of haploid strains with the alpha orientation or a orientation in liquid minimal medium without nitrogen resulted in the appearance of both MAT region orientations within 12 h (Fig. 4B). Because the short timeframe (H. polymorpha typically requires >24 h for mating) and agitation of the cultures on a shaker preventing prolonged contact between cells made mating events in these cultures unlikely, these data suggest that inversion of the MAT region occurs before the formation of diploids. The appearance of both orientations was suppressed by the addition of ammonium sulfate (Fig. 4B), indicating that inversion of the MAT region is induced by the same nitrogen-starvation conditions that induce mating. We suggest that inversion in a haploid cell leads to a change of expressed mating type, leading to mating. PCR analysis of 153 random spores formed by sporulation of a diploid strain that was made by crossing two alpha-orientation haploids showed that 84 (55%) were in the a orientation and 69 (45%) were in the alpha orientation, a frequency that is not significantly different from 50% (P = 0.26 by two-tailed binomial test).
Fig. 4.
Mating-type switching by inversion in H. polymorpha. (A) Crossing two strains in the same orientation results in diploids that are heterozygous for orientation. Haploid MATalpha (SHY2-3 and SHY2-24) and MATa (SHY2-4 and SHY2-28) strains with complementary auxotrophic mutations were obtained by random spore analysis of a sporulated diploid (SHY2) and were mated to produce diploids (SHY7 from alpha × alpha and SHY10 from a × a). (Left) Schematics indicate the MAT locus organization in the strains used in each cross and the resulting diploids. Purple ovals represent the centromere. The MAT genes closer to the centromere are silenced (pale colors). (Right) Gel images show amplification across the IR regions with the primer combinations indicated. (B) MAT locus inversion is induced by nitrogen limitation. Gel images show MAT locus organization for haploid MATalpha (SHY2-3) and MATa (SHY2-4) strains after growth for 12 h in YPD, minimal medium without nitrogen (NaKG), and NaKG supplemented with 40 mM ammonium sulfate as a nitrogen source.
P. pastoris also Has Two MAT-Like Loci at the Ends of an Invertible Region.
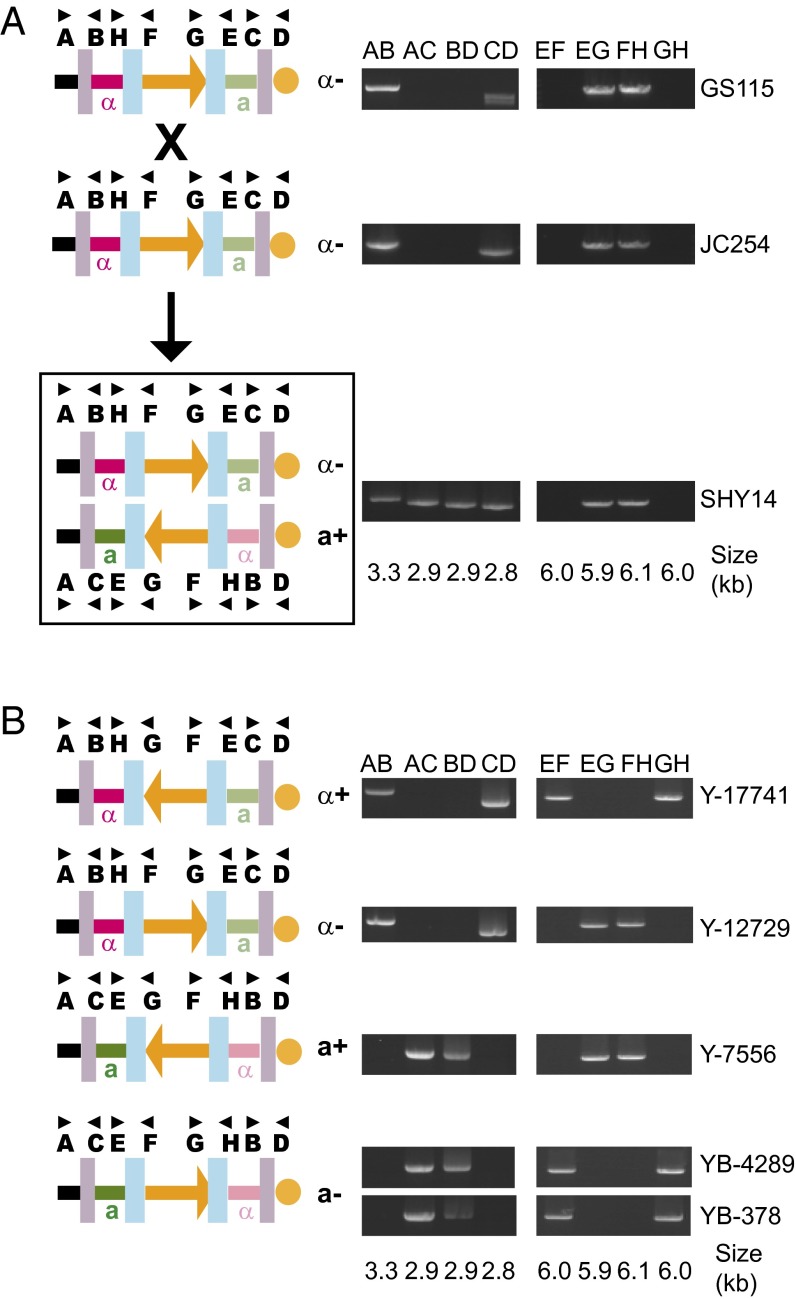
P. pastoris is a related homothallic methylotroph that is predominantly haploid (18, 41). The taxonomy of P. pastoris strains also has been revised recently, but all the strains we discuss here are from a single species that is properly called “Komagataella phaffii” (46). The genome sequence of strain CBS7435 (38) contains two MAT-like loci. One locus is close to the telomere of chromosome 4 and contains MATa1 and a2, whereas the other locus is 138 kb away and contains MATalpha1 and alpha2. These loci are flanked on one side by a 3-kb IR and on the other side by a 6-kb IR, which we refer to as the “outer” and “inner” IRs, respectively (Fig. 2). The reported genome sequence of P. pastoris strain GS115 (21), which was derived from strain CBS7435 by mutagenesis, terminates in the telomere-proximal copy of the inner IR and so does not include the second MAT-like locus, but we have confirmed by PCR that it is present in GS115 and is similar to the structure in CBS7435.
The organization of P. pastoris chromosome 4 suggested that the MAT-like locus close to the telomere might be silenced by telomere position effect (TPE) (47), making the telomere-distal MAT-like locus the active one, and that haploids could switch mating type by recombining the outer IRs to invert the whole 138-kb region. Indeed, haploid strains of P. pastoris differ as to whether the telomere-distal locus is MATalpha (designated as the alpha orientation) or MATa (designated as the a orientation). As in H. polymorpha, crossing two strains in the same orientation results in diploids that are heterozygous for orientation (Fig. 5A). Surprisingly, RT-PCR analysis indicates that haploid strains of both orientations transcribe all four MAT genes, as do diploids (SI Appendix, Fig. S3), and public RNA sequencing data from strain GS115 also show expression of all four genes (48). However, by quantitative RT-PCR (qRT-PCR) we find that haploid expression of MATalpha2 and MATa2 differs in the two orientations, with an alpha-orientation strain having 15-fold higher expression of MATalpha2 and fourfold lower expression of MATa2 than an a-orientation strain (Table 1 and SI Appendix, Table S1). For genes a2, alpha1, and alpha2, a diploid strain has a level of expression that is intermediate between the two haploids. MATa1 is highly expressed in both haploid orientations, at a level only slightly lower than the control gene ACT1 in our experiments, and is three- to fourfold lower in diploids. Southern blot analysis shows that a haploid P. pastoris culture grown in rich medium does not contain a mixture of chromosomal orientations, so the expression of both sets of MAT genes probably is caused by leaky silencing rather than by chromosomal rearrangement (SI Appendix, Fig. S4).
Fig. 5.
Mating-type switching by inversion in P. pastoris. (A) Crossing two strains in the same orientation results in diploids that are heterozygous for orientation. (Left) The schematic indicates chromosome 4 organization in the haploid parents and their diploid progeny. The orange arrow shows the orientation of the 123-kb region between the inner IRs. The MAT locus closer to the telomere (orange circle) is repressed (pale colors). Purple and blue boxes represent the outer and inner IRs, respectively. (Right) Gel images show amplification across the IRs using primer combinations as indicated. (B) All four possible structures of chromosome 4 are found among natural isolates of P. pastoris. The isolates are from tree fluxes and were obtained from the US Department of Agriculture Agricultural Research Service collection as Komagataella phaffii (SI Appendix, Table S2).
Table 1.
Fold change in expression of P. pastoris MAT loci determined by qRT-PCR
| Gene | Ratio GS190(a)/GS115(α)* | Ratio GS115(α)/GS190(a)* | Ratio GS190(a)/SHY15(a/α)* | Ratio GS115(α)/SHY15(a/α)* |
| a1 | 0.75 (0.57–0.97) | — | 3.36 (2.57–4.41) | 4.50 (3.86–5.24) |
| a2 | 4.38 (3.41–5.62) | — | 2.83 (2.20–3.63) | 0.65 (0.47–0.89) |
| α1 | — | 1.82 (1.19–2.77) | 0.66 (0.43–1.00) | 1.20 (1.03–1.39) |
| α2 | — | 15.03 (13.18–17.15) | 0.11 (0.06–0.20) | 1.61 (1.41–1.84) |
n = 2-ΔΔCt.
Values in parentheses show ± SD; see SI Appendix, Table S1 for details of calculations.
Four Isomers of P. pastoris Chromosome 4.
Because inversions could occur by recombination between the inner IRs, the outer IRs, or both, there are four possible isomeric structures for P. pastoris chromosome 4. We name these structures “alpha+,” “alpha−,” “a+,” and “a−” (Fig. 5B). The prefixes “alpha” and “a” indicate the mating type (the MAT genotype at the telomere-distal position), and the suffixes “+” and “−” indicate the orientation of the unique 123-kb region between the inner IRs relative to the telomere. PCR amplification of genomic DNAs with orientation-specific primer pairs showed that homothallic mating causes orientation alpha− to switch to a+ (Fig. 5A). Switching therefore occurs by recombination in the outer IRs, which is equivalent to swapping the locations of the telomere and the remaining 1.7 Mb of the chromosome. Telomeric repeats (TGCTGGAn) begin immediately beside the outer IR with no DNA between them (SI Appendix, Fig. S5).
Inversion across the inner IRs would not change the mating type but would make the two copies of chromosome 4 in a diploid collinear except at the MAT loci themselves. We guessed that such inversions might occur during meiosis, because otherwise any meiotic recombination within the 123-kb unique region would produce inviable chromosomes (SI Appendix, Fig. S6), but we found no evidence by random spore analysis to support this idea. However, all four possible chromosome structures are found among natural isolates of P. pastoris (Fig. 5B), indicating that inversion across the inner IR must occur at some rate.
Discussion
Our results show that homothallic methylotrophic yeast species switch mating types using a flip-flop inversion mechanism. In the late 1970s flip-flop inversion was discussed as a possible model for mating-type switching in several yeast species (49–54) and was found to be the mechanism of flagellar-phase variation in Salmonella (55). In a classic study in 1977, Hicks and Herskowitz (51) rejected the two-locus flip-flop model for S. cerevisiae in favor of the three-locus cassette model, which was proven soon after (56). In 1981, Tolstorukov and colleagues showed that the cassette model did not explain the pattern of switching in the methylotroph Pichia methanolica and proposed instead that this organism has a flip-flop system (52, 53). Our results now validate this proposal for methylotrophs. It remains to be determined whether the mechanism of inducible inversion involves a site-specific recombinase (such as Hin in phase variation or Flp in the yeast 2-μm plasmid) or uses the general recombination machinery. The methylotroph genomes do not have homologs of HO endonuclease.
Their conserved syntenic location shows clearly that the MAT loci of methylotrophs are orthologous to those of other yeasts in subphylum Saccharomycotina (Fig. 2). The gene SLA2, which is present in the H. polymorpha IR and the P. pastoris inner IR, also forms the Z region in many Saccharomycetaceae family yeasts (29) and is located beside the single MAT locus of many Pezizomycotina species (6). On the other side DIC1, which is located in the outer IR of P. pastoris and beside MATalpha in the 19-kb invertible region of H. polymorpha, was ancestrally beside the MAT locus in family Saccharomycetaceae and forms the X region in some species (29). Therefore, the MAT regions of methylotrophs and Saccharomycetaceae share a common ancestor.
Did the common ancestor of methylotrophs and Saccharomycetaceae switch mating types, and if so did it use a two-locus or a three-locus system? Parsimony suggests that it did switch, because a single loss (in the CTG clade) is more likely than two independent gains in Saccharomycetaceae and methylotrophs. We propose that the ancestor had a simple two-locus system with one IR, similar to H. polymorpha. Such a system can be formed spontaneously by DNA introgression between the MAT idiomorphs in a heterothallic fungus (SI Appendix, Fig. S7), as appears to have occurred very recently in Sclerotinia sclerotiorum (Pezizomycotina), which inverts part of its MAT locus in every meiotic generation (57).
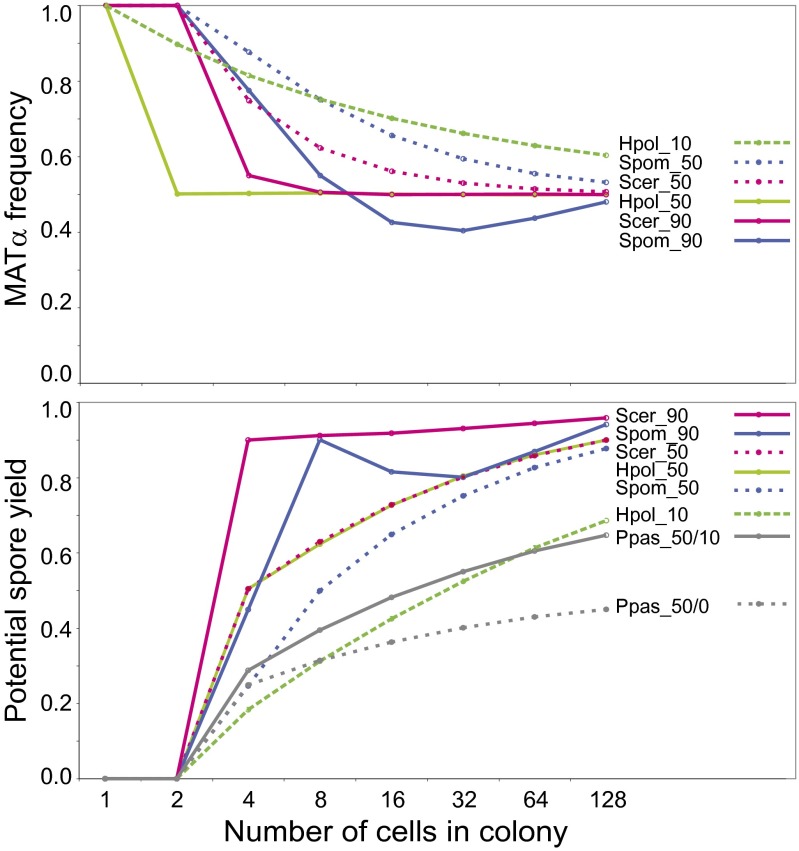
Further evidence for a two-locus ancestor comes from computer simulations (Fig. 6 and SI Appendix), which show that a simple two-locus system can evolve into a fully featured three-locus system as the result of continuous natural selection for a single trait—the ability of tiny colonies (4–16 cells) to produce the highest possible number of spores if nutrients run out. Selection for this trait would be expected if it was the reason that switching originated (29). Our simulations measured allele frequency and the potential yield of spores in colonies expanding from 1 to 128 haploid cells under different switching systems (Fig. 6). Taking the eight-cell stage as an example, potential spore yield from the colony would be maximized if it had four alpha cells and four a cells and so could make four diploids and hence 16 spores. However, no switching system can achieve a 4:4 ratio consistently in every eight-cell colony. In an H. polymorpha-like system, if inversion of the MAT region occurred in 10% of cells, simulations show that on average only five spores would be produced per colony instead of 16, so the potential spore yield is only 31% of the theoretical maximum (Hpol_10 in Fig. 6). As shown in Fig. 6, the maximum spore yield from eight-cell colonies switching by the H. polymorpha system is 62% (10 spores), which occurs when the MAT region inverts in 50% of cells (Hpol_50). In comparison, the highly efficient systems of S. cerevisiae (Scer_90) and S. pombe (Spom_90) achieve about 90% of theoretical maximum spore yield (>14 spores) at the eight-cell stage.
Fig. 6.
Simulations of changes in allele frequency and potential spore yield under different switching systems during growth from 1–128 haploid cells in a colony started by a germinating MATalpha spore. (Upper) S. cerevisiae (Scer) and S. pombe (Spom) systems were simulated at switching productivities of 90% (strong donor bias) and 50% (no bias), and H. polymorpha (Hpol) systems were simulated at productivities of 50% (all cells switch randomly) and 10% (fewer cells switch randomly). The productivity of switching is the proportion of cells that successfully change mating type when permitted to do so. Allele frequency curves for P. pastoris (not plotted) are identical to those for H. polymorpha. (Lower) The gray lines show the effect of allowing inversion in the inner IR of P. pastoris at a frequency of 10% (Ppas_50/10) compared with no inversion (Ppas_50/0), when the productivity of switching is 50%. Graphs are averages from 10,000 replicate colonies. Details of simulations are given in SI Appendix.
The simulations show that continued selection for increased spore yield can lead to a two-locus system becoming more complex. If the invertible region between the IRs in a two-locus system is large, meiotic recombination within this region becomes more probable and will decrease potential spore yield (SI Appendix, Fig. S6), but this decrease can be overcome by adding a second IR (compare the yield in Ppas_50/10 with that of Ppas_50/0 in Fig. 6, which shows the effect of allowing inversion at the inner IR in P. pastoris). Duplicating one set of MAT genes will convert a two-locus system into a three-locus system and form Z and X regions from the two previous IRs (Fig. 2). This step will either increase potential spore yield or leave it unchanged if the two-locus system already was at the maximum productivity of switching (the yields from Scer_50 and Hpol_50 are identical and higher than Hpol_10; Fig. 6). Once three loci are in place, the system can change from a conservative single Holliday junction mechanism to a nonconservative synthesis-dependent strand-annealing mechanism (3), which introduces the potential for biased choice of donor and increased efficiency (SI Appendix). When donor bias is possible, cell-lineage tracking becomes advantageous, because preventing switching in some cells while activating it in others can maximize potential spore yield in the important early generations of the colony (in Fig. 6, compare spore yields at the eight-cell stage in Scer_90 vs. Scer_50 and in Spom_90 vs. Spom_50, i.e., strong donor bias vs. unbiased switching in these species). Thus, an S. cerevisiae-like system with three MAT-like loci, Z and X guide sequences, HO, RE, and Ash1 can evolve step by step, starting from a simple H. polymorpha-like system, by continuous selection for one trait: increased spore yield from young colonies. Although haploidy and diploidy may each be beneficial in particular growth environments (58), and the ability to transition between them may be advantageous, selection for efficient spore production alone is sufficient to explain the origin and subsequent elaboration of the mating-type switching system.
What silencing mechanism was used in the ancestral system? It seems likely that the primordial system used either centromeric or telomeric heterochromatin to silence the nonexpressed MAT genes. However, all Saccharomycotina species, including H. polymorpha and P. pastoris, have lost the proteins (Clr4 and Swi6/HP1) that make the histone H3K9me modification that is characteristic of silent heterochromatin in most eukaryotes, including Schizosaccharomyces and Pezizomycotina (59). Instead, H. polymorpha may use Cse4-containing nucleosomes to silence its centromere-proximal MAT genes, similar to the mechanism proposed for neocentromeres in Candida albicans (60). It is less clear how expression differences between the MAT genes in P. pastoris haploids are achieved. TPE is a good candidate, but we see differential expression of only two of the four MAT genes, a result that is unexpected under TPE. TPE is a stochastic process that silences expression of telomere-linked genes in a proportion of cells in a population (47), possibly explaining why we see diminution rather than complete silencing of expression. It is unclear also how differential expression of only MATalpha2 and MATa2 could determine cell type under the current paradigm for the function of these genes (36).
Our results push back the origin of mating-type switching in the lineage leading to S. cerevisiae to before the common ancestor of methylotrophs and Saccharomycetaceae and hence almost to the base of subphylum Saccharomycotina. Consequently, they imply that the ability to switch has been lost in the CTG clade which includes the pathogen C. albicans. Genomic data indicate that, despite its benefits, switching also has been lost several other times including in the methylotrophs Kuraishia capsulata and Dekkera bruxellensis (22–24), and in the Saccharomycetaceae Lachancea kluyveri and Kazachstania africana (29, 61). Null alleles of HO are frequent among natural populations of S. cerevisiae (62). Losses of switching may simply reflect life in environments that do not require frequent spore formation for survival or dispersal. Studying the mechanisms of switching and silencing in deeper lineages of Saccharomycotina predating and postdating the loss of H3K9me heterochromatin will help us learn more about the origins of these systems and the evolutionary pressures that created them.
Materials and Methods
Yeast Strains and Culture Conditions.
Strains of H. polymorpha and P. pastoris used in this study are listed in SI Appendix, Table S2. Cultures were grown under standard rich-medium conditions. The medium used for the induction of mating in H. polymorpha was 2% malt extract and in P. pastoris was minimal medium without nitrogen (NaKG: 0.5% sodium acetate, 1% potassium chloride, 1% glucose). Isolation of spores from sporulating diploid populations was performed by random spore analysis. After growth on mating/sporulation medium for 4 d, diploids were suspended in diethyl ether for ∼1 h before plating on rich medium. Individual colonies were screened for MAT locus orientation by colony PCR (see below). To induce inversion of the MAT locus, overnight cultures of H. polymorpha grown in yeast extract/peptone/dextrose (YPD) were diluted to OD600 ∼0.5 in YPD, NaKG, or NaKG with 40 mM ammonium sulfate and were grown in an orbital shaker for 12 h at 30 °C with agitation at ∼200 cycles per min.
DNA Extraction and MAT Locus PCR.
DNA was harvested from stationary-phase cultures by homogenization with glass beads followed by phenol-chloroform extraction and ethanol precipitation. Primers used for MAT locus amplification are listed in SI Appendix, Table S3. Amplification was done using GoTaq (Promega) or Phusion Taq (New England Biolabs) polymerase for 25–35 cycles with an annealing temperature of 55–60 °C. Colony PCR was performed by heating cells in 10 μL water at 95 °C for 5 min followed by addition of PCR reagents.
RNA Extraction and RT-PCR.
RNA was extracted from log-phase cultures by either hot acid phenol-chloroform extraction or homogenization with glass beads followed by phenol-chloroform extraction and ethanol precipitation. Following DNase I (Invitrogen) treatment, cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen). Primers used for amplification of MAT gene cDNA are listed in SI Appendix, Table S3. For nonquantitative RT-PCR, amplification was performed using GoTaq (Promega) polymerase for 30 cycles with an annealing temperature of 55 °C. qRT-PCR was performed on an Agilent Mx3005P system using Sensifast Lo-ROX SYBR Green mix (Bioline) according to the manufacturer’s instructions.
Cse4 ChIP.
A synthetic construct containing the H. polymorpha CSE4 gene tagged at an internal site with 3HA and expressed from the S. cerevisiae TEF1 promoter was designed (SI Appendix, Fig. S8) and made by Integrated DNA Technologies. The construct was inserted into a panARS vector (63) containing a KanMX marker by restriction digestion and ligation and was transformed into H. polymorpha by electroporation as previously described (64). Log-phase cells were crosslinked using formaldehyde (1% final concentration) for 20 min followed by the addition of glycine to stop the reaction. Cells were harvested and lysed using glass beads followed by chromatin fragmentation by sonication with a Bioruptor Standard (Diagenode). Immunoprecipitation of chromatin fragments was performed with EZview Red Anti-HA Affinity Gel (Sigma-Aldrich) or mouse IgG1 (Cell Signaling Technology) as isotype control. After washes, bound DNA was eluted with HA peptide (Sigma-Aldrich), crosslinks were reversed, and the samples were purified by phenol-chloroform extraction. qPCR was performed on an Agilent Mx3005P system using Sensifast Lo-Rox SYBR Green mix (Bioline) according to the manufacturer’s instructions. Primers used for amplification of centromeric and MAT region DNA sequences are listed in SI Appendix, Table S3.
Localization of Centromeres in H. polymorpha Hi-C Data.
Using Hi-C data from the synthetic metagenomics experiment of Burton et al. (44), which included H. polymorpha, we aligned sequence reads to the NCYC495 genome with the Burrows–Wheeler alignment tool (65), requiring perfect full-length matches that were within 500 bp of HindIII or NcoI sites. Centromeres were localized from the interchromosomal contact map using a modification of the method of Marie-Nelly et al. (45). We integrated the diffuse signals from regional centromeres by counting the total number of interchromosomal contacts of each 1-kb interval on each chromosome. Expressed sequence tag data for H. polymorpha from the Joint Genome Institute (National Center for Biotechnology Information accession no. SRR346565) was aligned to the genome using STAR (66) and was visualized in JBrowse (67).
Genome Data.
Detailed views of the H. polymorpha and P. pastoris MAT regions are shown in SI Appendix, Fig. S5. The H. polymorpha NCYC495 leu1.1 (O. polymorpha) genome sequence was downloaded from the Joint Genome Institute (genome.jgi.doe.gov/Hanpo2/Hanpo2.home.html) and is used with permission. The genome sequence and our stock of NCYC495 are in the a orientation. Its MAT locus is located on chromosome 3, between positions 901 kb (MATalpha genes) and 920 kb (MATa genes). The two copies of the IR are at bases 899,527 to 901,575 (2,048 bp) and 920,077 to 922,124 (2,049 bp) and are identical except for a 1-bp indel. The IR includes the 3′ end of SLA2 and most of MATa1 except for the start codon. The centromeric region to the left of the MAT locus has no annotated genes between 887 and 899 kb, except for a Tpa5 (Ty5 family) retrotransposon (68).
In the H. polymorpha DL-1 (O. parapolymorpha) genome sequence (20) the MAT locus is on chromosome 5 (GenBank accession number AEOI02000008.1), which is the homolog of NCYC495 chromosome 3. This whole chromosome has been reported on the opposite strand in DL-1 relative to NCYC495. The genome sequence and our stock of DL-1 are in the alpha orientation. The DL-1 MAT locus (20) is between positions 426 kb (MATalpha genes) and 446 kb (MATa genes). The IRs are at bases 424,078 to 426,143 and 444,674 to 446,739 (2,066 bp), are identical to each other, and have 94% identity to the NCYC495 IRs. The centromere is to the right of the MAT locus at 447–459 kb (20).
The structure of P. pastoris chromosome 4 as represented in the genome sequences of CBS7435 (38) and GS115 (21) is alpha+ in the nomenclature used here. However, our laboratory stocks of CBS7435 and GS115 are both alpha− by PCR, indicating inversion or sequence misassembly across the inner IRs. In the sequence of CBS7435 chromosome 4 (GenBank accession no. FR839631.1), the outer IRs are at bases 127 to 2,703 and 140,670 to 143,246 (2,577 bp) and are identical, and the inner IRs are at bases 4,765 to 10,509 and 133,274 to 139,018 (5,745 bp with three differences). The MAT genes do not overlap with the IRs. The outer IR contains the complete DIC1 gene and the 5′ half of a hypothetical gene (PAS_chr4_0876). The inner IRs contain the complete CWC25 and SUI1 genes, the complete SLA2 gene except for the stop codon, and a pseudogene of alcohol dehydrogenase. Bases 1 to 126 of FR839631 are telomere repeats. The sequence of chromosome 4 from P. pastoris strain GS115 (GenBank accession no. FN392322.1) is incomplete and lacks the telomere-proximal copies of the outer IR and MAT locus (MATa genes) and most of the inner IR. It begins at the equivalent of base 10,302 of the CBS7435 sequence. The whole sequence of chromosome 4 also is on the opposite strand in FN392322 relative to FR839631.
Supplementary Material
Acknowledgments
We thank G. Jalowicki for assistance; K. Lahtchev, S. Harashima, I. van der Klei, K. de Schutter, J. Cregg, and I. Tolstorukov for strains; L. Schoenfeld for advice on Cse4 tagging; G. Butler and C. P. Kurtzman for comments; and A. A. Sibirny for permission to use the H. polymorpha NCYC495 sequence data that were produced by the US Department of Energy Joint Genome Institute (www.jgi.doe.gov) in collaboration with the user community. This study was supported by European Research Council Advanced Grant 268893 and Science Foundation Ireland Grant 13IA1910.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416014111/-/DCSupplemental.
References
- 1.Strathern JN, Herskowitz I. Asymmetry and directionality in production of new cell types during clonal growth: The switching pattern of homothallic yeast. Cell. 1979;17(2):371–381. doi: 10.1016/0092-8674(79)90163-6. [DOI] [PubMed] [Google Scholar]
- 2.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52(4):536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klar AJ. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu Rev Genet. 2007;41:213–236. doi: 10.1146/annurev.genet.39.073103.094316. [DOI] [PubMed] [Google Scholar]
- 5.Egel R. Fission yeast mating-type switching: Programmed damage and repair. DNA Repair (Amst) 2005;4(5):525–536. doi: 10.1016/j.dnarep.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Lin X, Heitman J. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism. In: Heitman J, Kronstad JW, Taylor JW, Cassleton LA, editors. Sex in Fungi. American Society for Microbiology; Washington, DC: 2007. pp. 35–57. [Google Scholar]
- 7.Lee SC, Ni M, Li W, Shertz C, Heitman J. The evolution of sex: A perspective from the fungal kingdom. Microbiol Mol Biol Rev. 2010;74(2):298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11(7):512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 9.Rusche LN, Rine J. Switching the mechanism of mating type switching: A domesticated transposase supplants a domesticated homing endonuclease. Genes Dev. 2010;24(1):10–14. doi: 10.1101/gad.1886310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knop M. Yeast cell morphology and sexual reproduction—a short overview and some considerations. C R Biol. 2011;334(8-9):599–606. doi: 10.1016/j.crvi.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Nagy LG, et al. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nat Commun. 2014;5:4471. doi: 10.1038/ncomms5471. [DOI] [PubMed] [Google Scholar]
- 12.Edskes HK, Wickner RB. The [URE3] prion in Candida. Eukaryot Cell. 2013;12(4):551–558. doi: 10.1128/EC.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naumov GI, Naumova ES, Marinoni G, Piskur J. Genetic analysis of the yeast Saccharomyces castellii, S. exiguus, and S. martiniae. Russ J Genet. 1998;34(4):457–460. [PubMed] [Google Scholar]
- 14.Watanabe J, Uehara K, Mogi Y. Diversity of mating-type chromosome structures in the yeast Zygosaccharomyces rouxii caused by ectopic exchanges between MAT-like loci. PLoS ONE. 2013;8(4):e62121. doi: 10.1371/journal.pone.0062121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barsoum E, Martinez P, Aström SU. Alpha3, a transposable element that promotes host sexual reproduction. Genes Dev. 2010;24(1):33–44. doi: 10.1101/gad.557310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Rienzi SC, et al. Genetic, genomic, and molecular tools for studying the protoploid yeast, L. waltii. Yeast. 2011;28(2):137–151. doi: 10.1002/yea.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gleeson MA, Sudbery PE. Genetic analysis in the methylotrophic yeast Hansenula polymorpha. Yeast. 1988;4(4):293–303. [Google Scholar]
- 18.Tolstorukov II, Cregg JM. 2007. Pichia Protocols. Classical Genetics. Methods in Molecular Biology, ed Cregg JM (Humana, Totowa, NJ), 2nd Ed, Vol 389, pp 189–201.
- 19.Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. Elsevier; Amsterdam: 2011. [Google Scholar]
- 20.Ravin NV, et al. Genome sequence and analysis of methylotrophic yeast Hansenula polymorpha DL1. BMC Genomics. 2013;14:837. doi: 10.1186/1471-2164-14-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Schutter K, et al. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27(6):561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- 22.Curtin CD, Borneman AR, Chambers PJ, Pretorius IS. De-novo assembly and analysis of the heterozygous triploid genome of the wine spoilage yeast Dekkera bruxellensis AWRI1499. PLoS ONE. 2012;7(3):e33840. doi: 10.1371/journal.pone.0033840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piškur J, et al. The genome of wine yeast Dekkera bruxellensis provides a tool to explore its food-related properties. Int J Food Microbiol. 2012;157(2):202–209. doi: 10.1016/j.ijfoodmicro.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Morales L, et al. Complete DNA sequence of Kuraishia capsulata illustrates novel genomic features among budding yeasts (Saccharomycotina) Genome Biol Evol. 2013;5(12):2524–2539. doi: 10.1093/gbe/evt201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunze G, et al. The complete genome of Blastobotrys (Arxula) adeninivorans LS3 - a yeast of biotechnological interest. Biotechnol Biofuels. 2014;7:66. doi: 10.1186/1754-6834-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443(7113):818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 27.Jakočiūnas T, Holm LR, Verhein-Hansen J, Trusina A, Thon G. Two portable recombination enhancers direct donor choice in fission yeast heterochromatin. PLoS Genet. 2013;9(10):e1003762. doi: 10.1371/journal.pgen.1003762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329(5987):82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon JL, et al. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc Natl Acad Sci USA. 2011;108(50):20024–20029. doi: 10.1073/pnas.1112808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knop M. Evolution of the hemiascomycete yeasts: On life styles and the importance of inbreeding. BioEssays. 2006;28(7):696–708. doi: 10.1002/bies.20435. [DOI] [PubMed] [Google Scholar]
- 31.Reuter M, Bell G, Greig D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr Biol. 2007;17(3):R81–R83. doi: 10.1016/j.cub.2006.11.059. [DOI] [PubMed] [Google Scholar]
- 32.Stefanini I, et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci USA. 2012;109(33):13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coluccio AE, Rodriguez RK, Kernan MJ, Neiman AM. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS ONE. 2008;3(8):e2873. doi: 10.1371/journal.pone.0002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butler G, et al. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc Natl Acad Sci USA. 2004;101(6):1632–1637. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabaldón T, et al. Comparative genomics of emerging pathogens in the Candida glabrata clade. BMC Genomics. 2013;14:623. doi: 10.1186/1471-2164-14-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 37.Butler G. Fungal sex and pathogenesis. Clin Microbiol Rev. 2010;23(1):140–159. doi: 10.1128/CMR.00053-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Küberl A, et al. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 2011;154(4):312–320. doi: 10.1016/j.jbiotec.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Sudbery PE, Gleeson MAG. Genetic manipulation of the methylotrophic yeasts. In: Walton EF, Yarranton GT, editors. Molecular and Cell Biology of Yeasts. Blackie; Glasgow: 1989. pp. 304–329. [Google Scholar]
- 40.Lahtchev K. In: Basic Genetics of Hansenula polymorpha. Hansenula polymorpha: Biology and Applications. Gellissen G, editor. Wiley-VCH; Weinheim: 2002. pp. 8–20. [Google Scholar]
- 41.Sreekrishna K, Kropp KE. In: Pichia pastoris. Nonconventional Yeasts in Biotechnology: A Handbook. Wolf K, editor. Springer; Berlin: 1996. pp. 203–253. [Google Scholar]
- 42.Kurtzman CP. A new methanol assimilating yeast, Ogataea parapolymorpha, the ascosporic state of Candida parapolymorpha. Antonie van Leeuwenhoek. 2011;100(3):455–462. doi: 10.1007/s10482-011-9603-0. [DOI] [PubMed] [Google Scholar]
- 43.Lynch DB, Logue ME, Butler G, Wolfe KH. Chromosomal G + C content evolution in yeasts: Systematic interspecies differences, and GC-poor troughs at centromeres. Genome Biol Evol. 2010;2:572–583. doi: 10.1093/gbe/evq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burton JN, Liachko I, Dunham MJ, Shendure J. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 (Bethesda) 2014;4(7):1339–1346. doi: 10.1534/g3.114.011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marie-Nelly H, et al. Filling annotation gaps in yeast genomes using genome-wide contact maps. Bioinformatics. 2014;30(15):2105–2113. doi: 10.1093/bioinformatics/btu162. [DOI] [PubMed] [Google Scholar]
- 46.Kurtzman CP. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J Ind Microbiol Biotechnol. 2009;36(11):1435–1438. doi: 10.1007/s10295-009-0638-4. [DOI] [PubMed] [Google Scholar]
- 47.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 48.Liang S, et al. Comprehensive structural annotation of Pichia pastoris transcriptome and the response to various carbon sources using deep paired-end RNA sequencing. BMC Genomics. 2012;13:738. doi: 10.1186/1471-2164-13-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egel R. “Flip-flop” control and transposition of mating-type genes in fission yeast. In: Bukhari AI, Shapiro JA, Adhya SL, editors. DNA Insertion Elements, Plasmids and Episomes. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1977. pp. 447–455. [Google Scholar]
- 50.Hicks JB, Strathern JN, Herskowitz I. The cassette model of mating-type interconversion. In: Bukhari AI, Shapiro JA, Adhya SL, editors. DNA Insertion Elements, Plasmids and Episomes. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1977. pp. 457–462. [Google Scholar]
- 51.Hicks JB, Herskowitz I. Interconversion of yeast mating types. II. Restoration of mating ability to sterile mutants in homothallic and heterothallic strains. Genetics. 1977;85(3):373–393. doi: 10.1093/genetics/85.3.373b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tolstorukov II, Benevolensky SV, Efremov BD. Mechanism of mating and self-diploidization in the haploid yeast Pichia pinus. VI. Functional organization of the mating type locus. Sov Genet. 1981;17(6):685–690. [Google Scholar]
- 53.Tolstorukov II, Benevolensky SV, Efremov BD. Genetic control of cell type and complex organization of the mating type locus in the yeast Pichia pinus. Curr Genet. 1982;5(2):137–142. doi: 10.1007/BF00365704. [DOI] [PubMed] [Google Scholar]
- 54.Oshima Y. Homothallism, mating-type switching, and the controlling element model. In: Hall MN, Linder P, editors. Saccharomyces cerevisiae. The Early Days of Yeast Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1993. pp. 291–304. [Google Scholar]
- 55.Silverman M, Simon M. Phase variation: Genetic analysis of switching mutants. Cell. 1980;19(4):845–854. doi: 10.1016/0092-8674(80)90075-6. [DOI] [PubMed] [Google Scholar]
- 56.Hicks J, Strathern JN, Klar AJ. Transposable mating type genes in Saccharomyces cerevisiae. Nature. 1979;282(5738):478–483. doi: 10.1038/282478a0. [DOI] [PubMed] [Google Scholar]
- 57.Chitrampalam P, Inderbitzin P, Maruthachalam K, Wu BM, Subbarao KV. The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation. PLoS ONE. 2013;8(2):e56895. doi: 10.1371/journal.pone.0056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zörgö E, et al. Ancient evolutionary trade-offs between yeast ploidy states. PLoS Genet. 2013;9(3):e1003388. doi: 10.1371/journal.pgen.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hickman MA, Froyd CA, Rusche LN. Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot Cell. 2011;10(9):1183–1192. doi: 10.1128/EC.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ketel C, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5(3):e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Souciet JL, et al. Génolevures Consortium Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009;19(10):1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katz Ezov T, et al. Heterothallism in Saccharomyces cerevisiae isolates from nature: Effect of HO locus on the mode of reproduction. Mol Ecol. 2010;19(1):121–131. doi: 10.1111/j.1365-294X.2009.04436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liachko I, Dunham MJ. An autonomously replicating sequence for use in a wide range of budding yeasts. FEMS Yeast Res. 2014;14(2):364–367. doi: 10.1111/1567-1364.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faber KN, Haima P, Harder W, Veenhuis M, Ab G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr Genet. 1994;25(4):305–310. doi: 10.1007/BF00351482. [DOI] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: A next-generation genome browser. Genome Res. 2009;19(9):1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neuvéglise C, Feldmann H, Bon E, Gaillardin C, Casaregola S. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 2002;12(6):930–943. doi: 10.1101/gr.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.