Significance
In higher plant Photosystem II, the PsbP and PsbQ proteins provide critical support for oxygen evolution at physiological calcium and chloride concentrations. The locations of these components within the photosystem, however, are unclear. Our findings that (i) the N terminus of PsbP, which is unresolved in the current high-resolution structure of this subunit, forms a compact structure and associates with the C-terminal domain of the protein and (ii) PsbP and PsbQ directly interact to form a framework for understanding the organization of these subunits within the higher plant photosystem.
Keywords: photosynthesis, Photosystem II, PsbP, PsbQ, mass spectrometry
Abstract
Protein cross-linking and radiolytic footprinting coupled with high-resolution mass spectrometry were used to examine the structure of PsbP and PsbQ when they are bound to Photosystem II. In its bound state, the N-terminal 15-amino-acid residue domain of PsbP, which is unresolved in current crystal structures, interacts with domains in the C terminus of the protein. These interactions may serve to stabilize the structure of the N terminus and may facilitate PsbP binding and function. These interactions place strong structural constraints on the organization of PsbP when associated with the Photosystem II complex. Additionally, amino acid residues in the structurally unresolved loop 3A domain of PsbP (90K–107V), 93Y and 96K, are in close proximity (≤11.4 Å) to the N-terminal 1E residue of PsbQ. These findings are the first, to our knowledge, to identify a putative region of interaction between these two components. Cross-linked domains within PsbQ were also identified, indicating that two PsbQ molecules can interact in higher plants in a manner similar to that observed by Liu et al. [(2014) Proc Natl Acad Sci 111(12):4638–4643] in cyanobacterial Photosystem II. This interaction is consistent with either intra-Photosystem II dimer or inter-Photosystem II dimer models in higher plants. Finally, OH• produced by synchrotron radiolysis of water was used to oxidatively modify surface residues on PsbP and PsbQ. Domains on the surface of both protein subunits were resistant to modification, indicating that they were shielded from water and appear to define buried regions that are in contact with other Photosystem II components.
Photosystem II (PS II) is a light-driven water-plastoquinone oxidoreductase that is found in all oxygenic photosynthetic organisms. This membrane protein complex contains at least 20 protein subunits, 17 of which are intrinsic membrane proteins. Higher plants contain three extrinsic proteins associated with the complex—PsbO, PsbP, and PsbQ—whereas cyanobacteria contain PsbO, PsbU, PsbV, and CyanoQ (a homolog of PsbQ). In higher plants the PsbO, PsbP, and PsbQ proteins are required for optimal rates of O2 evolution under physiological inorganic calcium and chloride concentrations (1, 2).
The PsbO protein appears to play a central role in the stabilization of the manganese cluster in all oxygenic photosynthetic organisms (3). In higher plants, PsbO and PsbP are required for photoautotrophic growth, PS II assembly, and the stabilization of PS II supercomplexes (4–9), with PsbP also being required for normal thylakoid assembly (6). Under low-light growth conditions, PsbQ is required for photoautotrophy (10).
Although the crystal structure of cyanobacterial PS II has been resolved to 1.9 Å (11), no crystal structures for higher plant PS II have been presented. The structure and interactions of PsbO with the intrinsic subunits have been approximated by analogy to the cyanobacterial photosystem. Although high-resolution crystal structures of isolated spinach PsbP [1.98 Å; Protein Data Bank (PDB) ID code 2VU4] (12) and PsbQ (1.49 Å; PDB ID code 1VYK) are available (13, 14), the domains of these components that interact with PS II are not well understood (2, 15).
In this communication, we have used protein cross-linking coupled with high-resolution mass spectrometry to identify internally cross-linked domains within the PsbP protein when it is bound to the photosystem. In its bound state, the N-terminal 15-amino-acid residue domain of PsbP, which is unresolved in current crystal structures, interacts with domains in the C terminus of the protein. Additionally, amino acid residues in a structurally unresolved loop domain of PsbP (90K–107V), 93Y and 96K, are in close proximity (≤11.4 Å) to the N-terminal 1E residue of PsbQ. This is the first report, to our knowledge, of an interaction between PsbP and PsbQ. Cross-linked domains within PsbQ were also identified, indicating that two PsbQ molecules can interact in a manner similar to that observed by Liu et al. (16). Finally, OH• produced by the synchrotron radiolysis was used to oxidatively modify surface residues on PsbP and PsbQ that were in contact with water. Domains on the surface of both proteins were resistant to modification, indicating that they were shielded from the bulk solvent and appear to define buried regions that are in contact with other components of PS II.
Results and Discussion
Protein Cross-Linking Reveals Inter- and Intraprotein Associations Within PS II.
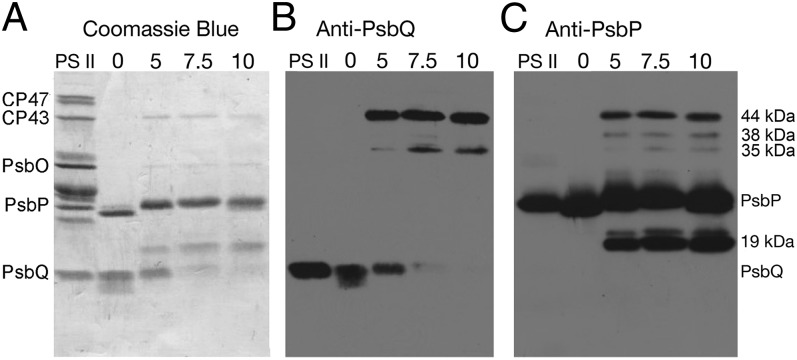
Fig. 1 illustrates the results observed upon treatment of PS II membranes with varying concentrations of the cross-linker bis-sulfosuccinimidyl suberate (BS3) followed by lithium dodecyl sulfate (LiDS)–PAGE and immunobloting. As expected, in the absence of BS3, no cross-linked products were observed, and only PsbP and PsbQ were removed from the membranes by salt-washing. Increasing concentrations of the cross-linker (5–10 mM) lead to increased accumulation of a number of putative cross-linked products (19, 35, 38, and 44 kDa), which were released from the PS II membrane by salt-washing. Immunoblot analysis indicated that the 19-kDa band reacted only with anti-PsbP, whereas the 35-, 38-, and 44-kDa bands reacted with both anti-PsbP and anti-PsbQ. The cross-linked products obtained by treatment with 5 mM BS3 were further analyzed. At 0 mM BS3, both PsbP and PsbQ appeared to exhibit proteolysis in the NaCl-wash extracts. It had been shown previously that both of these proteins are susceptible to proteolytic attack (17–19). The presence of the cross-linker BS3, however, appeared to suppress this endogenous proteolytic activity. The 19-kDa band was the result of intrachain protein cross-linking. Subsequent mass spectral analysis indicated that no proteolysis was evident, as we obtained complete mass spec coverage of both the N and C termini. The three higher mass bands contained PsbP and PsbQ and represent interchain cross-linked products. The 19- and 44-kDa cross-linked products were most abundant and were selected for further analysis.
Fig. 1.
Cross-linking of PsbP and PsbQ with BS3 within the PS II complex. Cross-linker concentrations were 0–10 mM BS3. Illustrated are “Western” blots of PS II membranes (PS II) and dialyzed 1.0 M NaCl extracts obtained after cross-linking of the PS II membranes at the indicated BS3 concentrations. (A) Overall protein profile of the samples. B and C identify the proteins that cross-react with anti-PsbQ and anti-PsbP, respectively. A number of PS II proteins are labeled to the left of A for reference. To the right of C, cross-linked products are labeled. Lanes were loaded with 10 μg of protein except the PS II lane, which was loaded with 10 μg of chl.
The cross-linker BS3 has been used extensively in protein cross-linking studies in a variety of systems. The N-hydroxysulfosuccidimyl leaving group reacts with primary amines and, at low pH, with the hydroxyl groups found in tyrosyl, threonyl, and seryl residues (20). The modification of threonyl and seryl hydroxyl groups is relatively inefficient and may exhibit lower stability (20, 21); consequently, in this study, we have considered only cross-links involving lysyl and tyrosyl residues and unblocked N termini.
In Table S1, we present the cross-linked residues identified in this study by mass spectrometry. Eleven intrachain cross-linked products were identified within PsbP, the vast majority of these being found in the 19-kDa cross-linked product. Two interchain cross-links involving PsbP and PsbQ were identified in the 44-kDa cross-linked product. Finally, three cross-linked residue pairs of PsbQ also were identified from the 44-kDa cross-linked product. All of the cross-linked peptides identified exhibited extremely low P values ranging from 2.5 × 10−4 to 5 × 10−16. The quality of the data used in this communication is illustrated in Fig. S1.
The N Terminus of PsbP Associates with C-Terminal Domains.
Elucidation of the structure of the N terminus of PsbP is critical for understanding the function of this component. The N terminus is required for the efficient binding of PsbP to the photosystem and for its function (22) in lowering the calcium and chloride requirement for oxygen evolution (23–25). Unfortunately, this important domain is not resolved in the current crystal structure of unbound PsbP (12) (PDB ID code 2VU4). Our observation that after cross-linking, PsbP, which normally migrates at 24 kDa, is observed to migrate at 19 kDa (Fig. 1) indicates that BS3 treatment prevents complete unfolding of the protein in the presence of LiDS and suggests that PsbP, when associated with the membrane, has a compact structure. Subsequent mass spectrometry data demonstrate that the N-terminal domain (1A–15N) of PsbP is closely associated with the C terminus (170K–186A) of the protein. Nine independent cross-linked products were identified that demonstrate this association (Table S1). All of the residue pairs identified to be cross-linked in these products are within 11.4 Å of each other within the PsbP subunit when it is bound to PS II. This information places strong constraints on the possible structures PsbP can assume in the bound state. It should also be noted that one pair of cross-linked residues, PsbP:40K and PsbP:155K, appears in the crystal structure of unbound PsbP to be separated by 14.3–16.3 Å (including rotamers) (12). The observed cross-linking of these residues with BS3 (span of ≤11.4 Å) when PsbP is bound to the photosystem indicates that a conformational change of 3–5 Å occurs in this region of the protein upon binding of the subunit to the PS II complex. Recently, Ido et al. (26) observed a similar apparent conformational change: PsbP:115E is in van der Waals contact with PsbP:173K, as these residues were cross-linked with 1-ethyl-3-(3-diethylaminopropyl) carbodiimide (EDC). This indicates that a conformational change of 7.4 Å occurs in this region of the protein when it associates with the photosystem.
Secondary structure analysis using the Genesilico Metaserver (27) of the unresolved N-terminal domain of PsbP indicates that it is likely that this region assumes either an α-helical or random coil secondary structure. Given the strong distance constraints provided by the observed interchain PsbP cross-linked products, molecular dynamic refinement can provide useful models for the structure of the N terminus of PsbP. The program MODELER (28) was used to provide the structures shown in Fig. 2. Illustrated in this figure are the 10 top models predicted for an α-helical N terminus (Fig. 2A) and an N terminus assuming a random coil configuration (Fig. 2B). In both instances, all 10 predicted structures exhibit similar low discrete optimized protein energy scores (∼–17,000). Importantly, regardless of the type of N terminus modeled (α-helical or random coil), the location of the N-terminal residue 1A is located in very similar positions. Our data indicate that PsbP assumes a compact structure when the protein is associated with the PS II complex. It had been suggested that the unresolved N terminus might form an extended structure when associated with the PS II core complex (2, 26); this, apparently, is not the case. Residue 1A has been shown to directly interact with PsbE:57E (26, 29); consequently, our results indicate that PsbP must be in close proximity to PsbE and CP47.
Fig. 2.
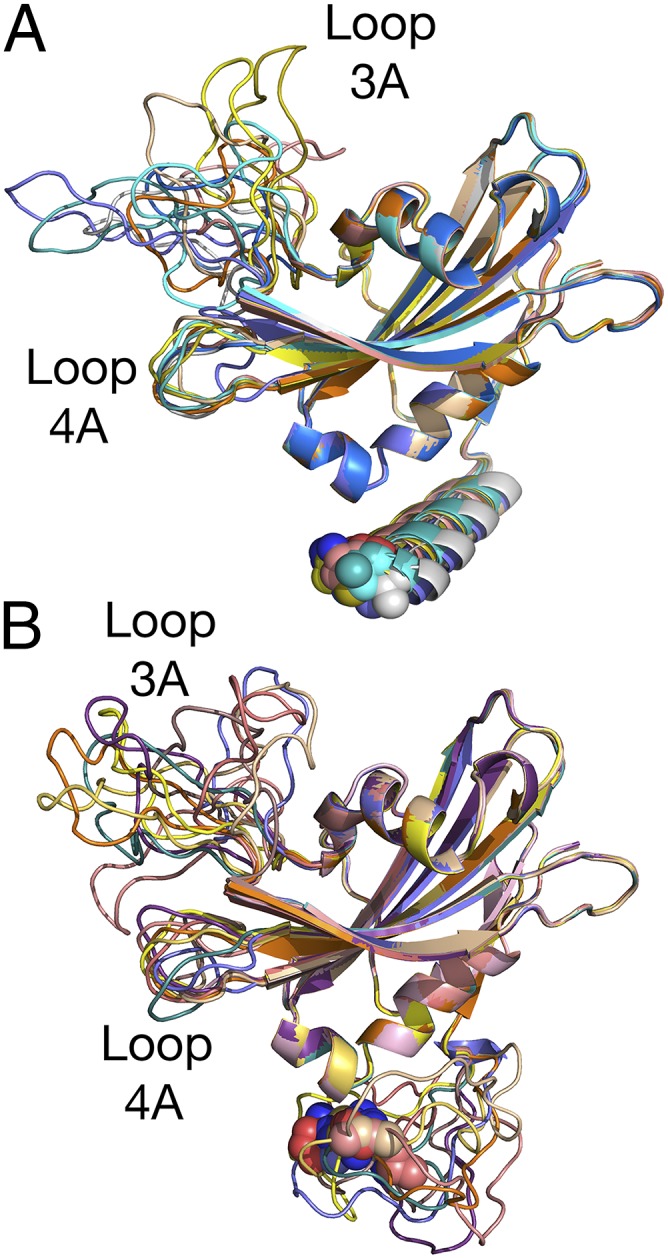
Distance-constrained molecular dynamic refinement for the N terminus of PsbP. Shown are the top 10 models for both (A) N-terminal α-helix or (B) random coil incorporating the distance constraints shown in Table S1. Residue 1A is shown as spheres. In both the α-helical or random coil models, 1A is located in very similar positions, indicating that the N terminus of PS II-bound PsbP does not assume an extended conformation.
PsbP and PsbQ Are Closely Associated.
Both PsbP and PsbQ bind to PS II with extremely high affinity (low nM Kds), and their removal requires high salt concentrations or markedly elevated pH conditions (30). The direct interaction between PsbP and PsbQ has been proposed by a number of investigators. Reconstitution studies have indicated that PsbQ cannot bind to the photosystem in the absence of PsbP (30, 31). PsbQ appears to stabilize the structural and functional interaction of PsbP to PS II. An N-terminal 19-amino-acid residue-truncated PsbP cannot bind to PS II in the absence of PsbQ but can bind in its presence. Importantly, in the presence of PsbQ, the function of the truncated PsbP is partially restored (32, 33).
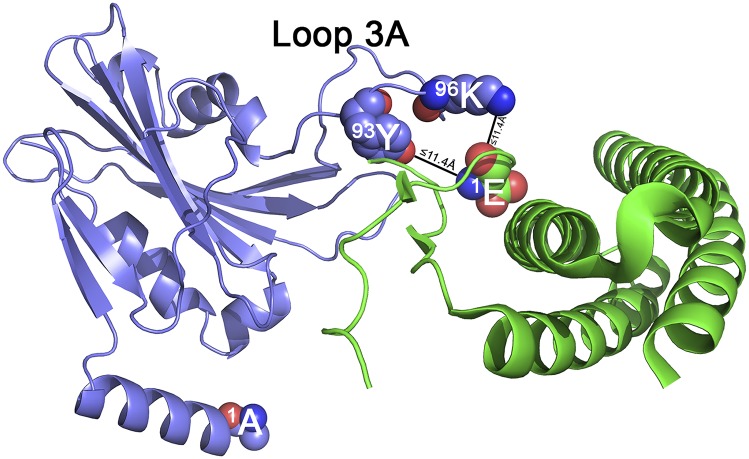
Analysis of the 44-kDa cross-linked band (Fig. 1) allowed the identification of an interacting domain between the PsbP and PsbQ subunits. Both PsbP:96K and PsbP:93Y can be cross-linked to PsbQ:1E using the cross-linker BS3, indicating that both PsbP residues are within 11.4 Å of the N terminus of PsbQ (Table S1). Fig. 3 illustrates one of the many possible models of this interaction. Both PsbP:93Y and PsbP:96K are located in the 17-residue loop 3A of PsbP (89G–105S). This loop is unresolved in the crystal structure of unbound PsbP. Because we lack intrachain cross-linked products involving this domain, our molecular dynamics refinements predict a wide variety of possible loop 3A structures (Fig. 2 A and B). Our identification of interacting domains between PsbP and PsbQ helps explain the observation that PsbQ assists in the stabilization of PsbP binding and function (32, 33). This observation may also explain why loop 3A is disordered and not resolved in the current PsbP crystal structure. In the absence of PsbQ, this domain may not exhibit any one preferred structure and may be quite mobile, which would preclude crystallographic structural assignment.
Fig. 3.
Interaction between PsbP and PsbQ. Schematic illustration of one possible model for BS3 cross-linking of PsbP and PsbQ. PsbP is shown in blue, and PsbQ is shown in green. Residue PsbQ:1E and its cross-linking partners PsbP:93Y and 96K are shown as spheres, as is PsbP:1A.
Observation of a PsbQ–PsbQ Dimer.
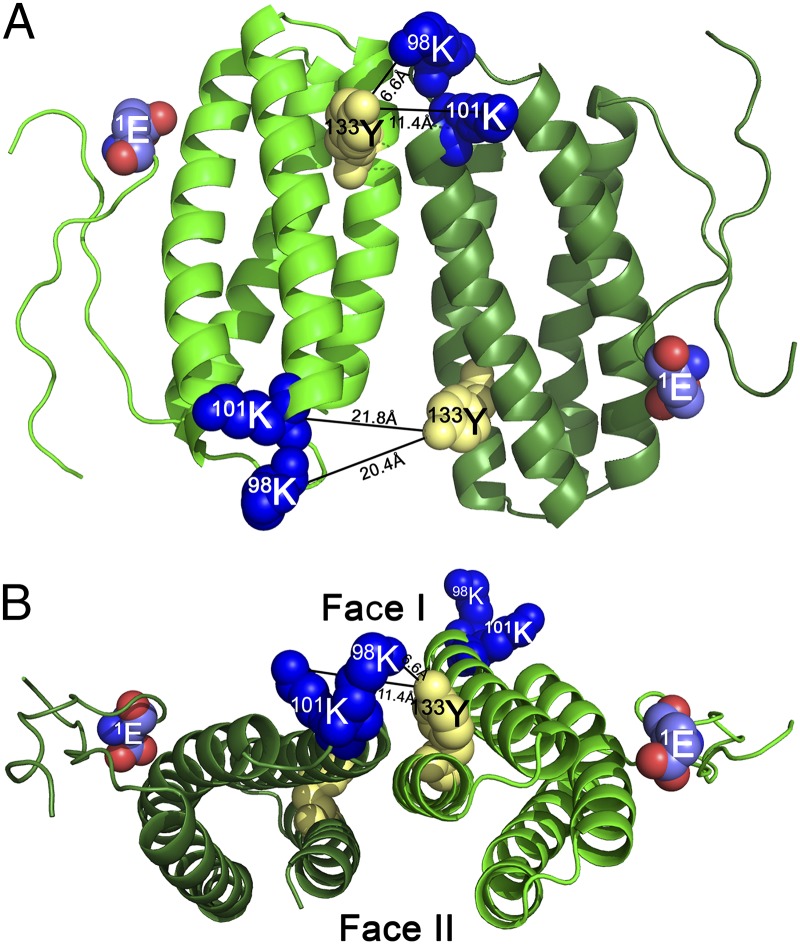
Additionally, three cross-linked products internal to PsbQ were observed in the 44-kDa band (Table S1). One of these, 53K–96K, is fully consistent with the unbound crystal structure of PsbQ, as these residues are located 8.5 Å apart. The other two cross-linked products observed, however, are significantly more difficult to explain. In the crystal structure of the unbound protein, the residue pairs 98K–133Y and 101K–133Y are located at a distance of 33.1 Å and 30 Å, respectively. The cross-linker BS3 cannot span this distance, and an unprecedented and massive conformational change in the PsbQ protein would be required to bring these residue pairs within the cross-linker’s active radius within a single PsbQ molecule. Consequently, we hypothesize that these observed cross-linked species represent interactions between two different PsbQ molecules.
GRAMM-X (34) was used to model this interaction using the PsbQ crystal structure and the distance constraints imposed by the cross-linker (≤11.4 Å). One possible model is illustrated in Fig. 4A. The two PsbQ monomers are shown to associate in an antiparallel configuration. In Fig. 4B, an end-on view of the modeled dimer is shown, with the cross-linked residues appearing in the foreground. In this view, the protein dimer is quite flattened and exhibits two faces, which we have designated face I and face II. Based on the radiolytic footprinting data shown in Fig. S2 we hypothesize that face I is oriented toward the lumen and, consequently, is more exposed to the bulk solvent, whereas face II is oriented toward the PS II core complex and is less exposed.
Fig. 4.
PsbQ–PsbQ interaction and one distance-constrained GRAMM-X model (34) for the PsbQ–PsbQ interaction. (A) View looking from the lumen toward face I of the dimer. (B) Edge view of the dimer. The two faces of the dimer—face I, which is oriented toward the lumen, and face II, which is oriented toward the intrinsic proteins of the PS II dimer—are indicated. The two interacting molecules of PsbQ are shown in light and dark green. Lysyl residues participating in this interaction are shown as blue spheres, and the tyrosyl residue is shown as yellow spheres.
Recently, Liu et al. (16) have proposed a similar model for the organization of the cyanobacterial PsbQ homolog, CyanoQ, in Synechocystis. These authors observed a cross-linked species involving the CyanoQ residues 120K and 96K, which are separated by 41 Å. Liu et al. (16) hypothesized that the cross-linked residues were on two different CyanoQ molecules, each of these associated with one PS II monomer of the dimeric PS II complex. They placed this antiparallel homodimer at the lumenal interface of the PS II complex dimer. Cross-linked products were identified between CyanoQ and PsbO (CyanoQ:120K-PsbO:59K and CyanoQ:180K-PsbO:180) and CyanoQ and CP47 (CyanoQ:102K-CP47:440D), supporting this assignment.
Ido et al. (26) have presented a markedly different model for the placement of PsbQ in higher plant PS II. In their report, PsbQ is placed on the periphery of the PS II complex adjacent to CP43 and near the lumenal surface of the light-harvesting chlorophyll-protein II-S trimer. Cross-linked products were observed between PsbQ and CP43 and between PsbQ and CP26; however, the cross-linking sites were not identified. No cross-linked products were identified between PsbP and PsbQ. The models proposed by Liu et al. (16) and Ido et al. (26) appear to be mutually exclusive.
Our identification of a cross-linked species between two different PsbQ molecules appears to support the assignment and location of CyanoQ presented by Liu et al. (16). Another possibility exists, however; the CyanoQ–CyanoQ interaction presented by Liu et al. (16) is modeled as an intra-PS II dimer interaction, with the CyanoQ of one monomer interacting with the CyanoQ of the second monomer at the lumenal surface of the monomer–monomer interface. In this study, protein cross-linking was performed on dodecyl-maltoside PS II particles (35). In our study, and that of Ido et al. (26), protein cross-linking was performed on higher plant PS II membranes. In this membrane environment, it is possible that protein cross-linking between two different PS II dimer complexes could occur. Such inter-PS II dimer interactions might be facilitated by the high concentration of PS II complexes within PS II membranes. Consequently, our observation of the interaction between PsbP and PsbQ appears to be consistent with either the model presented by Liu et al. (16) or that of Ido et al. (26).
Radiolytic Footprinting Identifies Shielded Domains on PsbP and PsbQ.
The use of radiolytic footprinting to identify protein–protein interactions is a developing technique that allows the identification of residues exposed to the bulk solvent (36, 37). The OH• that is produced during synchrotron radiolysis is extremely reactive and can modify at least 14 residues that are identifiable by mass spectrometry (36).
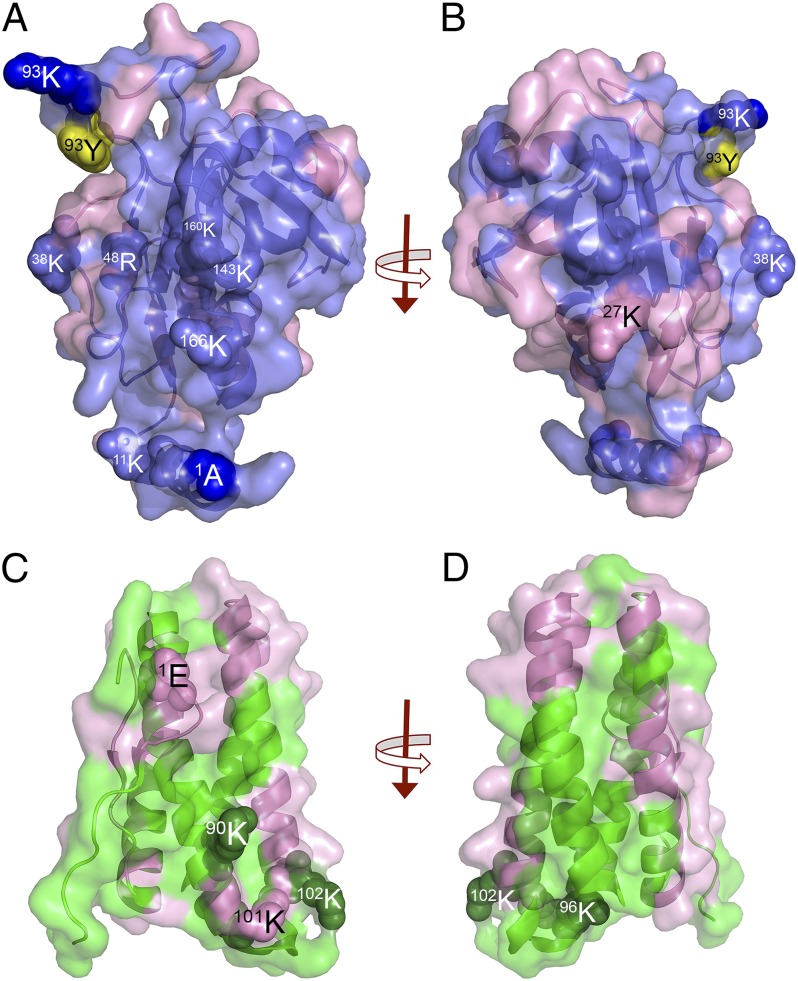
Fig. 5 and Table S2 present the results from the radiolytic footprinting of the PsbP and PsbQ subunits that are associated with PS II membranes. Mass spectrometry coverage for the PsbP and PsbQ proteins was 97% and 100%, respectively, when integrated over all of the time points (0, 4, 8, and 16 s irradiation) examined. Fig. 5 represents the union of all of the identified oxidatively modified residues observed in the experiment. The residues modified at each time point are summarized in Table S2.
Fig. 5.
Radiolytic mapping of PsbP and PsbQ. (A) View of the face of the PsbP protein proposed to face the core of PS II. (B) Rotation of A by 180°, showing the face of PsbP proposed to be exposed to the bulk solvent and PsbR. Oxidatively modified residues are shown in pink, and unmodified residues are shown in blue or yellow. (C) View of monomeric PsbQ. (D) Rotation of C by 180°. Oxidatively modified residues are shown in pink, and unmodified residues are shown in green. Several residues are shown as spheres.
The radiolytic modification of PsbP is shown in Fig. 5 A and B and Table S2. These two faces of the protein exhibit markedly different radiolytic labeling patterns. The face of the protein shown in Fig. 5A has essentially no radiolytic modifications except at the periphery of the protein. This indicates that few of the residues located on this face are exposed to the bulk solvent. Residue 1A, which earlier had been identified as interacting with PsbE:57E (26, 29), and residues 96K and 93Y, which we have identified as interacting with PsbQ:1E, are all unmodified. Interestingly, the three residues that have recently been identified by Nishimura et al. (38) (48R, 143K, and 160K) as being required for PsbP binding to PS II are unmodified surface residues that lie on this face of the protein. Earlier, Tohri et al. (39) also identified a group of conserved higher plant PsbP residues (11K, 13K, 33K, 38K, 143K, 166K, 170K, and 174K) whose modification with N-succinimidyl propionate + EDC prevented the association of PsbP with PS II. None of these residues, except 174K, were radiolytically modified, and all of these unmodified residues are also located on this face of PsbP. It should be noted that none of the interacting partners for any of the basic residues identified in these studies (38, 39) have been located. Our finding that virtually all of the residues identified by Nishimura et al. (38) and Tohri et al. (39) are located on this face of PsbP indicates that the unmodified domain of PsbP, which is illustrated in Fig. 5A, forms a major interacting interface of PsbP with the intrinsic components of PS II.
A 180° rotation of PsbP is shown in Fig. 5B. Many more radiolytically modified residues are present on this face of the protein, indicating that these are exposed to the bulk solvent. Interestingly, a domain of unmodified residues is also evident, indicating that residues in this region are not exposed. This unmodified domain is immediately adjacent to 27K, which is cross-linked to PsbR:22D with EDC (26). Although 27K is radiolytically modified, we hypothesize that the adjacent unmodified domain may interact closely with PsbR and may form the interaction interface between PsbP and PsbR.
It should be noted that no radiolytic modification was observed for the N-terminal 13 amino acid residues (1AYGEAANVFGKP13K) even though we observed 100% mass spectrometry coverage in this domain. These residues are not resolved in the current PsbP crystal structure (12). Our results indicate that these residues are not exposed to the bulk solvent. We hypothesize that they are buried at the interface of PsbP with PsbE and PsbR.
The radiolytic modification of PsbQ is shown in Fig. 5 C and D and Table S2. Monomeric PsbQ is shown in Fig. 5C. In this view and the 180° rotation of the monomer (Fig. 5D), it is clear that large domains on both of the illustrated faces of the subunit are protected from radiolytic modification. This indicates that significant portions of the surface of PsbQ are inaccessible to the bulk solvent, presumably due to intimate interaction with other PS II components. Fig. S2 illustrates the radiolytic labeling of PsbQ within the context of the putative dimer illustrated in Fig. 5. Face I of the dimer (Fig. S2A) exhibits moderate labeling, with two domains that are unmodified. We hypothesize that these interact with other yet unidentified PS II components. Intriguingly, face II of the dimer (Fig. S2B) exhibits little oxidative labeling except at the periphery of the dimer. We hypothesize that this face interacts with PS II core complex components possibly in a manner similar to that suggested by Liu et al. (16).
Earlier, we had demonstrated that NHS-biotin modification of four lysyl residues (90K, 96K, 101K, and 102K) prevented efficient binding of this component to the photosystem (40). Three of these residues (90K, 96K, and 102K) were not observed to be radiolytically modified. These residues cluster in a domain that bridges the end of helix II and the start of helix III. Our radiolytic footprinting data support and extend our hypothesis that these residues form an interacting domain for PsbQ to the photosystem (40).
Conclusions
Our results significantly expand our understanding of the structural organization of PsbP and PsbQ within higher plant PS II. The observation that the N-terminal domain of PsbP interacts directly with residues at the C terminus of the protein places strong constraints on the location of PsbP within the photosystem and appears to preclude the positioning for this component recently suggested by Ido et al. (26). Additionally, our identification of interacting domains between PsbP and PsbQ provides a framework for understanding the structural and functional interactions between these two subunits. Finally, our observation that PsbQ forms putative dimers in higher plants, similar to dimers observed for CyanoQ in cyanobacteria (16), must be taken into account when proposing global models for extrinsic protein interactions within the photosystem. Two of the many possible models for the global interaction of PsbP and PsbQ with PS II are presented in Fig. S3. The first (Fig. S3 A and B) is analogous to the model presented by Liu et al. (16) for localization of CyanoQ within cyanobacterial PS II. The second model (Fig. S3 C and D) is based on a model presented by Ido et al. (26) for the organization of PsbP and PsbQ in the higher plant photosystem. The position of PsbQ in our model required significant adjustment to fulfill the distance constraints that we observe. Ongoing experimentation will provide additional structural constraints that will allow the differentiation between these and other possible models for the organization of the extrinsic proteins associated with higher plant PS II.
Materials and Methods
PS II Membrane Isolation and Protein Cross-Linking.
PS II membranes were isolated from market spinach by the method of Berthold et al. (41). Chlorophyll (Chl) concentration was determined by the method of Arnon (42). Oxygen evolution rates were >400 mmoles O2·mg chl–1·h−1. After isolation, the PS II membranes were suspended at 2 mg chl per mL in 50 mM Mes-NaOH, pH 6.0, 300 mM sucrose, 15 mM NaCl (SMN) buffer and frozen at –80 °C until use. Protein cross-linking was performed using BS3 (ProteoChem, Inc.). BS3 can cross-link primary amino groups and, at low pH, threonyl, tyrosyl, and possibly seryl residues (20, 43–45). PS II membranes were suspended at a chl concentration of 100 μg/mL in SMN buffer and treated with various concentrations of BS3 (0–10 mM) for 1 h at room temperature and in the dark. The reaction was quenched by bringing the reaction mixture to 30 mM ammonium bicarbonate and incubating it for 20 min at room temperature. The membranes were harvested by centrifugation for 25 min at 39,000 × g, and the final pellet was resuspended in 1.0 M NaCl for 1 h at 4 °C to release any PsbP and PsbQ proteins not cross-linked to intrinsic membrane protein or PsbO. The PS II membranes were pelleted by centrifugation for 25 min at 39,000 × g, and the supernatant, which contained free PsbP, free PsbQ, and PsbP–PsbQ cross-linked products, was collected. The salt-extracted sample was then dialyzed overnight against 10 mM Mes-NaOH, pH 6.0, using a 6–8-kDa membrane (Spectrum Laboratories, Inc.), centrifuged for 25 min at 39,000 × g, and proteins were concentrated by ultrafiltration using a 10-kDa cutoff membrane (Millipore Co.). Protein concentrations were determined using the BCA protein assay (46).
Synchrotron Radiolysis.
Synchrotron radiolysis was performed as described previously (47). Briefly, PS II membranes were prepared as described above. Radiolysis was performed on the XLRM2 beamline of The J. Bennett Johnston, Sr. Center for Advanced Microstructures & Devices synchrotron. Samples (200 μL at 2 mg chl per mL) were exposed for various lengths of time (0, 4, 8, and 16 s) at room temperature in a multichannel Plexiglas chamber. After exposure, the samples were immediately removed from the chamber and held on ice until being stored at –80 °C before further analysis.
Electrophoresis and Protein Digestion.
For the cross-linking experiments, the protein samples were resolved on a standard acrylamide 12.5–20% LiDS–PAGE gradient gel (48). For the radiolytic experiments, however, the proteins were resolved on 12.5–20% LiDS–PAGE gradient using a nonoxidizing gel system (49, 50). After electrophoresis, the gels were stained with Coomassie Blue, destained, and protein bands of interest were excised. These were then processed for protease digestion (trypsin or trypsin + Lys-C) using standard protocols. In some cases, the mass spectrometry-compatible ProteaseMax (Promega) was included during digestion. After digestion, the proteolytic peptides were processed using a C18 ZipTip before mass analysis.
Reversed-phase chromatography and mass spectrometry were performed as described previously (50). Briefly, peptides were separated by reversed-phase chromatography using a Water’s X-Bridge C18 3.5-μm 2.1 × 100 mm column. Mass spectrometry was performed on a Thermo Scientific linear-trap quadrupole–Fourier transform mass spectrometer, a hybrid instrument consisting of a linear ion trap and a Fourier transform ion cyclotron resonance mass spectrometer.
Identification and analysis of peptides containing cross-linked products or oxidative mass modifications were performed using the MassMatrix online search engine (51, 52). A FASTA library containing PsbO, PsbP, PsbQ, and PsbR proteins was searched, as was a decoy library that contained the same proteins but with reversed amino acid sequences. For the identification of cross-linked products, peptides were selected if their P value was ≤0.001. For the identification of oxidative modifications, a more stringent P value (≤0.00001) was used. In both instances, the peptides were required to exhibit 0% hits to the decoy library for further consideration.
Protein Modeling.
Secondary structure analysis for the N terminus of PsbP was performed using the online Genesilico Metaserver (27), which provides the prediction using 16 different secondary structure prediction algorithms. Eight of these predicted a random coil structure, and eight predicted an α-helical architecture for the N terminus of PsbP. Molecular dynamics refinement of the PsbP protein was carried out using the program MODELER (28). The random coil and α-helix models of PsbP were compared with the X-ray structure rmsd values. Values for all of the models presented were in the range of 0.20–0.28. The modeled protein structures were further validated for their stereochemistry using the program PROCHECK (53) to obtain Ramachandran plots. All of our models satisfy Ramachandran plot statistics, with 0% of the residues in the random coil models falling in disallowed regions and 0.6% of the residues of α-helix models in disallowed regions. ProSA-web (54, 55) was used to determine the Z scores for all of our modeled structures. It was found that all of the α-helix and random coil models had Z scores very close to that reported for the actual X-ray crystal structure of PsbP. Distance-constrained models for the PsbQ–PsbQ interaction were generated by the program GRAMM-X (34).
Supplementary Material
Acknowledgments
We thank Dr. Johnna L. Roose for her critical reading of the manuscript. This work was supported by US Department of Energy, Office of Basic Energy Sciences Grant DE-FG02-09ER20310 (to T.M.B. and L.K.F.) and National Institutes of Health Grants RR019900 and GM58843 (to P.A.L.). H.D.B. is supported by the Louisiana Governor’s Biotechnology Initiative.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415165111/-/DCSupplemental.
References
- 1.Roose JL, Wegener KM, Pakrasi HB. The extrinsic proteins of Photosystem II. Photosynth Res. 2007;92(3):369–387. doi: 10.1007/s11120-006-9117-1. [DOI] [PubMed] [Google Scholar]
- 2.Bricker TM, Roose JL, Fagerlund RD, Frankel LK, Eaton-Rye JJ. The extrinsic proteins of Photosystem II. Biochim Biophys Acta. 2012;1817(1):121–142. doi: 10.1016/j.bbabio.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Bricker TM, Frankel LK. The structure and function of the 33 kDa extrinsic protein of Photosystem II. A critical review. Photosynth Res. 1998;56(2):157–173. [Google Scholar]
- 4.Yi X, McChargue M, Laborde S, Frankel LK, Bricker TM. The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J Biol Chem. 2005;280(16):16170–16174. doi: 10.1074/jbc.M501550200. [DOI] [PubMed] [Google Scholar]
- 5.Yi X, Hargett SR, Liu H, Frankel LK, Bricker TM. The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J Biol Chem. 2007;282(34):24833–24841. doi: 10.1074/jbc.M705011200. [DOI] [PubMed] [Google Scholar]
- 6.Yi X, Hargett SR, Frankel LK, Bricker TM. The PsbP protein, but not the PsbQ protein, is required for normal thylakoid architecture in Arabidopsis thaliana. FEBS Lett. 2009;583(12):2142–2147. doi: 10.1016/j.febslet.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 7.Ido K, et al. Knockdown of the PsbP protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochim Biophys Acta. 2009;1787(7):873–881. doi: 10.1016/j.bbabio.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Bricker TM, Frankel LK. Auxiliary functions of the PsbO, PsbP and PsbQ proteins of higher plant Photosystem II: A critical analysis. J Photochem Photobiol B. 2011;104(1-2):165–178. doi: 10.1016/j.jphotobiol.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Allahverdiyeva Y, et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013;75(4):671–684. doi: 10.1111/tpj.12230. [DOI] [PubMed] [Google Scholar]
- 10.Yi X, Hargett SR, Frankel LK, Bricker TM. The PsbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. J Biol Chem. 2006;281(36):26260–26267. doi: 10.1074/jbc.M603582200. [DOI] [PubMed] [Google Scholar]
- 11.Umena Y, Kawakami K, Shen J-R, Kamiya N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature. 2011;473(7345):55–60. doi: 10.1038/nature09913. [DOI] [PubMed] [Google Scholar]
- 12.Kopecky V, Jr, et al. Raman spectroscopy adds complementary detail to the high-resolution x-ray crystal structure of photosynthetic PsbP from Spinacia oleracea. PLoS ONE. 2012;7(10):e46694. doi: 10.1371/journal.pone.0046694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balsera M, Arellano JB, Revuelta JL, de las Rivas J, Hermoso JA. The 1.49 A resolution crystal structure of PsbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. J Mol Biol. 2005;350(5):1051–1060. doi: 10.1016/j.jmb.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 14.Ristvejová J, et al. Structure and dynamics of the N-terminal loop of PsbQ from photosystem II of Spinacia oleracea. Biochem Biophys Res Commun. 2006;345(1):287–291. doi: 10.1016/j.bbrc.2006.04.087. [DOI] [PubMed] [Google Scholar]
- 15.Ifuku K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol Biochem. 2014;81:108–114. doi: 10.1016/j.plaphy.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Liu H, et al. MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc Natl Acad Sci USA. 2014;111(12):4638–4643. doi: 10.1073/pnas.1323063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwabara T, Murata T, Miyao M, Murata N. Partial degradation of the 18 kDa of the photosynthetic oxygen-evolving complex: A study of a binding site. Biochim Biophys Acta. 1986;850(1):146–155. [Google Scholar]
- 18.Miyao M, Fujimura Y, Murata N. Partial degradation of the extrinsic 23 kDa protein of the Photosystem II complex of spinach. Biochim Biophys Acta. 1988;936(3):465–474. [Google Scholar]
- 19.Kuwabara T, Shinohara K. Association of extrinsic proteases with spinach Photosystem II membranes. In: Baltscheffsky M, editor. Current Research in Photosynthesis. Vol III. Kluwer; Dordrecht, The Netherlands: 1990. pp. 743–746. [Google Scholar]
- 20.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25(4):663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 21.Hermanson GT. Bioconjugate Techniques. Academic; San Diego, CA: 1996. [Google Scholar]
- 22.Ifuku K, Sato F. Importance of the N-terminal sequence of the extrinsic 23 kDa polypeptide in Photosystem II in ion retention in oxygen evolution. Biochim Biophys Acta. 2001;1546(1):196–204. doi: 10.1016/s0167-4838(01)00139-x. [DOI] [PubMed] [Google Scholar]
- 23.Ghanotakis DF, Topper JN, Babcock GT, Yocum CF. Water-soluble 17 and 23 kDa polypeptide restore oxygen evolution by creating a high-affinity site for Ca2+ on the oxidizing side of Photosystem II. FEBS Lett. 1984;170(1):169–173. [Google Scholar]
- 24.Miyao M, Murata N. Calcium ions can be substituted for the 24 kDa polypeptide in photosynthetic oxygen evolution. FEBS Lett. 1984;168(1):118–120. [Google Scholar]
- 25.Ghanotakis DF, Babcock GT, Yocum CF. On the role of water-soluble polypeptides (17,23 kDa) calcium and chloride in photosynthetic oxygen evolution. FEBS. 1985;192(1):1–3. [Google Scholar]
- 26.Ido K, et al. Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant photosystem II supercomplex. J Biol Chem. 2014;289(29):20150–20157. doi: 10.1074/jbc.M114.574822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31(13):3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Prot Sci. 2007;50:2.9.1–2.9.31. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 29.Ido K, et al. The conserved His-144 in the PsbP protein is important for the interaction between the PsbP N-terminus and the Cyt b559 subunit of photosystem II. J Biol Chem. 2012;287(31):26377–26387. doi: 10.1074/jbc.M112.385286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyao M, Murata N. The mode of binding of three extrinsic proteins of 33 kDa, 23 kDa and 18 kDa in the Photosystem II complex of spinach. Biochim Biophys Acta. 1989;977(3):315–321. [Google Scholar]
- 31.Kavelaki K, Ghanotakis DF. Effect of the manganese complex on the binding of the extrinsic proteins (17, 23 and 33 kDa) of Photosystem II. Photosynth Res. 1991;29(3):149–155. doi: 10.1007/BF00036218. [DOI] [PubMed] [Google Scholar]
- 32.Ifuku K, Sato F. A truncated mutant of the extrinsic 23-kDa protein that absolutely requires the extrinsic 17-kDa protein for Ca2+ retention in photosystem II. Plant Cell Physiol. 2002;43(10):1244–1249. doi: 10.1093/pcp/pcf136. [DOI] [PubMed] [Google Scholar]
- 33.Kakiuchi S, et al. The PsbQ protein stabilizes the functional binding of the PsbP protein to photosystem II in higher plants. Biochim Biophys Acta. 2012;1817(8):1346–1351. doi: 10.1016/j.bbabio.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Tovchigrechko A, Vakser IA. Development and testing of an automated approach to protein docking. Proteins. 2005;60(2):296–301. doi: 10.1002/prot.20573. [DOI] [PubMed] [Google Scholar]
- 35.Bricker TM, Morvant J, Masri N, Sutton HM, Frankel LK. Isolation of a highly active photosystem II preparation from Synechocystis 6803 using a histidine-tagged mutant of CP 47. Biochim Biophys Acta. 1998;1409(1):50–57. doi: 10.1016/s0005-2728(98)00148-0. [DOI] [PubMed] [Google Scholar]
- 36.Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- 37.Orban T, Gupta S, Palczewski K, Chance MR. Visualizing water molecules in transmembrane proteins using radiolytic labeling methods. Biochemistry. 2010;49(5):827–834. doi: 10.1021/bi901889t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura T, et al. Identification of the basic amino acid residues on the PsbP protein involved in the electrostatic interaction with photosystem II. Biochim Biophys Acta. 2014;1837(9):1447–1453. doi: 10.1016/j.bbabio.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 39.Tohri A, et al. Identification of domains on the extrinsic 23 kDa protein possibly involved in electrostatic interaction with the extrinsic 33 kDa protein in spinach photosystem II. Eur J Biochem. 2004;271(5):962–971. doi: 10.1111/j.1432-1033.2004.03998.x. [DOI] [PubMed] [Google Scholar]
- 40.Meades GD, Jr, et al. Association of the 17-kDa extrinsic protein with photosystem II in higher plants. Biochemistry. 2005;44(46):15216–15221. doi: 10.1021/bi051704u. [DOI] [PubMed] [Google Scholar]
- 41.Berthold DA, Babcock GT, Yocum CF. A highly resolved oxygen-evolving Photosystem II preparation from spinach thylakoid membranes. FEBS Lett. 1981;134(2):231–234. [Google Scholar]
- 42.Arnon DI. Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 1949;24(1):1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swaim CL, Smith JB, Smith DL. Unexpected products from the reaction of the synthetic cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate), DTSSP with peptides. J Am Soc Mass Spectrom. 2004;15(5):736–749. doi: 10.1016/j.jasms.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Kalkhof S, Sinz A. Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal Bioanal Chem. 2008;392(1-2):305–312. doi: 10.1007/s00216-008-2231-5. [DOI] [PubMed] [Google Scholar]
- 45.Mädler S, Bich C, Touboul D, Zenobi R. Chemical cross-linking with NHS esters: A systematic study on amino acid reactivities. J Mass Spectrom. 2009;44(5):694–706. doi: 10.1002/jms.1544. [DOI] [PubMed] [Google Scholar]
- 46.Smith PK, et al. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 47.Frankel LK, et al. Radiolytic mapping of solvent-contact surfaces in Photosystem II of higher plants: Experimental identification of putative water channels within the photosystem. J Biol Chem. 2013;288(32):23565–23572. doi: 10.1074/jbc.M113.487033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delepelaire P, Chua NH. Lithium dodecyl sulfate/polyacrylamide gel electrophoresis of thylakoid membranes at 4 degrees C: Characterizations of two additional chlorophyll a-protein complexes. Proc Natl Acad Sci USA. 1979;76(1):111–115. doi: 10.1073/pnas.76.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun G, Anderson VE. Prevention of artifactual protein oxidation generated during sodium dodecyl sulfate-gel electrophoresis. Electrophoresis. 2004;25(7-8):959–965. doi: 10.1002/elps.200305800. [DOI] [PubMed] [Google Scholar]
- 50.Frankel LK, Sallans L, Limbach PA, Bricker TM. Identification of oxidized amino acid residues in the vicinity of the Mn(4)CaO(5) cluster of Photosystem II: Implications for the identification of oxygen channels within the Photosystem. Biochemistry. 2012;51(32):6371–6377. doi: 10.1021/bi300650n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133–137. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Freitas MA. MassMatrix: A database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9(6):1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26(2):283–291. [Google Scholar]
- 54.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17(4):355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 55.Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.