Significance
Macrolide antibiotics inhibit translation by binding in the ribosomal nascent peptide exit tunnel. It was believed that macrolides interfere with protein synthesis by obstructing the egress of nascent proteins. In contrast to this view, the results of ribosome profiling analysis suggest that the main mode of macrolide action is context-specific inhibition of peptide bond formation. The ribosome with a macrolide molecule bound in the tunnel is impaired in catalysis of peptide bond formation between specific combinations of the peptidyl donors and aminoacyl acceptors, leading to interruption of translation when such problematic substrates are encountered. These findings underscore the existence of a link between the ribosomal tunnel and the peptidyl transferase center and pave the way for development of superior antibiotics.
Keywords: ribosome, antibiotics, macrolides, translation, peptidyl transferase
Abstract
Macrolides are clinically important antibiotics thought to inhibit bacterial growth by impeding the passage of newly synthesized polypeptides through the nascent peptide exit tunnel of the bacterial ribosome. Recent data challenged this view by showing that macrolide antibiotics can differentially affect synthesis of individual proteins. To understand the general mechanism of macrolide action, we used genome-wide ribosome profiling and analyzed the redistribution of ribosomes translating highly expressed genes in bacterial cells treated with high concentrations of macrolide antibiotics. The metagene analysis indicated that inhibition of early rounds of translation, which would be characteristic of the conventional view of macrolide action, occurs only at a limited number of genes. Translation of most genes proceeds past the 5′-proximal codons and can be arrested at more distal codons when the ribosome encounters specific short sequence motifs. The problematic sequence motifs are confined to the nascent peptide residues in the peptidyl transferase center but not to the peptide segment that contacts the antibiotic molecule in the exit tunnel. Therefore, it appears that the general mode of macrolide action involves selective inhibition of peptide bond formation between specific combinations of donor and acceptor substrates. Additional factors operating in the living cell but not functioning during in vitro protein synthesis may modulate site-specific action of macrolide antibiotics.
Macrolide antibiotics are among the most successful antibacterials and have been widely used for the treatment of serious infections. These drugs stop bacterial growth by inhibiting protein synthesis. Macrolides bind to the ribosome in the nascent peptide exit tunnel (NPET), a narrow conduit that the polypeptides assembled in the peptidyl transferase center (PTC) pass through on their way out of the ribosome (1–3). Binding of a macrolide molecule in the NPET at a short distance from the PTC obstructs the passage of the nascent peptides. Treatment of sensitive cells with macrolide antibiotics leads to a rapid decline of the overall protein synthesis and accumulation of peptidyl-tRNA (4). In the cell-free translation system, these drugs were shown to inhibit the synthesis of model polypeptides and cause early peptidyl-tRNA drop-off (5, 6). Based on these observations, it was largely assumed that macrolides indiscriminately inhibit the production of all cellular polypeptides by preventing the nascent chain egress and blocking translation at its early rounds (7).
This general view, which has prevailed over decades, has been challenged by several recent findings. Crystallographic studies have shown that macrolides, although significantly narrowing the NPET, nevertheless still leave sufficient room for the growing nascent protein chain, thereby raising the possibility that some proteins may bypass the constriction created by the drug (3). Indeed, progression of the drug-bound ribosome along mRNAs can be halted in vitro at codons distal to the beginning of the ORF (8). Some biochemical evidence argues that the action of macrolide antibiotics depends on the properties of the polypeptide being synthesized by the drug-bound ribosome as revealed by differential inhibition of in vitro translation of certain model and natural mRNA templates (8–10). Consistent with these observations, the sites of macrolide-induced programmed ribosome stalling at the regulatory leader ORFs, which control expression of the macrolide resistance genes, are defined by the sequence of the encoded nascent peptide (11–15). Importantly, the protein-specific action of macrolides is readily observed in vivo, because treatment of sensitive cells with even very high concentrations of antibiotics allows the continued production of a subset of cellular polypeptides (8).
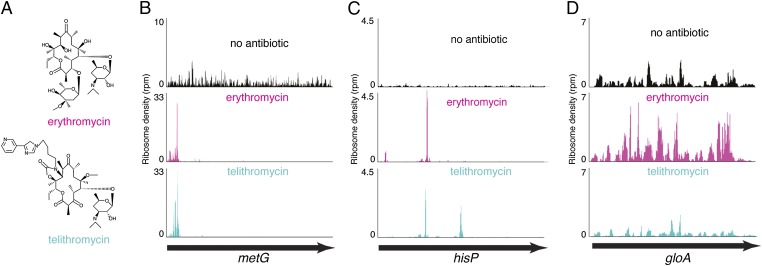
Despite the growing evidence that macrolides may affect protein synthesis in a sequence-specific manner, the general mode of action of these antibiotics remains obscure. Therefore, to understand the key principles of inhibition of translation by macrolides, we carried out genome-wide ribosome profiling analysis in Escherichia coli cells treated with two types of antibiotics and monitored drug-induced changes in the translation of individual genes. Erythromycin (ERY), one of the drugs used in our study, represents the prototype macrolide antibiotic, whereas the second drug, telithromycin (TEL), belongs to the newest generation of macrolides, called ketolides (Fig. 1A). The results of the ribosome profiling experiments and in vitro biochemical testing allowed us to define some of the amino acid sequence motifs that are difficult to synthesize for drug-bound ribosomes and to propose a generalized model for the action of macrolide antibiotics, alleviating discrepancies that confounded the previous models.
Fig. 1.
Gene-specific action of macrolide antibiotics. (A) Chemical structures of ERY and TEL antibiotics used in the profiling experiments. (B–D) Three common patterns of macrolide action observed in ribosome profiling analysis: (B) inhibition of translation at early stages, (C) translation arrest at the internal codons of the gene, and (D) translation through the entire length of the gene.
Results
Gene-Specific and Global Effects of Macrolide Antibiotics.
We used the ribosome profiling technique (16, 17) to analyze macrolide-induced redistribution of ribosomes on the cellular mRNAs. The exponentially growing cells of the antibiotic hypersusceptible E. coli strain (8) were exposed to ERY or TEL at concentrations exceeding the minimal inhibitory concentrations (MICs) by 100-fold. Under these conditions, the maximal inhibition of translation is achieved within the first 5 min upon addition of the drugs (8). However, we extended the incubation time with antibiotics for a total of 15 min to ensure that the drug-bound ribosomes reach the sites of translation arrest, which in some mRNAs could be hundreds of codons away. After rapid cell harvesting and lysis, the polysomal mRNA was digested by nuclease treatment, and the ribosome-protected mRNA fragments were subjected to next-generation sequencing (16, 17). The resulting reads, representing ribosomal “footprints” on mRNAs, were mapped to the E. coli genome, and distribution of ribosomes on individual ORFs was computed.
Antibiotic treatment resulted in a notable redistribution of ribosomes along most of the ORFs. However, the changes in ribosome density patterns differed significantly between individual ORFs. The genes could be loosely grouped into three major categories based on the changes in the ribosome pattern in response to macrolides (Fig. 1 B–D). One class included ORFs with an increase in ribosome occupancy of the 5′-terminal codons (Fig. 1B). This pattern, which was observed more frequently in the ERY-treated cells but was seen only rarely in cells treated with TEL, is reflective of either stalling of the ribosome at the initial rounds of translation or early peptidyl-tRNA drop-off and thus generally conforms to the conventional model of macrolide action. ORFs in the second category were characterized by the presence of one or several peaks of ribosome density within the internal codons, reflective of site-specific arrest of translation after the nascent peptide’s N terminus has bypassed the NPET-bound antibiotic (Fig. 1C). The distinct sharpness of the peaks readily attests to the context-specific nature of these “late” translation arrest events. Finally, the third category included the genes actively translated in the drug-treated cells. Such ORFs were characterized by continuous ribosome occupancy throughout their entire length, albeit with certain changes in the ribosome profiles as a result of the drug treatment (Fig. 1D). The conventional model of macrolide action fails to explain the ribosome profile patterns of the second and third categories.
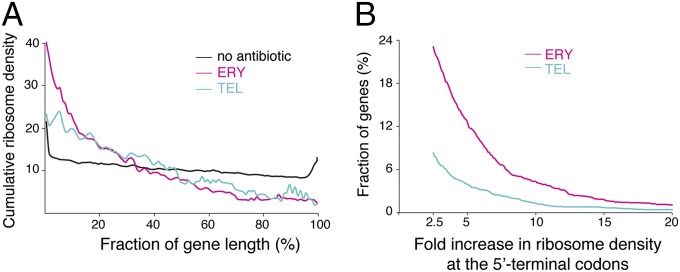
For obtaining a more general view of the global macrolide-induced redistribution of ribosomes along the ORFs, we carried out metagene analysis. To reduce the experimental noise imposed by the stochastic occurrence of ribosome footprints at the poorly translated genes, the metagene analysis was limited to the highly expressed ORFs in the drug-free and drug-treated samples (n = 1,081) (see Materials and Methods for details). The length of each gene was split into 100 segments and the cumulative relative ribosome density was calculated for each segment across all of the ORFs (Fig. 2A). The observed general pattern was dramatically different from what would be expected based on the conventional view of macrolide action, which presumes that these drugs act primarily at the early stages of translation. Although we did observe somewhat increased ribosome occupancy of the early codons, more pronounced in the ERY compared with the TEL sample, the ribosome density continued to remain high through the extended fraction of the ORF lengths, indicating that translation of many genes continues past the initial codons. A continuous steady decrease in ribosome occupancy through the ORF length (Fig. 2A) shows that a large number of proteins evade macrolide inhibition at the early rounds of their synthesis, but their translation is then inhibited at later stages, with the sites of translation arrest scattered throughout the length of the genes. A more rapid drop of the ERY curve compared with TEL suggests that the ERY-bound ribosomes more frequently encounter the sites where inhibition of translation takes place.
Fig. 2.
Metagene analysis. (A) Global redistribution of ribosome density through the length of the genes in macrolide-treated cells. Highly expressed genes were length-normalized, split into an equal number of length segments, and ribosome density within each segment was computed as a fraction of the total density within the gene. Cumulative ribosome density for each segment across all of the ORFs was plotted against normalized gene length. (B) A limited number of genes show increased ribosome occupancy of the early codons in macrolide-treated cells. The combined ribosome density within codons 2–15 of common highly expressed genes in the ERY/control (n = 2,056) or TEL/control (n = 1,922) samples was computed, and the fraction of genes (%) showing a specific increase in the ratios between antibiotic-treated and drug-free samples was plotted.
We assessed the fraction of proteins inhibited at the early stages of their synthesis by computing the number of ORFs showing an increase in relative ribosome occupancy of the 5′-proximal codons across common well-expressed genes in the ERY/untreated or TEL/untreated pairs of samples. Only 480 out of ∼2,000 genes showed a 2.5-fold or higher increase in ribosome density within the codons 2–15 in ERY-treated cells; even fewer genes (173 out of ∼1,900) in TEL-treated cells exhibited such a trend (Fig. 1B). Thus, the conventional model of macrolide action that presumes early interruption of protein synthesis fails to account for the effects observed with the majority of actively expressed E. coli genes, where the macrolide-bound ribosome continues translation beyond the 5′-terminal codons.
Macrolides Induce Context-Specific Ribosome Stalling.
The high peaks of ribosome occupancy at the defined codons of many ORFs reflect site-specific arrest of translation. To systematically identify sites at which such ribosome stalling takes place, we selected sites exhibiting significantly increased occupancy in drug-treated samples. Even with conservative cutoff values for gene-wide and codon-specific ribosome density, we identified 1,117 (ERY) and 1,057 (TEL) codons with at least 20-fold higher relative ribosome occupancy. To correlate the density peaks with the position of the corresponding mRNA codon in the ribosome, we had to take into account a known ambiguity in assigning the ribosome placement on the basis of ribosome profiling data in bacteria (17, 18). By analyzing the profiling signals in the drug-free sample at the termination codons and at the known stalling sites in the genome (e.g., the secM gene; Fig. S1), we concluded that in our experiments the assignment algorithm (17) identifies the mRNA codons located either in the P or A sites of the ribosome. To simplify the discussion, but bearing this ambiguity in mind, in the following sections we will consider the peaks in ribosome density to represent the P-site codons.
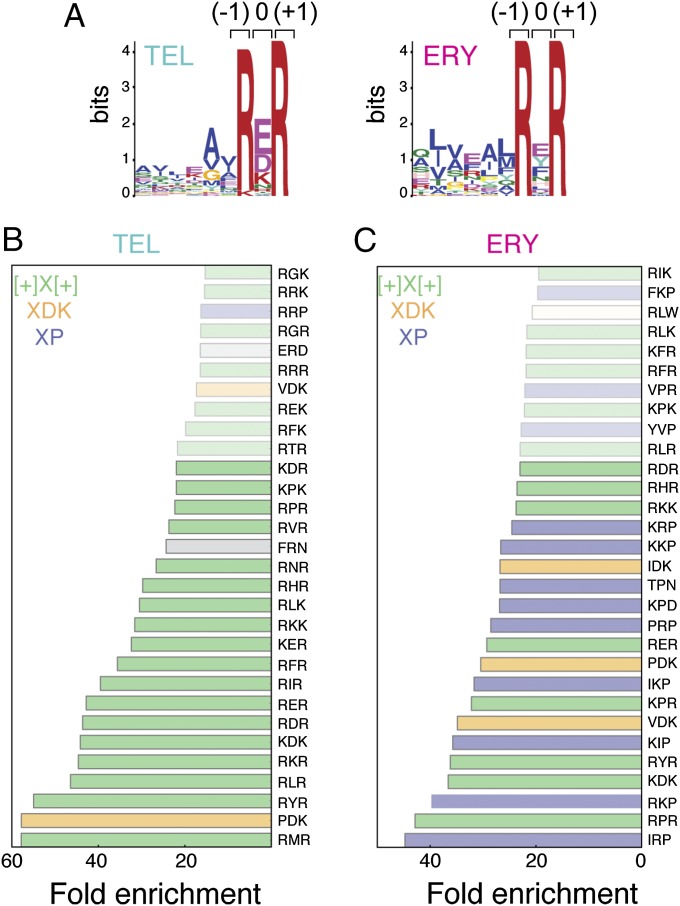
Previous analysis of the regulatory leader peptides of erythromycin resistance rRNA methyltransferase (erm) genes showed that macrolide-dependent programmed translation arrest depends on the sequence of the nascent chain preceding the stall site (8, 12–15, 19). Recent structural studies revealed that out of the C-terminal amino acids of the stalled nascent peptide, residues −8 to −2 (if the C-terminal residue is assigned position number 0) are in close proximity to the NPET-bound macrolide antibiotic (14, 20). Consequently, we anticipated that this segment of the nascent polypeptide chain that interacts with the drug would define the sites of macrolide-induced ribosome stalling. In search for the translation arrest signal, we compared the sequences of the nascent peptide segments in the vicinity of the drug-induced stalling sites. Specifically, we analyzed 11 amino acid-long sequences including 10 C-terminal amino acids of the stalled nascent peptide and the amino acid residue specified by the codon following the peak of the ribosome density corresponding to the incoming acceptor amino acid. Strikingly, comparison of the sequences did not show any significant conservation within the first eight positions of the analyzed segment. However, the last three residues, which could be tentatively assigned to the two C-terminal amino acids of the nascent chain and the incoming A-site amino acid, showed notable enrichment in specific residues. The conservation was especially pronounced at the penultimate (–1) position of the nascent peptide and the incoming aminoacyl acceptor (position +1) (Fig. 3A).
Fig. 3.
Conservation of amino acid sequences at the sites of macrolide-induced translation arrest. (A) MEME Suite-based search for consensus sequences within the 11 amino acid segments derived from the strong arrest sites showed enrichment of specific amino acid residues only in the immediate proximity to the site of arrest. The shown plots represent the [+]X[+] consensus found in many sites of TEL- and ERY-induced arrest. The main peak of the ribosome density in the profiling data corresponded to the codon specifying the amino acid residue at position 0 tentatively located in the PTC P site. (B and C) Triamino acid motifs enriched in the sites of strong arrest induced by TEL (B) or ERY (C). The top 30 tripeptide motifs are shown; motifs showing at least 20-fold enrichment are shown in bold.
Having realized that macrolide-induced late translation arrest heavily relies on the triamino acid sequence centered at the peak of the ribosome density, we reanalyzed the strong arrest sites, computing the extent of enrichment of individual triamino acid sequences. The combined results of 11 and 3 amino acid sequence enrichment analysis showed that the most prevalent motif at the sites of ERY- and TEL-induced translation arrest conformed to the consensus [R/K]X[R/K] around the codon of the drug-induced high ribosome density. Within this motif, the first amino acid residue (presumably occupying the penultimate position of the nascent chain) and the incoming amino acids are Arg or Lys, whereas the nature of the nascent peptide C-terminal residue (“X”) residing in the P site of the PTC is less conserved. Because Arg and Lys are positively charged at neutral pH, we designate this motif as [+]X[+].
More than 80% of the top TEL stall sites conform to the [+]X[+] consensus (Fig. 3B). Among 80 possible amino acid combinations conforming to the [+]X[+] motif, some could direct macrolide-induced ribosome stalling more readily than others. Thus, triamino acid sequences containing Arg at positions –1 and +1 are more frequent at the sites of TEL-induced stalling than those containing Lys, whereas the charged (Asp, Glu, Lys) or nonpolar (Ile, Leu, Phe, Pro) amino acids are overrepresented at the C termini of the nascent peptides (X in the [+]X[+] motif) in the sites of arrest (Fig. 3 B and C). Although the [+]X[+] motif is more prevalent in the TEL arrest sites, it also accounts for ∼50% of the sites of significant ribosome stalling in ERY-treated cells (Fig. 3C). Other motifs, including those ending with Asp–Lys (XDK) or containing proline (XP), are found in many of the remaining strong ERY-induced stalling sites. It is noteworthy that a much more diverse array of motifs cause the ERY-bound ribosome to halt translation compared to the motifs that stall the TEL-occupied ribosome.
Stalling Motifs Define the Combinations of PTC Substrates Problematic for the Macrolide-Bound Ribosome.
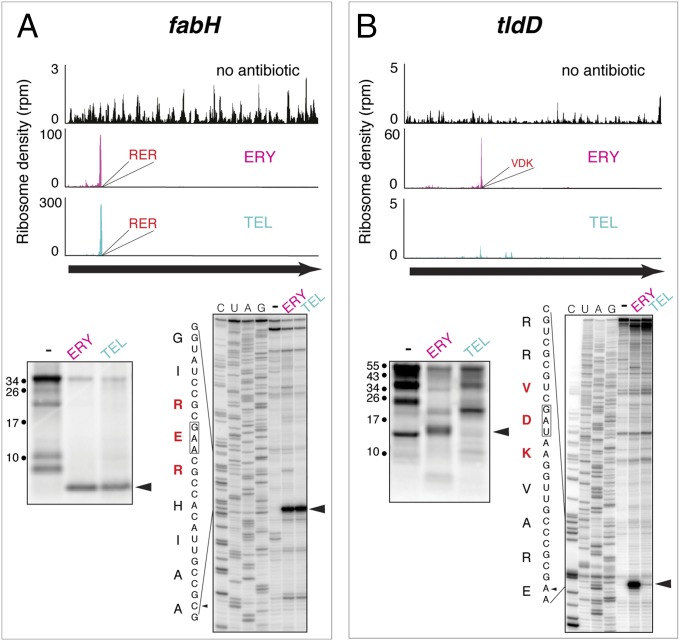
Due to the ambiguity of assigning exact ribosome placement on mRNA based on the profiling data, we wanted to define more accurately the position of the drug-stalled ribosome and thus better understand how the identified stalling motifs are related to the nature of the donor and acceptor substrates of the PTC. We chose several genes where well-defined peaks of drug-induced ribosome density were observed in the profiling experiment and analyzed antibiotic-dependent ribosome stalling in the cell-free translation system using the primer extension inhibition (toe-printing) technique (21). According to the profiling data, treatment of E. coli with ERY and TEL stalls the ribosomes at the RE41R site in the fabH ORF (Fig. 4A). When FabH was translated in the E. coli S30 cell-free system in the presence of ERY or TEL, accumulation of a truncated polypeptide with the expected size of ca. 4.5 kDa was observed. Toe-printing analysis unambiguously placed the fabH Glu41 codon in the P site of the stalled ribosome (Fig. 4A), indicating that antibiotics prevented the transfer of the 41 amino acid-long FabH nascent chain ending with Arg–Glu41 to the incoming Arg–tRNA. Similarly, a strong peak of ribosome density is present at the Glu358 codon of fusA ORF in the TEL-treated cells (Fig. S2). In our previous studies, we showed that the cell-free translation of fusA in the presence of TEL is arrested when the codon Glu358 enters the ribosomal P site (8), confirming that the tripartite [+]X[+] motif represents the last two amino acids of the nascent chain and the incoming aminoacyl acceptor.
Fig. 4.
Defining the exact sites of macrolide-induced ribosome stalling. Drug-dependent translation arrest within the [+]X[+] (RER) motif in the fabH ORF (A) or XDK (VDK) motif in tldD ORF (B). In each panel, the top images represent the ribosome density within the corresponding ORFs in the ribosome profiling experiments, the gel on the left shows fractionation of the [35S]-labeled protein products of translation of the ORF in the S30 cell-free system, and sequencing gel on the right shows the results of toe-printing analysis of ribosome stalling during translation in the in vitro system composed of purified components. Arrowheads show the main translation products and the main toe-printing bands corresponding to the peaks of the ribosome density in the profiling experiments.
The XDK motif is enriched at the sites of ERY-induced ribosome stalling (Fig. 3 B and C), and a strong peak of ribosome density is observed in the ERY-treated cells at the Asp142 codon, specifying the middle residue of the VDK sequence of the protein TldD (Fig. 4B). Toe-printing analysis placed the tldD Asp142 codon in the P site of the stalled ribosome, showing that ERY renders the catalysis of peptide bond formation between the nascent chain ending with Asp142 and the incoming Lys–tRNA highly inefficient.
For testing stalling at the XP motif, we chose the gene rseA, whose translation is inhibited in vivo by both ERY and TEL at Pro96 located within the RPW sequence (Fig. S3). Toe-printing analysis confirmed the placement of the Pro96 codon in the P site of the ERY- and TEL-carrying ribosome and thus established the inhibition of the reaction of transpeptidation between the P-site nascent chain ending with Pro96 and the A-site Trp–tRNATrp.
Although we verified in vitro the sites of drug-induced ribosome stalling for only several genes, combined with the previously published data (8, 14, 15), these results uniquely define the placement of the identified macrolide stalling motifs within the ribosome. An unexpected conclusion that emerges from these data is that the sites of macrolide-dependent translation arrest are defined primarily not by the sequence of the nascent chain juxtaposed with the antibiotic in the NPET but by the nature of the amino acid residues in the PTC.
Additional Cellular Factors Might Influence Macrolide-Induced Translation Arrest.
With many genes, the site-specific in vivo inhibitory action of macrolides could be readily reproduced in vitro. However, translation of a few other genes responded differently to macrolide presence in the living cell or in the cell-free translation system. For example, TEL caused ribosome stalling at the PD266K motif of the rfaD ORF in vivo, but no accumulation of the expected ∼30-kDa truncated polypeptide was noted in vitro (Fig. S4A). ERY and TEL cause strong ribosome stalling at the RL99R motif of clpX in vivo (Fig. S4B), but the corresponding ∼10-kDa truncated polypeptide was not observed during cell-free translation, perhaps due to inhibition of the clpX in vitro synthesis at the earlier codons. Similarly, although in vivo resistance to macrolides of some genes could be recapitulated in the E. coli S30 translation system, (e.g., yciT; Fig. S4C), some other tested proteins exhibited much higher sensitivity to the antibiotic in vitro (e.g., tpx; Fig. S4D).
A seeming discrepancy between the in vivo and in vitro data observed for some genes indicate that additional factors may stimulate or interfere with the ability of macrolide antibiotics to inhibit translation. The rate of ribosome progression along mRNA, protein secretion, or interaction of the nascent peptide with the chaperones or other proteins could be among the parameters that may influence antibiotic action in the living cell.
Discussion
The results of the genome-wide ribosome profiling analysis significantly redefine our understanding of the mode of action of macrolide antibiotics. In contrast to the previously prevailing view, and in agreement with the more recent proteomic and biochemical experiments, ribosome profiling clearly reveals these antibiotics as context- and thus protein-specific inhibitors of translation. Synthesis of the protein is interrupted not simply when the nascent chain reaches the site of the drug binding in the NPET but also when the drug-bound ribosome comes across a sequence of codons specifying specific combinations of amino acids.
A striking finding that emerged from the results of the profiling analysis and biochemical experiments is that the sites of macrolide-dependent translation arrest are defined primarily not by the sequence of the nascent chain juxtaposed with the antibiotic in the NPET but by the nature of the amino acid residues in the PTC: specifically, the C-terminal amino acids of the peptidyl donor and the incoming aminoacyl-tRNA acceptor (Figs. 3 and 4 and Fig. S3). Our previous studies have shown that binding of macrolide antibiotics to the ribosome alters the PTC structure and may interfere with peptide bond formation (12–15). Our new data expand these observations, indicating that macrolides, in general, inhibit translation by making certain combinations of the PTC substrates problematic for the drug-bound ribosome and arresting translation when such substrates are encountered. Combined, these results suggest that the primary mode of macrolide action may not be the obstruction of the NPET per se but context-specific interference with peptide bond formation. Such selective inhibition of the PTC activity can be promoted by the allosteric effects of the NPET-bound antibiotics upon the PTC and additionally assisted by constraining the placement of the nascent chain in the drug-obstructed tunnel.
The view of macrolides as selective PTC inhibitors helps to rationalize a number of previously reported results as well as observations that originated from ribosome profiling (this study) and proteomic experiments (8). If the antibiotic-bound ribosome encounters problematic PTC substrates during the early steps of translation when the nascent peptide is very short, the peptidyl-tRNA will be prone to dissociation, leading to peptidyl-tRNA drop-off (4–6). Some short nascent chain sequences may have higher affinity for the NPET, leading to early stalling of the ribosome similar to that observed at the leader ORFs of macrolide resistance genes (11, 13, 15). The combination of these scenarios would account for the increase in the relative ribosome occupancy of the early codons of some ORFs seen in ERY-treated cells (and to a lesser extent in the TEL-treated cells) (Fig. 2). Noteworthy, the studies of the early peptidyl-tRNA drop-off (7–9) have documented the phenomenon, but did not quantitatively analyze its contribution to the overall inhibition of production of individual proteins by macrolide antibiotics. Additional yet unknown factors influencing the initial threading of the N terminus of the nascent chain through the drug-obstructed NPET may contribute to the macrolide effects at the early stages of translation. Identifying these factors will require thorough structural, bioinformatic, and biochemical analyses.
In the absence of arrest sequences near the 5′ end of the ORF, threading of the nascent chain through the drug-obstructed NPET would stabilize the association of peptidyl-tRNA with the ribosome, so that if a problematic sequence is encountered at the later stages of translation, the ribosome will stall and remain stably associated with the mRNA, resulting in high peaks of ribosome occupancy at the internal codons. It is unclear whether peptidyl-tRNA drop-off or premature peptide release could contribute to the resolution of such stalled complexes. If none of the stalling sequences were encountered in the gene, the ribosome would successfully produce a complete protein.
The structure of the NPET-bound antibiotic defines the type and variety of the challenging PTC substrates (Fig. S5). Binding of a ketolide (TEL) antibiotic significantly inhibits peptide bond formation primarily (although not exclusively) between the [R/K]-X and the [R/K] substrates (the [+]X[+] motif; Figs. 3B and 4A). However, binding of ERY makes the PTC more restrictive by also slowing the peptidyl transfer reaction that involves the Asp donor and the Lys acceptor (XDK motif; Fig. 3C), or the reactions involving proline (the XP motif; Fig. S3). Because of a broader variety of the ERY arrest motifs, the ERY-bound ribosome stalls more frequently than the TEL-bound ribosome. As a result, the overall ribosome density is skewed more strongly toward the 5′ portions of the ORFs in ERY-treated cells, and fewer ERY-bound ribosomes reach the 3′-terminal segments of the ORFs (Fig. 2A). This result is also in line with our previous observation that more full-size proteins are synthesized in TEL-treated bacteria compared with those synthesized in cells exposed to ERY (8).
Our analysis was focused on the sites of the most severe macrolide-induced arrest and thus could detect only the “worst” combinations of the PTC substrates. It is likely that macrolides could also create a challenge for some other donor–acceptor pairs, with a range in the severity of the effects. This would lead to transient ribosome pausing at the corresponding sites and could account for redistribution of ribosome density seen on most of the genes in antibiotic-treated cells (Fig. 2C and Fig. S4 C and D). At the moment, it is unclear whether macrolide antibiotics simply accentuate the intrinsic difference in reactivity of the PTC substrates or impose an “unnatural” selectivity upon the PTC active site. Peptide bond formation is not rate-limiting for most of the ribosomal substrates during in vivo translation. In the absence of macrolides, the difference in reactivity of various donor–acceptor pairs would be obscured by slower steps of the elongation cycle. By making the catalysis of peptide bond formation generally less efficient, macrolide antibiotics may sufficiently slow the reaction so that it may become rate limiting for some combination of substrates. In this regards, it is worth noting that the frequent presence of proline residues in the ERY arrest sites (Fig. 3B) is reminiscent of proline motifs that are poorly translated even by the drug-free ribosomes (22, 23). Like with macrolide-dependent arrest, ribosome stalling at the proline motifs is influenced by the nascent chain residues proximal to the C terminus (24, 25). On the other hand, some other short motifs, which are known to stall drug-free ribosomes (26), were not enriched in the sites of macrolide-dependent stalling.
Although the amino acid residues at the very C terminus of the donor substrate and the aminoacyl-tRNA acceptor seem to play the primary role in macrolide-induced translation arrest, more distal segments of the nascent chain may modulate the efficiency of stalling. In some ORFs, arrest at even a strong stalling motif could be rather ineffective, allowing translation to continue. For example, TEL-bound ribosomes readily bypass the “best” TEL arrest sequence RM192R in fadH and stall at the subsequent arrest site (RE204R) instead (Fig. S6). Thus, although the “stalling” short sequence motif at the PTC of the translating ribosome is the primary determinant of macrolide-induced translation arrest, the local context possibly influences the interactions of the nascent chain with the NPET and antibiotic and may affect the drug action.
The context specificity of macrolide action revealed by ribosome profiling provides insights into the origin and evolution of inducible macrolide resistance controlled by programmed translation arrest of the regulatory ORFs of the resistance genes. Strikingly, many such ORFs contain the motifs that, according to our analysis, are the most problematic for the drug-bound ribosome. For example, the RLR sequence, which is among the top stalling motifs for the TEL- or ERY-bound ribosome (Fig. 3 B and C), is the site of translation arrest at ermDL and several other regulatory ORFs controlling macrolide resistance genes (15, 27). Translation of another leader ORF, ermBL, is halted when the ribosome reaches the VDK sequence, which matches one of the main ERY stalling motifs, XDK (Fig. 3C) (14). The proline-containing sequences, corresponding to the ERY motif XP, are found at the sites of stalling in erm leader peptides of the “RP” class (27). Therefore, it appears that regulatory ORFs of many macrolide resistance genes have evolved by incorporating the most efficient ribosome stalling motifs allowing for robust induction of resistance when cells are exposed to macrolides. Curiously, however, the IFVI and IAVV motifs found in the well-characterized ErmCL and ErmAL leader peptides do not appear among the ERY stalling sequences, nor did we observe ERY-dependent stalling at these motifs in cellular proteins. It is possible that the structural context more significantly affects drug-induced stalling at these sequences compared with the other motifs.
In conclusion, ribosome profiling, which provided a genome-wide view of translation inhibition by macrolides, yielded important insights into the mode of action of these antibiotics. Applying a similar genome-wide analysis to other protein synthesis inhibitors may lead to a more accurate understanding of the mechanisms of translation inhibition and provide new approaches to develop superior antibiotics.
Materials and Methods
Ribosome Profiling.
An antibiotic hypersusceptible E. coli strain, BWDK (8), was used for the ribosome profiling experiments. Exponentially growing E. coli cells were exposed to 100-fold MICs of ERY (100 μg/mL) or TEL (50 μg/mL) for 15 min, and the ribosome-protected mRNA fragments were isolated and sequenced using previously published protocols (17, 18). The resulting reads were mapped to the E. coli MG1655 genome, and the nucleotide-level ribosomal density was assigned using the described algorithm (17). More than 11 million reads have been mapped to the protein coding genes in each experimental sample.
Metagene Analysis.
Genes containing at least 16 codons and with a minimum gene-wide ribosome density of 50 in the three samples (ERY, TEL, and “no antibiotic”) were used for the metagene analysis (n = 1,081). Each gene was computationally split into 100 segments, and the relative ribosome occupancy at each segment was calculated by normalizing segment-specific density against the total ribosome density for the entire ORF. This value was added across the same segment in all 1,081 genes, resulting in a cumulative value of normalized ribosome density for each segment in every sample. The plot shown in Fig. 1B was then generated using moving average curve smoothing.
To identify the number of genes with increased ribosomal occupancy of the 5′-proximal codons, the combined ribosome density at codons 2–15 was calculated as a fraction of the total ribosome density within the entire ORF. Such relative ribosome occupancy of codons 2–15 was calculated for all well-expressed genes shared between ERY-treated and untreated cells or TEL-treated and untreated cells. The ratios of the values for the drug-exposed and untreated samples were computed and plotted.
Identifying Sites of ERY- and TEL-Induced Ribosome Stalling.
To identify codons with a significantly increased ribosome occupancy in ERY- and TEL-exposed cells, we used ORFs with a minimum ribosome density of 100 and, within these ORFs, selected codons with the minimum ribosome density of 5. “Codon density” was calculated by summing up the ribosome densities at each nucleotide position of the codon. To avoid bias resulting from higher ribosome density around the start and stop codons, each gene was divided into three segments: 5′-proximal 10 codons, 3′-proximal 10 codons, and the remainder of the gene. Codon density was converted into relative ribosome density per codon by normalizing against the total ribosome density in the gene segment to which the codon belongs (5′-terminal, 3′-terminal, or the rest of the gene). Ratios of the normalized ribosome density values (fold-increase/decrease) were calculated by comparing relative ribosome density per codon for ERY versus untreated or TEL versus untreated samples.
Identifying Amino Acid Context at the Site of Macrolide-Induced Translation Arrest.
Eleven amino acid segments, including the sequence encoded in the 10 codons up to the site of translation arrest and one codon following, were computationally extracted for the codons, showing a minimum of a 20-fold increase in relative ribosome density in antibiotic-treated samples. MEME Suite (28) was used for searching the conserved motifs, with a minimum cutoff of 50 occurrences of each motif.
To identify amino acid triplets enriched at the sites of antibiotic-induced stalling, the number of occurrences of every possible triamino acid motif in the five amino acid segment spanning positions –3 to +1 around the stall codons was calculated. This value was normalized against the frequency of occurrence of the triamino acid sequence in the E. coli proteome, resulting in the fold enrichment of individual motifs at the codons showing at least 20-fold increased ribosome density in the ERY or TEL samples.
In Vitro Translation.
The E. coli S30 cell-free transcription–translation system (Promega) was programmed with 0.5 pmol of PCR template carrying the gene of interest under the Ptac promoter. The 5-μL reactions were supplemented with 2 μCi [35S] Met (1,175 Ci/ mmol) (MP Biomedicals) and either no antibiotic or 50 μM of ERY or TEL. The reactions were incubated at 37 °C for 30–45 min and terminated by addition of 0.5 μg RNase A (Sigma-Aldrich) for 5 min at 37 °C. The translation products were precipitated with four volumes of acetone and resolved on a 16.5% (wt/vol) Tris⋅Tricine gel.
Primer extension inhibition analysis (toe-printing) was carried out as described in refs. 12, 29. The PCR-generated templates were transcribed and translated for 30 min at 37 °C in the PURExpress system (New England Biolabs) followed by primer extension.
Supplementary Material
Acknowledgments
We thank Nora Vazquez-Laslop for help, advice, and critical reading of the manuscript and Elizabeth Woods for proofreading. This work was supported by the Howard Hughes Medical Institute (J.S.W.) and National Institutes of Health Grant R01GM106386 (to A.S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61619).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417334111/-/DCSupplemental.
References
- 1.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107(40):17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlünzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413(6858):814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 3.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121(2):257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Menninger JR. Functional consequences of binding macrolides to ribosomes. J Antimicrob Chemother. 1985;16(Suppl A):23–34. doi: 10.1093/jac/16.suppl_a.23. [DOI] [PubMed] [Google Scholar]
- 5.Otaka T, Kaji A. Release of (oligo) peptidyl-tRNA from ribosomes by erythromycin A. Proc Natl Acad Sci USA. 1975;72(7):2649–2652. doi: 10.1073/pnas.72.7.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330(5):1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 7.Vazquez D. Inhibitors of Protein Biosynthesis. Springer-Verlag; New York: 1979. [DOI] [PubMed] [Google Scholar]
- 8.Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151(3):508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Hardesty B, Picking WD, Odom OW. The extension of polyphenylalanine and polylysine peptides on Escherichia coli ribosomes. Biochim Biophys Acta. 1990;1050(1-3):197–202. doi: 10.1016/0167-4781(90)90166-y. [DOI] [PubMed] [Google Scholar]
- 10.Starosta AL, et al. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol. 2010;17(5):504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39(4):797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30(2):190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Ramu H, et al. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41(3):321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Arenz S, et al. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sothiselvam S, et al. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA. 2014;111(27):9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh E, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147(6):1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484(7395):538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayford M, Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989;206(1):69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- 20.Arenz S, et al. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 22.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339(6115):85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 23.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339(6115):82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 24.Elgamal S, et al. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 2014;10(8):e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starosta AL, et al. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014;42(16):10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woolstenhulme CJ, et al. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110(10):E878–E887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(web server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orelle C, et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013;41(14):e144. doi: 10.1093/nar/gkt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.