Abstract
The AAA+ protease ClpAP, consisting of the ClpA chaperone and the ClpP protease, processively unfolds and translocates its substrates into its proteolytic core, where they are cleaved. Unfolding and efficient translocation require ATP-dependent conformational changes in ClpA’s D2 loop, where the conserved GYVG motif resides. To explore the role of the essential tyrosine of this motif, we investigated how two mutations at this residue (Y540C and Y540A) affect the rate at which the enzyme processes unstructured substrates. The mutations decrease ClpA’s ability to process unfolded or unstable protein substrates, but have only mild effects on the rates of ATP hydrolysis or hydrolysis of small peptide substrates. The mutants’ substrate binding properties were also characterized, using single molecule fluorescence microscopy. The single-molecule studies demonstrate that the conserved tyrosine is essential for formation of the pre-hydrolytic, high substrate affinity conformation observed in wild-type ClpA. Together, the results support a model in which destabilization of the high substrate affinity conformation of ClpA makes translocation less efficient and uncouples it from ATP hydrolysis.
AAA+ proteins are a ubiquitous set of ATP-dependent molecular machines that participate in multiple processes including unwinding, disassembly, denaturation, and proteolysis of macromolecule substrates (1). Members of this large superfamily contain one or two AAA+ modules, each of which incorporates Walker A and B motifs that are responsible for ATP binding and hydrolysis and various other elements that sense the nucleotide occupancy of the ATP-binding site (1, 2). The Clp/Hsp100 subfamily of AAA+ proteins is responsible for ATP-dependent protein unfolding and proteolysis in bacteria and eukaryotes (3, 4). Like other members of the AAA+ superfamily, Clp/Hsp100 family members use the energy of ATP hydrolysis to perform mechanical work. This energy conversion requires ClpA/P conformational changes, but the exact mechanisms of these changes are not yet clear.
ClpAP is a Clp/Hsp100 barrel-shaped protease consisting of the serine protease ClpP flanked by two hexameric ClpA rings (5). Each ClpA monomer subunit contains two AAA+ modules in addition to a flexible, largely helical N-domain. The ClpA hexamer binds its substrates and processively unfolds and translocates them into ClpP's central cavity, where they are proteolyzed (6, 7). Addition of the ssrA undecapaptide (AANDENYALAA) to a protein is sufficient for substrate recognition and subsequent degradation by ClpAP (8, 9), and the ssrA peptide is by itself a ClpAP substrate (10). Binding of ATP or the non-hydrolyzable analog ATPγS at the first of ClpA's two AAA+ modules allows for self-assembly of the ClpA hexamer, but active ATP hydrolysis at the second AAA+ module is required for substrate unfolding and translocation (11).
Previous single-molecule studies have shown that ClpA switches between a pre-hydrolytic conformation with high affinity for substrate and a post-hydrolytic low-affinity conformation (10). Recent structural evidence suggests that one difference between these two conformations involves the central pore D2 loop of the second AAA+ domain, consisting of residues 520-542. Hexameric models based on crystallographic studies of ADP-bound ClpA show the D2 loop in a "down" position (possibly stabilized by a bound magnesium ion), pointing towards the ClpP binding face (12, 13). Synchrotron protein footprinting characterization of ATPγS-bound ClpA suggests this loop takes on an alternate "up" position, perhaps while interacting with a sensor region in the first AAA+ domain (14). Repetitive cycling between “up” and “down” conformations has been hypothesized to mediate substrate translocation by ClpA (13). Similar nucleotide-dependent conformational changes have been reported in other ATP-dependent proteases (15). The D2 loop in question contains a highly conserved GYVG motif also present in other AAA+ unfoldases (16). In the case of ClpA, substrates interact directly with the D2 loop, and two non-conservative mutations at the tyrosine in this motif, Y540C and Y540A, cause severe defects in unfolding and translocation but allow binding of an ssrA-tagged substrate (13). Mutations at the equivalent site in other Clp/Hsp100 family members, including ClpX (Y153) (17), HslUV (Y91) (18), and ClpB (Y653A) (19) cause similar phenotypes, although it is still unclear how this conserved aromatic residue carries out its essential role in substrate denaturation and translocation.
We further investigated how ClpA's D2 loop and Y540 in particular contribute to translocation by examining the ability of the Y540C and Y540A mutants to process unfolded and poorly structured protein substrates as well as short peptides. We then used single-molecule experiments to demonstrate that the conserved tyrosine is essential for stable formation of the observed pre-hydrolytic high-affinity state. The enzyme’s ability to process folded substrates is thus correlated with the ability to stabilize this ATP-bound high substrate affinity state.
MATERIALS AND METHODS
Preparation of Protein Samples
Wild-type ClpA-FLAG, K501R-FLAG ClpA and ClpP-His6 (referred to here as ClpP) were prepared as previously described (10, 20). Standard PCR techniques were used to prepare plasmids containing the Y540A and Y540C mutations, with the primers 5'-T ATT GGT GCG CCT CCG GGA GCC GTT GGT TTT GAT CAG-3' and 5'-GGT GCG CCT CCG GGA TGC GTT GGT TTT GAT-3', respectively. DNA sequencing was used to verify the resulting constructs' identities in each case. As the FLAG tag (DYKDDDDKI) is present at ClpA’s N-terminus, the mutated tyrosine is the 549th residue, but we have maintained the Y540A and Y540C nomenclature to maintain consistency with previous published reports of these mutant ClpA proteins. The FLAG-tagged ClpA Y540A and Y540C proteins were each prepared in the same manner as the wild-type and the K501R proteins.
Concentrations of ClpA are expressed as concentrations of hexamers, while concentrations of ClpP are expressed as concentrations of tetradecamers.
Gel-based Assays
Gel-based assays were conducted to determine the extent of ClpA autodegradation and of casein degradation. ClpP (0.7 µM), ATP (10 mM), phosphocreatine (30 mM), and creatine phosphokinase (0.05 units/mL) were mixed in a Buffer A (sodium chloride (100 mM) magnesium chloride (20 mM), glycerol (5% w/v), and HEPES (50 mM, pH 7.5)) at 37°C. To determine the extent of the reaction in the presence of a non-hydrolyzable ATP analog, 3 mM ATPγS was used in lieu of ATP and phosphocreatine. α-Casein (25 µM) was used for caseinolysis assays. All reactions were initiated with addition of ClpA (0.7 µM). At specified time points, samples were removed from the reaction mixture, mixed with an equal volume of 2× SDS buffer (0.1M TrisCl/SDS, pH 6.8, 20% (w/v) glycerol, 4% (w/v) SDS, 3.1% (w/v) DTT, and 0.001% (w/v) bromphenol blue), and immediately placed in a boiling water bath for five minutes both to quench the reaction and to prepare the sample for gel electrophoresis. Samples were run on 8% bis/acrylamide gels at 30 mA for approximately one hour. Gels were stained with Coomassie Brilliant Blue R and destained with a mixture of 7% (v/v) acetic acid and 5% (v/v) methanol in water. Gel images were collected digitally using a CCD camera (AlphaImager 2200).
BODIPY Fluoroscein Casein Degradation Assays
To measure the kinetics of casein degradation, BODIPY FL-labeled casein (Invitrogen) was used as a substrate in a fluorescence-based assay. The substrate was incubated with ATP in Buffer A at 37°C for five minutes; a mixture of ClpA and ClpP in buffer A was incubated separately at the same temperature for the same period of time. To initiate the reaction, the two mixtures were combined for a final composition of 20 mM ATP, 200 nM ClpA, 100 nM ClpP, 20 mM magnesium chloride, 5% (w/v) glycerol, and 100 mM sodium chloride, buffered with 50 mM HEPES at pH 7.5, with concentrations of the labeled casein substrate ranging from 0 to 250 µg/mL. The fluorescence of the individual BODIPY FL moieties is quenched when the casein molecule is intact, but upon cleavage of the casein substrate, fluorescence is detectable. The excitation and detection wavelengths used in the assays were 485 nm and 530 nm, respectively. Apparent kcat and KM values were determined by measuring rates of fluorescence increase over the first 60 seconds after reaction initiation. For assays determining the caseinolytic activity of partially autodegraded ClpAP, the reactions were carried out in the same manner, but 20 mM ATP was incubated with the ClpA and ClpP components prior to addition of the substrate/ATP mixture. For these assays, the BODIPY FL-casein substrate concentration was 250 µg/mL (approximately 6-fold greater than the apparent KM).
ATPase Assays
ATPase activities of wild-type and mutant ClpA-FLAG were measured with a standard coupled enzyme assay. Wild-type or mutant ClpA (100 nM) was combined with phosphoenolpyruvate (7.5 mM), reduced β-nicotinamide adenine dinucleotide (NADH, 0.2 mM), ATP (5 mM), pyruvate kinase (9.3 units/mL), and lactate dehydrogenase (10.7 units/mL) in a buffer consisting of 50 mM HEPES at pH 7.5, 300 mM potassium chloride, 40 mM magnesium chloride, 0.1% Nonidet P40 (CAS 9016-45-9), and 10% (w/v) glycerol. Assays measuring the ATPase rate after partial degradation of ClpA also contained 50 nM ClpP. Formation of ADP is coupled to conversion of NADH to its oxidized form, resulting in a loss of absorbance at 340 nm. Enzymatic ATPase rates were calculated from the change in absorbance.
Mass Spectrometric Analysis of Autodegradation Product
Partially autodegraded Y540A ClpA samples were prepared and run on an 8% bis/acrylamide SDS gel and Coomassie stained as described above. The apparent autodegradation product and the apparent intact ClpA bands were cut from the gel and submitted to the MIT Center for Cancer Research Proteomics Core Facility for enzymatic digestion and LC-MS/MS analysis. Separate analyses after a trypsin digest and after a GluC digest provided reasonable coverage of the intact ClpA sample (59%).
Peptidolysis Assays
ClpP cleavage rates of the fluorogenic substrate succinyl-Leu-Tyraminomethylcoumarin (succ-LY-AMC) were measured before and after ClpA partial autodegradation. ClpP (400 nM), ClpA (1 mM), ATP (10 mM), phosphocreatine (30 mM), and creatine phosphokinase (0.05 units/mL) were mixed in Buffer A. The succ-LY-AMC substrate was added (final concentration of 1 mM) simultaneously with the ClpA for one set of samples and five minutes after addition of ClpA for the second set of samples. Fluorescence readings (excitation and detection wavelengths 345 and 440 nm, respectively) were taken at 10-second intervals.
TIRF Experiments
Single-molecule total internal reflection fluorescence (TIRF) studies were conducted on the wild-type and mutant ClpA proteins to determine residence time distributions of a sample substrate. The substrate in these studies, Cy3-ssrA, consisted of the ssrA peptide (AANDENYALAA) with a Cy3 fluorescent dye moiety appended to its N-terminus. This substrate was prepared as described previously (10). The microscope set-up was designed and constructed in the laboratory of Professor Jeff Gelles (Brandeis University), who generously provided access to it for these experiments. The set-up (21) allows for the detection of fluorescence generated by TIR excitation of fluorophores near the slide surface. The slide functionalization and data collection and analyses were performed as described previously (10). Briefly, wild-type or mutant FLAG-tagged hexameric ClpA was immobilized on a glass surface using anti-FLAG antibodies in the presence of ATPγS. The Cy3ssrA substrate molecule was perfused onto the surface in the presence of an oxygen-scavenging system (consisting of glucose, glucose oxidase, catalase, and β-mercaptoethanol) used to decrease photobleaching. Video images were taken at 35 ± 1 ms/frame and then analyzed using publicly available scripts developed in the laboratory of Prof. David Weitz (Harvard University) (http://www.seas.harvard.edu/projects/weitzlab/matlab/latest_code/) and home-written software to determine the length of residence time of single fluorophores. The resulting distributions were fitted using a maximum likelihood fitting algorithm in the QuB software suite (22). Using the data and a user-defined model, this algorithm finds the values of the rate constants in the model that are most likely to produce the observed data. This type of fitting algorithm is commonly used in analysis of single molecule data and is especially useful in determining whether an additional state should be added to a model in order to obtain a better fit to the observed data.
Bulk Cy3-ssrA Degradation Assay
An HPLC-based assay was used to measure the steady state rates of Cy3-ssrA degradation. Reaction mixtures contained ClpA (200 nM), ClpP (100 nM), ATP (10 mM), Cy3-ssrA (1 µM), aminomethylcoumarin (AMC, 25 µM, serving as an internal standard detectable via absorption at 344 nm), DTT (0.5 mM), sodium chloride (300 mM), magnesium chloride (20 mM), glycerol (10% w/v), and HEPES (50 mM, pH 7.5). Samples were taken from the reaction mixtures at various time points and quenched with guanidinium hydrochloride, to a final concentration of 3 M; the quenched samples were then analyzed by reverse-phase C4 chromatography. Initial enzymatic rates (vo) were calculated based on the appearance of the major product peak, which was quantified using its integrated absorbance at 550 nm and normalized to the integrated absorption of the AMC peak at 344 nm. Values for apparent kcat/KM were calculated as kcat/KM = vo/([ClpA6]i[Cy3-ssrA]i).
RESULTS
Enzymatic Properties of D2 Mutants
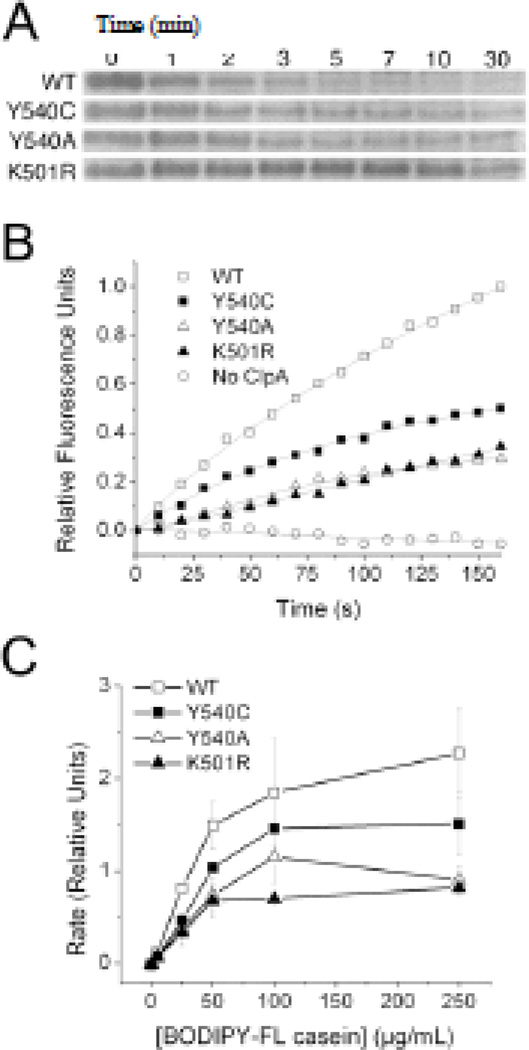
We prepared N-terminally FLAG-tagged versions of ClpA containing the Y540A and Y540C D2 loop mutations, as well as a wild-type FLAG-tagged ClpA. Addition of the FLAG peptide at the N-terminus had no significant effect on ATPase activity of wild-type ClpA (data not shown). Previous studies showed that these mutants bind but are unable to unfold GFP-ssrA (13), a highly stable substrate that wild-type ClpA readily processes. To determine whether these mutants' catalytic defects are less severe with less stable substrates, we tested their activity on casein, a ClpA substrate that lacks a stable tertiary structure (23). The D2 mutants were able to proteolyze α-casein, albeit at a much slower rate than wild-type ClpA, as qualitatively determined by gel electrophoresis data (Figure 1a).
Figure 1.
Enzymatic Properties of ClpA D2 Loop Mutants. A) Casein bands from gel electrophoresis samples show that wild-type ClpAP degrades α-casein more rapidly than ClpA(Y540A/C)P mutants, which in turn process the substrate slightly faster than ClpA(K501R)P. Numbers above gel lanes indicate time in minutes at which samples were quenched. B) Fluorescence emission at 530 nm for reaction mixtures containing 250 µg/mL BODIPY FL casein; fluorescence increases upon substrate cleavage by ClpAP. The D2 loop mutants degrade the BODIPY FL casein substrate at a slower rate than does wild-type ClpA. C) Plot of rate as a function of substrate concentration for BODIPY FL casein degradation by wild-type and mutant ClpAP. Error bars are the standard error in the mean for three trials.
To determine the kinetics of caseinolysis by the ClpA D2 mutants more quantitatively, we used a fluorogenic BODIPY FL casein assay. The BODIPY FL casein packaged in the commercially available assay is heavily labeled with BODIPY FL moieties, the fluorescence of which is quenched when the casein molecule is intact. Proteolytic cleavage of the labeled casein molecule reduces intramolecular quenching, resulting in increased fluorescence (24). The ClpA D2 mutants were able to process BODIPY FL casein, but at a slower rate than the wild-type ClpA (Figure 1b). Using this assay, we determined the steady-state kinetic parameters of the ClpA mutants' processing of BODIPY FL casein (Figure 1c). We also determined the ability of a K501R mutant to process the BODIPY FL casein substrate. The K501R mutation has previously been shown to have a dramatically reduced ATP hydrolysis rate (11), and any caseinolytic ability this mutant possesses is assumed to stem from ClpPpore opening rather than active translocation. Because the Y540A mutant's Vmax (0.93 ± 0.13 relative fluorescence units per second (RFU/s)) is not significantly greater than that of the K501R mutant (0.78 ± 0.15 RFU/s), we conclude that the majority of the Y540A mutant's ability to process the BODIPY FL casein substrate stems from its ability to open the ClpP pore and provide greater access to the ClpP proteolytic active sites. The Y540C mutant (Vmax = 1.49 ± 0.15 RFU/s), on the other hand, while still processing the BODIPY FL casein substrate more slowly than wild-type (Vmax = 2.53 ± 0.53 RFU/s), proteolyzes its substrate two-fold faster than the K501R mutant, suggesting that it retains at least some active translocation ability. This result is not surprising given that the Y540C mutant has some ability to unfold and proteolyze GFPssrA (13). The apparent Kms are the same within the experimental uncertainty for the wild-type (39 ± 16 µg substrate/mL) and mutant enzymes (K501R, 32 ± 16 µg substrate/mL; Y540C, 39 ± 7 µg substrate/mL; Y540A, 32 ± 8 µg substrate/µL). The lack of an observable effect on apparent Km indicates that the mutants have a relatively small net effect on the overall substrate affinity as averaged over all of the conformational states in the catalytic cycle, but does not rule out substantial effects on individual conformational states.
D2 loop mutants maintain ATPase activity
To evaluate whether the defects in substrate processing by the ClpA mutants were related to altered ATPase activity, we tested the mutants' ability to hydrolyze ATP in the presence or absence of substrate. In the absence of added substrate, the Y540C and Y540A mutants' ATPase rates (2.22 and 2.07 µmol/min/mg ClpA, respectively) are not significantly different from that of wild-type ClpA (2.31 µmol/min/mg ClpA). The ATPase rate of wild-type ClpA upon addition of the eleven amino acid ssrA peptide substrate is 91 ± 10% (s.e.m) of its rate in the absence of the substrate. This small reduction has been noted for other peptide substrates of the protein (25). Similarly, the Y540C and Y540A mutants' ATPase activities are also mildly reduced by the addition of the ssrA peptide substrate, to 76 ± 4% (s.e.m) and 75 ± 4% (s.e.m) of the rates in absence of substrate, respectively.
Incomplete autodegradation of D2 mutants
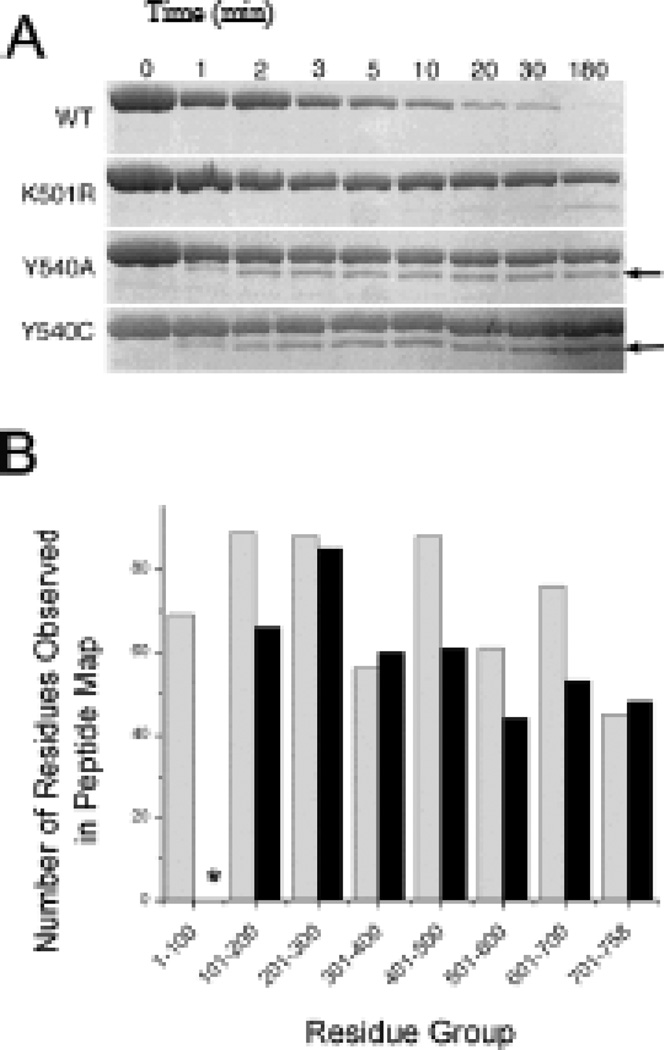
As an additional test of how the D2 mutations affect substrate processing, we examined autodegradation by these mutants. When paired with ClpP, wild-type ClpA has been reported to undergo ATP-dependent autodegradation (26, 27), and gel based assays confirm that wildtype ClpA is almost completely degraded within 5 minutes at 37° C (Figure 2a). Based on the results of casein and GFPssrA processing experiments, we expected to see low levels of autoprocessing of ClpA in the Y540C mutant and no activity in the Y540A mutant. Instead, both mutants begin to autodegrade, then stop. Concurrent with this initial loss of intact ClpA is the formation of an apparent autodegradation product of approximately 70 kDa (Figure 2a); this product does not form when the reaction is carried out with ATPγS (Supporting Information). The amount of autodegradation product formed increases for the first two minutes of the reaction, with no further change over the subsequent three hours. A band corresponding to the intact protein is present throughout, indicating that not all of the ClpA in the reaction mixture is partially degraded. Densitometric analysis of sequential reaction time points on Coomassie-stained gels indicates that the sum of the apparent autodegradation product and the intact mutant ClpA stays relatively constant over the course of the reaction (data not shown), consistent with the hypothesis that the product is derived from the intact enzyme.
Figure 2.
Autodegradation of ClpA D2 loop mutants. A) ClpA and autodegradation product bands from gel electrophoresis samples. Numbers above the gel lanes indicate time in minutes at which samples were quenched. ClpA runs at ~80 kDa; the autodegradation product formed by the Y540 mutants (indicated by arrows) runs at ~70 kDa. B) Bar graph indicating the number of residues observed in various sections of the peptide maps for an intact ClpA control (gray) and the autodegradation product (black). The asterisk indicates that no residues are represented in the autodegradation product’s 1–100 residue group.
Peptide mapping was used to identify the apparent autodegradation product after Edman sequencing failed to yield detectable signals. Samples of the apparent autodegradation product and the intact mutant ClpA-FLAG bands were analyzed by mass spectrometry after tryptic or Glu-C digestion. The two peptide maps yielded 59% coverage for the intact 767-amino acid ClpA-FLAG molecule (Supporting Information). The C terminus of the intact ClpA molecule was well-represented in the peptide map, with the peptide detected closest to the C-terminus being LTYGFQSAQKHKAE (residues 742-755). Likewise, the first N-terminal peptide of the intact ClpA control sample detected was HLLLALLSNPSARE (residues 29-42); this residue and other peptide residues near the N-terminus were matched with high confidence. The autodegradation product's C-terminus was also well-represented, with the same LTYGFQSAQKHKAE peptide being detected with high confidence. The initial N-terminal peptide detected (VTGANVLVAIFSEQE), however, corresponds to amino acid residues 103-117 in the intact protein. Thus, the autodegradation product appears to consist of the intact ClpA molecule minus approximately 100 N-terminal residues (Figure 2b, Supporting Information). Western blot analysis confirmed that the FLAG-tagged N-terminus was missing from the autodegradation product (data not shown).
Enzymatic activity of partially degraded D2 mutants
The observation that the partial autodegradation of the ClpA D2 mutants stops after the first few minutes of the reaction suggests that partial autoprocessing inhibits further degradation of substrates. The cessation of autodegradation after the first few minutes does not appear to be due to the decrease in ClpA concentration, since at the point where the rate has dropped to an undetectable level, the ClpA concentration has only decreased approximately two-fold. One possible mechanism of inactivation is simple pore blockage by a partially processed ClpA subunit, preventing degradation of other ClpA subunits. To investigate whether this might be the case, we probed the enzymatic activity of partially autodegraded ClpA towards other substrates.
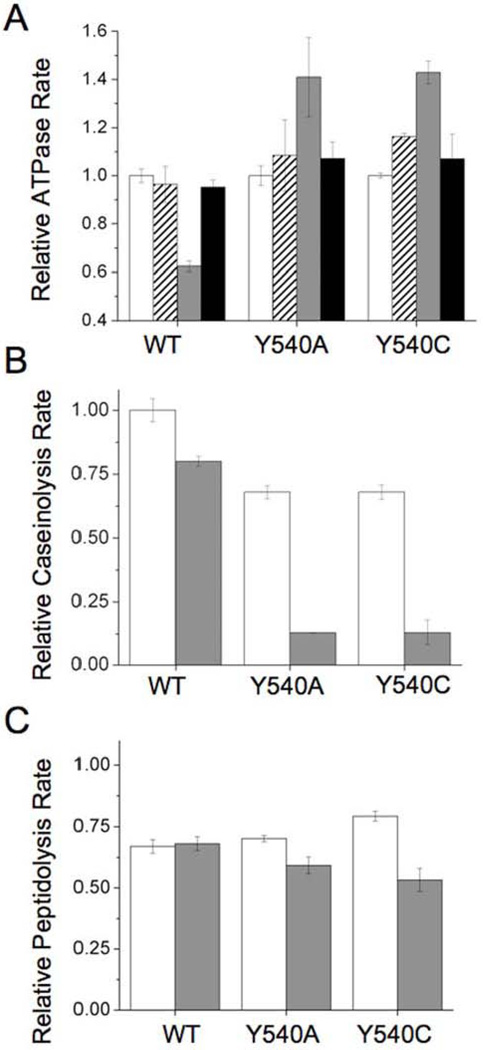
We first verified that partially autodegraded ClpA mutants maintain their ATPase activities. Addition of ClpP to ClpA does not immediately alter the ATPase activity of wild-type ClpA (Figure 3a). Upon incubation of ClpAP at 37°C for five minutes, wild-type ClpA's ATPase activity is reduced to ~60% of its original level; this is expected, as the wild-type ClpA undergoes extensive autodegradation during this time period. To verify that this loss of activity is ClpP-dependent, we tested the ATPase activity of ClpA incubated at 37°C in the absence of ClpP. The resulting value is not significantly different from ClpA's activity prior to incubation, suggesting that the loss observed in the presence of ClpP is indeed caused by proteolytic degradation of ClpA molecules. The Y540A and Y540C mutants behave somewhat differently under these conditions. Addition of ClpP initially marginally increases the ATPase activity; after five minutes, this increase becomes substantial, at ~140% of the pre-ClpP levels. Thus, partial autodegradation of ClpA D2 mutants slightly increases, rather than decreases, the protein's ATPase activity. As observed for wild-type ClpA, the activity of the mutants in the absence of ClpP remains constant over the course of the five-minute incubation period.
Figure 3.
Relative enzymatic rates before and after partial ClpA autodegradation. Error bars are the standard error in the mean for three trials. A) ATPase rates, normalized to the rate of ClpA at t=0. White bars: ATPase rate of ClpA alone in the absence of ClpP at t=0. Dashed bars: ATPase rate of ClpA upon addition of ClpP. Gray bars: ATPase rate of ClpA after a five minute-incubation at 37°C with ClpP. Black bars: ATPase rate of ClpA after a five minute-incubation at 37°C in absence of ClpP. Note that the minimum relative ATPase rate on this plot is 0.4. B) Caseinolysis rates, normalized to the rate of wild-type ClpA prior to autodegradation. White bars: caseinolysis rate of ClpP immediately after addition of ClpA. Gray bars: caseinolysis rate of ClpP after five minutes incubation with ClpA at 37°C. C) Peptidolysis rates, normalized to the rate of ClpP in the absence of ClpA. White bars: peptidolysis rate of ClpP immediately after addition of ClpA. Gray bars: peptidolysis rate of ClpP after a five minute-incubation with ClpA at 37°C.
We next investigated the protease activity of partially degraded ClpA mutants towards the less tightly structured α-casein substrate. Using the BODIPY FL casein assay described above, we tested the caseinolytic activity of ClpAP before and after a five-minute incubation at 37°C with ATP. Wild-type ClpAP was found to have 79 ± 2% of its caseinolytic activity after this incubation (Figure 3b). This preservation of activity is consistent with the significant reduction in ClpA autodegradation in the presence of casein (Figure S-3), presumably due to competition between free ClpA and casein for ClpA/P binding sites. In contrast, when paired with ClpP, the ClpA D2 mutants Y540A and Y540C have only 19.4 ± 0.3% and 18 ± 6%, respectively, of their caseinolytic activity after incubation period, despite the fact that the majority of the protein is not proteolyzed (Figure S-3). This result is consistent with a model whereby the ClpA entrance pore is blocked to some extent by partially degraded ClpA molecules, leaving little room for access of the unfolded substrate casein while completely preventing further proteolysis of stably folded ClpA. Previous work on ClpXP (28) suggests that simultaneous processing of multiple polypeptide chains is feasible in the Clp proteases. In the case of ClpAP, simultaneous processing of more than one polypeptide chain would allow for slow caseinolysis even in the presence of partially degraded ClpA.
Another possible explanation for this result is blockage not of the ClpA pore, but of the ClpP proteolytic active sites. To determine whether the ClpP active sites are accessible in the postautodegradation state, we tested the peptidolytic activity of ClpAP before and after partial autodegradation had occurred. ClpP readily cleaves small peptides in the presence or absence of ClpA, and previous studies have suggested that ClpP equitorial pores are large enough to allow passage of small peptides (29), providing access to proteolytic sites even if the ClpA cavity is blocked. Peptidolysis activity towards the well-studied fluorogenic succinyl-Leu-Tyr-AMC substrate for wild-type ClpAP remains the same before and after autodegradation (Figure 3c). Peptidolytic rates for the D2 loop mutants Y540A and Y540C decrease to 84% and 67% of their initial levels, respectively, after partial autodegradation; this decrease is significant but not as great as that seen in caseinolytic rates.
These data are consistent with a model of mutant ClpAP inactivation by partial blockage of both the ClpA entrance pore and ClpP proteolytic active sites.
D2 loop mutations destabilize peptide-enzyme interactions in the pre-hydrolytic state
High affinity substrate binding associated with the ATP-bound state of wild-type ClpA was previously observed using single-molecule total internal reflectance fluorescence (TIRF) experiments (10). We used the same method to investigate whether high affinity substrate binding was possible in translocation-deficient D2 loop mutants. We studied a substrate composed of the ssrA peptide (AANDENYALAA) with the fluorophore Cy3 covalently tagged to its N-terminus (Cy3-ssrA).
We first compared the steady-state kinetic parameters of Cy3-ssrA degradation by ClpP when associated with wild-type or mutant ClpA. Relative to wild-type ClpA, the apparent kcat/KM values for degradation by the ATPase-deficient K501R and the Y540A and Y540C variants are reduced (23 ± 1%, 10 ± 1%, and 10 ± 1% of wild-type values, respectively). These results suggest that wild-type ClpAP degradation of Cy3-ssrA occurs by active translocation, rather than by simple pore opening.
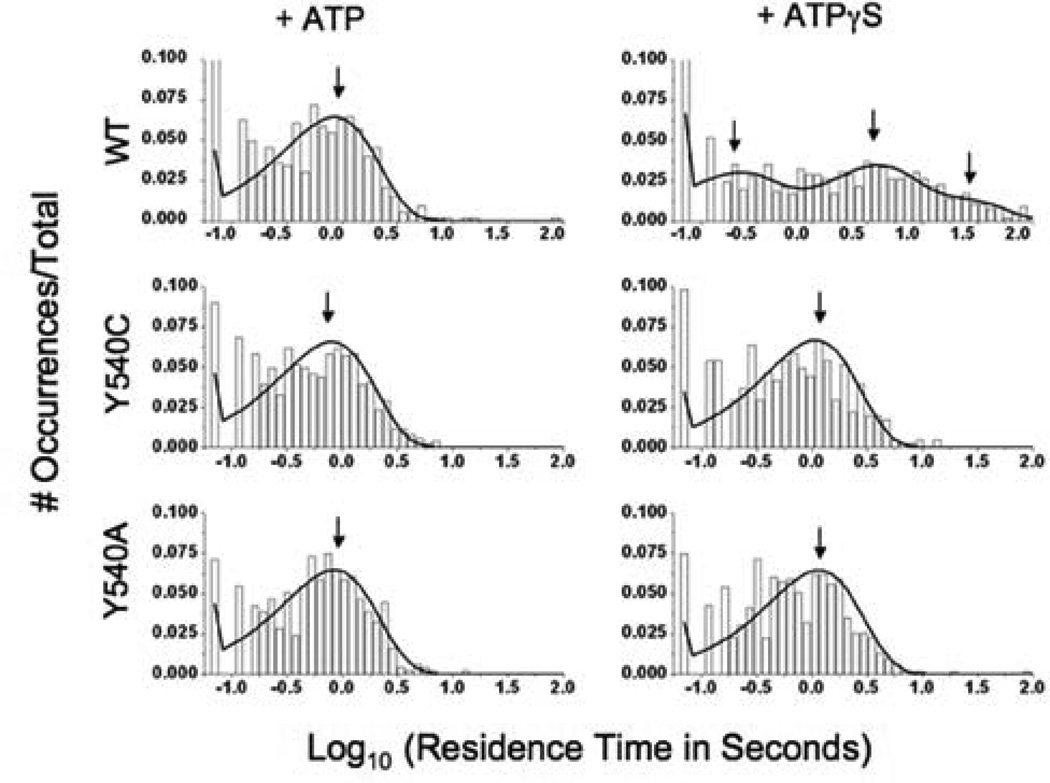
After verifying the difference in efficiencies between mutant and wild-type ClpAP in processing Cy3-ssrA, we turned to a single molecule TIRF assay to determine whether the mutations affect the stability of the enzyme-substrate complex. In this assay, wild-type or mutant FLAG-tagged ClpA is immobilized on a coated glass surface in the presence of the poorly hydrolyzable ATP analogue ATPγS; the Cy3-ssrA substrate and either ATP or ATPγS are then added to the slide. The previous study (10) demonstrated that photobleaching and non-specific substrate binding are minimal under the experimental conditions. Using a maximum likelihood fitting method (22), we found the residence time distributions of substrate on Y540A and Y540C to be best fitted by single exponential terms with lifetimes of 730 ± 40 ms and 770 ± 40 ms, respectively, in the presence of ATP (Figure 4), where the post-hydrolytic form of ClpA is expected to predominate in the steady-state. These lifetimes are only slightly longer than that of wild-type ClpA (650 ± 40 ms (10)), despite the wild-type protein's advantage in processing efficiency. We also measured the residence time distribution for these mutants in the presence of ATPγS to mimic the pre-hydrolytic form of the enzyme. As observed in the presence of ATP, the distributions in the presence of ATPγS were characterized by single kinetic components; the lifetimes of these components for the Y540A and Y540C mutants were 1090 ± 40 ms and 970 ± 50 ms, respectively. Thus, the mutants' affinity for substrate in the pre-hydrolytic state is only slightly greater than in the post-hydrolytic state. In contrast, addition of ATPγS to wild-type ClpA drives the enzyme into a number of states, two of which are much longer-lived (lifetimes of 3200 ± 490 ms and 20700 ± 2800 ms) than the enzyme-substrate complex observed in the post-hydrolytic state (10). These results suggest that the long-lived high affinity state observed in the wild-type is destabilized in the D2 loop mutants.
Figure 4.
Histograms indicating Cy3-ssrA residence times on wild-type (above), Y540C (middle), and Y540A (below) ClpA in the presence of ATP (left column) and ATPγS (right column). Maximum likelihood fits to the data are shown as black curves. Arrows indicate the average lifetimes of the major components of these fits.
DISCUSSION
Impaired Ability of ClpA Pore Loop Mutants to Degrade Unfolded/Disordered Protein Substrates
ClpA pore loop mutants have defects in their ability to process partially unfolded or unstructured protein substrates, consistent with the proposed role of this loop in substrate translocation. The ClpA pore loop mutants Y540C and Y540A process casein, a protein substrate with little or no stable tertiary structure, but at slower rates than the wild-type enzyme (Figure 1). Although unfolding of the substrate is not required for processive proteolysis of casein, processive translocation of the unfolded substrate is required for degradation of this substrate. These results thus support the hypothesis that substrate translocation activity is slower, but not eliminated, in the Y540C and Y540A mutants.
The phenotype of Y540C/A mutants in autodegradation is also consistent with the previously proposed roles of these mutants in protein unfolding and translocation (13). Neither of the Y540 mutants is capable of complete autodegradation. Both mutants are, however, capable of translocating and proteolyzing their N-termini to produce a ~70 kDa partial autodegradation product (Figure 2). The crystal structure of ClpA (12) shows that although the N-terminal 150 amino acids of ClpA have a well-defined tertiary structure, this region of the protein is composed of a series of helices and loops that are relatively loosely packed. This N-domain, which has functional roles in ClpP binding (13) and substrate binding/processing (30), has been shown to be highly flexible (31). Matouschek et al. have demonstrated that ClpA can process α-helices and surface loops much more readily than other elements of secondary structure (7). The facile processing of the ClpA N-domain by the ClpA pore loop mutants contrasts with the inability of these mutants to process stable substrates such as ssrA-tagged GFP (13). The observed partial autodegradation of the mutants is thus consistent with at least partial maintenance of the substrate translocation activity. The mutants’ inability to degrade ClpA past the N-domain may be due to the intrinsically more stable structure of the rest of polypeptide and/or to effects of removing the N-domain on the rest of the structure. These results also provide evidence that ClpA autodegradation proceeds from its N-terminus.
Evidence for Uncoupling of ATPase/Protease Activities from Substrate Translocation
Several alternative mechanisms might account for the role of Y540 in translocation and processive proteolysis. This residue might participate in activation or deactivation of the ClpA ATPase active site (with possible indirect effects on the ClpP protease active sites). Alternatively, this residue might be required for proper coupling of the ATPase and/or protease catalytic cycles to substrate translocation, without having any direct effect on the ATPase or protease activities themselves.
To help distinguish between these alternative models for the role of Y540 in translocation, we investigated the ability of Y540C and Y540A mutants to catalyze ATP hydrolysis and to support amide bond hydrolysis by ClpP. Y540C and Y540A have near wild-type levels of ATPase activity and peptidolysis activity (using the small substrate succinyl-Leu-Tyr-AMC, which does not require active translocation (25) (Figure 3)). The observations that the Y540 mutants retain wild-type levels of ATPase and peptidolysis activities but exhibit reduced proteolysis activities indicate that the peptidolytic and ATP hydrolytic activities are intact, but suggest that they have become decoupled from translocation.
Kinetic characterization of D2 mutants during autodegradation is also consistent with uncoupling of ATP hydrolysis and processive proteolysis. The ClpA mutants lose most of their ability to degrade casein when autodegradation can occur, suggesting that ClpA's axial pores are blocked by ClpA subunits bound as substrate (Figure 3b). However, under the same conditions, the mutants' ability to digest succinyl-Leu-Tyr-AMC is only slightly impaired and their ATPase activity is slightly enhanced (Figures 3c and 3a, respectively). These observations suggest that the mutants remain bound to substrate ClpA subunits after degradation of the relatively unstable N-terminus, and that the bound complexes continue to be competent for ATP hydrolysis and amide bond hydrolysis even though they are not competent for further autodegradation or caseinolysis.
Effects of Y540 Mutations on the Kinetic Stability of the Pre-Hydrolytic High-Affinity Substrate Binding Conformation
The previously proposed affinity-switch mechanism for translocation (10) suggests how the ATPase catalytic cycle might become uncoupled from translocation in the ClpA pore loop mutants. In this mechanism, the protein or peptide substrate binds to the D2 loop with high affinity at the apical ClpA pore in the pre-hydrolytic state, but with low affinity after the loop/substrate complex has moved through the pore following ATP hydrolysis. The motion of the substrate is therefore coupled to the ATPase catalytic cycle. However, if mutation of Y540 disrupts the loop-substrate interaction in the pre-hydrolytic state, efficient translocation will be impaired. Disruption of the high-affinity state would increase the probability that the substrate is released without being translocated, uncoupling ATP hydrolysis from substrate translocation.
Single-molecule fluorescence experiments support the hypothesis that mutations at Y540 destabilize the high-affinity, pre-hydrolytic loop-substrate complex. The Y540A and Y540C mutants in the ATPγS-bound state (mimicking the pre-hydrolytic conformation) exhibit substrate association times that are much shorter than those exhibited by wild-type ClpA. Faster dissociation of the substrate from the loop during loop movement would reduce the efficiency of translocation (defined as ktransloc/(ktransloc+kdissoc), where ktransloc is the translocation rate constant and kdissoc is the dissociation rate constant), thereby uncoupling active translocation from ATP hydrolysis. It is likely that substrate dissociation from the Y540 mutants in the active, nucleotide-hydrolyzing condition also occurs from the low-affinity ADP-bound state. However, the uncoupling of active translocation from ATP hydrolysis in the mutants suggests that the substrate often dissociates from the mutants prior to being moved through the ClpA central channel. As described in previous studies (10), rates of substrate binding to ClpA appear to be similar for ATP- and ATPγS-bound complexes, suggesting that the differences in kinetic stabilities observed are also correlated to differences in thermodynamic affinities.
The Y540A and Y540C mutations may also interfere with the D2 loop's ability to form an initial, pre-bound complex with substrate. Many biological binding reactions can be described as a pre-equilibration to form an initial, low-affinity complex, followed by a conformational change to form a stable complex (32, 33). Such a two-state binding mechanism has been suggested for ClpX, for which a metastable, short-lived intermediate has been observed during the processing of ssrA-tagged substrates (34). In the previous study on wild-type ClpA binding to Cy3-ssrA (10), a very short-lived state (τ = 170 ± 40 ms) was observed in the presence of ATPγS (but not ATP) and proposed to represent a low-affinity complex that equilibrates with the stable high-affinity complex. This short-lived state is not observed for the Y540C and Y540A mutants. (We note that some binding events take place on this time scale, but the overall distributions for both mutants are fitted best to single-exponential curves with τ ~ 1000 ms.). It therefore seems likely that the conserved tyrosine residue of the GYVG pore motif (Y153 in ClpX, Y540 in ClpA) contributes to formation of the short-lived pre-bound complex. Formation of a short-lived pre-bound complex may help the enzyme achieve a high rate of association with its substrate; if so, the inability of the loop mutants to form this complex would also contribute to their deficits in processing substrates.
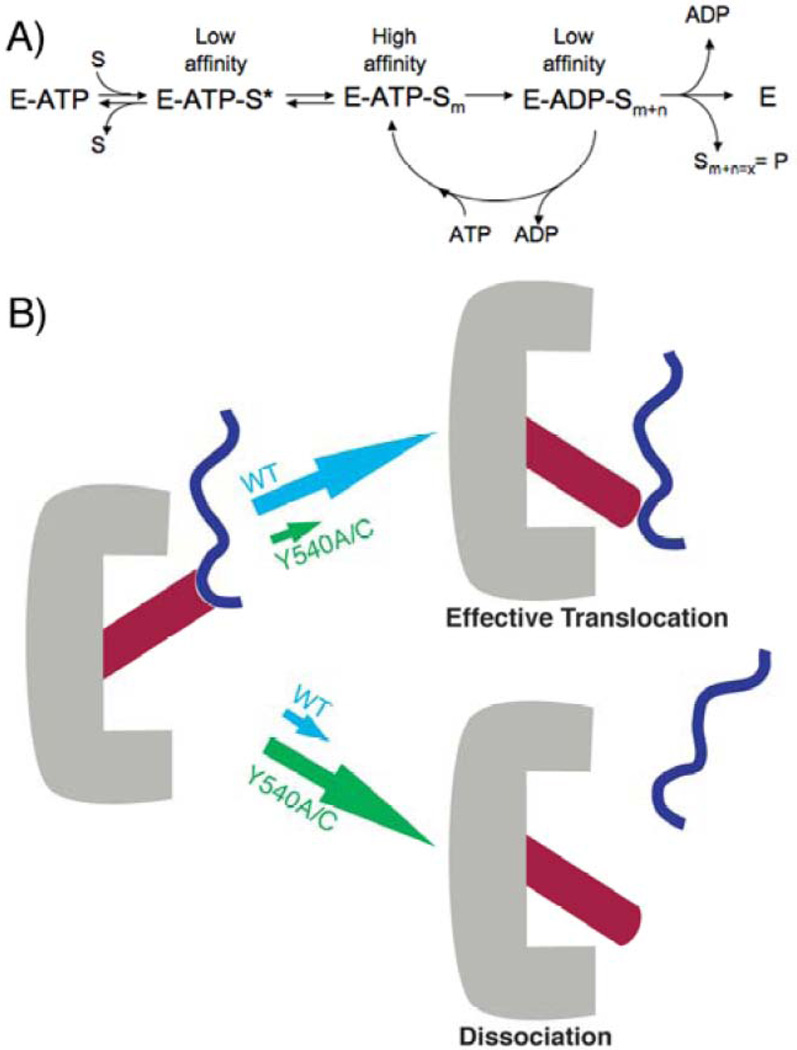
A Model for the Effects of Y540 Mutations on Translocation
The previous observation of wild-type ClpA's substrate affinity switch helped to explain its ability to processively unfold and translocate protein substrates. Here we have demonstrated that this affinity switch is perturbed in Y540 pore mutants, significantly destabilizing the pre-hydrolytic state. We hypothesize that this destabilization effectively uncouples ATPase activity from proteolysis (Figure 5). As in the wild-type ClpA, ATP hydrolysis in the mutants fuels the conformational changes between the states, but these conformational changes are decoupled from effective translocation due to a weaker "grip" in the pore loop mutants, thus increasing the probability of non-productive ATP hydrolysis. This model is also consistent with the observation that mutation of Y540 does not impair ATP hydrolysis and, under active turnover conditions, can even accelerate it.
Figure 5.
Model for substrate processing. A) The enzyme (E) binds ATP and then substrate to form a putative pre-binding complex E-ATP-S* that is in equilibrium with a higher affinity, more tightly bound complex E-ATP-Sm. Active ATP hydrolysis drives an iterative cycling process. Some cycles will result in an increase in the position ‘m’ (i.e., n>0), translocating the substrate partially through the central pore. Other cycles will result in ineffective ATP hydrolysis, such that n=0 and the position m does not change (i.e., the substrate is not moved with respect to the pore). Product is released when the substrate has been completely translocated through the central pore. B) Cartoon depiction of loop movement in ClpA. The loop movement conformational change associated with ATP hydrolysis may be coupled to an effective translocation step for substrate, or the substrate may dissociate before the conformational change, preventing effective translocation. Partitioning between these two possibilities is a function of the stability of the pre-hydrolytic state (i.e., how tightly the loop holds on to substrate in the ATP-bound state).
This model for the role of Y540 in translocation further suggests that interactions between Y540 and other regions of ClpA are important in maintaining the kinetic stability of the pre-hydrolytic complex. One possible region of interaction is the D1 sensor region, which has also been implicated as a substrate-binding region (13). Further structural and functional studies will be required to test this hypothesis. Because the catalytic cycle of ClpAP includes a large number of coordinated catalytic steps, mutations in the D2 loop may well have effects on microscopic steps throughout the catalytic cycle in addition to those observed so far. The D2 loop mutants may therefore be generally useful in studying how ClpA-substrate interactions function in the complex catalytic mechanism of ClpAP.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Prof. Jeff Gelles (Brandeis University) for use of the TIRF laser set-up and to Ioannis Papayannopoulos at MIT for helpful analysis and discussion of mass spectrometric data.
ABBREVIATIONS USED
- AAA+
ATPases associated with various cellular activities
- ATP
adenosine 5´-triphosphate
- ATPγS
adenosine 5’-O-(3-thiotriphosphate)
- ssrA
small stable RNA
Footnotes
- autodegradation data in the presence of ATPγS
- peptide maps of intact ClpA Y540A and its autodegradation product
- complete gel images for partial gels shown in manuscript
- sample video data for TIRF experiments This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:7–43. [PubMed] [Google Scholar]
- 2.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure, diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 3.Schirmer EC, Glover JR, Singer MA, Lindquist S. Hsp100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 4.Gottesman S, Maurizi MR, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 5.Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. Homology in structural organization between Escherichia coli ClpAP protease and the eukaryotic 26S proteasome. J. Mol. Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa T, Beuron F, Kessel M, Wickner S, Maurizi MR, Steven AC. Translocation pathway of protein substrates in ClpAP protease. Proc. Natl. Acad. Sci USA. 2001;98:4328–4333. doi: 10.1073/pnas.081543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 10.Farbman ME, Gershenson A, Licht S. Single-molecule analysis of nucleotide-dependent substrate binding by the protein unfoldase ClpA. J. Am. Chem. Soc. 2007;129:1237812379. doi: 10.1021/ja074168x. [DOI] [PubMed] [Google Scholar]
- 11.Singh SK, Maurizi MR. Mutational analysis demonstrates different functional roles for the 2 ATP-binding sites in ClpAP protease from Escherichia coli. J. Biol. Chem. 1994;269:29537–29545. [PubMed] [Google Scholar]
- 12.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 13.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Bohon J, Jennings LD, Phillips CM, Licht S, Chance MR. Synchrotron protein footprinting supports substrate translocation by ClpA via ATP-induced movements of the D2 loop. Structure. 2008 doi: 10.1016/j.str.2008.04.016. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Licht S, Lee I. Molecular machines: from conformational changes to substrate translocation and processivity. Biochem., 2008;47:3595–3605. doi: 10.1021/bi800025g. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Song JJ, Franklin MC, Kamtekar CS, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU protease-ATPase complex releal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 17.Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park EY, Rho YM, Koh OJ, Ahn SW, Seong IS, Song JJ, Bang O, Seol JH, Wang JM, Eom SH, Chung CH. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J. Biol, Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 19.Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 20.Choi KH, Licht S. Control of peptide product sizes by the energy-dependent protease ClpAP. Biochem. 2005;44:13921–13931. doi: 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- 21.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Susi H, Timasheff SN, Stevens L. Infrared spectra and protein conformations in aqueous solutions: the amide I band in H2O and D2O solutions. J. Biol. Chem. 1967;242:5460–5466. [PubMed] [Google Scholar]
- 24.Jones LJ, Upson RH, Haugland RP, Panchuk-Voloshina N, Zhou M. Quenched BODIPY dye-labeled casein substrates for the assay of protease activity by direct fluorescence measurement. Anal. Biochem. 1997;251:144–152. doi: 10.1006/abio.1997.2259. [DOI] [PubMed] [Google Scholar]
- 25.Maurizi MR, Thompson MW, Singh SK, Kim SH. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Meth. Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 26.Gottesman S, Clark WP, Maurizi MR. The ATP-dependent Clp protease of Escherichia coli : sequence of ClpA and identification of a Clp-specific substrate. J. Biol. Chem. 1990;265:7886–7893. [PubMed] [Google Scholar]
- 27.Seol JH, Yoo SJ, Kim KI, Kang MS, Ha DB, Chung CH. The 65-kDa protein derived from the internal translation iniation site of the ClpA gene inhibits the ATP-dependent protease Ti in Escherichia coli. J. Biol. Chem. 1994;269:29468–29473. [PubMed] [Google Scholar]
- 28.Burton RE, Siddiqui SM, Kim YI, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J., 2001;20:3091–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE. Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. USA. 2005;102:16678–16683. doi: 10.1073/pnas.0507370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo JH, Baker TA, Sauer RT. Characterization of the N-terminal repeat domain of Escherichia coli ClpA – A class I Clp/Hsp100 ATPase. Protein Science. 2001;10:551–559. doi: 10.1110/ps.41401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa T, Maurizi MR, Steven AC. The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J. Struct. Biol. 2004;146:180–188. doi: 10.1016/j.jsb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. The Enzymes. 1992;20:1–61. [Google Scholar]
- 33.Fersht A. Measurement and magnitude of enzymatic rate constants, in. Enzyme Structure and Mechanism. (2nd ed) 1985:121–154. [Google Scholar]
- 34.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.