Abstract
Our understanding of the antiviral actions of IFIT1, one of the most strongly induced interferon stimulated genes (ISGs), has advanced remarkably within the last few years. This review focuses on the recent cellular, biochemical, and structural discoveries that have provided new insight as to how IFIT1 functions as both a sensor and effector molecule of the cellular innate immune system. IFIT1 can detect viral RNA lacking 2′-O methylation on their cap structures or displaying a 5′-triphosphate moiety and inhibit their translation or sequester them from active replication. Because of these inhibitory actions, many viruses have evolved unique mechanisms to evade IFIT1 to facilitate replication, spread of infection, and disease pathogenesis.
Abbreviations: IFN, interferon; ISG, interferon-stimulated gene; IFIT, interferon-induced protein with tetratricopeptide repeats; PAMP, pathogen-associated molecular pattern; PRR, pathogen recognition receptor; TLR, toll-like receptor; IFNAR, interferon α/β receptor; TPR, tetratricopeptide repeat; IRF, interferon regulatory factor; ISRE, interferon stimulated response element; VSV, vesicular stomatitis virus; IAV, influenza A virus; EMSA, electrophoretic mobility shift assay; WNV, West Nile virus; JEV, Japanese encephalitis virus; DENV, dengue virus; SARS-CoV, Severe acute respiratory syndrome coronavirus; eiF4, eukaryotic initiation factor 4; HCV, hepatitis C virus; RVFV, Rift Valley Fever virus; 5′-ppp, 5′-triphosphate; IRES, internal ribosome entry site; VPg, viral protein genome-linked
Keywords: Interferon, Antiviral, 2′-O methylation, Immune evasion, Cap structure
1. Introduction
After virus infection, most mammalian cells develop an antiviral response that is triggered by detection of pathogen-associated molecular patterns (PAMPs), including single-stranded and double-stranded viral nucleic acids. Viral PAMPs are detected by specific host pattern recognition receptors (PRRs) including Toll-like receptors (TLR3, TLR7, TLR8 and TLR9), RIG-I-like receptors (MDA5 and RIG-I) and DNA sensors (cGAS, DAI, IFI16, DHX9, and DHX36) in the endosome and within the cytoplasm [1], [2], [3]. Binding of viral PAMPs to PRRs triggers signaling pathways that induce the expression of virus-responsive genes and antiviral cytokines (e.g., type I interferon (IFN)), which limit virus replication and shape adaptive immunity.
Type I IFNs comprise a family of functionally and genetically related cytokines, with IFNα and IFNβ the most extensively studied [4]. Type I IFN signaling is mediated through the heterodimeric IFNα/β receptor (IFNAR), which is composed of IFNAR1 and IFNAR2 subunits [5]. Signal transduction following the binding of type I IFN to IFNAR occurs via Janus kinase (JAK) and Signal transducer and activator of transcription (STAT) proteins and results in nuclear translocation of the transcription factor complex IFN-stimulated gene factor 3 (ISGF3, which is comprised of IFN regulatory factor 9 (IRF9) and phoshorylated STAT1 and STAT2), and induction of hundreds [6], [7], [8], [9] of different IFN-stimulated genes (ISGs). These ISGs encode distinct proteins with diverse biological functions that block multiple stages of the viral lifecycle including entry, translation, replication, assembly and spread. Additionally, some ISGs have immunomodulatory activity including effects on leukocyte recruitment and priming of adaptive immunity.
Until recently, most effort was focused on defining the mechanism of action of a very limited number of antiviral ISGs with well-established antiviral phenotypes (e.g., PKR, RNAse L, Mx1, and OAS). Recent ectopic expression and gene silencing screens [9], [10], [11], [12], [13] have identified many novel antiviral ISGs with inhibitory activity against different families of RNA and DNA viruses (reviewed in [14], [15]). Here, I describe the advances in our understanding of the broad-spectrum antiviral activity of one ISG, IFIT1, and how it functions both as a sensor and effector molecule to inhibit the disease pathogenesis of several virus families.
2. IFIT1 is a member of a family of inhibitory ISGs
2.1. IFIT1 gene family
IFIT1 is a member of a family of related genes that arose by gene duplication and are induced after type I IFN treatment or viral infection [16]. Four IFIT family members have been characterized extensively in humans (IFIT1 (also known as ISG56), IFIT2 (ISG54), IFIT3 (ISG60) and IFIT5 (ISG58)) and are localized to chromosome 10q23. Three members are expressed in mice: Ifit1 (Isg56), Ifit2 (Isg54), Ifit3 (Isg49) and are located on chromosome 19qC1. Additional less characterized yet highly related IFIT genes (IFIT1B (human) and Ifit1b, Ifit1c and Ifit3b (mouse)) in syntenic regions of the chromosome exist, although their functional significance remain undefined. A non-transcribed IFIT1-related pseudogene is present on human chromosome 13 [17]. IFIT genes share a similar genomic structure with most composed of two exons, with the second exon containing the vast majority of the coding sequence. IFIT1 gene homologs exist in other mammalian species as well as birds, fish and amphibians (reviewed in [18]).
IFIT1 proteins localize within the cytoplasm and lack any enzymatic domains or activity. They contain multiple tetratricopeptide repeats (TPR); this motif is composed of 34 amino acids that adopt a helix–turn–helix structure and mediate protein–protein interactions. Proteins containing TPR motifs regulate the cell cycle, transcription, protein transport, and protein folding [19]. Different IFIT family members have distinct numbers and arrangements of TPR motifs (Fig. 2B), which likely dictate unique functions; for example, IFIT1 has six and IFIT2 has four. Human and mouse IFIT1 share a sequence identity of 53% at the amino acid level, and have varied relatedness (∼38–57%) to other IFIT homologs within a given species. Although the sequence similarity between the different IFIT proteins within a species is high [18], their distinct effects on replication of individual viruses suggest they serve non-redundant functions in the host response to viral infections [20], [21], [22].
Fig. 2.
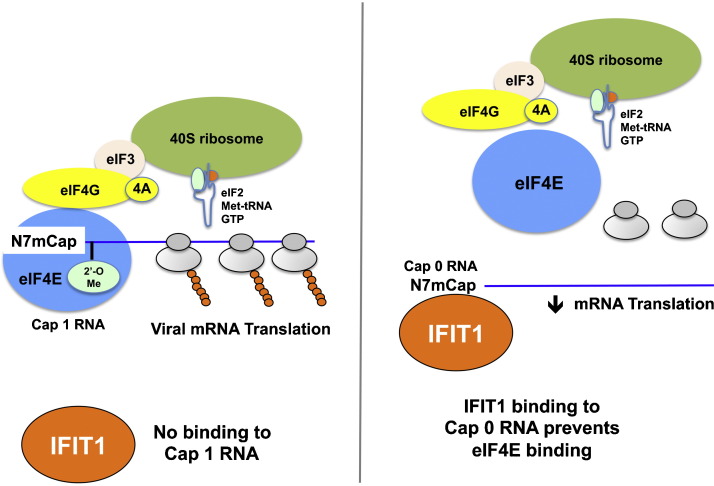
IFIT1 inhibits translation initiation by competing with eIF4E for binding to viral mRNA lacking 2′-O methylation. Assembly of the ribosome initiation complex at the 5′ end of a viral mRNA. eIF4E, as part of the eIF4F complex, binds the m7G-cap structure. eIF4G binds eIF3, which recruits the 40S ribosomal subunit and its associated ternary complex (eIF2-Met-tRNA-GTP). (Left) Viral mRNA with cap 1 structures (N-7 and 2′-O methylation) recruit eIF4E and initiates translation and polyribosome formation. IFIT1 binds poorly to cap 1 mRNA. (Right) Viral mRNA with cap 0 structures lacking 2′-O methylation are recognized by preferentially by IFIT1, which prevents binding of eIF4E and efficient translation. This Figure was adapted from published models [54], [85].
2.2. Expression pattern of IFIT proteins
Although most cell types do not express IFIT proteins under basal conditions, they are induced rapidly and to high levels in many cells following virus infection [23]. This expression pattern is determined in part by the upstream promoter regions of IFIT genes, which contain IFN-stimulated response elements (ISRE) [24], [25], [26]. Ifit1 and Ifit2 are induced within two hours of exogenous IFNα treatment [25]. In some cells, subsets of IFIT genes are induced selectively after stimulation with type I IFN or viral infection [27]. Cell-type and tissue-specific kinetics of expression of individual IFIT genes [20], [21], [28], [29] may contribute to the distinctive antiviral functions that have been observed in vivo [22], [30], [31], [32], [33].
IFIT gene expression also can be triggered independently of type I IFN, through signals generated directly after the ligation of PRRs (such as TLR3, TLR4, MDA5, RIG-I, and cGAS) by PAMPs (such as double-stranded RNA, DNA, and lipopolysaccharide (LPS)). IFIT genes were described as viral stress-inducible genes [23] and are induced at the transcriptional level directly by IRF3 [34], [35], which is activated soon after viral infection (via a MAVS or STING-dependent signal), often prior to the induction of type I IFN. Other IRF proteins (such as IRF1, IRF5, and IRF7) can induce the expression of IFIT genes directly [36], [37], although these pathways remain less well defined. Some IFIT genes, including human IFIT1B, lack ISRE-containing promoters and presumably are not induced by type I IFN or IRF-dependent signals [38]. Human IFIT genes also are induced by retinoic acid [39], though the kinetics are slower and might be regulated by IFNα induction [37].
2.3. Structure and RNA binding activity of IFIT proteins
Although an atomic structure of a full-length mouse or human IFIT1 has not been described, four studies have reported high-resolution X-ray crystallographic structures of other IFIT family members, including human IFIT2 [40] and IFIT5 [41], [42], [43]. In the 2.8 Å resolution IFIT2 structure, monomers of IFIT2 had nine TPR motifs and formed domain-swapped homodimers. IFIT2 had an extensively positively charged C-terminal region that supported RNA binding with or without 5′ triphosphorylation (5′-ppp) [40]. Mutation or deletion of charged residues in this region that altered RNA binding to IFIT2 negatively affected antiviral activity against Newcastle disease and Sendai viruses when these IFIT2 variants were expressed ectopically in 293T cells [40]. This study also suggested that IFIT2 binds to RNA containing adenylate uridylate (AU)-rich elements. These are found in mRNA of some genes that encode cytokines or apoptotic factors and their targeting could contribute to how IFIT2 regulates inflammatory responses [44], [45].
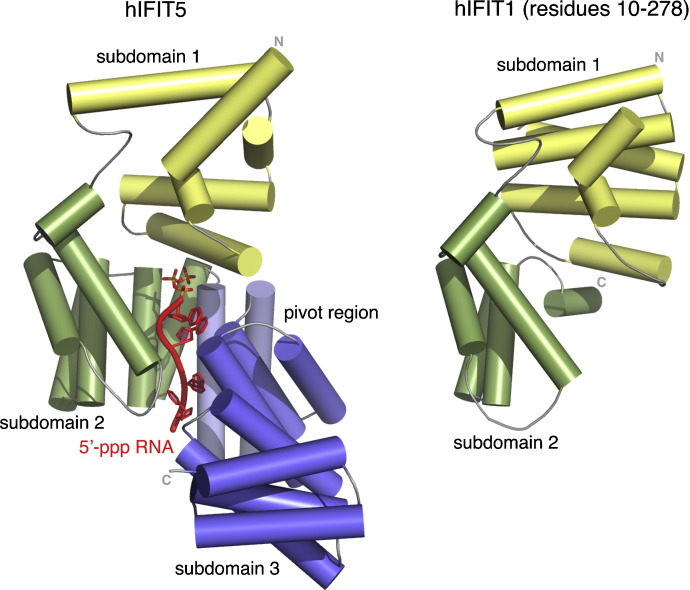
Abbas et al. described the crystal structures of IFIT5 alone or in complex with 5′-ppp RNA as well as a separate structure of the N-terminal, protease resistant fragment (amino acid residues 10–279) of human IFIT1 [41]. In IFIT5, 18 of its 24 α-helices form canonical TPRs with the remaining helices intervening between TPRs. This results in the formation of three distinct bundles of TPRs oriented in a V-like configuration to give the overall protein a clamp-shaped structure (Fig. 1 ). Structural analysis also revealed a helical domain containing a positively charged cavity that engaged a single-stranded 5′-ppp RNA. Binding of 5′-ppp RNA to IFIT1 and IFIT5 occurs in a non-sequence specific manner and requires an overhang of five and three 5′ nucleotides, respectively. Mutation of residues that altered 5′-ppp RNA binding to IFIT5 reduced antiviral activity against vesicular stomatitis (VSV) or influenza (IAV) viruses when variant IFIT5 proteins were expressed ectopically in 293T cells.
Fig. 1.
Structure of IFIT5 and N-terminal protease resistant fragment of IFIT1. Cartoon diagram of the structure of the IFIT5 monomer (PDB 4HOR) and N-terminal protease resistant fragment of IFIT1 (PDB 4HOU). α-helical structural elements are shown as cylinders. The subdomains are labeled: subdomain 1 (yellow), subdomain 2 (green), pivot region (light blue), and subdomain 3 (blue). In the IFIT5 structure, the 5′-ppp RNA is shown in red with the phosphate atoms in orange. The Figure was prepared with PyMOL (http://pymol.org/) and is adapted from the original publication [41].
IFIT5 also may bind endogenous 5′-monophosphate-capped RNA including tRNA and other single-stranded RNA moieties. Katibah et al. reported that RNA recognition by IFIT5 requires a convoluted intramolecular fold of the TPRs, which scaffolds additional helices to form an RNA binding cleft [42]. Analogous to studies with IFIT2, IFIT5 co-purification of cellular RNA or binding to tRNA was reduced by C-terminal truncation and mutation of positively charged residues in the cleft also diminished RNA binding activity [42]. Feng et al. described electrophoresis mobility shift assay (EMSA) experiments in which IFIT5 bound with high-affinity (10–100 nM) to poly A, poly U, and poly C single-stranded RNA and AT-rich double-stranded DNA [43]. C-terminal truncations and site-specific mutations of positively charged residues abolished the poly A single-stranded RNA and AT-rich double-stranded DNA binding activity of IFIT5. Collectively, these structural and biochemical studies show that IFIT5 binds to several types of RNA, and that its RNA binding ability contributes to its antiviral activity.
3. IFIT1 is a sensor and recognizes 2′-O unmethylated RNA
3.1. IFIT1 restricts infection of viruses lacking 2′-O methylation of viral RNA
The cellular mRNA of higher eukaryotes and many viral RNAs are methylated at the N-7 and 2′-O positions of the 5′ guanosine cap by host methyltransferases. Whereas N-7 methylation is essential for RNA translation and stability, until recently, the function of 2′-O methylation was uncertain [46], [47]. My group showed that a West Nile virus (WNV) mutant in the viral NS5 gene (WNV-NS5-E218A) lacking 2′-O methyltransferase activity [48] was attenuated in wild type cells and mice but was pathogenic in the absence of Ifit1 expression [30], [49]. The mutant virus lacking 2′-O methyltransferase activity showed increased replication in peripheral and central nervous system tissues of Ifit1 −/− mice compared with wild type mice. 2′-O methylation of viral RNA did not affect IFNβ induction in WNV-infected cells but instead modulated the antiviral activity of IFIT genes including IFIT1.
This finding has been confirmed with several related flaviviruses. Two groups made identical or analogous mutations in the NS5 gene (NS5-E218A or NS5-K61A) of Japanese encephalitis virus (JEV), which resulted in abrogation of 2′-O methylation, attenuation in wild type mice, and enhanced sensitivity and restriction by type I IFN and IFIT1 [50], [51]. Another report describes similar mutations in Dengue virus serotype 1 (DENV-1) and serotype 2 (DENV-2) in which substitutions were engineered (NS5-E216A (DENV-1) and NS5-E217A and NS5-K61A (DENV-2)) that abrogate 2′-O methyltransferase activity [52]. DENV strains lacking 2′-O methylation were attenuated in mice and macaques and more sensitive to the antiviral effects of IFIT1. As both the mutant JEV and DENV retained immunogenicity in animal models, 2′-O-methyltransferase mutant flaviviruses have been proposed as novel live-attenuated vaccines that are self-limited due to the enhanced restriction by IFIT1 and the host innate immune response.
Coronavirus mutants lacking 2′-O methyltransferase activity also show enhanced sensitivity to the antiviral actions of IFIT proteins [49], [53], [54], [55]. Human and mouse coronavirus nsp16 mutants (e.g., mouse hepatitis virus (MHV) nsp16-D130A) lacking 2′-O methylation induced higher levels and were more sensitive to type I IFN. The absence of 2′-O-methylation on MHV-D130A viral RNA preferentially activated the MDA5-MAVS signaling pathway and also resulted in enhanced sensitivity to IFIT1 [53]. IFN-α treatment preferentially inhibited infection of a human coronavirus (strain 229E) 2′-O methyltransferase mutant (nsp16 D129A) in HeLa cells, and this restriction was linked to IFIT1 expression [54]. Consistent with this cell culture data, growth of MHV-D130A was partially restored in Ifit1 −/− animals. In studies with an analogously mutated SARS coronavirus (SARS-CoV), an nsp16-D130A mutation showed attenuated replication in human airway epithelial cells and wild type mice but was virulent in Ifit1 −/− mice [55]. These studies establish that IFIT1 has a central role in restraining the growth of 2′-O methyltransferase-deficient flaviviruses and coronaviruses (Table 1 ).
Table 1.
Viruses lacking 2′-O methylation of mRNA are restricted by IFIT1.
| Virus | Family | Mutationa | References |
|---|---|---|---|
| West Nile virus | Flaviviridae | NS5-E218A | [30], [49] |
| Dengue virus serotype-1 | Flaviviridae | NS5-E216A | [52] |
| Dengue virus serotype-2 | Flaviviridae | NS5-E217A; NS5-K61A | [52] |
| Japanese encephalitis virus | Flaviviridae | NS5-E218A; NS5-K61A | [50], [51] |
| Mouse hepatitis virus | Coronaviridae | nsp16-D130A | [49], [53], [54] |
| Human 229E Coronavirus | Coronaviridae | nsp16-D129A | [54] |
| SARS-CoV | Coronaviridae | nsp16-D130A | [55] |
| Vaccinia virus | Poxviridae | J3-K175R | [49] and unpublished results |
| Venezuelan equine encephalitis virus (strain TC-83) | Togaviridae | Nucleotide 3 (G → A), 5′-UTR | [56] |
| Sindbis virus | Togaviridae | Nucleotide 8 (G → U), 5′-UTR | [56] |
| Hepatitis C virus | Flaviviridae | [64], [65] |
The indicated mutation enhances the sensitivity of the virus to IFIT1-dependent restriction.
3.2. IFIT1 binds mRNA lacking 2′-O methylation
Studies have begun to define how IFIT1 inhibits viruses that lack 2′-O methylation on their 5′ cap structures. EMSA experiments revealed that IFIT1 retarded the electrophoretic mobility of viral RNA displaying cap 0 (without 2′-O methylation) but not cap 1 (with 2′-O methylation) structures [51], [56]. The preferential binding of IFIT1 to cap 0 RNA was corroborated by RNA immunoprecipitation experiments [51]. A separate group ectopically expressed IFIT proteins and performed affinity purifications using capped RNA. Among the IFIT family members only human and murine IFIT1 were detected when capped RNA was used as bait [54]. Using affinity purification and mass spectrometry, this group showed that IFIT1 bound efficiently to cap 0 RNA but poorly to cap 1 RNA [54]. Primer extension assays showed that recombinant human and rabbit IFIT1 and IFIT1B bind cap-proximal regions of cap 0 mRNAs with high affinity (K 1/2,app 9–23 nM). Methylation at the 2′-O position abrogated IFIT1-mRNA interaction, whereas IFIT1B retained the ability to bind cap 1 mRNA with reduced affinity (K 1/2,app ∼ 450 nM) [57]. These data suggest that IFIT1 can sense the methylation state of capped RNA.
4. IFIT1 is an effector molecule that restricts viral translation
4.1. IFIT1 blocks translation of viral RNA lacking 2′-O methylation
After establishing that IFIT1 preferentially binds cap 0 RNA lacking 2′-O methylation, four groups have shown independently that IFIT1 mediates its antiviral effect by inhibiting viral RNA translation [51], [54], [56], [57]. Kimura et al. and Hyde et al. transfected luciferase reporter gene RNA with different 5′ cap structures into IFN-primed wild type or Ifit1 −/− MEFs and measured luicferase activity. Whereas cap 0 RNA showed reduced translation in wild type compared to Ifit1 −/− cells, addition of 2′-O methylation resulted in equivalent levels of translation in both wild type and Ifit1 −/− cells [51]. Habjan et al. used pulsed stable isotope labeling in cell culture to assess the impact of IFIT1 on global translation. Ifit1 +/+ and Ifit1 −/− macrophages were infected with either MHV-WT or MHV-nsp16-D130A and analyzed by whole-proteome shotgun mass spectrometry. Translation of viral proteins was reduced selectively in Ifit1 +/+ cells infected with MHV-nsp16-D130A [54]. Kumar et al. investigated the influence of IFIT1 on translation initiation using an in vitro reconstituted translation system and a toe-printing assay. 48S ribosomal complexes were assembled from individual purified 40S subunits, Met-tRNA, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B and eIF4F on cap 0 and cap 1 mRNA in the presence and absence of different IFIT proteins. At 800 nM, IFIT1 nearly abrogated 48S complex formation on cap 0 mRNA, whereas IFIT2, IFIT3 and IFIT5 did not affect 48S complex formation. Titration experiments showed that 48S complex formation on cap 0 mRNA was sensitive even to low (∼50 nM) concentrations of IFIT1 [57]. Collectively, these data indicate that synthesis of proteins encoded by viral RNA lacking 2′-O methylation is inhibited by IFIT1.
4.2. IFIT1 competes with eIF4E and eIF4F for binding to cap 0 mRNA
Further studies have defined how IFIT1 restricts translation of cap 0 RNA. As translation of cellular mRNA requires binding of the cap-binding protein eIF4E [58], initial experiments tested whether IFIT1 could compete with eIF4E for binding to cap 0 mRNA as a means of selectively inhibiting translation. Cap 0 and cap 1 mRNA were coupled to beads and assayed for binding to recombinant eIF4E in the presence or absence of recombinant IFIT1. eIF4E interaction with bead-bound cap 0 but not cap 1 RNA was reduced by IFIT1, suggesting that the two proteins compete for the RNA target [54]. A subsequent study confirmed these results as IFIT1 binding to cap 0 mRNA was unaffected by a 10-fold excess of eIF4E or eIF4F [57]. Thus, the affinity of IFIT1 for cap 0 RNA enables it to outcompete eIF4E and eIF4F for binding and selectively prevents translation initiation (Fig. 2 ).
4.3. IFIT proteins inhibit other steps in translation initiation
Eukaryotic initiation factor 3 (eIF3) is a multi-subunit protein complex that functions in translation initiation at several steps including formation of the 43S pre-initiation complex, mRNA recruitment to the 43S pre-initiation complex, and scanning of the mRNA for AUG (start codon) recognition (reviewed in [59]). Biochemical studies suggest that some IFIT family members reduce the efficiency of cap-dependent protein translation more generally by binding subunits of the eIF3 translation initiation complex [60]. Human IFIT1 and IFIT2 reportedly blocks binding of eIF3-mediated recruitment of the eIF2–GTP–Met-tRNA ternary complex to 40S ribosomes by interacting with eIF3e. In addition, human IFIT2, and mouse IFIT1 and IFIT2 reportedly inhibit the formation of the 48S pre-initiation complex by binding to eIF3c [20], [60], [61]. Recently, some of these results were questioned as addition of human IFIT1 failed to affect 43S complex formation irrespective of whether Met-tRNA, 40S subunits, or Met-tRNA and eIF2 were pre-incubated with excess IFIT1 [57].
Hepatitis C virus (HCV), a positive-stranded RNA virus, contains an internal ribosome entry site (IRES), which regulates the assembly of cap-independent translation initiation complexes on viral mRNA by a sequential pathway requiring eIF3 [62]. Type I IFN inhibits HCV infection in part, by blocking its translation [63], [64]. Examination of the cellular proteins associated with HCV-translation complexes in IFN-treated human cells showed that human IFIT1 is an eIF3-associated factor that fractionates with the initiator ribosome-HCV RNA complex [64]. IFIT1 suppressed IRES-dependent translation of HCV, whereas a mutant IFIT1 protein reportedly lacking eIF3-binding activity failed to inhibit HCV. Moreover, ectopic expression of IFIT1 decreased HCV infection in human hepatocytes [65]. Thus, IFIT1 may block HCV replication by targeting eIF3-dependent steps in the viral RNA translation initiation process; these include HCV IRES-dependent recognition of the 43S pre-initiation complex and assembly of the 43S-mRNA complex.
5. IFIT1 is a sensor and recognizes 5′-ppp RNA
Human IFIT1 also can function as a sensor for viral RNA by recognizing an uncapped 5′-ppp and sequestering it from the actively replicating pool [32]. Using a proteomics approach with 5′-ppp RNA as bait, mass spectrometry analysis identified IFIT1 as a primary binding partner. Subsequent experiments showed that IFIT1 and IFIT5 interact directly with 5′-ppp on RNA, whereas IFIT2 and IFIT3 form a complex with IFIT1 that may be required for its function [41]. These IFIT-dependent interactions were relevant against RNA viruses displaying a 5′-ppp, as silencing of IFIT1, IFIT2 and IFIT3 in HeLa cells to varying degrees enhanced replication of the negative strand Rift Valley fever virus (RVFV), VSV, and IAV. Studies with Ifit1 −/− mouse fibroblasts and myeloid cells also showed enhanced replication of VSV despite wild-type production levels of type I IFN and other inflammatory cytokines. In vivo, Ifit1 −/− mice were more vulnerable to infection with VSV, with higher virus-induced mortality observed. This result, however, has been questioned, as experiments by a second group with the same VSV strain but an independently generated Ifit1 −/− mouse revealed no difference in mortality compared with wild type mice over a wide range of VSV doses [22].
Biochemical and structural studies have begun to elucidate the characteristics of IFIT1 binding to 5′-ppp RNA. EMSA studies revealed that IFIT1 could bind 5′-ppp RNA in a non-sequence specific manner with a requirement of at least a five-nucleotide single-stranded overhang [41]. The affinity of IFIT1 for 5′-ppp RNA has been measured at ∼250–500 nM by surface plasmon resonance or filter binding assays [32], [56], which is at least 10-fold lower than that observed for cap 0 RNA, although one group failed to measure stable binding of IFIT1 to 5′-ppp mRNA using a primer extension technique even at higher (1 μM) concentrations [57]. IFIT1 is hypothesized to have antiviral activity against viruses that generate 5′-ppp RNA through translation-independent mechanisms including sequestration of RNA from the replicating pool [32].
6. Mechanisms of viral evasion of IFIT1
The cap structure of host mRNA is formed through a canonical series of sequential enzymatic reactions that occur in the nucleus [66], [67]: (i) an RNA triphosphatase removes the ©-phosphate from the 5′-ppp end of the nascent RNA to generate 5′-diphosphate RNA (ppN-RNA); (ii) an RNA guanylyltransferase transfers the GMP moiety from GTP to ppN-RNA to yield the core cap structure (GpppN-RNA); and (iii) an RNA guanine-N-7-methyltransferase methylates the guanine at the N-7 position to produce a cap 0 structure (m7GpppN-RNA). In higher eukaryotes, m7GpppN-RNA also is methylated at the ribose 2′-O position of the nascent mRNA by a 2′-O methyltransferase to form cap 1 (m7GpppNm) and cap 2 (m7GpppNmNm) structures. Host capping of mRNA at the 5′ end allows for efficient mRNA translation, directs pre-mRNA splicing and mRNA export from the nucleus, limits mRNA degradation by cellular exonucleases, and allows recognition of foreign RNA as ‘non-self’ [68]. As IFIT1 is one of the most strongly induced genes by the cell-intrinsic innate immune responses, “successful” viruses infecting vertebrate cells have evolved specific and efficient mechanisms to overcome its inhibitory action (Fig. 3 ).
Fig. 3.
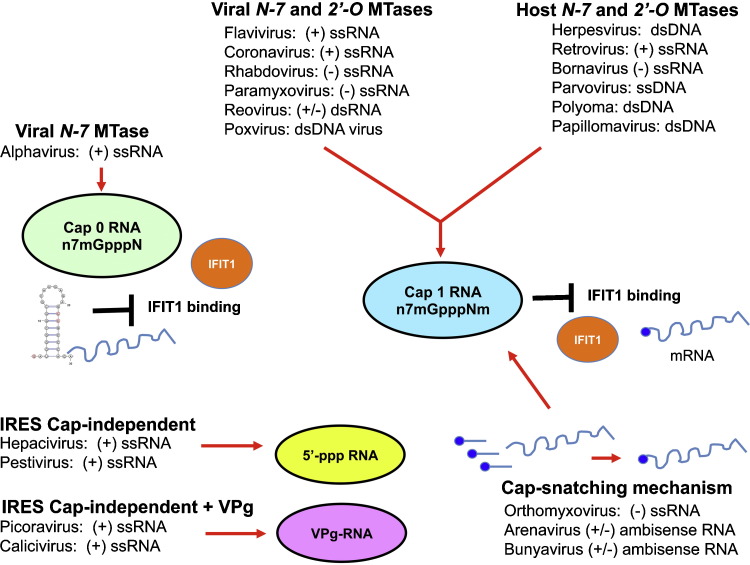
Mechanisms of evasion of IFIT1 by viruses. Different families of RNA and RNA viruses use distinct mechanisms to evade IFIT1-dependent restriction. These include: (i) viral-encoded N-7 and 2′-O methyltranferases (MTase) to generate a cap 1 structure on their mRNA (flavivirus, coronavirus, rhabdovirus, paramyxovirus, reovirus, and poxvirus), which prevents IFIT1 binding; (ii) use of host-encoded N-7 and 2′-O methyltranferases (MTase) in the nucleus to generate a cap 1 structure on their mRNA (herpesvirus, retrovirus, bornavirus, parvovirus, polyomavirus, and papillomavirus); (iii) ‘cap-snatching’ mechanisms to cleave cap 1 structures from host mRNA (10–20 nucleotides). The capped leader RNA is used to prime transcription on the viral genome, which leads to the synthesis of capped, translatable viral mRNAs (orthomyxovirus, arenavirus, and bunyavirus); (iv) RNA structural elements at the 5′-end of type 0 capped alphavirus RNA antagonize IFIT1 binding and function; and (v) some positive sense RNA viruses utilize IRES elements to mediate cap-independent translation. For hepaciviruses and pestiviruses the 5′-end remains as an uncapped 5′-ppp RNA. Picornaviruses and caliciviruses covalently attach a viral protein (VPg) to the 5′-end. This figure was adapted from a published model [68].
6.1. Viral 2′-O methytransferases
Several families of DNA and RNA viruses that replicate in the cytoplasm have evolved their own capping machinery to carry out N-7 and 2′-O methylation of viral RNA. This includes DNA (poxviruses), positive-sense (flaviviruses and coronaviruses), negative-sense (rhabdoviruses and paramyxoviruses) and double-stranded (reovirus) RNA viruses. The capping of viral mRNA can be classified as ‘conventional’, when it follows the enzymatic steps of the mRNA-capping pathways used by host or ‘non-conventional’, when it does not (reviewed in [67], [68]). Poxviruses (e.g., vaccinia virus) encode a ‘conventional’ capping system, which contains a multifunctional mRNA cap-synthesizing enzyme (D1 gene) that includes an RNA triphosphatase, guanylyltransferase, and N-7 methyltransferase. After the N-7 methylguanosine type 0 cap is added, the cap 1 structure is completed by the activity of a separate viral protein (VP39 encoded by J3 gene) that adds a 2′-O-methyl group. The cap formation of flavivirus RNA also follows the conventional pathway of mRNA cap formation through sequential activities of the viral RNA triphosphatase (NS3 gene), guanylyltransferase (NS5 gene) and N-7 and 2′-O methyltransferases (NS5 gene). Coronaviruses also are believed to produce mRNAs with a cap 1 structure through the “conventional” capping pathway although all of the component enzymes have not been defined. The viral Nsp14 and nsp16 genes encode the N-7 and 2′-O coronavirus methyltransferases, respectively.
Some virus families (e.g., Rhabdoviridae) form RNA cap structures through a ‘non-conventional’ mechanism that is distinct from the cellular mRNA capping pathway. During the mRNA cap formation of VSV, an unknown NTPase hydolyzes GTP to GDP. Monophosphorylated viral mRNA is transferred to GDP by a polyribonucleotidyltransferase, which then generates the Gpp-pA-RNA cap structure. This RNA cap is methylated sequentially by the L protein of VSV first at the ribose-2′-O position and then at the guanine-N-7 position, resulting in m7GpppAm-RNA cap 1 structure [69].
6.2. Cap-snatching
As an alternative to encoding capping machinery, several negative-strand (Orthomyxoviridae) and ambisense (Arenaviridae and Bunyaviridae) RNA viruses acquire cap structures by ‘stealing’ them from cellular mRNA. This ‘cap snatching’ mechanism was first identified in influenza virus [70], [71] and is performed by two of its polymerase subunits (PB2 and PA). PB2 binds to the 5′ end of capped cellular mRNA and then the endonuclease activity of PA cleaves the cellular RNA 10–13 nucleotides downstream of the cap structure. The released short, capped mRNAs that are released are then used as primers by the viral polymerase to synthesize nascent viral mRNA [72]. The sequence, length and structure of the 5′ end of the mRNA that comes with the cap varies among virus families. Most sequences are 15–20 nucleotides long but arenaviruses may use shorter primers [73].
6.3. Cap-independent translation
Viruses have developed alternative cap-independent translation programs, which to some extent avoid IFIT1-mediated restriction. Hepaciviruses and pestiviruses of the Flaviviridae family of RNA viruses lack a cap structure at their 5′ end but encode for an IRES to initiate viral translation in a cap-independent manner. The HCV IRES directly recruits the 40S ribosome subunit to the translation initiator codon of the genome and does not require interactions with eukaryotic initiation factors eIF1, 1A, 4A, 4B and 4E [74]. Thus, in theory, HCV could still be translated even if IFIT1 were bound to the 5′ end of the genomic RNA. However, IFIT1 may block HCV translation through an independent mechanism by targeting eIF3-dependent steps in the viral RNA translation initiation process (see Section 4.3). Although other RNA viruses (e.g., picornaviruses) encode IRES elements, there are differences in function and requirements for eukaryotic translation initiation factors (reviewed in [75]). Indeed, in contrast to HCV, the IRES-containing encephalomyocarditis virus (a picornavirus) is resistant to the antiviral effects of IFIT1 [32], [49].
As another possible means to prevent immune molecules including IFIT1 from recognizing non-2′-O methylated or non-capped mRNA, Picornaviridae and Caliciviridae positive-sense RNA viruses covalently attach a small basic protein VPg (viral protein genome-linked) to the 5′ end of viral RNA [76], [77]. For viruses of the Picornaviridae family, the VPg ‘cap’ does not function in translation but instead serves as a primer to synthesize RNA by the viral polymerase and protects RNA from exonuclease degradation. In comparison, the calicivirus VPg protein acts as a ‘cap substitute’ and interacts directly with the cap-binding protein eIF4E to facilitate viral mRNA translation [78].
6.4. Use of host methyltransferases in the nucleus
Viruses that synthesize their mRNA using cellular RNA polymerase II use the host capping machinery to generate cap 1 structures on their 5′ end. This occurs for most nuclear DNA viruses (e.g., Herpesviruses, Polyomaviruses, and Papillomaviruses) and for selected RNA viruses including those belonging to the Retroviridae and Bornaviridae families [79].
6.5. Antagonism of IFIT1 by viral RNA structural elements
Alphaviruses are positive sense RNA viruses that replicate in the cytoplasm, translate via a cap-dependent mechanism, and yet lack a virally encoded 2′-O-methyltransferase or cap-snatching mechanism, and thus should be restricted by IFIT1. Alphaviruses have a defined cap 0 structure lacking 2′-O methylation on the 5′ end of their viral genomic and subgenomic RNA [80], [81], which is synthesized through a non-conventional mechanism [67]. A recent study shows that alphaviruses use a stable stem-loop structure in their 5′-untranslated region (UTR) to antagonize IFIT1 binding and antiviral activity [56]. Mutations within the 5′-UTR that affect stable RNA structural elements enabled restriction by or antagonism of Ifit1 in vitro and in vivo. This phenomenon explains why some alphavirus strains are more sensitive to the antiviral effects of type I IFN [82], and links the phenotype to single nucleotide changes in the 5′-UTR of the alphavirus RNA [83]. Thus, structural elements at the 5′ end of alphavirus genome can function to evade IFIT1-dependent restriction of non-2′-O methylated viral RNA.
7. Conclusions
Triggering an effective intrinsic cellular antiviral response is essential for the host to eliminate invading pathogens. To promote their own survival, viruses have developed strategies to escape host recognition by interfering with antiviral detection pathways, signaling cascades, and effector mechanisms. IFIT1 is an IFN-induced highly expressed protein that functions in the cytoplasm as a dual sensor and effector molecule to restrict virus infection. IFIT1 preferentially recognizes non-2′-O methylated or uncapped non-self viral mRNA and suppresses translation initiation or sequesters the RNA from active replication. IFIT1 likely contributes to a species barrier that puts evolutionary pressure on viruses to generate mRNA with host cap structures. ‘Successful’ or pathogenic viruses have evolved mechanisms to produce mRNA with 5′-ends that mimic host cellular mRNAs, including viral RNA with N-7 and 2′-O methylation through several independent strategies (encoding capping machinery, ‘cap-snatching’, or using host capping machinery). Viruses also can avoid or attenuate IFIT1 restriction by using cap-independent translation mechanisms, covalent binding of viral proteins to the 5′-end of the RNA, or evolving secondary structures at their 5′-end that inhibit IFIT1 binding and function. As both host and many viral mRNA share 2′-O methylation, higher eukaryotic cells may be trapped into retaining this modification, despite its relative lack of protective effect against viral pathogens [84]. A deeper mechanistic understanding of IFIT1 biology could facilitate the development of novel antiviral agents that target viral methyltransferases or other proteins to re-sensitize pathogenic viruses to the inhibitory activities of IFIT1.
Conflict of interest
The author has no financial conflict to disclose.
Acknowledgements
NIH grants U19 AI083019, R01 AI104972, and R01 AI104002 supported this work. The author gratefully acknowledges J. Hyde, S. Austin, and J. White for critical comments on the manuscript and help with figure preparation.
Biography

Michael S. Diamond received his M.D. and Ph.D. from Harvard University, and his post-doctoral and clinical training in infectious diseases and virology from the University of California, Berkeley and the University of California, San Francisco. He is currently a Professor of Medicine, Molecular Microbiology, Pathology & Immunology at Washington University School of Medicine and the Associate Director of the Center for Human Immunology and Immunotherapy Programs at Washington University. Michael Diamond's research focuses on the interface between viral pathogenesis and the host immune response with a primary focus on flaviviruses and alphaviruses. His group has studied how novel innate immune sensor and effector molecules restrict infection of multiple families of pathogenic human viruses.
References
- 1.Kawai T., Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 2.Keating S.E., Baran M., Bowie A.G. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Cai X., Chiu Y.H., Chen Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 4.Theofilopoulos A.N., Baccala R., Beutler B., Kono D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 5.Pestka S., Krause C.D., Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 6.Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Veer M.J., Holko M., Frevel M., Walker E., Der S, Parajape J.M. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 8.Lanford R.E., Guerra B., Lee H., Chavez D., Brasky K.M., Bigger C.B. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 9.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karki S., Li M.M., Schoggins J.W., Tian S., Rice C.M., Macdonald M.R. Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PLoS ONE. 2012;7:e37398. doi: 10.1371/journal.pone.0037398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J., Ding S.C., Cho H., Chung B.C., Gale M., Jr., Chanda S.K. A short hairpin RNA screen of interferon-stimulated genes identifies a novel negative regulator of the cellular antiviral response. MBio. 2013;4:e003853-13. doi: 10.1128/mBio.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schoggins J.W., Dorner M., Feulner M., Imanaka N., Murphy M.Y., Ploss A. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proc Natl Acad Sci USA. 2012;109:14610–14615. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins J.W. Interferon-stimulated genes: roles in viral pathogenesis. Curr Opin Virol. 2014;6C:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen G.C., Sarkar S.N. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 17.Wathelet M.G., Clauss I.M., Content J., Huez G.A. The IFI-56K and IFI-54K interferon-inducible human genes belong to the same gene family. FEBS Lett. 1988;231:164–171. doi: 10.1016/0014-5793(88)80724-5. [DOI] [PubMed] [Google Scholar]
- 18.Fensterl V., Sen G.C. The ISG56/IFIT1 gene family. J Interferon Cytokine Res. 2011;31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D’Andrea L.D., Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Terenzi F., Hui D.J., Merrick W.C., Sen G.C. Distinct induction patterns and functions of two closely related interferon-inducible human genes, ISG54 and ISG56. J Biol Chem. 2006;281:34064–34071. doi: 10.1074/jbc.M605771200. [DOI] [PubMed] [Google Scholar]
- 21.Terenzi F., White C., Pal S., Williams B.R., Sen G.C. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J Virol. 2007;81:8656–8665. doi: 10.1128/JVI.00322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fensterl V., Wetzel J.L., Ramachandran S., Ogino T., Stohlman S.A., Bergmann C.C. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarkar S.N., Sen G.C. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Levy D., Larner A., Chaudhuri A., Babiss L.E., Darnell J.E., Jr. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc Natl Acad Sci USA. 1986;83:8929–8933. doi: 10.1073/pnas.83.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bluyssen H.A., Vlietstra R.J., Faber P.W., Smit E.M., Hagemeijer A., Trapman J. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics. 1994;24:137–148. doi: 10.1006/geno.1994.1591. [DOI] [PubMed] [Google Scholar]
- 26.de Veer M.J., Sim H., Whisstock J.C., Devenish R.J., Ralph S.J. IFI60/ISG60/IFIT4, a new member of the human IFI54/IFIT2 family of interferon-stimulated genes. Genomics. 1998;54:267–277. doi: 10.1006/geno.1998.5555. [DOI] [PubMed] [Google Scholar]
- 27.Fensterl V., White C.L., Yamashita M., Sen G.C. Novel characteristics of the function and induction of murine p56 family proteins. J Virol. 2008;82:11045–11053. doi: 10.1128/JVI.01593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusari J., Sen G.C. Regulation of synthesis and turnover of an interferon-inducible Mrna. Mol Cell Biol. 1986;6:2062–2067. doi: 10.1128/mcb.6.6.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wacher C., Muller M., Hofer M.J., Getts D.R., Zabaras R., Ousman S.S. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol. 2007;81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szretter K.J., Daniels B.P., Cho H., Gainey M.D., Yokoyama W.M., Gale M., Jr. 2′-O methylation of the viral mRNA cap by West Nile virus evades Ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8:e1002698. doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho H., Shrestha B., Sen G.C., Diamond M.S. A role for Ifit2 in restricting West Nile virus infection in the brain. J Virol. 2013;87:8363–8371. doi: 10.1128/JVI.01097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichlmair A., Lassnig C., Eberle C.A., Gorna M.W., Baumann C.L., Burkard T.R. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- 33.Butchi N.B., Hinton D.R., Stohlman S.A., Kapil P., Fensterl V., Sen G.C. Ifit2 deficiency results in uncontrolled neurotropic coronavirus replication and enhanced encephalitis via impaired alpha/beta interferon induction in macrophages. J Virol. 2014;88:1051–1064. doi: 10.1128/JVI.02272-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grandvaux N., Servant M.J., tenOever B., Sen G.C., Balachandran S., Barber G.N. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa S., Lozach J., Benner C., Pascual G., Tangiraia R.K., Westin S. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes B.J., Richards J., Mancl M., Hanash S., Beretta L., Pitha P.M. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem. 2004;279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 37.Lou Y.J., Pan X.R., Jia P.M., Li D., Xiao S., Zhang Z.L. IRF-9/STAT2 [corrected] functional interaction drives retinoic acid-induced gene G expression independently of STAT1. Cancer Res. 2009;69:3673–3680. doi: 10.1158/0008-5472.CAN-08-4922. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Zhang Y.B., Liu T.K., Gui J.F. Lineage-specific expansion of IFIT gene family: an insight into coevolution with IFN gene family. PLoS ONE. 2013;8:e66859. doi: 10.1371/journal.pone.0066859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu M., Tong J.H., Mao M., Kan L.X., Lui M.M., Sun Y.W. Cloning of a gene (RIG-G) associated with retinoic acid-induced differentiation of acute promyelocytic leukemia cells and representing a new member of a family of interferon-stimulated genes. Proc Natl Acad Sci USA. 1997;94:7406–7411. doi: 10.1073/pnas.94.14.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z., Liang H., Zhou Q., Li Y., Chen H., Ye W. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 2012;22:1328–1338. doi: 10.1038/cr.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbas Y.M., Pichlmair A., Gorna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katibah G.E., Lee H.J., Huizar J.P., Vogan J.M., Alber T., Collins K. tRNA binding, structure, and localization of the human interferon-induced protein IFIT5. Mol Cell. 2013;49:743–750. doi: 10.1016/j.molcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng F., Yuan L., Wang Y.E., Crowley C., Lv Z., Li J. Crystal structure and nucleotide selectivity of human IFIT5/ISG58. Cell Res. 2013;23:1055–1058. doi: 10.1038/cr.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stawowczyk M., Van Scoy S., Kumar K.P., Reich N.C. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem. 2011;286:7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reich N.C. A death-promoting role for ISG54/IFIT2. J Interferon Cytokine Res. 2013;33:199–205. doi: 10.1089/jir.2012.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei C.M., Gershowitz A., Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- 47.Wei C.M., Moss B. Methylated nucleotides block 5′-terminus of vaccinia virus messenger RNA. Proc Natl Acad Sci USA. 1975;72:318–322. doi: 10.1073/pnas.72.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y., Ray D., Zhao Y., Dong S., Ren Z., Li Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S.H., Dong H., Li X.F., Xie X., Zhao H., Deng Y.Q. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J Virol. 2013;87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura T., Katoh H., Kayama H., Saiga H., Okuyama M., Okamoto T. Ifit1 inhibits JEV replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol. 2013;87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zust R., Dong H., Li X.F., Chang D.C., Zhang B., Balakrishnan T. Rational design of a live attenuated dengue vaccine: 2′-o-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog. 2013;9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zust R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B.W., Ziebuhr J. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Habjan M., Hubel P., Lacerda L., Benda C., Holze C., Eberl C.H. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013;9:e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menachery V.D., Yount B.L., Jr., Josset L., Gralinski L.E., Scobey T., Agnihothram S. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J Virol. 2014;88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hyde J.L., Gardner C.L., Kimura T., White J.P., Lui G., Trobaugh D.W. A viral RNA structural element alters host recognition of nonself RNA. Science. 2014;343:783–787. doi: 10.1126/science.1248465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar P., Sweeney T.R., Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp-mRNAs. Nucleic Acids Res. 2014;42:3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furuichi Y., Shatkin A.J. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinnebusch A.G. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 60.Guo J., Peters K.L., Sen G.C. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology. 2000;267:209–219. doi: 10.1006/viro.1999.0135. [DOI] [PubMed] [Google Scholar]
- 61.Hui D.J., Bhasker C.R., Merrick W.C., Sen G.C. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP. Met-tRNAi. J Biol Chem. 2003;278:39477–39482. doi: 10.1074/jbc.M305038200. [DOI] [PubMed] [Google Scholar]
- 62.Otto G.A., Puglisi J.D. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 63.Sumpter R., Jr., Wang C., Foy E., Loo Y.M., Gale M., Jr. Viral evolution and interferon resistance of hepatitis C virus RNA replication in a cell culture model. J Virol. 2004;78:11591–11604. doi: 10.1128/JVI.78.21.11591-11604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang C., Pflugheber J., Sumpter R., Jr., Sodora D.L., Hui D., Sen G.C. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898–3912. doi: 10.1128/JVI.77.7.3898-3912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Raychoudhuri A., Shrivastava S., Steele R., Kim H., Ray R., Ray R.B. ISG56 and IFITM1 proteins inhibit hepatitis C virus replication. J Virol. 2011;85:12881–12889. doi: 10.1128/JVI.05633-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 67.Dong H., Fink K., Zust R., Lim S.P., Qin C.F., Shi P.Y. Flavivirus RNA methylation. J Gen Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 68.Decroly E., Ferron F., Lescar J., Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogino T., Banerjee A.K. An unconventional pathway of mRNA cap formation by vesiculoviruses. Virus Res. 2011;162:100–109. doi: 10.1016/j.virusres.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouloy M., Plotch S.J., Krug R.M. Globin mRNAs are primers for the transcription of influenza viral RNA in vitro. Proc Natl Acad Sci USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Plotch S.J., Bouloy M., Ulmanen I., Krug R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 72.Dias A., Bouvier D., Crepin T., McCarthy A.A., Hart D.J., Baudin F. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 73.Raju R., Raju L., Hacker D., Garcin D., Compans R., Kolakofsky D. Nontemplated bases at the 5′ ends of Tacaribe virus mRNAs. Virology. 1990;174:53–59. doi: 10.1016/0042-6822(90)90053-t. [DOI] [PubMed] [Google Scholar]
- 74.Niepmann M. Hepatitis C virus RNA translation. Curr Top Microbiol Immunol. 2013;369:143–166. doi: 10.1007/978-3-642-27340-7_6. [DOI] [PubMed] [Google Scholar]
- 75.Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 76.Flanegan J.B., Petterson R.F., Ambros V., Hewlett N.J., Baltimore D. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc Natl Acad Sci USA. 1977;74:961–965. doi: 10.1073/pnas.74.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaffer F.L., Ehresmann D.W., Fretz M.K., Soergel M.I. A protein, VPg, covalently linked to 36S calicivirus RNA. J Gen Virol. 1980;47:215–220. doi: 10.1099/0022-1317-47-1-215. [DOI] [PubMed] [Google Scholar]
- 78.Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labue L. Calicivirus translation initiation requires an interaction between VPg and eIF 4E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pyper J.M., Clements J.E., Zink M.C. The nucleolus is the site of Borna disease virus RNA transcription and replication. J Virol. 1998;72:7697–7702. doi: 10.1128/jvi.72.9.7697-7702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hefti E., Bishop D.H., Dubin D.T., Stollar V. 5′ nucleotide sequence of sindbis viral RNA. J Virol. 1975;17:149–159. doi: 10.1128/jvi.17.1.149-159.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pettersson R.F., Soderlund H., Kaariainen L. The nucleotide sequences of the 5′-terminal T1 oligonucleotides of Semliki-Forest-virus 42-S and 26-S RNAs are different. Eur J Biochem. 1980;105:435–443. doi: 10.1111/j.1432-1033.1980.tb04518.x. [DOI] [PubMed] [Google Scholar]
- 82.Jahrling P.B., Navarro E., Scherer W.F. Interferon induction and sensitivity as correlates to virulence of Venezuelan encephalitis viruses for hamsters. Arch Virol. 1976;51:23–35. doi: 10.1007/BF01317831. [DOI] [PubMed] [Google Scholar]
- 83.Kinney R.M., Chang G.J., Tsuchiya K.R., Sneider J.M., Roehrig J.T., Woodward T.M. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol. 1993;67:1269–1277. doi: 10.1128/jvi.67.3.1269-1277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daugherty M.D., Malik H.S. Rules of engagement: molecular insights from host–virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 85.Hay N., Sonenberg N. Upstream and downstream of Mtor. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]