Abstract
Here, we review the genetic risk factors for late onset Alzheimer's disease (AD) and their role in AD pathogenesis. Recent advances in our understanding of the human genome, namely technological advances in methods to analyze millions of polymorphisms in thousands of subjects, have revealed new genes associated with AD risk: ABCA7, BIN1, CASS4, CD33, CD2AP, CELF1, CLU, CR1, DSG2, EPHA1, FERMT2, HLA-DRB5-DBR1, INPP5D, MS4A, MEF2C, NME8, PICALM, PTK2B, SLC24H4 RIN3, SORL1, ZCWPW1. Emerging technologies to analyze the entire genome in large datasets have also revealed coding variants that increase AD risk: PLD3 and TREM2. We review the relationship between these AD risk genes and the cellular and neuropathological features of AD. Together, understanding the mechanisms underlying the association of these genes with risk for disease will provide the most meaningful targets for therapeutic development to date.
Keywords: Alzheimer's disease, amyloid precursor protein, genome wide association studies, endocytosis, immune response, cholesterol metabolism
Alzheimer's Disease
Alzheimer's disease is pathologically defined by extensive neuronal loss and the accumulation of intracellular neurofibrillary tangles and extracellular amyloid plaques in the brain. Genetic, biochemical, and neuropathological data suggest that A aggregation is central to initiating AD pathogenesis (1). Neurofibrillary pathology strongly correlates with neuronal dysfunction and progression of the clinical phase of AD (2). The clinical phase of AD is also marked by synaptic loss, selective neuronal death, neurotransmitter loss, and neuroinflammation (2).
Emerging Genetics
Dominantly inherited, early onset AD is associated with classical Mendelian patterns of inheritance with age-dependent penetrance. Late onset AD (LOAD) also has a strong genetic component. The identification of novel loci that affect LOAD risk is critical to our understanding of the underlying etiology of AD. Genome wide associated studies (GWAS) have identified polymorphisms in or near several genes that are associated with AD risk: ABCA7, CLU, CR1, CD33, CD2AP, EPHA1, BIN1, PICALM, MS4A (3-7) (Fig. 1). Additional loci were identified in a meta analysis of these large LOAD consortium datasets: CASS4, CELF1, DSG2, FERMT2, HLA DRB5 DBR1, INPP5D, MEF2C, NME8, PTK2B, SLC24H4 RIN3, SORL1, ZCWPW1 (6). The identification of common variants that have small effects on AD risk has begun to create a broader picture of the processes and pathways involved in AD risk. Variants in genes involved in lipid metabolism, the inflammatory response, and endocytosis have been identified through these GWAS.
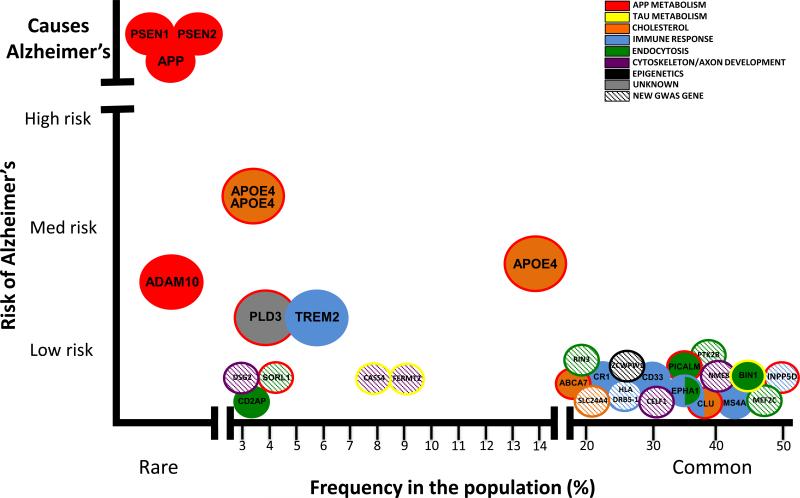
Figure 1. Rare and common variants contribute to Alzheimer's disease risk.
Figure updated and modified from (149).
Although large datasets with whole genome or exome sequencing are being generated, these approaches in smaller datasets have yielded evidence of rare coding variants in two genes with moderate to large effects on LOAD risk: PLD3 and TREM2 (8-11) (Fig. 1). The identification of rare variants in the population that have moderate to large effects on AD risk will be valuable in identifying pathways that are central to disease pathogenesis. In contrast to the GWAS, sequencing studies have identified variants within the coding sequence that can be more easily examined in in vitro and in vivo model systems. These methods may provide the most meaningful targets for therapeutic development.
In complex, heterogeneous diseases like AD, novel approaches to integrate genetic, expression, and epigenetic into organized molecular networks may facilitate our understanding of the underlying disease pathogenesis. AD likely arises from a complex interplay between genetic susceptibility and downstream molecular pathways. A recent study constructed gene-regulatory networks from 1,647 AD and control brain samples to demonstrate that networks involved in immune-and microglia-specific modules are disrupted in AD brains (12). TYROBP was identified as a key regulator in a module of genes involved in pathogen phagocytosis (12). Interestingly, TYROBP, a.k.a. DAP12, is key signaling molecule for TREM2, another recently identified AD risk gene. Thus, these methods are useful in developing integrated models of the molecular pathways disrupted in AD.
Alternative AD Phenotypes
The majority of AD risk genes affect Aβ production and clearance, highlighting the importance of this pathway in AD pathogenesis. This is likely the result of the methods by which the genes were identified, in studies testing for association with AD case control status (3-7, 13). Using alternative AD phenotypes may reveal additional genes that modify particular aspects of the disease. Use of biomarkers as quantitative endophenotypes has led to the identification of additional genes that modify tau and Aβ metabolism in CSF and neuroimaging phenotypes (14-21). Using biomarkers as quantitative endophenotypes in populations who are tracked over the course of disease will give us more information regarding genes that influence disease onset and progression (14). Additional risk alleles may modify tau metabolism and impact AD progression; however, these studies are still on going.
APP, PSEN1, and PSEN2
Dominantly inherited mutations in β-amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) cause early onset Alzheimer's disease (AD) (2, 22). Sequential cleavage of APP, a transmembrane neuronal protein, by β-secretase and then by γ-secretase produces Aβ (23). PSEN1 and PSEN2 are critical components of the γ-secretase complex. The amyloid cascade hypothesis posits that changes in APP and/or Aβ homeostasis lead to the aggregation of Aβ and deposition in plaques and that these events are sufficient to initiate the cascade of pathologic abnormalities associated with AD (1). APP proteolysis by secretase results in cleavage within the Aβ domain generating non-amyloidogenic fragments that are reported to possess neurotrophic and neuroprotective properties (24, 25).
Increasing evidence suggests that there are additional variants in APP and APP-modifying genes that alter AD risk in LOAD cases. Novel, rare variants in APP, PSEN1, PSEN2 and ADAM10 have been identified in large, LOAD families (26-28). Segregation data and bioinformatic analysis suggests that these rare variants in APP may increase (e.g.: APP N660Y), decrease (e.g.: APP A673T), or have no effect on AD risk (e.g.: APP E599K) (26, 29). A polymorphism in PSEN1, PSEN1 E318G, is associated with a 10-fold increase in LOAD risk in APOE4 carriers (27). Additionally, rare coding variants in ADAM10, the major α-secretase involved in shedding of the APP ectodomain (30), co-segregate in seven LOAD families (8, 31). ADAM10 risk variants, Q170H and R181G, increase Aβ levels in vitro (8). In Tg2576 AD mice, ADAM10 Q170H and R181G disrupt α-secretase activity and shift APP processing toward amyloidogenic cleavage, yielding increased plaque load (31). Together, these findings illustrate that variants in APP and APP-modifying genes (e.g.: PSEN1, PSEN2, ADAM10) can cause early onset AD or alter risk for LOAD.
CHOLESTEROL METABOLISM
APOE genotype is the strongest risk factor for LOAD. Its central role in cholesterol metabolism implicates this pathway in AD pathogenesis. In recent LOAD GWAS, variants in several genes were identified that are involved in cholesterol metabolism: CLU, ABCA7, SORL1 (3-6, 13).
APOE
Apolipoprotein E (APOE) is the strongest risk factor for LOAD. APOE is located on chromosome 19q13.2. APOE encodes three common alleles (ε2, ε3, ε4). APOEε4 is associated with increased AD risk (32, 33): one APOEε4 allele increases AD risk 3 fold and two APOEε4 alleles increases AD risk by 12 fold. APOEε4 is also associated with a dose dependent decrease in age at onset. Conversely, APOEε2 is associated with decreased risk for AD and later age at onset (32, 33).
APOE is a regulator of lipoprotein metabolism (34). APOE plays several important roles in the central nervous system: cholesterol transport, neuroplasticity and inflammation (35). APOE binds to Aβ and influences the clearance of soluble Aβ and the Aβ aggregation (35, 36). APOE also regulates Aβ metabolism indirectly by interacting with receptors such as LRP1 (37). In APP transgenic mice, APOE influences the amount and structure of intraparenchymal Aβ deposits in an isoform specific manner (38-41). Neuropathological and neuroimaging studies demonstrate that APOEε4 carriers exhibit accelerated and more abundant Aβ deposition than APOEε4 negative individuals (42-44). APOEεgenotype is also associated with CSF Aβ42 and tau levels (15, 16, 43). Thus, genetic, cellular, animal and human studies demonstrate that APOE is a risk factor for LOAD and modifies AD pathogenesis via an APP-dependent manner.
CLU
Clusterin (CLU) is an apolipoprotein. Clusterin is a stress-activated chaperone protein that functions in apoptosis, complement regulation, lipid transport, membrane protection, and cell-cell interactions (45).
CLU is located on chromosome 8p21.1 and encodes 3 alternative transcripts (46). Several single nucleotide polymorphisms (SNPs) have been identified in CLU that confers protection against LOAD: rs11136000, rs9331888, rs2279590, rs7982, and rs7012010 (3-5, 13). Lambert et al reported an association of CLU rs9331896 with LOAD in 74,046 individuals (6). The functional impact of these polymorphisms is poorly understood. Rs9331888 is associated with expression of an alternative splice variant (36), while rs9331888 and rs11136000 are associated with plasma clusterin levels (47-49). Elevated clusterin plasma levels are also associated with brain atrophy, disease severity, and disease progression (50-52).
Prior to the identification of risk alleles in LOAD, clusterin was implicated in AD pathogenesis. Clusterin mRNA expression is elevated in AD brains (53, 54) and is detected in amyloid plaques (55, 56). Purified clusterin interacts with Aβ and influences fibril formation in vitro (57-59). Clusterin-deficient APP transgenic mice have reduced fibril formation, fewer dystrophic neurites, and altered soluble Aβ levels (60). Thus, clusterin likely influences Aβ clearance, amyloid deposition, and neuritic toxicity. APOE-and Clusterin deficient APP transgenic mice exhibit earlier and more extensive Aβ deposition than in control mice (61).
Clusterin is also associated with the complement system. Clusterin modulates the membrane attack complex, where it inhibits the inflammatory response associated with complement activation (45). Because neuroinflammation is a hallmark of AD, SNPs that alter clusterin expression or its functions as an amyloid response agent could impact AD pathogenesis and downstream effects.
ABCA7
ATP-binding cassette transporter A7 (ABCA7) is a member of the ABC transporter superfamily, where it functions to transport substrates across cell membranes (62). ABCA7 is located on chromosome 19p13.3 and can undergo alternate splicing to generate two transcripts, both of which are expressed in the brain (63).
GWAS in LOAD identified several SNPs near ABCA7 as risk alleles, including rs3764650 (3-6) and rs4147929, which was identified in a meta analysis of 74,046 individuals (6). Polymorphisms in this region increase LOAD risk. However, the impact of these polymorphisms on ABCA7 function and in AD is poorly understood (53, 64).
ABCA7 is expressed in hippocampal CA1 neurons and at 10-fold higher levels in microglia (65). rs3764650 in ABCA7 is associated with neuritic plaque burden in AD brains (20). ABCA7 mRNA expression in autopsy brain tissue is also associated with advanced cognitive decline (53, 64).
ABCA7 functions in the efflux of lipids from cells into lipoprotein particles. ABCA7-deficient mice exhibit only modest effects on lipid homeostasis compared with ABCA1-eficient mice (66, 67), suggesting that ABCA7 is not essential. In vitro, ABCA7 stimulates cholesterol efflux and inhibits Aβ secretion (68). ABCA7 also modulates phagocytosis of apoptotic cells by macrophages via the C1q complement pathway (69). Increasing ABCA7 expression also increases microglial phagocytosis of apoptotic cells, synthetic substrates, and Aβ (67, 69-71). APP transgenic mice that are ABCA7-deficient have increased Aβ deposition compared to the singly transgenic animals (67). Thus, ABCA7 may influence AD risk via cholesterol transfer to APOE or by clearing Aβ aggregates (67, 68, 72).
Immune Response
Neuroinflammation and dysregulation of the immune response is a central feature of AD (2). Common variants have been identified in several genes that are associated with LOAD in GWAS: CR1, CD33, MS4A, CLU, ABCA7, and EPHA1 (3-7, 13). Additionally, rare, coding variants were identified in TREM2 in sequencing studies of LOAD cohorts (9, 10).
CR1
Complement receptor 1 (CR1) encodes the CR1 protein. CR1 is a component of the complement response. CR1 is located on chromosome 1q32 in a cluster of complement-related proteins. CR1 encodes 4 isoforms that differ based on genetic duplication and deletions (73). CR1 expression on phagocytic cells, such as erythrocytes, results in the ingestion and removal of complement activated particles (74).
SNPs in CR1 were identified in GWAS in LOAD (3-6, 13). The SNP rs6656401 tags several SNPs that are strongly associated with AD risk. A second SNP, rs3818361, is associated with LOAD risk in APOEε4 carriers (13). Variants in the CR1 locus are also associated with neuroimaging measures associated with AD (19) and neuritic plaque burden in AD brains (20). CR1 mRNA expression in autopsy brain tissue is also associated with advanced cognitive decline (53).
CR1 is an interesting AD risk gene, as expression of complement factors are reportedly upregulated in affected regions of AD brains (75, 76). Neurons and glia are sources of complement in the brain (77-79). Additionally, material isolated from neurofibrillary tangles and amyloid plaques activates the complement system (80, 81).
CR1 encodes high expression and low expression alleles (74). Individuals who are homozygous for the low expression CR1 allele have fewer than 200 copies of CR1 per cell, while individuals who are homozygous for the high expression allele express nearly 1,400 copies per cell (73). Higher CR1 protein expression is associated with a higher clearance rate of immune complexes (82, 83). Clearance of plasma Aβ42 is dependent on C3b binding to CR1 (84). It is also hypothesized that Aβ42 activates the complement system (85). Because elevated complement cascade activity could exacerbate AD pathology, individuals with CR1 variants that dampen the complement response may be at lower risk of developing AD pathology.
CD33
CD33 is a member of the sialic acid-binding Ig-like lectin family of receptors and is expressed on myeloid cells and microglia (86-88). Sialic acid binding activates CD33, leading to monocyte inhibition via immunoreceptor tyrosine-based inhibitory motif (ITIM) domains (89). CD33 is also reported to play a role in clathrin-independent receptor-mediated endocytosis (90).
CD33 is located on chromosome 19q13.3. SNPs proximal to CD33 (e.g. rs3865444) were identified in LOAD GWAS that reduce LOAD risk (4, 5, 7). Rs3865444 is associated with an increase in CD33 lacking exon 2 (87) and rs12459419 modulates exon 2 splicing efficiency (87). Splicing of CD33 influences microglial activation (87). Rs3865444 failed to reach genome wide significance in the most recent study of 74,046 individuals; however, the strength of the biological findings makes CD33 an interesting player in AD (6).
CD33 mRNA expression is specifically increased in microglia and expression in autopsy brain tissue is associated with more advanced cognitive decline (53, 88). Aβ phagocytosis is inhibited in immortalized microglial cells expressing CD33, and this effect is abolished in cells expressing CD33 lacking exon 2 (88). The minor allele of rs3865444 is associated with reduced CD33 mRNA expression and insoluble Aβ42 in AD brains (88). CD33 positive immunoreactive microglia are also positively correlated with insoluble Aβ42 and plaque burden in AD brains (88). Thus, CD33 may play an important role in Aβ clearance and other neuroinflammatory pathways mediated by microglia in the brain.
MS4A
MS4A is a locus that contains several genes associated with the inflammatory response: MS4A4A, MS4A4E, MS4A6E. While this gene family is poorly characterized, MS4A is structurally similar to CD20 (91). CD20 regulates calcium influx following the activation of B-cell antigen receptor (92). MS4A genes are expressed in myeloid cells and monocytes.
GWAS in LOAD identified a SNP rs983392 (near MS4A6A) and rs670139 (near MS4A4E) as AD risk alleles (4-6). rs983392 is associated with reduced LOAD risk, while rs670139 is associated with increased LOAD risk. MS4A6E mRNA expression and rs670139 are associated with more advanced Braak tangle and plaque stages in AD brain tissue (53). However, the functional SNP(s) in this region has yet to be identified.
TREM2
TREM2 is a receptor expressed on microglia that stimulates phagocytosis and suppresses inflammation (93). TREM2 is located on chromosome 6q21.1 and occur as 3 transcripts. The longest transcript is a transmembrane protein that is trafficked to the cell surface where it interacts with DAP12 (a.k.a. TYROBP) and binds with several ligands (94). The transmembrane domain is missing from the shorter transcripts. While these transcripts have not been experimentally verified, they are predicted to be secreted.
Homozygous mutations in TREM2 are associated with autosomal recessive forms of dementia with bone cysts and fractures (95). Autosomal recessive mutations in TREM2 were also identified in a family with frontotemporal dementia (FTD)-like syndrome without bone involvement (96). Recently rare, missense mutations in TREM2 have been reported to increase LOAD risk. Gene based burden tests suggest that multiple rare, coding variants in TREM2 make increase risk for disease. The most common variant in European-descent populations, R47H (rs75932628), is reported to increase LOAD risk approximately two fold (9, 10, 97-99). However, there is some debate regarding the degree to which carrying TREM2 R47H increases AD risk: studies report a range of 1.7-3.4-fold increased AD risk in TREM2 R47H carriers (100, 101). TREM2 R47H is also associated with increased risk for Parkinson's disease, FTD and ALS (96, 97, 102, 103).
TREM2 mutation carriers with AD have more extensive brain atrophy than non-carriers with AD (104). Variants in the TREM2 region are also associated with CSF tau levels (15). Interestingly, after trafficking to the cell surface, TREM2 is cleaved by γ-secretase (105). TREM2 may play an important role in neurodegeneration, possibly in clearance of protein aggregates or in neuroinflammatory mechanisms.
Endocytosis
Endocytosis is critical for normal processing of APP, which is central to AD pathogenesis. Furthermore, synaptic activity and neurotransmitter release is disrupted in AD (2). Genes associated with endocytosis and synaptic function were identified in several GWAS of LOAD risk: BIN1, PICALM, CD2AP, EPHA1, SORL1 (3-6, 13).
BIN1
Bridging integrator 1 (BIN1) is involved in regulating endocytosis and trafficking, immune response, calcium homeostasis and apoptosis. BIN1 is located on chromosome 2q14.3 and is differentially spliced to 7 major transcripts (106). BIN1 interacts with clathrin and AP2/α-adaptin (107, 108) and binds to lipid membranes and induce membrane curvature (109).
GWAS identified SNPs in BIN1 increase risk for LOAD (3, 4). The most significant SNPs, rs744373 and rs7561528, are located more than 25kB upstream from the BIN1 coding region. The most recent LOAD GWAS of 74,046 individuals identified rs6733839 (6). Rs7561528 is associated with entorhinal cortical thickness and temporal pole cortical thickness (19). Rs59335482, in linkage disequilibrium with rs744373, is associated with elevated BIN1 mRNA expression and tau loads but not tangles in AD brains (110). BIN1 protein levels are altered in aged mice, AD mouse models and human AD brains (110, 111). In AD brains, elevated BIN1 mRNA expression levels are associated with delayed disease onset and shorter disease duration (53).
BIN1 may play a role in tau processing. BIN1 interacts with another microtubule associated protein (CLIP 170) (112). BIN1 and tau interact in neuroblastoma cells and mouse brains (110). BIN1 knockdown suppresses tau induced toxicity in a Drosophila model of AD (110).
BIN1 is also implicated in clathrin-mediated endocytosis and intracellular endosome trafficking, where it could modify APP trafficking (23, 113). BIN1 binds to GTPase dynamin (114). BIN1-deficient mice have impaired endocytic protein scaffolds and synaptic vesicle recycling (115).
BIN1 also plays an important role in senescence and apoptosis (116, 117). BIN1 is implicated in phagocytosis by macrophages and binds α-integrins, which regulate the immune response (118). Thus, while BIN1 has functions relevant to several aspects of disease pathogenesis, the exact role of BIN1 in disease and the functional variant associated with disease risk remains to be resolved.
PICALM
Phosphatidylinositol binding clathrin assembly protein (PICALM) encodes a protein involved in clathrin assembly. PICALM is located on chromosome 11q14 and results in 23 alternative transcripts. PICALM is predominantly expressed in neurons (119). SNPs 5’ to PICALM rs3851179 and rs541458 are associated with reduced LOAD risk (3, 6, 13). However, the functional effects of these SNPs remain to be determined.
PICALM recruits clathrin and adaptor protein complex 2 (AP2) to the cell membrane, where it plays a role in determining the amount of membrane to be recycled by regulating clathrin cage size (120). PICALM also plays an essential role in synaptic vesicle fusion to the presynaptic membrane via VAMP2 trafficking (121). Deletion of the PICALM homolog AP180 in Drosophila and yeast result in impaired clathrin-mediated endocytosis (122, 123). PICALM-deficient mice have no overt neurological phenotypes but display abnormal iron metabolism, which has been implicated in APP processing (124).
PICALM co-localizes with APP in vitro and in vivo (119). Disrupting PICALM expression alters APP trafficking in vitro, and overexpression of PICALM in vivo increases plaque deposition in AD transgenic mice (119). PICALM modulates Aβ-induced toxicity in a yeast model (125). PICALM binds to autophagosomes, suggesting that PICALM may also play a role in autophagy mediated Aβ clearance (126). Thus, targeting PICALM-mediated Aβ generation and clearance may influence accumulation of Aβ in AD brains.
CD2AP
CD2 associated protein (CD2AP) is a scaffolding protein that is involved in cytoskeletal reorganization and intracellular trafficking (127). CD2AP is located on chromosome 6q12. SNPs in CD2AP rs9296559 and rs9349407 are associated with increased LOAD risk (4, 5). CD2AP rs9349407 is associated with neuritic plaque burden in AD brains (20). Rs10948363 was most recently identified in a meta-analysis of 74,046 individuals (6). However, the putative functional SNP remains undetermined, and CD2AP mRNA expression is not altered in AD brains (53). Knockdown of a CD2AP fly ortholog in a Drosophila model of AD enhances tau neurotoxicity (128).
CD2AP is required for synapse formation (127), where it associates with Cbl, enodophilin, and synaptojanin. Lysosomal function is also impaired in cells from CD2AP-deficient mice, suggesting that CD2AP is a critical regulator of vesicular trafficking to the lysosome (129).
EPHA1
EPHA1 is a member of the ephrins family of tyrosine kinase receptors that binds to membrane-bound ephrins-A ligands on adjacent cells. This interaction leads to contact-dependent, bidirectional signaling to adjacent cells (130). EPHA1 is located on chromosome 7q34. A SNP near EPHA1, rs11767557, is associated with reduced LOAD risk (4, 5). Rs11771145 was associated with reduced LOAD risk in the largest GWAS study to date (6). However, there is no evidence that EPHA1 mRNA expression is altered in AD brains (53).
EPHA1 plays roles in cell and axonal guidance and synaptic plasticity (131, 132). EPHA1 is expressed by CD4-positive T lymphocytes and monocytes (133). Although the most strongly associated SNP is close to EPHA1, there are several other genes within the region of linkage disequilibrium defined by this SNP and thus the functional SNP could be in or affect expression of one of these neighboring genes.
SORL1
Sortilin related receptor L (SORL1) is involved in vesicle trafficking from the cell surface to the Golgi-endoplasmic reticulum. SORL1 is a member of the Vsp10p domain receptor family and is comprised of five type I transmembrane receptors.
SORL1 is located on chromosome 11q23.2. SORL1 was originally identified as an AD risk gene in candidate based approaches (134, 135). A recent GWAS in 74,046 individuals revealed that rs11218343 near SORL1 is associated with reduced AD risk (6).
SORL1 directs APP to endocytic pathways for recycling (134) and plays an important role in Aβ generation (136-138). SORL1-deficient mice have elevated Aβ levels (139). SORL1 mRNA expression is reduced in AD brains (140 142). SORL1 is also a receptor that binds lipoproteins, including APOE-containing particles, and mediates their uptake via endocytotic pathways (134). Thus, the role of SORL1 in controlling APP cleavage and APOE uptake may be critical to maintaining signaling functions in the brain.
Unknown
PLD3
Phospholipase D3 (PLD3) is a poorly characterized “non classical” member of the PLD protein family with no reported catalytic activity (143). PLD3 is located at chromosome 19q13.2 and is alternatively spliced into 25 predicted transcripts. Whole exome sequencing in LOAD families was coupled with genotyping in large case-control series to identify PLD3 V232M as an AD risk factor (11).
Classical PLD proteins catalyze the hydrolysis of phophatidylcholine to generate phosphatidic acid, which acts as an effector for clathrin mediated endocytosis (144) and have been implicated in AD pathogenesis (145-148). PLD3 is highly expressed in neurons in the hippocampus, entorhinal cortex, and frontal cortex. In vitro, co-expression of PLD3 with APP produces significantly lower extracellular Aβ levels by a mechanism that remains unknown (11).
New LOAD Risk Genes
Additional loci were identified in the largest LOAD GWAS to-date: CASS4, CELF1, DSG2, FERMT2, HLA-DRB5-DBR1, INPP5D, MEF2C, NME8, PTK2B, SLC24H4-RIN3, ZCWPW1 (6). Much less is known of the role of these genes in AD; however, many of these genes fit into known pathways that are altered in AD. HLA-DRB5-DRB1 and INPP5D are involved in the immune response. MEF2C is involved in the immune response and in synaptic function. PTK2B is involved in cell migration and synaptic function. CELF1, NME8, and CASS4 are involved in cytoskeletal function and axonal transport. CASS4 is implicated in APP and tau metabolism. FERMT2 is also implicated in tau metabolism (6). Importantly, several of these susceptibility loci occur in gene-dense regions; so, it remains unclear which gene is responsible for the association.
Conclusions
The identification of common and rare variants that contribute to AD risk has provided new opportunities to understand the mechanisms underlying AD. The majority of the genes recently identified affect Aβ production and clearance, highlighting the importance of this pathway in AD pathogenesis. As whole genome and exome sequencing studies in large datasets are completed, it is very likely that many more genes will be added to this list. It remains to be seen whether additional pathways are identified or whether most genes will fall into the already identified pathways and cellular mechanisms.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Dr. Karch and Dr. Goate report no biomedical financial interests or potential conflicts of interest.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013 doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, et al. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, et al. Potential late onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate {alpha} secretase activity. Hum Mol Genet. 2009;18:3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, et al. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature. 2013 doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 14.Cruchaga C, Kauwe JSK, Mayo K, Spiegel N, Bertelsen S, Nowotny P, et al. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer's disease. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer's disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauwe JSK, Cruchaga C, Bertelsen S, Mayo K, Latu W, Nowotny P, et al. Validating predicted biological effects of Alzheimer's disease associated SNPs using CSF biomarker levels. J Alzheimers Dis. 2010;21:833–842. doi: 10.3233/JAD-2010-091711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauwe JSK, Cruchaga C, Karch CM, Sadler B, Lee M, Mayo K, et al. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer's disease. PLoS One. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauwe JSK, Cruchaga C, Mayo K, Fenoglio C, Bertelsen S, Nowotny P, et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proceedings of the National Academy of Sciences. 2008;105:8050–8054. doi: 10.1073/pnas.0801227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9:e111–194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerreiro RJ, Gustafson DR, Hardy J. The genetic architecture of Alzheimer's disease: beyond APP. PSENs and APOE. Neurobiol Aging. 2012;33:437–456. doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thinakaran G, Koo EH. Amyloid precursor protein trafficking, processing, and function. J Biol Chem. 2008;283:29615–29619. doi: 10.1074/jbc.R800019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattson MP, Cheng B, Culwell AR, Esch FS, Lieberburg I, Rydel RE. Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the beta-amyloid precursor protein. Neuron. 1993;10:243–254. doi: 10.1016/0896-6273(93)90315-i. [DOI] [PubMed] [Google Scholar]
- 25.Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, et al. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS One. 2012;7:e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benitez BA, Karch CM, Cai Y, Jin SC, Cooper B, Carrell D, et al. The PSEN1, p.E318G variant increases the risk of Alzheimer's disease in APOE-epsilon4 carriers. PLoS Genet. 2013;9:e1003685. doi: 10.1371/journal.pgen.1003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin SC, Pastor P, Cooper B, Cervantes S, Benitez BA, Razquin C, et al. Pooled-DNA sequencing identifies novel causative variants in PSEN1, GRN and MAPT in a clinical early-onset and familial Alzheimer's disease Ibero-American cohort. Alzheimers Res Ther. 2012;4:34. doi: 10.1186/alzrt137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 30.Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. Embo J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, et al. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80:385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 33.Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, et al. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci U S A. 2013;110:E1807–1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, et al. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 39.Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, et al. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25:2803–2810. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, Holtzman DM. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer's disease. Neurobiol Dis. 2002;9:305–318. doi: 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- 42.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer's disease: allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 43.Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones SE, Jomary C. Clusterin. Int J Biochem Cell Biol. 2002;34:427–431. doi: 10.1016/s1357-2725(01)00155-8. [DOI] [PubMed] [Google Scholar]
- 46.Rizzi F, Caccamo AE, Belloni L, Bettuzzi S. Clusterin is a short half-life, poly-ubiquitinated protein, which controls the fate of prostate cancer cells. J Cell Physiol. 2009;219:314–323. doi: 10.1002/jcp.21671. [DOI] [PubMed] [Google Scholar]
- 47.Szymanski M, Wang R, Bassett SS, Avramopoulos D. Alzheimer's risk variants in the clusterin gene are associated with alternative splicing. Transl Psychiatry. 2011;1 doi: 10.1038/tp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xing YY, Yu JT, Cui WZ, Zhong XL, Wu ZC, Zhang Q, et al. Blood clusterin levels, rs9331888 polymorphism, and the risk of Alzheimer's disease. J Alzheimers Dis. 2012;29:515–519. doi: 10.3233/JAD-2011-111844. [DOI] [PubMed] [Google Scholar]
- 49.Schurmann B, Wiese B, Bickel H, Weyerer S, Riedel-Heller SG, Pentzek M, et al. Association of the Alzheimer's disease clusterin risk allele with plasma clusterin concentration. J Alzheimers Dis. 2011;25:421–424. doi: 10.3233/JAD-2011-110251. [DOI] [PubMed] [Google Scholar]
- 50.Thambisetty M, Simmons A, Velayudhan L, Hye A, Campbell J, Zhang Y, et al. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch Gen Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. Jama. 2011;305:1322–1326. doi: 10.1001/jama.2011.381. [DOI] [PubMed] [Google Scholar]
- 52.Kiddle SJ, Sattlecker M, Proitsi P, Simmons A, Westman E, Bazenet C, et al. Candidate Blood Proteome Markers of Alzheimer's Disease Onset and Progression: A Systematic Review and Replication Study. J Alzheimers Dis. 2013 doi: 10.3233/JAD-130380. [DOI] [PubMed] [Google Scholar]
- 53.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.May PC, Lampert-Etchells M, Johnson SA, Poirier J, Masters JN, Finch CE. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer's disease and in response to experimental lesions in rat. Neuron. 1990;5:831–839. doi: 10.1016/0896-6273(90)90342-d. [DOI] [PubMed] [Google Scholar]
- 56.Calero M, Rostagno A, Matsubara E, Zlokovic B, Frangione B, Ghiso J. Apolipoprotein J (clusterin) and Alzheimer's disease. Microsc Res Tech. 2000;50:305–315. doi: 10.1002/1097-0029(20000815)50:4<305::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 57.Matsubara E, Frangione B, Ghiso J. Characterization of apolipoprotein J-Alzheimer's A beta interaction. J Biol Chem. 1995;270:7563–7567. doi: 10.1074/jbc.270.13.7563. [DOI] [PubMed] [Google Scholar]
- 58.Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1 42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136:22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 59.Matsubara E, Soto C, Governale S, Frangione B, Ghiso J. Apolipoprotein J and Alzheimer's amyloid beta solubility. Biochem J. 1996;316(Pt 2):671–679. doi: 10.1042/bj3160671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Demattos RB, O'Dell MA, Parsadanian M, Taylor JW, Harmony JAK, Bales KR, et al. Clusterin promotes amyloid plaque formation and is critical for neuritic toxicity in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA. 2002;99:10843–10848. doi: 10.1073/pnas.162228299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeMattos RB, Cirrito JR, Parsadanian M, May PC, O'Dell MA, Taylor JW, et al. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- 62.Kim WS, Weickert CS, Garner B. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J Neurochem. 2008;104:1145–1166. doi: 10.1111/j.1471-4159.2007.05099.x. [DOI] [PubMed] [Google Scholar]
- 63.Ikeda Y, Abe-Dohmae S, Munehira Y, Aoki R, Kawamoto S, Furuya A, et al. Posttranscriptional regulation of human ABCA7 and its function for the apoA-I-dependent lipid release. Biochem Biophys Res Commun. 2003;311:313–318. doi: 10.1016/j.bbrc.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 64.Vasquez JB, Fardo DW, Estus S. ABCA7 expression is associated with Alzheimer's disease polymorphism and disease status. Neurosci Lett. 2013 doi: 10.1016/j.neulet.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim WS, Guillemin GJ, Glaros EN, Lim CK, Garner B. Quantitation of ATP-binding cassette subfamily-A transporter gene expression in primary human brain cells. Neuroreport. 2006;17:891–896. doi: 10.1097/01.wnr.0000221833.41340.cd. [DOI] [PubMed] [Google Scholar]
- 66.Kim WS, Fitzgerald ML, Kang K, Okuhira K, Bell SA, Manning JJ, et al. Abca7 null mice retain normal macrophage phosphatidylcholine and cholesterol efflux activity despite alterations in adipose mass and serum cholesterol levels. J Biol Chem. 2005;280:3989–3995. doi: 10.1074/jbc.M412602200. [DOI] [PubMed] [Google Scholar]
- 67.Kim WS, Li H, Ruberu K, Chan S, Elliott DA, Low JK, et al. Deletion of Abca7 increases cerebral amyloid-beta accumulation in the J20 mouse model of Alzheimer's disease. J Neurosci. 2013;33:4387–4394. doi: 10.1523/JNEUROSCI.4165-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan SL, Kim WS, Kwok JB, Hill AF, Cappai R, Rye KA, et al. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem. 2008;106:793–804. doi: 10.1111/j.1471-4159.2008.05433.x. [DOI] [PubMed] [Google Scholar]
- 69.Jehle AW, Gardai SJ, Li S, Linsel-Nitschke P, Morimoto K, Janssen WJ, et al. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J Cell Biol. 2006;174:547–556. doi: 10.1083/jcb.200601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka N, Abe-Dohmae S, Iwamoto N, Fitzgerald ML, Yokoyama S. HMG-CoA reductase inhibitors enhance phagocytosis by upregulating ATP-binding cassette transporter A7. Atherosclerosis. 2011;217:407–414. doi: 10.1016/j.atherosclerosis.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka N, Abe-Dohmae S, Iwamoto N, Yokoyama S. Roles of ATP-binding cassette transporter A7 in cholesterol homeostasis and host defense system. J Atheroscler Thromb. 2011;18:274–281. doi: 10.5551/jat.6726. [DOI] [PubMed] [Google Scholar]
- 72.Wildsmith KR, Holley M, Savage JC, Skerrett R, Landreth GE. Evidence for impaired amyloid beta clearance in Alzheimer's disease. Alzheimers Res Ther. 2013;5:33. doi: 10.1186/alzrt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krych-Goldberg M, Moulds JM, Atkinson JP. Human complement receptor type 1 (CR1) binds to a major malarial adhesin. Trends Mol Med. 2002;8:531–537. doi: 10.1016/s1471-4914(02)02419-x. [DOI] [PubMed] [Google Scholar]
- 74.Liu D, Niu ZX. The structure, genetic polymorphisms, expression and biological functions of complement receptor type 1 (CR1/CD35). Immunopharmacol Immunotoxicol. 2009;31:524–535. doi: 10.3109/08923970902845768. [DOI] [PubMed] [Google Scholar]
- 75.Eikelenboom P, Stam FC. Immunoglobulins and complement factors in senile plaques. An immunoperoxidase study. Acta Neuropathol. 1982;57:239–242. doi: 10.1007/BF00685397. [DOI] [PubMed] [Google Scholar]
- 76.Shen Y, Li R, McGeer EG, McGeer PL. Neuronal expression of mRNAs for complement proteins of the classical pathway in Alzheimer brain. Brain Res. 1997;769:391–395. doi: 10.1016/s0006-8993(97)00850-0. [DOI] [PubMed] [Google Scholar]
- 77.Terai K, Walker DG, McGeer EG, McGeer PL. Neurons express proteins of the classical complement pathway in Alzheimer disease. Brain Res. 1997;769:385–390. doi: 10.1016/s0006-8993(97)00849-4. [DOI] [PubMed] [Google Scholar]
- 78.Gasque P, Ischenko A, Legoedec J, Mauger C, Schouft MT, Fontaine M. Expression of the complement classical pathway by human glioma in culture. A model for complement expression by nerve cells. J Biol Chem. 1993;268:25068–25074. [PubMed] [Google Scholar]
- 79.Hosokawa M, Klegeris A, Maguire J, McGeer PL. Expression of complement messenger RNAs and proteins by human oligodendroglial cells. Glia. 2003;42:417–423. doi: 10.1002/glia.10234. [DOI] [PubMed] [Google Scholar]
- 80.McGeer PL, Akiyama H, Itagaki S, McGeer EG. Activation of the classical complement pathway in brain tissue of Alzheimer patients. Neurosci Lett. 1989;107:341–346. doi: 10.1016/0304-3940(89)90843-4. [DOI] [PubMed] [Google Scholar]
- 81.Shen Y, Lue L, Yang L, Roher A, Kuo Y, Strohmeyer R, et al. Complement activation by neurofibrillary tangles in Alzheimer's disease. Neurosci Lett. 2001;305:165–168. doi: 10.1016/s0304-3940(01)01842-0. [DOI] [PubMed] [Google Scholar]
- 82.Schifferli JA, Paccaud JP. Two isotypes of human C4, C4A and C4B have different structure and function. Complement Inflamm. 1989;6:19–26. doi: 10.1159/000463068. [DOI] [PubMed] [Google Scholar]
- 83.Gibson NC, Waxman FJ. Relationship between immune complex binding and release and the quantitative expression of the complement receptor, type 1 (CR1, CD35) on human erythrocytes. Clin Immunol Immunopathol. 1994;70:104–113. doi: 10.1006/clin.1994.1017. [DOI] [PubMed] [Google Scholar]
- 84.Rogers J, Li R, Mastroeni D, Grover A, Leonard B, Ahern G, et al. Peripheral clearance of amyloid beta peptide by complement C3-dependent adherence to erythrocytes. Neurobiol Aging. 2006;27:1733–1739. doi: 10.1016/j.neurobiolaging.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 85.Velazquez P, Cribbs DH, Poulos TL, Tenner AJ. Aspartate residue 7 in amyloid beta-protein is critical for classical complement pathway activation: implications for Alzheimer's disease pathogenesis. Nat Med. 1997;3:77–79. doi: 10.1038/nm0197-77. [DOI] [PubMed] [Google Scholar]
- 86.Crocker PR, Hartnell A, Munday J, Nath D. The potential role of sialoadhesin as a macrophage recognition molecule in health and disease. Glycoconj J. 1997;14:601–609. doi: 10.1023/a:1018588526788. [DOI] [PubMed] [Google Scholar]
- 87.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, et al. CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, et al. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Linnartz B, Neumann H. Microglial activatory (immunoreceptor tyrosine-based activation motif)- and inhibitory (immunoreceptor tyrosine-based inhibition motif)-signaling receptors for recognition of the neuronal glycocalyx. Glia. 2013;61:37–46. doi: 10.1002/glia.22359. [DOI] [PubMed] [Google Scholar]
- 90.Tateno H, Li H, Schur MJ, Bovin N, Crocker PR, Wakarchuk WW, et al. Distinct endocytic mechanisms of CD22 (Siglec-2) and Siglec-F reflect roles in cell signaling and innate immunity. Mol Cell Biol. 2007;27:5699–5710. doi: 10.1128/MCB.00383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Howie D, Nolan KF, Daley S, Butterfield E, Adams E, Garcia-Rueda H, et al. MS4A4B is a GITR-associated membrane adapter, expressed by regulatory T cells, which modulates T cell activation. J Immunol. 2009;183:4197–4204. doi: 10.4049/jimmunol.0901070. [DOI] [PubMed] [Google Scholar]
- 92.Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5:e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rohn TT. The triggering receptor expressed on myeloid cells 2: “TREMming” the inflammatory component associated with Alzheimer's disease. Oxid Med Cell Longev. 2013;2013:860959. doi: 10.1155/2013/860959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colonna M. TREMs in the immune system and beyond. Nat Rev Immunol. 2003;3:445–453. doi: 10.1038/nri1106. [DOI] [PubMed] [Google Scholar]
- 95.Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, et al. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet. 2002;71:656–662. doi: 10.1086/342259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guerreiro RJ, Lohmann E, Bras JM, Gibbs JR, Rohrer JD, Gurunlian N, et al. Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 2013;70:78–84. doi: 10.1001/jamaneurol.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giraldo M, Lopera F, Siniard AL, Corneveaux JJ, Schrauwen I, Carvajal J, et al. Variants in triggering receptor expressed on myeloid cells 2 are associated with both behavioral variant frontotemporal lobar degeneration and Alzheimer's disease. Neurobiol Aging. 2013;2077;34:e2011–2078. doi: 10.1016/j.neurobiolaging.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bertram L, Parrado AR, Tanzi RE. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1565. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 99.Benitez BA, Cruchaga C. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1567–1568. doi: 10.1056/NEJMc1306509#SA4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pottier C, Wallon D, Rousseau S, Rovelet-Lecrux A, Richard AC, Rollin Sillaire A, et al. TREM2 R47H variant as a risk factor for early-onset Alzheimer's disease. J Alzheimers Dis. 2013;35:45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- 101.Guerreiro R, Hardy J. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1569–1570. doi: 10.1056/NEJMc1306509. [DOI] [PubMed] [Google Scholar]
- 102.Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley WW, et al. TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson's disease. Mol Neurodegener. 2013;8:19. doi: 10.1186/1750-1326-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, et al. TREM2 Variant p.R47H as a Risk Factor for Sporadic Amyotrophic Lateral Sclerosis. JAMA Neurol. 2014 doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rajagopalan P, Hibar DP, Thompson PM. TREM2 and neurodegenerative disease. N Engl J Med. 2013;369:1565–1567. doi: 10.1056/NEJMc1306509#SA3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wunderlich P, Glebov K, Kemmerling N, Tien NT, Neumann H, Walter J. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) by ectodomain shedding and gamma-secretase dependent intramembranous cleavage. J Biol Chem. 2013 doi: 10.1074/jbc.M113.517540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ren G, Vajjhala P, Lee JS, Winsor B, Munn AL. The BAR domain proteins: molding membranes in fission, fusion, and phagy. Microbiol Mol Biol Rev. 2006;70:37–120. doi: 10.1128/MMBR.70.1.37-120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramjaun AR, McPherson PS. Multiple amphiphysin II splice variants display differential clathrin binding: identification of two distinct clathrin-binding sites. J Neurochem. 1998;70:2369–2376. doi: 10.1046/j.1471-4159.1998.70062369.x. [DOI] [PubMed] [Google Scholar]
- 108.McMahon HT, Wigge P, Smith C. Clathrin interacts specifically with amphiphysin and is displaced by dynamin. FEBS Lett. 1997;413:319–322. doi: 10.1016/s0014-5793(97)00928-9. [DOI] [PubMed] [Google Scholar]
- 109.Tsutsui K, Maeda Y, Seki S, Tokunaga A. cDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem Biophys Res Commun. 1997;236:178–183. doi: 10.1006/bbrc.1997.6927. [DOI] [PubMed] [Google Scholar]
- 110.Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang S, Liu T, Li S, Zhang X, Ding Q, Que H, et al. Comparative proteomic analysis of brains of naturally aging mice. Neuroscience. 2008;154:1107–1120. doi: 10.1016/j.neuroscience.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 112.Meunier B, Quaranta M, Daviet L, Hatzoglou A, Leprince C. The membrane-tubulating potential of amphiphysin 2/BIN1 is dependent on the microtubule-binding cytoplasmic linker protein 170 (CLIP-170). Eur J Cell Biol. 2009;88:91–102. doi: 10.1016/j.ejcb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/Ehd1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wigge P, McMahon HT. The amphiphysin family of proteins and their role in endocytosis at the synapse. Trends Neurosci. 1998;21:339–344. doi: 10.1016/s0166-2236(98)01264-8. [DOI] [PubMed] [Google Scholar]
- 115.Di Paolo G, Sankaranarayanan S, Wenk MR, Daniell L, Perucco E, Caldarone BJ, et al. Decreased synaptic vesicle recycling efficiency and cognitive deficits in amphiphysin 1 knockout mice. Neuron. 2002;33:789–804. doi: 10.1016/s0896-6273(02)00601-3. [DOI] [PubMed] [Google Scholar]
- 116.Galderisi U, Di Bernardo G, Cipollaro M, Jori FP, Piegari E, Cascino A, et al. Induction of apoptosis and differentiation in neuroblastoma and astrocytoma cells by the overexpression of Bin1, a novel Myc interacting protein. J Cell Biochem. 1999;74:313–322. [PubMed] [Google Scholar]
- 117.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wixler V, Laplantine E, Geerts D, Sonnenberg A, Petersohn D, Eckes B, et al. Identification of novel interaction partners for the conserved membrane proximal region of alpha-integrin cytoplasmic domains. FEBS Lett. 1999;445:351–355. doi: 10.1016/s0014-5793(99)00151-9. [DOI] [PubMed] [Google Scholar]
- 119.Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, et al. Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baig S, Joseph SA, Tayler H, Abraham R, Owen MJ, Williams J, et al. Distribution and expression of picalm in Alzheimer disease. J Neuropathol Exp Neurol. 2010;69:1071–1077. doi: 10.1097/NEN.0b013e3181f52e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Harel A, Wu F, Mattson MP, Morris CM, Yao PJ. Evidence for CALM in directing VAMP2 trafficking. Traffic. 2008;9:417–429. doi: 10.1111/j.1600-0854.2007.00694.x. [DOI] [PubMed] [Google Scholar]
- 122.Wendland B, Emr SD, Riezman H. Protein traffic in the yeast endocytic and vacuolar protein sorting pathways. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 123.Zhang B, Koh YH, Beckstead RB, Budnik V, Ganetzky B, Bellen HJ. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
- 124.Duce JA, Tsatsanis A, Cater MA, James SA, Robb E, Wikhe K, et al. Iron-export ferroxidase activity of β-amyloid precursor protein is inhibited by zinc in Alzheimer's disease. Cell. 2010;142:857–867. doi: 10.1016/j.cell.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Treusch S, Hamamichi S, Goodman JL, Matlack KE, Chung CY, Baru V, et al. Functional links between Abeta toxicity, endocytic trafficking, and Alzheimer's disease risk factors in yeast. Science. 2011;334:1241–1245. doi: 10.1126/science.1213210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer's APP CTF for terminal degradation via autophagy. Proc Natl Acad Sci U S A. 2013;110:17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 128.Shulman JM, Chipendo P, Chibnik LB, Aubin C, Tran D, Keenan BT, et al. Functional screening of Alzheimer pathology genome-wide association signals in Drosophila. Am J Hum Genet. 2011;88:232–238. doi: 10.1016/j.ajhg.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cormont M, Meton I, Mari M, Monzo P, Keslair F, Gaskin C, et al. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 2003;4:97–112. doi: 10.1034/j.1600-0854.2003.40205.x. [DOI] [PubMed] [Google Scholar]
- 130.Yamazaki T, Masuda J, Omori T, Usui R, Akiyama H, Maru Y. EphA1 interacts with integrin-linked kinase and regulates cell morphology and motility. J Cell Sci. 2009;122:243–255. doi: 10.1242/jcs.036467. [DOI] [PubMed] [Google Scholar]
- 131.Martinez A, Otal R, Sieber BA, Ibanez C, Soriano E. Disruption of ephrin-A/EphA binding alters synaptogenesis and neural connectivity in the hippocampus. Neuroscience. 2005;135:451–461. doi: 10.1016/j.neuroscience.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 132.Lai KO, Ip NY. Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Curr Opin Neurobiol. 2009;19:275–283. doi: 10.1016/j.conb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 133.Sakamoto A, Sugamoto Y, Tokunaga Y, Yoshimuta T, Hayashi K, Konno T, et al. Expression profiling of the ephrin (EFN) and Eph receptor (EPH) family of genes in atherosclerosis-related human cells. J Int Med Res. 2011;39:522–527. doi: 10.1177/147323001103900220. [DOI] [PubMed] [Google Scholar]
- 134.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70:887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spoelgen R, von Arnim CA, Thomas AV, Peltan ID, Koker M, Deng A, et al. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, et al. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 139.Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, Peng J, et al. Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer's disease. J Neurosci. 2008;28:12877–12886. doi: 10.1523/JNEUROSCI.4582-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, et al. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- 141.Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, et al. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62:640–647. doi: 10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Munck A, Bohm C, Seibel NM, Hashemol Hosseini Z, Hampe W. Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum. Febs J. 2005;272:1718–1726. doi: 10.1111/j.1742-4658.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 144.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 145.Cai D, Zhong M, Wang R, Netzer WJ, Shields D, Zheng H, et al. Phospholipase D1 corrects impaired betaAPP trafficking and neurite outgrowth in familial Alzheimer's disease-linked presenilin-1 mutant neurons. Proc Natl Acad Sci U S A. 2006;103:1936–1940. doi: 10.1073/pnas.0510710103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cai D, Netzer WJ, Zhong M, Lin Y, Du G, Frohman M, et al. Presenilin-1 uses phospholipase D1 as a negative regulator of beta-amyloid formation. Proc Natl Acad Sci U S A. 2006;103:1941–1946. doi: 10.1073/pnas.0510708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jin JK, Ahn BH, Na YJ, Kim JI, Kim YS, Choi EK, et al. Phospholipase D1 is associated with amyloid precursor protein in Alzheimer's disease. Neurobiol Aging. 2007;28:1015–1027. doi: 10.1016/j.neurobiolaging.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 148.Oliveira TG, Chan RB, Tian H, Laredo M, Shui G, Staniszewski A, et al. Phospholipase d2 ablation ameliorates Alzheimer's disease-linked synaptic dysfunction and cognitive deficits. J Neurosci. 2010;30:16419–16428. doi: 10.1523/JNEUROSCI.3317-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Guerreiro R, Bras J, Hardy J. SnapShot: genetics of Alzheimer's disease. Cell. 2013;155:968, 968–e961. doi: 10.1016/j.cell.2013.10.037. [DOI] [PubMed] [Google Scholar]