Abstract
5-hydroxymethylcytosine (5hmC), a novel modified cytosine, is oxidized from 5-methylcytosine (5mC) by the ten-eleven translocation (Tet) protein family. The specific distribution of 5hmC in mammalian brain and its roles in gene regulation suggest that 5hmC is important in brain development. 5hmC may also contribute to the mechanisms underlying neurological diseases. Here, we summarize the current knowledge of 5hmC, with an emphasis on its roles in neurodevelopmental and neurodegenerative disorders.
Keywords: 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), Neurodevelopmental disorder, Neurodegenerative disease, Ten-eleven translocation (Tet) protein
Introduction
“Epigenetics,” a term originally proposed by Conrad Waddington 75 years ago, is defined as the mitotically and/or meiotically stable, heritable changes in gene expression that are caused by mechanisms other than alterations in the genetic sequence (Berger et al., 2009; Dupont et al., 2009; Waddington, 1939). There are three widely recognized primary epigenetic marks: posttranslational modification of histones, cytosine modifications, and non-coding RNAs, all of which play crucial roles in many aspects of mammalian development, including stem cell self-renewal and differentiation and neurodevelopment (Davis et al., 2008; Li, 2002; Li and Zhao, 2008; Smith and Meissner, 2013; Yao and Jin, 2014). Among these epigenetic marks, methylation of the fifth position of cytosine (5-methylcytosine, 5mC), which exists symmetrically at CpG dinucleotides, is one of the best characterized and is involved in the regulation of gene transcription (Smith and Meissner, 2013). Methylation to 5mC is catalyzed by the “writers,” DNA methyltransferases (DNMTs), which either maintain cytosine methylation during DNA replication (e.g., DNMT1) or methylate DNA de novo during development (e.g., DNMT3A and DNMT3B). 5mC is then recognized by the “readers,” methyl-CpG binding protein 2 (MeCP2) and methyl-CpG-binding domain proteins 1–4 (MBD1–4) (Bogdanovic and Veenstra, 2009; Hendrich and Bird, 1998; Jones and Liang, 2009; Okano et al., 1999).
For decades, 5mC was thought to be the only epigenetic mark on DNA. Despite being reported decades earlier in bacteria (Wyatt and Cohen, 1953) and mammals (Penn et al., 1972), the importance of 5-hydroxymethylcytosine (5hmC), another modified DNA base, went unrecognized until 2009, when two independent research teams demonstrated the presence of 5hmC in mouse Purkinje neurons and embryonic stem cells (ESCs) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Conversion of 5mC to 5hmC is catalyzed by the 5hmC “writers,” the novel ten-eleven translocation (Tet) protein family (Ito et al., 2011). 5hmC is also recognized by its own dynamic binding proteins (5hmC “readers”) (Spruijt et al., 2013). 5hmC plays an essential role in DNA demethylation since it can be further oxidized by Tet proteins to produce 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), which are quickly removed from the genome by thymine-DNA glycosylase (TDG) to initiate base excision repair (BER) (He et al., 2011; Ito et al., 2011; Maiti and Drohat, 2011).
5hmC is highly enriched in the central neural system (CNS). For example, 5hmC levels in the brain are approximately ten-fold higher than in ESCs (Globisch et al., 2010). Using the 5hmC-selective chemical-labeling approach, we and others have determined the unique, conserved patterns of 5hmC expression in the mouse cerebellum and hippocampus. We found that 5hmC is depleted along the X chromosome, but accumulates at intragenic- and exon-enriched loci, and that regulation of 5hmC may play a role in neurological disorders (Song et al., 2011; Szulwach et al., 2011). 5hmC is now recognized as an important epigenetic mark in developmental and neurological disease and is the subject of many new studies. Here, we summarize recent progress in 5hmC research and discuss the role of 5hmC in neurodevelopmental and neurodegenerative disorders.
Generation, distribution and roles of 5hmC
Tet proteins generate 5hmC
The Tet protein family (the 5hmC “writers”) consists of three proteins: Tet1, Tet2, and Tet3. These are 2-oxoglutatate (2OG)- and Fe(II)-dependent dioxygenases that oxidize 5mC to 5hmC using alpha-ketoglutarate as a cosubstrate (Ito et al., 2010; Pastor et al., 2013; Tahiliani et al., 2009). Tet1 and Tet2 are highly expressed in ESCs, while Tet3 is present mostly in oocytes (Gu et al., 2011; Koh et al., 2011; Moran-Crusio et al., 2011). The Tet proteins share a conserved catalytic cysteine-rich domain in the C-terminus, while Tet1 and Tet3, but not Tet2, also contain a CXXC zinc finger domain in the N-terminus (Wu and Zhang, 2011). Although CXXC domains in DNMT1 and MBD1 are known to bind to unmethylated CpGs, the function of Tet CXXC domains is more complex (Frauer et al., 2011; Xu et al., 2011; Zhang et al., 2010). The CXXC domain of Tet3 has been found to be critical for targeting Tet3 to specific gene promoters, where Tet3 regulates the 5mC/5hmC status (Xu et al., 2012). In contrast, due to a gene triplication and subsequent inversion, the Tet2 gene has been split into two segments, such that the CXXC domain is encoded by a distinct gene, IDAX (also known as CXXC4) (Iyer et al., 2011; Iyer et al., 2009). In this case, IDAX binds unmethylated CpGs and recruits Tet2 to DNA (Ko et al., 2013).
Cell- and tissue-specific levels of 5hmC
In the initial studies of 5hmC in 2009, which used thin-layer chromatography and mass spectrometry, the total amount of 5hmC was estimated to account for 0.6% of all nucleotides in Purkinje neurons (40% of 5mC levels) and 0.03% in mouse ESCs (7% of 5mC levels) (Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). Since then, several new approaches for detecting 5hmC have been developed, allowing our lab and others to investigate the precise genome-wide distribution of 5hmC (Pastor et al., 2011; Song et al., 2011; Wu et al., 2011). These analyses have revealed that, unlike the relatively equal distribution of 5mC between different cell types, the overall 5hmC level varies widely between cell types and tissues (Nestor et al., 2012; Pastor et al., 2011). In addition, certain tissue-specific stem cells dynamically acquire 5hmC and 5mC during differentiation. For example, 26,044 dynamically hydroxymethylated loci (DhMLs) and 16,123 dynamically methylated loci (DMLs) were uncovered during the differentiation of human ESCs into neuronal progenitor cells (NPCs) (Kim et al., 2014).
Genomic distribution of 5hmC
The distribution of 5hmC across the genome in mouse brain has been explored by our group and others. It is particularly abundant in synaptic genes with a tissue-specific differential distribution at exon-intron boundaries, pointing to a possible additional role for 5hmC in RNA splicing in brain (Khare et al., 2012). In human and mouse ESCs, on the other hand, 5hmC is located mostly in gene bodies, promoters, and enhancers (Ficz et al., 2011; Wu et al., 2011). Enrichment of 5hmC in gene promoters is generally associated with high gene expression, which may be due to the function of 5hmC in overcoming 5mC-mediated gene silencing (Ficz et al., 2011). However, it has been pointed out that the 5hmC levels in gene bodies of ESCs are not simply correlated with gene expression levels; for instance, some genes (e.g., housekeeping genes) with very low 5hmC levels in promoters and gene bodies are highly expressed (Xu et al., 2011). Base-resolution 5hmC profile methods (oxBS-Seq and Tet-assisted bisulfite sequencing) have provided more detailed information about the location of 5hmC in ESCs and shown that 5hmC is enriched around, but not within, transcription factor consensus motifs (Booth et al., 2012; Yu et al., 2012). Moreover, 5hmC is consistently found to be enriched at euchromatin in both mouse ESCs and neuronal cells, while 5mC gradually accumulates in the heterochromatin (Chen et al., 2014; Ficz et al., 2011; Szulwach et al., 2011). Finally, 5hmC is preferentially distributed at cis-regulatory elements, and its enrichment is more significant in human ESCs than in mouse ESCs (Pastor et al., 2011; Szulwach et al., 2011; Wu et al., 2011).
Molecular functions of 5hmC: demethylation and recruitment of chromatin binding proteins
Because 5hmC is converted from 5mC and can subsequently be oxidized to 5fC and 5caC by Tet proteins, 5hmC has been recognized as an intermediate in the process of demethylation. 5hmC mediates demethylation by three distinct mechanisms: passive demethylation, DNA repair-based demethylation, and DNMT3-mediated demethylation. Passive demethylation is the result of poor binding between 5hmC and UHRF1 (ubiquitin-like, containing PHD and RING finger domain 1). Together, DNMT1 and its partner, UHRF1, are responsible for DNA methylation maintenance (Bostick et al., 2007; Sharif et al., 2007). The presence of 5hmC interferes with the maintenance of DNA methylation patterns due to the poor binding efficiency between 5hmC and UHRF1 relative to that between 5mC and UHRF1; this may result in a failure of DNMT1 recruitment (Frauer et al., 2011; Hashimoto et al., 2012) (Fig. 1a). 5hmC is also a critical component of DNA repair-based DNA demethylation. 5fC and 5caC, which are derived from 5hmC, can be excised by TDG, triggering DNA base excision repair (BER) to generate an unmethylated cytosine (He et al., 2011; Ito et al., 2011; Maiti and Drohat, 2011). Alternatively, 5hmC can be deaminated to 5-hydroxyuracil (5hmU) by AID (activation-induced cytidine deaminase) and APOBEC (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide), and subsequently removed by SMUG1 (single strand-selective monofunctional uracil DNA glycosylase 1) or TDG-mediated BER (Fig. 1b) (Cortellino et al., 2011). Finally, a recent study found that DNMT3A and DNMT3B have a dehydroxymethylation function in vitro (Chen et al., 2012a). Whether this activity occurs in vivo and, if it does occur, how DNMT3-mediated DNA demethylation affects gene regulation, are still unknown.
Figure 1. Biological roles of 5hmC in DNA demethylation and gene regulation.
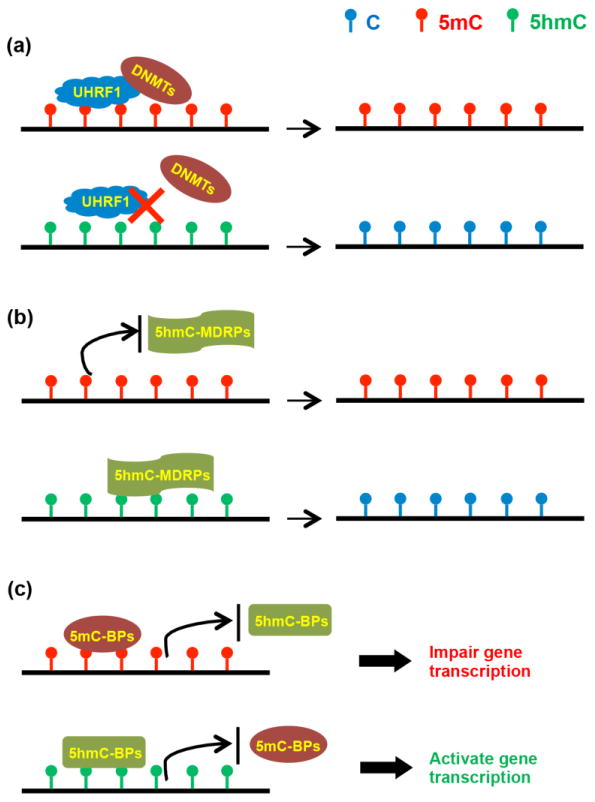
(a) The presence of 5hmC prevents the binding of UHRF1, inhibiting the recruitment of DNMTs, and eventually leading to passive demethylation. (b) 5hmC can be further oxidized or deaminated in the process of active demethylation with the help of 5hmC-mediated demethylation related proteins (5hmC-MDRPs). (c) The levels of 5mC and 5hmC affect which readers bind to DNA. 5mC-binding proteins (5mC-BPs) usually impair gene transcription, while enriched 5hmC improves gene expression by modulating the binding between 5hmC-binding proteins (5hmC-BPs) and chromatin.
Given the fact that 5hmC is found at such high levels in brain and that dynamic 5hmC readers have been identified, 5hmC is thought to be not merely an intermediate in DNA demethylation. In support of this, it has been observed that once established, 5hmC in mature neurons is maintained throughout adulthood (Chen et al., 2014). Data suggest that 5hmC plays an important role in the recruitment of chromatin-binding proteins and gene expression. A key function of 5hmC in gene regulation is the modulation of the binding between protein complexes and chromatin (Fig. 1c) (Spruijt et al., 2013). For example, methyl-CpG-binding domain protein 3 (MBD3), which binds 5mC poorly, is reported to bind 5hmC-enriched regions in mouse ESCs (Yildirim et al., 2011). Binding of MBD3 to chromatin is dependent on 5hmC; MBD3 appears to be important for the establishment or maintenance of global 5hmC through an interaction with Tet proteins (Yildirim et al., 2011).
The role of 5hmC in neuronal development
As has previously been reviewed, 5mC plays an essential role in neurogenesis in both developing and adult brains (Hirabayashi and Gotoh, 2010; Ma et al., 2010). However, less is known about the functions of 5hmC. The very high 5hmC levels in brain suggest that 5hmC is essential for proper neurodevelopment and that disruption of this system could play a role in neurological diseases. Indeed, we previously reported the marked acquisition of 5hmC in mouse neuronal cells from postnatal neurodevelopment through adulthood (Szulwach et al., 2011). The subsequent genome-wide analyses of 5hmC distribution in human cerebellum further revealed conserved characteristics of 5hmC in mammals (Szulwach et al., 2011). Furthermore, we identified fetus-specific and adult-specific DhMLs in the human cerebellum, which were enriched within exons and 5′ UTR regions; interestingly, fetus-specific DhMLs associated more strongly with TSSs than the adult-specific DhMLs (Wang et al., 2012). Specifically, we found that during development, DhMLs were highly enriched in genes encoding mRNAs that are regulated by fragile X mental retardation protein (FMRP), a RNA-binding protein that can inhibit mRNA translation and is associated with fragile X syndrome (FSX) and autism spectrum disorder (ASD). We have also reported an enrichment of intragenic 5hmC and an age-dependent acquisition of this modification in genes linked to neurodegenerative disease (Song et al., 2011; Szulwach et al., 2011). In addition, a recent study found that the distribution and localization of 5mC and 5hmC within chromatin, as well as interactions with 5mC- and 5hmC-binding proteins, changed throughout development and that these changes parallel neuronal differentiation and development (Chen et al., 2014). Consistent with the idea that 5hmC contributes to neuronal differentiation, a subset of 5hmC-binding proteins are specifically expressed in NPCs or brain, suggesting that 5hmC-mediated effects on gene regulation may be cell type- and tissue-specific (Spruijt et al., 2013). Taken together, these data suggest that the patterns of 5hmC acquired throughout development are critical for normal neurodevelopment and neurological function in the adult brain and that dysregulation of 5hmC may contribute to neurodevelopmental disorders or neurodegenerative diseases. In the sections below, we summarize and discuss the roles of 5hmC in selected neurodevelopmental and neurodegenerative disorders.
Roles of 5hmC in neurodevelopmental disorders
Rett syndrome
Rett syndrome (RTT) is a progressive neurodevelopmental disorder that causes mental retardation that affects females almost exclusively, with an incidence of 1/10,000 to 1/15,000 (Amir et al., 1999). In 1999, mutation of the gene MeCP2, which encodes X-linked methyl-CpG binding protein 2 (MeCP2), was revealed to be the primary cause of RTT. Because this is X-linked, mutation of MeCP2 in males is usually lethal (Amir et al., 1999). RTT was once thought to be an irreversible neurodevelopmental disease; however, activation of MeCP2 expression has successfully rescued the RTT phenotype in mice, suggesting that this is a feasible therapeutic target for the treatment of RTT in people (Castro et al., 2013; Giacometti et al., 2007; Guy et al., 2007).
As its name implies, MeCP2 binds 5mC (Lewis et al., 1992); interestingly, MeCP2 also binds 5hmC (Mellen et al., 2012). Mutation of MeCP2 interferes with normal regulation of 5mC and 5hmC. For example, our lab has previously found that the gene dosage of MeCP2 in mouse cerebellum is negatively correlated with the amount of global 5hmC and that complete loss of MeCP2 increased intragenic and transposable element–associated 5hmC (Szulwach et al., 2011). However, loss of MeCP2 leads to a specific reduction of 5hmC at DhMLs, pointing to different MeCP2 mechanisms at epigenetically dynamic and stable regions (Szulwach et al., 2011). Mellen et al. (2012) proposed a new MeCP2-5hmC-mediated model of cell-specific regulation of chromatin structure and gene expression. This model requires a high 5hmC:5mC ratio within the bodies of the expressed genes and occupation of 5hmC-binding sites by MeCP2 (Table 1). The specific contribution of these factors varyies between cell types, indicating MeCP2-5hmC-mediated gene regulation could be cell- and tissue-specific. These observations clearly indicate there is an interaction between MeCP2 and 5hmC that likely plays an important role in Rett syndrome.
Table 1. Roles of 5hmC in neurodevelopmental and neurodegenerative disorders.
| Disease | Epigenetic effect |
|---|---|
| Rett syndrome | Disruption of MeCP2-5hmC binding (Mellen et al., 2012) |
| Autism spectrum disorders | Altered 5hmC in and downregulation of GAD67 and RELN (Zhubi et al., 2014) |
| Schizophrenia | Increased genome-wide 5hmC and downregulation of GAD67 and BDNF (Dong et al., 2012) |
| Fetal alcohol spectrum disorders | Delayed acquisition of 5hmC and 5mC during development in the hippocampus (Chen et al., 2013) |
| Alzheimer's disease (AD) | Increased 5hmC in AD brains (Coppieters et al., 2013) |
| Fragile X-associated tremor/ataxia syndrome | Enrichment of 5hmC specifically at cerebellar-specific enhancers (Yao et al., 2014) |
| Huntington's disease | Reduced 5hmC in 5′UTR of ADORA2A, possibly effecting splicing (Villar-Menendez et al., 2013); downregulation of Tet proteins (Wang et al., 2013) |
Further study is required to answer questions raised by this MeCP2-5hmC model of RTT. First, RTT patients carrying different mutations in MeCP2 present with a varying severity of a subset of symptoms. For example, RTT patients with mutation R133C, which preferentially impairs the binding between MeCP2 and 5hmC, present with a milder form characterized by delayed onset of regression, milder speech and motor deficits, and less severe feeding difficulties compared to patients with other mutations (Bebbington et al., 2008). It is not clear how the functional differences caused by these mutations result in these varied clinical features. Second, the binding of MeCP2 to 5hmC is recognized as an important step in decoding 5hmC in the CNS (Mellen et al., 2012). MeCP2 interacts with both 5mC and 5hmC, and it is still unclear how these two pathways interact to affect gene regulation. Analyzing the relationships between MeCP2, 5mC, and 5hmC in glial cells, which were recently found to play important roles in mouse models of RTT (Derecki et al., 2012; Lioy et al., 2011), may help to clarify the mechanism.
Autism spectrum disorders
Autism spectrum disorders (ASDs) comprise a clinically heterogeneous group of disorders that share the common features of impaired social relationships, impaired language and communication, and a limited range of interests and behavior (Kelleher and Bear, 2008). Although there is significant overlap between autism and other developmental disorders, the deficient social relationships distinguish ASDs from other developmental disorders (Rapin and Tuchman, 2008).
Only a few studies have explored the role of 5hmC in autism. Our lab examined 5hmC levels in the developing human cerebellum and found that overall 5hmC levels increased during development (Wang et al., 2012). Comparisons of 5hmC enrichment on all UCSC RefSeq genes, fragile X mental retardation protein (FMRP) target genes, and autism candidate genes revealed that, during development, DhMLs are highly enriched in genes regulated by FMRP or disrupted in autism, demonstrating that genes involved in autism are normally regulated by 5hmC (Wang et al., 2012). In addition, a recent study reported evidence of a MeCP2-5hmC-mediated pathway in ASD (Zhubi et al., 2014). This study found enrichment of 5hmC and increased recruitment of MeCP2 at the promoters but not the gene bodies of GAD67 (glutamic acid decarboxylase 67) and RELN (reelin), two genes downregulated in ASD, resulting in downregulation of these two genes in human cerebellar samples (Table 1). These findings support a MeCP2-5hmC-mediated pathway in ASD and underscore the importance of 5hmC in neurodevelopmental disorders.
Schizophrenia and psychotic disorders
Schizophrenia affects about 1% of the population; symptoms include psychosis, loss of drive and volition, and neurocognitive deficits and the precise diagnosis (schizophrenia or psychotic disorders) depends on the combination of symptoms present (van Os and Kapur, 2009). Diagnosis typically occurs in the 20s, with males presenting with diagnostic symptoms about five years earlier than females (Lewis and Levitt, 2002; van Os and Kapur, 2009). While many environmental and genetic factors have been identified, the specific causes of this disorder remain unknown. Recently, schizophrenia has been recognized as a neurodevelopmental disorder that begins earlier than the presentation of diagnosable clinical symptoms (Lewis and Levitt, 2002).
In recent years, a role of epigenetic modifications in schizophrenia has been recognized (Grayson and Guidotti, 2013; Labrie et al., 2012). Recently, Dong et al. (2012) observed an increase of Tet1 (but not Tet2 or Tet3) mRNA and protein in the parietal cortices of psychotic patients (schizophrenia and bipolar disorder), which accompanied an increase in genome-wide 5hmC and in the promoters of GAD67 and BDNF (brain-derived neurotrophic factor), two genes known to be downregulated in schizophrenia (Table 1). They also reported decreased expression of APOBEC3A and APOBEC3C in the cortex of psychotic patients, suggesting that the impairment of the demethylation pathway may cause the observed increase in genome-wide 5hmC in schizophrenia (Dong et al., 2012). Dysregulation of the demethylation pathways is supported by the finding that GADD45b, which is thought to coordinate the demethylation process by recruiting deaminases and glycosylases to promoters, is increased in the parietal cortex of psychotic subjects (Barreto et al., 2007; Cortellino et al., 2011; Gavin et al., 2012; Rai et al., 2008; Schmitz et al., 2009). These studies highlight a previously unrecognized role for hydroxymethylation and the demethylation pathway in the etiology of schizophrenia and psychotic disorders. Further study is needed to expand on these findings.
Fetal alcohol spectrum disorder
Maternal consumption of alcohol during pregnancy severely affects the developing fetus, leading to various degrees of developmental deficits and growth retardation, including but not limited to fetal alcohol syndrome (FAS). The prevalence of FAS is between 0.5 and 2 per 1,000 live births worldwide (May and Gossage, 2001). Children with FAS suffer from a range of neurodevelopmental deficits, including memory impairment, learning deficits, and affective disorders (Kodituwakku, 2009). Recently, FAS has been re-categorized into a broader class, termed fetal alcohol spectrum disorders (FASD) (Kodituwakku, 2009).
Alcohol directly influences DNA methylation by inhibiting synthesis of methionine, resulting in the decreased production of SAM (S-adenosylmethionine), the cosubstrate for DNMT-mediated methylation of cytosine (Bonsch et al., 2006; Resendiz et al., 2013). A recent immunocytochemistry-based study of the normal pattern of DNA methylation and hydroxymethlation throughout development found that alcohol exposure can delay the acquisition of proper patterns of these DNA modifications (Chen et al., 2013) (Table 1). Alteration of these DNA methylation programs by alcohol interferes with hippocampal neuronal differentiation and maturation and these changes are correlated with developmental retardation (Chen et al., 2013). Together, these results indicate that alcohol affects 5mC and 5hmC during neuronal differentiation, although further studies are required to investigate the mechanism by which alcohol affects these DNA methylation programs in FASD.
Roles of 5hmC in neurodegenerative disorders
Alzheimer's disease
Alzheimer's disease (AD), a neurodegenerative disease characterized by a progressive decline in cognitive functions, is the most common neurodegenerative disease, with more than 15 million people affected worldwide (Blennow et al., 2006). Loss of neurons and synapses is typically observed in the cerebral cortex and certain subcortical regions in AD patient brains, and this loss results in severe atrophy of the affected regions, including degeneration in the temporal/parietal lobe, and parts of the frontal cortex and cingulate gyrus (Wenk, 2003). More than 90% of AD is late onset, with an age of onset of 60-65 years or older, and most of these cases are sporadic (Bekris et al., 2010). Early-onset AD begins before the age of 60, and most early-onset AD cases are familial (Bekris et al., 2010). The exact causes of sporadic AD are unknown, but it has been suggested that alterations in epigenetic processes caused by environmental risk factors could be involved in its pathophysiology (Bihaqi et al., 2012; Irier and Jin, 2012).
As discussed above, we have reported a potential role for 5hmC in age-related disease (Song et al., 2011; Szulwach et al., 2011). Interestingly, despite the fact that many changes in the brain associated with aging are due to oxidative stress, the aging-associated increase of hippocampal 5hmC in mice appears to be independent of oxidative stress and may be caused by changes in the activity, but not expression levels, of Tet proteins (Chen et al., 2012b). An earlier study suggested that a variety of epigenetic marks, including 5mC, and DNMT1 are reduced in AD brains (Mastroeni et al., 2010). However, more recently, Coppieters et al. (2013) found a significant increase in 5mC and 5hmC in the middle frontal gyrus and middle temporal gyrus of AD brains (Table 1). In addition, these increased global levels of 5mC and 5hmC were positively correlated with each other and with markers of AD, including amyloid beta, tau, and ubiquitin load. Furthermore, a recent study ascertained epigenetic changes during AD progression by analyzing the hippocampus and parahippocampal gyrus of preclinical AD and late-stage AD patients and found significantly increased levels of Tet1, 5mC, and 5hmC, but decreased 5fC and 5caC levels (Bradley-Whitman and Lovell, 2013). The reasons for the differences among these studies are not clear. One potential explanation is that different brain regions were used in these studies and the levels of 5mC and 5hmC could vary between brain regions. These altered patterns of methylation in vulnerable brain regions prior to the onset of clinical symptoms lend further support to a role for DNA methylation in general, and 5hmC specifically, in the pathogenesis of AD.
Fragile X-associated tremor/ataxia syndrome
Clinically distinct from fragile X syndrome (FXS), fragile X-associated tremor/ataxia syndrome (FXTAS) is a late-onset (50-70 years old) neurodegenerative disorder associated with deficits in movement, memory, and the autonomic nervous system (Hagerman et al., 2004; Hagerman et al., 2001). Individuals with premutation alleles of the fragile X mental retardation-1 (FMR1) gene (55–200 CGG repeats) do not develop fragile X syndrome; however, a subset of male carriers develops FXTAS (Hagerman et al., 2004; Jacquemont et al., 2004).
Much of our current understanding of epigenetic mechanisms in FXTAS comes from the rCGG mouse model; these mice overexpress human rCGG repeats in the 5′ UTR of the FMR1 gene in Purkinje cells, causing intranuclear inclusions in Purkinje cells, Purkinje neuron cell death, and associated behavioral deficits (Hashem et al., 2009). Our lab recently investigated genome-wide 5hmC in this mouse model (Yao et al., 2014). Our analysis revealed that rCGG mice at 16 weeks of age showed an overall reduction in genome-wide 5hmC levels compared with age-matched wild-type littermates. In contrast, we identified regions enriched for 5hmC in these mice, specifically in cerebellum-specific, but not general, enhancers (Table 1). Furthermore, the DhMLs identified were highly correlated with genes and transcription factors important in neuronal development and neuronal function, including XBP1, AhR, and USF1/2 (Chen et al., 2003; Hayashi et al., 2007; Williamson et al., 2005). Accordingly, it seems that the presence or absence of 5hmC contributes to FXTAS pathogenesis by directly affecting these transcription factor binding sites. However, we still do not know the precise mechanism by which specific genes gain or lose 5hmC, and further research is needed to uncover these mechanisms.
Huntington's disease
Huntington's disease (HD) is an autosomal dominant, progressive neurodegenerative disorder characterized by chorea and dystonia, loss of coordination, cognitive decline, and behavioral problems, with a typical onset in middle age (Walker, 2007). The disease is caused by an expanded trinucleotide (CAG) repeat in the huntingtin gene that encodes a polyglutamine tract in the N-terminus of the huntingtin protein, resulting in a toxic gain of function that leads to HD (Tobin and Signer, 2000). Studies have explored epigenetic mechanisms in HD in the ADORA2A gene. ADORA2A encodes the adenosine A2A receptor (A2AR), a G-protein-coupled receptor that is highly expressed in basal ganglia; expression levels of A2AR are severely reduced in HD (Glass et al., 2000). A recent study found that reduced expression of the A2AR receptor in the putamen of HD patients was associated with reduced 5hmC levels, but increased 5mC levels, in the 5′UTR region of ADORA2A (Villar-Menendez et al., 2013) (Table 1). The same study found slightly different results in the R6 mouse model of HD, which is transgenic for the 5' end of the huntingtin gene, with approximately 120 +/- 5 repeat expansions (Mangiarini et al., 1996). In these mice, the striatal expression of A2AR receptor was also reduced; there was no change in 5mC or 5hmC in the 5′UTR region of ADORA2A in old mice, but 5hmC was reduced in younger mice. Reductions in 5hmC were also noted in exon m2. This location spans an exon-intron boundary, suggesting that 5hmC changes may be affecting A2AR mRNA splicing, leading to reduced expression (Khare et al., 2012).
Wang et al. (2013) also reported a genome-wide loss of 5hmC in striatum and cortex of the YAC128 (yeast artificial chromosome transgene with 128 CAG repeats) HD mouse model. The authors speculated that significant downregulation of the 5hmC writers, Tet2 and Tet3, and upregulation of the 5hmC reader, MeCP2, could be responsible for this loss of 5hmC. Significant loss of these proteins was seen in striatum, but not cortex, pointing to tissue-specific mechanisms of 5hmC reduction. A possible alternative mechanism is suggested by the decreased Tet1 expression in both striatum and cortex. Further, the expanded huntingtin protein is known to bind DNA directly (Benn et al., 2008), raising the possibility that it can act directly in the epigenetic modification process. Together, these data from mouse and humans implicate 5hmC in HD, but further research is required to determine the mechanisms behind this relationship.
Conclusions and outlook
Rapid advances in techniques for the detection of 5hmC have enabled us and others to determine its precise global distribution, providing essential information for analysis of the biological functions of 5hmC. These techniques have uncovered strong evidence for the critical function of 5hmC in brain development and related neurological disorders. Not only does 5hmC act as an intermediate in the DNA demethylation process, 5hmC-mediated pathways are critical for gene regulation. Many studies have begun to investigate the role of 5hmC in neurodevelopmental and neurodegenerative disorders; however, mechanistic studies of 5hmC-mediated processes are necessary. Moreover, there are other common neurodevelopmental and neurodegenerative disorders, such as Down's syndrome and Parkinson's disease, in which 5hmC has not been studied to date. Clearly, these ongoing and future studies have great potential to improve our understanding of these neurodevelopmental and neurodegenerative disorders.
Acknowledgments
The authors would like to thank Cheryl Strauss for critical reading of the manuscript. The authors are supported by grants from Natural Science Foundation of China (31329004) and in part by the National Institutes of Health (NS05163, NS079625 and HD073162).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bebbington A, Anderson A, Ravine D, Fyfe S, Pineda M, de Klerk N, Ben-Zeev B, Yatawara N, Percy A, Kaufmann WE, Leonard H. Investigating genotype-phenotype relationships in Rett syndrome using an international data set. Neurology. 2008;70:868–875. doi: 10.1212/01.wnl.0000304752.50773.ec. [DOI] [PubMed] [Google Scholar]
- Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. Journal of geriatric psychiatry and neurology. 2010;23:213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, Clark TW, Bouzou B, Cha JH. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. The Journal of neuroscience. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes & development. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi SW, Schumacher A, Maloney B, Lahiri DK, Zawia NH. Do epigenetic pathways initiate late onset Alzheimer disease (LOAD): towards a new paradigm. Current Alzheimer research. 2012;9:574–588. doi: 10.2174/156720512800617982. [DOI] [PubMed] [Google Scholar]
- Blennow K, de Leon MJ, Zetterberg H. Alzheimer's disease. Lancet. 2006;368:387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. Journal of neural transmission. 2006;113:1299–1304. doi: 10.1007/s00702-005-0413-2. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bradley-Whitman MA, Lovell MA. Epigenetic changes in the progression of Alzheimer's disease. Mechanisms of ageing and development. 2013;134:486–495. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro J, Mellios N, Sur M. Mechanisms and therapeutic challenges in autism spectrum disorders: insights from Rett syndrome. Current opinion in neurology. 2013;26:154–159. doi: 10.1097/WCO.0b013e32835f19a7. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. The Journal of biological chemistry. 2012a;287:33116–33121. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Dzitoyeva S, Manev H. Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restorative neurology and neuroscience. 2012b;30:237–245. doi: 10.3233/RNN-2012-110223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. The Journal of neuroscience. 2003;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Damayanti NP, Irudayaraj J, Dunn K, Zhou FC. Diversity of two forms of DNA methylation in the brain. Frontiers in genetics. 2014;5:46. doi: 10.3389/fgene.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PloS one. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiology of aging. 2013 doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derecki NC, Cronk JC, Lu Z, Xu E, Abbott SB, Guyenet PG, Kipnis J. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Translational psychiatry. 2012;2:e159. doi: 10.1038/tp.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Armant DR, Brenner CA. Epigenetics: definition, mechanisms and clinical perspective. Seminars in reproductive medicine. 2009;27:351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, Wang M, Qin W, Soding J, Spada F, Leonhardt H. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PloS one. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M, Dragunow M, Faull RL. The pattern of neurodegeneration in Huntington's disease: a comparative study of cannabinoid, dopamine, adenosine and GABA(A) receptor alterations in the human basal ganglia in Huntington's disease. Neuroscience. 2000;97:505–519. doi: 10.1016/s0306-4522(00)00008-7. [DOI] [PubMed] [Google Scholar]
- Globisch D, Munzel M, Muller M, Michalakis S, Wagner M, Koch S, Bruckl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS one. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38:138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Leavitt BR, Farzin F, Jacquemont S, Greco CM, Brunberg JA, Tassone F, Hessl D, Harris SW, Zhang L, Jardini T, Gane LW, Ferranti J, Ruiz L, Leehey MA, Grigsby J, Hagerman PJ. Fragile-X-associated tremor/ataxia syndrome (FXTAS) in females with the FMR1 premutation. American journal of human genetics. 2004;74:1051–1056. doi: 10.1086/420700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Human molecular genetics. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic acids research. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi A, Kasahara T, Iwamoto K, Ishiwata M, Kametani M, Kakiuchi C, Furuichi T, Kato T. The role of brain-derived neurotrophic factor (BDNF)-induced XBP1 splicing during brain development. The Journal of biological chemistry. 2007;282:34525–34534. doi: 10.1074/jbc.M704300200. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Molecular and cellular biology. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nature reviews. Neuroscience. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Irier HA, Jin P. Dynamics of DNA methylation in aging and Alzheimer's disease. DNA and cell biology. 2012;31(Suppl 1):S42–48. doi: 10.1089/dna.2011.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Progress in molecular biology and translational science. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Hagerman RJ, Leehey MA, Hall DA, Levine RA, Brunberg JA, Zhang L, Jardini T, Gane LW, Harris SW, Herman K, Grigsby J, Greco CM, Berry-Kravis E, Tassone F, Hagerman PJ. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature reviews. Genetics. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, Tice R, Tochigi M, Morera S, Nazarians A, Belsham D, Wong AH, Blencowe BJ, Wang SC, Kapranov P, Kustra R, Labrie V, Klimasauskas S, Petronis A. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nature structural & molecular biology. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Park YK, Kang TW, Lee SH, Rhee YH, Park JL, Kim HJ, Lee D, Lee D, Kim SY, Kim YS. Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Human molecular genetics. 2014;23:657–667. doi: 10.1093/hmg/ddt453. [DOI] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, Hogan PG, Aravind L, Rao A. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodituwakku PW. Neurocognitive profile in children with fetal alcohol spectrum disorders. Developmental disabilities research reviews. 2009;15:218–224. doi: 10.1002/ddrr.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie V, Pai S, Petronis A. Epigenetics of major psychosis: progress, problems and perspectives. Trends in genetics. 2012;28:427–435. doi: 10.1016/j.tig.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Levitt P. Schizophrenia as a disorder of neurodevelopment. Annual review of neuroscience. 2002;25:409–432. doi: 10.1146/annurev.neuro.25.112701.142754. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews. Genetics. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao X. Epigenetic regulation of mammalian stem cells. Stem cells and development. 2008;17:1043–1052. doi: 10.1089/scd.2008.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, Mandel G. A role for glia in the progression of Rett's syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nature neuroscience. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer's disease: decrements in DNA methylation. Neurobiology of aging. 2010;31:2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, Gossage JP. Estimating the prevalence of fetal alcohol syndrome. A summary. Alcohol research & health. 2001;25:159–167. [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, Perna F, Pandey S, Madzo J, Song C, Dai Q, He C, Ibrahim S, Beran M, Zavadil J, Nimer SD, Melnick A, Godley LA, Aifantis I, Levine RL. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome research. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews. Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. The Biochemical journal. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapin I, Tuchman RF. Autism: definition, neurobiology, screening, diagnosis. Pediatric clinics of North America. 2008;55:1129–1146. viii. doi: 10.1016/j.pcl.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Resendiz M, Chen Y, Ozturk NC, Zhou FC. Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics. 2013;5:73–86. doi: 10.2217/epi.12.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Molecular cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature reviews. Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature biotechnology. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, Munzel M, Wagner M, Muller M, Khan F, Eberl HC, Mensinga A, Brinkman AB, Lephikov K, Muller U, Walter J, Boelens R, van Ingen H, Leonhardt H, Carell T, Vermeulen M. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, Vasanthakumar A, Godley LA, Chang Q, Cheng X, He C, Jin P. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin AJ, Signer ER. Huntington's disease: the challenge for cell biologists. Trends in cell biology. 2000;10:531–536. doi: 10.1016/s0962-8924(00)01853-5. [DOI] [PubMed] [Google Scholar]
- van Os J, Kapur S. Schizophrenia. Lancet. 2009;374:635–645. doi: 10.1016/S0140-6736(09)60995-8. [DOI] [PubMed] [Google Scholar]
- Villar-Menendez I, Blanch M, Tyebji S, Pereira-Veiga T, Albasanz JL, Martin M, Ferrer I, Perez-Navarro E, Barrachina M. Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A2AR levels in Huntington's disease. Neuromolecular medicine. 2013;15:295–309. doi: 10.1007/s12017-013-8219-0. [DOI] [PubMed] [Google Scholar]
- Waddington CH. An introduction to modern genetics. G. Allen & Unwin; London: 1939. [Google Scholar]
- Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- Wang F, Yang Y, Lin X, Wang JQ, Wu YS, Xie W, Wang D, Zhu S, Liao YQ, Sun Q, Yang YG, Luo HR, Guo C, Han C, Tang TS. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington's disease. Human molecular genetics. 2013;22:3641–3653. doi: 10.1093/hmg/ddt214. [DOI] [PubMed] [Google Scholar]
- Wang T, Pan Q, Lin L, Szulwach KE, Song CX, He C, Wu H, Warren ST, Jin P, Duan R, Li X. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Human molecular genetics. 2012;21:5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk GL. Neuropathologic changes in Alzheimer's disease. The Journal of clinical psychiatry. 2003;64(Suppl 9):7–10. [PubMed] [Google Scholar]
- Williamson MA, Gasiewicz TA, Opanashuk LA. Aryl hydrocarbon receptor expression and activity in cerebellar granule neuroblasts: implications for development and dioxin neurotoxicity. Toxicological sciences. 2005;83:340–348. doi: 10.1093/toxsci/kfi031. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes & development. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. The Biochemical journal. 1953;55:774–782. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, Min J, Nicholson T, Chen T, Xu G, Shi Y, Zhang K, Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, Diao J, Wu F, He HH, Cui Q, Clark E, Ma C, Barbara A, Veenstra GJ, Xu G, Kaiser UB, Liu XS, Sugrue SP, He X, Min J, Kato Y, Shi YG. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Jin P. Cytosine modifications in neurodevelopment and diseases. Cellular and molecular life sciences. 2014;71:405–418. doi: 10.1007/s00018-013-1433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Lin L, Street RC, Zalewski ZA, Galloway JN, Wu H, Nelson DL, Jin P. Genome-wide alteration of 5-hydroxymethylcytosine in a mouse model of fragile X-associated tremor/ataxia syndrome. Human molecular genetics. 2014;23:1095–1107. doi: 10.1093/hmg/ddt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell research. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Dong E, Cook EH, Guidotti A, Grayson DR. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD) cerebellum. Translational psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]


