Abstract
The potential of PACE4 as a pharmacological target in prostate cancer has been demonstrated as this proprotein convertase is strongly overexpressed in human prostate cancer tissues and its inhibition, using molecular or pharmacological approaches, results in reduced cell proliferation and tumor progression in mouse tumor xenograft models. We developed a PACE4 high-affinity peptide inhibitor, namely, the multi-leucine (ML), and sought to determine whether this peptide could be exploited for the targeting of prostate cancer for diagnostic or molecular imaging purposes. We conjugated a bifunctional chelator 1,4,7-triazacyclononane-1,4,7- triacetic acid (NOTA) to the ML peptide for copper-64 (64Cu) labeling and positron emission tomography (PET)– based prostate cancer detection. Enzyme kinetic assays against recombinant PACE4 showed that the NOTA-modified ML peptide displays identical inhibitory properties compared to the unmodified peptide. In vivo biodistribution of the 64Cu/NOTA-ML peptide evaluated in athymic nude mice bearing xenografts of two human prostate carcinoma cell lines showed a rapid and high uptake in PACE4-expressing LNCaP tumor at an early time point and in PACE4-rich organs. Co-injection of unlabeled peptide confirmed that tumor uptake was target-specific. PACE4-negative tumors displayed no tracer uptake 15 minutes after injection, while the kidneys, demonstrated high uptake due to rapid renal clearance of the peptide. The present study supports the feasibility of using a 64Cu/NOTA-ML peptide for PACE4-targeted prostate cancer detection and PACE4 status determination by PET imaging but also provides evidence that ML inhibitor–based drugs would readily reach tumor sites under in vivo conditions for pharmacological intervention or targeted radiation therapy.
Abbreviations: ACN, acetonitrile; Ac, acetyl; 64Cu, copper-64; DCM, dichloromethane; [18F]-FDG, [18F]-fluoro-2-deoxy-d-glucose; HR-MS, high-resolution mass spectrometry; Ki, inhibitory constant; ML, multi-leucine; DMF, N,N′-dimethylformamide; NOTA, 1,4,7-triazacyclononane-1,4,7-triacetic acid; PC, proprotein convertase; PET, positron emission tomography
Introduction
Prostate cancer is the most prevalent cancer among men and is the second leading cause of cancer deaths in North American men [1]. Like the detection of primary tumors and subsequent metastases, the lack of novel approaches to improve aggressive prostate adenocarcinoma diagnosis is part of the important challenge that needs to be overcome for effective prostate cancer management. In the last two decades, diagnostic imaging using positron emission tomography (PET) has proven itself to be a useful tool for tumor and metastasis detection and assessment of treatment responses. Such modalities are met with the widely used radiolabeled metabolic PET tracer [18F]-fluoro-2-deoxy-d-glucose ([18F]-FDG) [2]. However, for several indications including prostate cancer, [18F]-FDG does not provide sufficient limit of detection due to low tumor avidity for this tumor type, which renders imaging difficult and furthermore does not allow tumor grading or discrimination of benign prostate hyperplasia [3], [4], [5], [6]. The use of target-specific PET imaging is an emerging way to overcome these issues and to generate novel radiopharmaceuticals for prostate cancer detection but also for molecular diagnostic as target-specific probe allows tumor target status determination [7], [8]. Still new target discovery remains a challenge, and the use of overexpressed proteins documented as pharmacological targets offers interesting possibilities for molecular imaging probe development, especially when inhibitors are available. In this context, molecular imaging also broadens the array of information as it can encompass tumor target status and define further treatment possibilities for each specific case analyzed, such as in personalized medicine approaches.
We recently reported the overexpression of the protease PACE4 in prostate cancer specimens [9]. Additionally, Oncomine (Compendia Bioscience, Ann Arbor, MI) resources, as well as two recent studies [10], [11], shows that PACE4 overexpression is observed in 90% to 95% of examined prostate tumors, correlating with aggressiveness. PACE4 is a member of the mammalian proprotein convertase (PC) family of enzymes that includes eight proteolytically active members, namely, furin, PC1/3, PC2, PC4, PC5/6, PC7, PACE4, and SKI-1/S1P [12], [13], [14]. PCs carry out their proteolytic processing functions in cell secretory pathways and are reported for their central role in cancer as their substrates include many cancer-related proteins that require an activation step before gaining their full biologic functions [15], [16]. PACE4 localizes both within and outside the cells, either in the secretory pathways [17], [18] or at the cell surface within the extracellular matrix [19], [20]. Through molecular silencing studies we demonstrated the importance of PACE4 in prostate cancer progression, especially through its role in cancer cell proliferation, growth factor activation, tumor growth, and neovascularization, making the case for PACE4 as a pharmacological target in prostate cancer [9], [21]. These findings led our group to the development of a high-affinity octapeptide inhibitor targeting PACE4 with considerable selectivity, namely, the multi-leucine (ML) peptide, Ac-LLLLRVKR-NH2 [22]. ML treatment phenocopies PACE4 knockdown leading to reduced proliferation and G0/G1 blockade of cell cycle progression in prostate cancer cells [12]. As small peptides are extremely valuable scaffolds for the generation of bioconjugated imaging probes [23], we hypothesized that this high-affinity inhibitor could be suitable for prostate cancer tumor detection as its target is highly expressed in prostate neoplasia, especially in basal and stromal cells [11], [24].
We therefore generated a PET suitable radiolabeled form of the ML peptide by the conjugation of a 1,4,7-triazacyclononane-1,4,7-triacetic acid (NOTA) chelating group at the N-terminal extremity that can further be quickly and efficiently used to generate radionuclide complexes such as with copper-64 (64Cu). 64Cu is a positron emitter with a low positron energy (Eβ +) of 656 keV that ensures high-resolution imaging [25] and a half-life of 12.7 hours. 64Cu can also be readily produced using a medical cyclotron, thus combining multiple advantages for molecular imaging [26]. In the present study, we demonstrate that the NOTA-conjugated PACE4 inhibitor maintains its affinity toward PACE4 enzyme using kinetic assays and distributes in PACE4-rich organs and within LNCaP prostate cancer xenografts, as supported by competition studies with unlabeled peptide co-injections. This supports the potential of PACE4 as a reachable target for prostate cancer detection but also for pharmacological intervention with such inhibitory compounds. The ability of the peptide to target the tumor could also provide novel information on tumor status, such as to profile tumor targets for availability in theranostic or personalized medicine applications.
Materials and Methods
Materials and Reagents
All solvents used were HPLC grade and used without further purification. TentaGel S RAM resin (0.22 mmol/g) was purchased from Rapp Polymere (Tübingen, Germany). All the amino acid derivatives and coupling reagents were purchased from ChemImpex International (Wood Dale, IL). Piperidine was obtained from A&C American Chemicals Ltd (Saint-Laurent, Québec). 1,4,7-Triazacyclononane was purchased from CheMatech (Dijon, France). All other reagents were purchased from Sigma-Aldrich (St Louis, MO). Preparative HPLC was carried out using a Waters Delta Prep 3000 module with a Gemini C18 (10 μm, 250 × 30 mm) column. HPLC–mass spectrometry (MS) purity and characterization analyses were performed on a Waters Alliance 2695 separation module coupled with a Micromass ZQ 2000 mass spectrometer equipped with an analytical column XBD-Eclipse plus C18 (1.8 μm, 3 × 50 mm). UPLC analyses were carried out on Acquity UPLC CLASS (Waters, Milford, MA) using an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) with either an ELSD detector or a radioactive detector (Bioscan photomultiplicator tube BFC-3200). The column was heated at 50°C and the analysis method used was a 15-minute gradient from 0% to 100% acetonitrile (ACN; 0.5 ml/min; 0.025% acetic acid). High-resolution MS (HR-MS; TripleTOF 5600; ABSciex, Foster City, CA) was also performed to confirm the identity of the pure products. According to HPLC, MS, and HR-MS, the purity of peptides exceeded 97%. Their physicochemical properties are presented in Table S1.
Peptide Synthesis
The peptide synthesis was performed on an automated system using Tentagel S RAM resin. Unless otherwise stated, all reactions were performed under nitrogen atmosphere. Water-sensitive reactions were performed in anhydrous solvents. N,N′-dimethylformamide (DMF) was dried over molecular sieves for 1 week before use. The resin was first loaded in the reaction column of the automatic peptide synthesizer (Pioneer Peptide Synthesis System; Applied Biosystems, Foster City, CA), and Fmoc amino acids were added in a three-fold excess using HATU in the presence of N,N-diisopropylethylamine. A deprotection step was performed using 20% piperidine in DMF. The NOTA chelate unit was synthesized using the procedure described by our group [27]. The N-terminal Fmoc was cleaved from the peptidyl resin using 50% piperidine for 30 minutes, followed by successive washing with DMF twice, dichloromethane (DCM) thrice, isopropanol, DCM, isopropanol, DCM, isopropanol, and DCM thrice. The resin was then treated with 2.5-fold equivalent of bromoacetic anhydride [preformed in DCM with diisopropylcarbodiimide (2.5 equivalents) and bromoacetic acid (5 equivalents), 15 minutes] for 30 minutes and then was washed as before. The resin was then suspended in DCM, and five-fold excess of 1,4,7-triazacyclononane was added followed by shaking for 3 hours before washing and suspending in N-methyl-2-pyrrolidone (NMP). t-Butyl bromoacetate (3 equivalents) was added and shaken for 2 hours before washing. The peptide was cleaved from the polymer solid support using a mixture of trifluoroacetic acid (TFA)/H2O/triisopropylsilane (95/2.5/2.5, vol/vol/vol) and stirred for 3 hours. The mixture was filtered, and the filtrate was precipitated in diethyl ether. After centrifugation and solvent decantation, the crude peptide was dissolved in a mixture of water and ACN, filtered, diluted in water, lyophilized, and then purified using preparative HPLC. The identity of the peptide was confirmed by MS and HR-MS.
Peptide Radiolabeling
The EBC-TR-19 cyclotron facility provides 64Cu isotope on a routine basis for research purposes, using a target system developed in collaboration with Advanced Cyclotron Systems Inc (Richmond, British Columbia). 64Cu was prepared following the 64Ni(p,n)64Cu reaction using an enriched 64Ni target electroplated on a rhodium disk [28]. [64Cu]CuCl2 was recovered from the target following the procedure described in McCarthy et al. [29] and converted to [64Cu]Cu[II] acetate ([64Cu]Cu(OAc)2) by dissolving the [64Cu]CuCl2 in ammonium acetate (0.1 M; pH 5.5). Peptide was labeled with 64Cu following conditions optimized in our laboratory [30]. Briefly, the peptide (5 μg) was dissolved in ammonium acetate buffer (0.1 M, pH 5.5) with [64Cu]Cu-(OAc)2 (300-370 MBq; 8-10 mCi) in a total volume of 300 to 350 μl. The resulting solution was incubated at 95°C for 10 minutes. The material was further diluted with water (5 ml) and applied on a C18 Sep-Pack Column (Waters) to eliminate unchelated 64Cu. Radiopeptide preparation was washed with 10 ml of water and eluted with 3 ml of ACN containing 0.025% TFA. The peptide fraction was collected, evaporated, and counted in a Capintec radioisotope calibrator (Capintec, Inc, Ramsey, NJ) to calculate the specific activity of the product. The resulting 64Cu/NOTA-ML. peptide was reconstituted in phosphate-buffered saline (PBS) at pH 7.4. Full peptide labeling was routinely assessed. Fluorine-18 was prepared by the 18O(p,n)18 F reaction on 18O enriched water as target material using the EBC-TR-19 cyclotron facility following established procedures and used for [18F]-FDG synthesis as described in Hamacher et al. [31].
In Vitro Kinetic Assays
PACE4 enzyme kinetic assays were performed using the fluorogenic substrate Pyr-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide (100 μM; Bachem, Torrance, CA.) in 20 mM bis-Tris (pH 6.5), 1 mM CaCl2, and 1.8 mg/ml BSA. Purified human recombinant PACE4 was obtained as described previously in Fugère et al. [32]. Assays were carried out at 37°C during 60 minutes, and real-time fluorescence was measured using a Gemini EM 96-well spectrofluorometer (Molecular Devices, Sunnyvale, CA; λEM = 370 nm; λEX = 460 nm; cutoff = 435 nm) as described in Levesque et al. [22]. Increasing concentrations of peptide were assayed to perform a competitive inhibition assay measured by substrate cleavage inhibition. Kinetics assays were analyzed using SoftMax Pro 5, and inhibitory constant (Ki) values were determined from IC50 using Cheng and Prusoff equation with 3.5 μM as PACE4 Km.
Cell Culture and Human Tumor Xenograft Models
LNCaP and PC3 human prostate cancer cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and were maintained in Roswell Park Memorial Institute medium (RPMI 1640) supplemented with 10% FBS (Wisent Bioproducts, St Bruno, Québec). PACE4 knockdown LNCaP were cultivated as described previously [21]. Cells were grown at 37°C in a water-saturated atmosphere with 5% CO2. Cells were grown in complete media and harvested at their exponential growing state. Four- to 6-week-old male athymic nude mice (Nu/Nu; Charles River Laboratories, LaSalle, Québec) were inoculated subcutaneously on the shoulders with a suspension of 1 × 106 PC3 cells in 100 μl of PBS or on the hips with 2.5 × 106 LNCaP cells in 200 μl of ice-cold PBS/Matrigel (1:1; BD Biosciences, Bedford, MA). Mice were housed under pathogen-free conditions and the inoculations were done under isoflurane anesthesia in a sterile laminar flow hood. Tumors were given a minimum of 4 weeks to grow to a minimal diameter of 5 mm before being used for biodistribution or PET imaging studies. All animal studies were conducted in compliance with the Canadian Council on Animal Care guidelines and with the approval of the Animal Care Committee of the Université de Sherbrooke.
Cell Uptake
For cell uptake experiments, 2.5 × 105 cells were seeded into 12-well plates 48 hours before the experiments were performed. Culture medium was replaced by fresh serum-free medium, and 180 to 200 kBq (5 μCi; 25 μl) of 64Cu/NOTA-ML were added to each well. After incubation, cells were washed three times with PBS and lysed using 10% sodium dodecyl sulfate. Cell lysate radioactive content was measured in a gamma counter (Cobra II auto-gamma counter; Packard, Minneapolis, MN). Precise cell counts (Trypan blue stain) were determined on mirror plates following the washing procedure. The results were expressed as percentage of the peptide dose per 106 cells.
In Vivo Stability Studies
For stability studies, the 64Cu-radiolabeled peptide reconstituted in PBS [20-30 MBq (500-800 μCi); 100 μl] was injected to air/isoflurane-anesthetized CD1 (Charles River Laboratories) mice through the caudal vein. Fifteen and 30 minutes following injection, blood was collected and centrifuged, and plasma protein precipitation with ACN (two volumes: 70% final concentration) followed. Samples were analyzed by UPLC with a radioactivity detector and retention times (RTs) were compared to the original radiolabeled peptide to monitor peptide integrity. For radio-thin layer chromatography (radio-TLC), blood was spotted directly on C18 TLC plates and sodium citrate buffer (0.1 M, pH 5.5) was used as a developing solvent. Image capture was performed on an Instant Imager scanner (Bioscan, DC Washington).
In Vivo Biodistribution
Biodistribution studies were performed by injecting 400 to 900 kBq (10-25 μCi; 100 μl) of 64Cu-NOTA-ML to isoflurane-anesthetized mice (either CD1 or tumor-bearing Nu/Nu mice) through the caudal vein. Doses were prepared and measured with a CRC-35R dose calibrator (Capintec, Inc). Fifteen minutes post-injection, the animals were killed by CO2 inhalation. Organs of interest were further collected, washed, weighed, and measured in a gamma counter. The results were expressed as percentage of the injected dose per gram of tissue (%ID/g). Target competition studies were performed with co-injections of 0.1 μmol of unlabeled peptide (without 64Cu chelated) to evaluate target-specific accumulation of the peptide. Experiments were realized with a minimum of three mice per group.
PET Imaging
PET scans were performed using a LabPET8 (Gamma Medica-IDEAS Inc., Sherbrooke, Quebec, Canada) small animal scanner with an axial field of view of 7.5 cm. Xenografted Nu/Nu mice were injected with 3.7 to 7.4 MBq (100-200 μCi; 100 μl) of 64Cu-NOTA-LLLLRVKR-NH2 (with or without co-injections of 0.1 μmol of unlabeled peptide) through the caudal vein under isoflurane anesthesia. Animal temperature was stabilized using a heated bed and monitored using a rectal probe. For [18F]-FDG tumor uptake assessment, 15 MBq (400 μCi; 100 μl) was injected. Each animal had a 20-minute dynamic scan immediately after compound injection. For cold peptide competition by co-injection, the same mice were reimaged after total radioactivity decay with both labeled and unlabeled peptides (0.1 μmol) injected together as a co-injection. The images were reconstructed by a three-dimensional MLEM algorithm implementing an analytically derived system matrix [33]. Regions of interest were traced for organs of interest and the respective activity was derived and reported to the injected dose per cubic centimeter of tissue for percentage calculation.
Statistical Analysis
All experiments were repeated at least three times (n = 3), and the results were expressed as mean ± SEM if not stated otherwise. Statistical analyses were done using Student’s t test to calculate P values.
Results
Peptide Synthesis
The ML peptide was synthesized by solid-phase automated synthesis with Fmoc amino acids followed by manual synthesis of the NOTA chelating moiety as described previously [27]. After purification (> 97% purity), peptide identity was confirmed by MS and HR-MS. Information regarding the analytical characterization of the obtained peptide is provided in Table S1. N-terminal extremity was favored on the basis of the previous observations that N-terminal modifications of the ML peptide (e.g., fluorescein isothiocyanate addition, alkylation, or PEGylation) were previously done without affecting the inhibitor potency toward PACE4 [22]. Using these fluorescent trackable analogs, peptide entry within cells was evaluated and found to readily penetrate within cells by its hydrophobic nature. The peptide (Figure 1A; MW calculated = 1294, found = 1294 g/mol) was purified by HPLC to a purity of > 97% and further used as starting material for each labeling with 64Cu.
Figure 1.
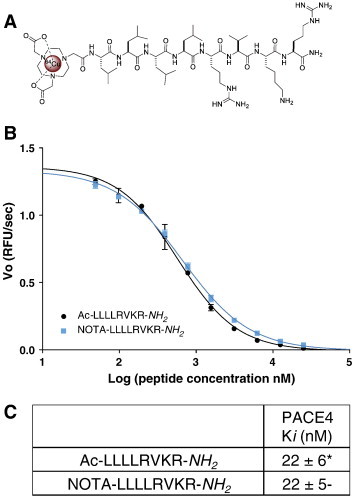
NOTA-LLLLRVKR-NH2 in vitro affinity toward PACE4. (A) Structure of the 64Cu-NOTA-LLLLRVKR-NH2. (B) PACE4 competitive enzyme kinetic assay with the cleavable fluorogenic substrate; Pyr-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide using increasing doses of both NOTA-ML and Ac-ML inhibitors. The presented plot is a representative experiment where Vo is the velocity of reaction in RFU/s. (B) Peptide Ki against recombinant PACE4. Data are mean ± SD (n = 3). *Data from Levesque et al. [22].
NOTA-Coupled ML Peptide Retains Original PACE4 Affinity and Is Retained in Prostate Cancer Cells Expressing PACE4
Unlabeled NOTA-ML peptide was first assayed against recombinant human PACE4 to assess that peptide intrinsic affinity for PACE4 was maintained despite the addition of the chelating group at the N-terminal extremity. Enzyme kinetic assay monitored by the cleavage of the fluorogenic substrate Pyr-Arg-Thr-Lys-Arg-methyl-coumaryl-7-amide (Figure 1, B and C) shows that both acetylated (Ac) and NOTA-coupled forms of the peptide display identical inhibition potencies (Ki = 22 ± 6 and 22 ± 5 nM, respectively) toward recombinant PACE4. To assess cell entry of the peptide in cells in vitro, cell uptake assays were performed on LNCaP cells and their counterparts knockdowned for PACE4 (shPACE4) [21] and compared to the PACE4-lacking PC3 cells. Uptake was performed on 2.5 × 105 cells up to 1 hour following addition of 64Cu/NOTA-ML. Radioactive peptide uptakes were measured within the cells following washes (Figure 2A). Whereas PC3 cell uptake reached a plateau within the first 20 minutes, LNCaP cells kept retaining the peptide over the full experiment as shown by the increase of the percentage of uptake over time. PACE4 knockdown LNCaP cells displayed a reduced uptake coherent with their attenuated levels of PACE4, as shown in Figure 2B. The difference between the uptake of LNCaP shPACE4 and PC3 cells is most likely attributable to the level of PACE4 distinguishing both cell lines, the shPACE4 cells still having considerable PACE4 compared to the PACE4-null PC3 cells.
Figure 2.
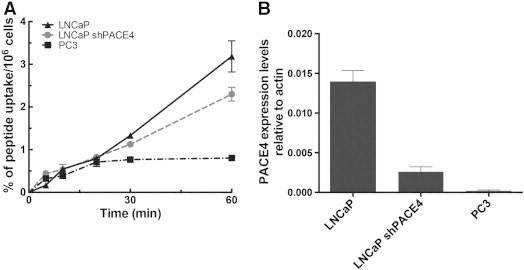
Peptide uptake within prostate cancer cells. (A) Time course of peptide uptake in LNCaP, PACE4 knockdown LNCaP (shPACE4), and PC3 cells. Percentage of the peptide dose measured in the cells was reported over cell number. Data are mean ± SD (n = 3). (B) Expression levels of PACE4 in the three cell lines determined by quantitative reverse transcription–polymerase chain reaction (data from [21]).
64Cu-NOTA-ML In Vivo Stability
NOTA-ML peptide was successfully radiolabeled with 64Cu with a yield not decay corrected greater than 95% and a specific activity of 74 to 93 TBq/mmol (2000 to 2500 Ci/mmol). As the peptide is strictly composed of natural amino acids, in vivo stability studies were first conducted to validate that imaging and biodistribution studies would be performed in an appropriate time range for the tracking of intact radiolabeled molecule instead of potential derived metabolites. CD1 mice were intravenously injected with 20 to 30 MBq of 64Cu-NOTA-ML through the tail vein. After 15 and 30 minutes, mouse blood samples were collected from the back paw and peptide integrity was analyzed by UPLC coupled to a radioactivity detector (Figure 3) and by radio-TLC (data not shown). At 15 minutes post-injection, circulating peptide was still uncleaved (RT = 5.1 minutes; Figure 3), whereas after 30 minutes, radioactive metabolites (RT = 3.1 and 3.9 minutes) were the main radioactive products detected in circulation. Another interesting observation is the absence of free 64Cu in mouse circulating blood and the apparition of radiolabeled metabolites, coherent with tight Cu chelation and the expected C-terminal degradation observed in mouse plasma ex vivo in Kwiatkowska et al. [34]. As C-terminal degradation would directly interfere with the peptide capability to recognize PACE4 by altering the PC recognition motif RVKR, molecular imaging beyond this window was avoided to avoid imaging metabolites.
Figure 3.
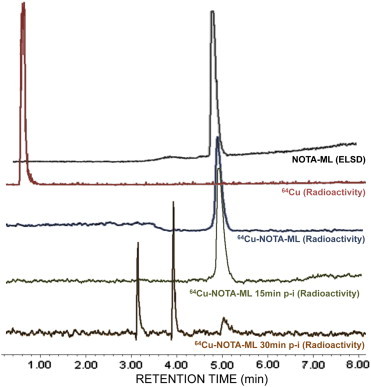
Radiolabeled peptide integrity analysis in vivo. UPLC analysis showing ELSD chromatogram for pure unlabeled compound (black trace) and radiometric profiles of free 64Cu (red) and radiolabeled compound after labeling (blue). The chromatograms of plasma taken from mice 15 minutes (green) and 30 minutes (orange) post-injection are also shown.
64Cu-NOTA-ML Distribution Is Specific to PACE4-Expressing Tissues In Vivo
Consistent with peptide stability, biodistribution studies were carried out on healthy CD1 mice 15 minutes post-injection, where circulating 64Cu-radiolabeled peptide is still unaltered. Following tail vein radiolabeled peptide injection, mice were sacrificed after 15 minutes and organs were collected to monitor radioactivity distribution. 64Cu/NOTA-ML peptide had a very high kidney uptake (50 ± 20%ID/g) 15 minutes post-injection. To distinguish target-specific uptake in organs from unspecific uptake, competitive biodistribution studies were realized with and without simultaneous injection of 0.1 μmol of unlabeled peptide together with the radiolabeled compound. Specific uptake can thereby be evaluated by comparing the difference between both conditions as unlabeled peptide molar ratio in the co-injection is greater than 500:1 (cold/radioactive). Figure 4A presents the %ID/g determined in each analyzed organ for both conditions in healthy mice. The data indicate that specific accumulation is significantly higher in the liver (13 ± 2% when unblocked vs 7 ± 1% when blocked), heart (7.4 ± 0.4% vs 4.3 ± 0.7%), and lungs (3.7 ± 0.5% vs 1.7 ± 0.2%), three known PACE4-rich organs (see Table 1). Moderate, but still significant, target-specific uptake was also observable in the pancreas (3.3 ± 0.1% vs 1.9 ± 0.2%), spleen (2.80 ± 0.06% vs 1.9 ± 0.2%), and bones (2.65 ± 0.09% vs 1.9 ± 0.2%).
Figure 4.
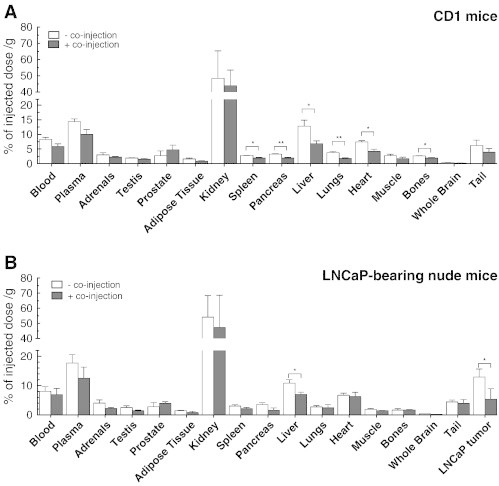
Radiopeptide biodistribution. Biodistribution at 15 minutes post-injection (A) in male CD1 mice and (B) in LNCaP xenograft-bearing Nu/Nu mice with or without co-injection of competitive non-radiolabeled peptide. Specific uptake can be visualized as the difference between the unblocked (− co-injection) and blocked (+ co-injection) conditions. Data are mean ± SEM (n = 3). *P < .05, **P < .01.
Table 1.
Comparison between 64Cu-NOTA-ML Target-Specific Distribution and PACE4 Reported Expression Pattern
| Organs | Specific Accumulation⁎ | Reported PACE4 Levels† | References |
|---|---|---|---|
| Liver | +++++ | ++++ | [47], [52], [53] |
| Heart | ++++ | ++++ | [47], [48] |
| Lungs | +++ | +++ | [47], [49] |
| Pancreas | ++ | +/− | [54] |
| Spleen | ++ | ++ | [47] |
| Bones | ++ | +/− | [55], [56] |
| Muscle | + | +/− | [57] |
| Adrenals | + | − | [47] |
| Adipose tissues | + | +/− | [58] |
| Testis | − | − | [47] |
Qualitative scoring based on co-injection blocking yields (difference in percentages between blocked and unblocked conditions).
Qualitative scoring based on reported tissue distribution; levels were estimated when possible by direct comparison of side-by-side results. “+/−” indicates data hardly comparable.
64Cu-NOTA-ML Targets LNCaP Prostate Cancer Xenografts and ML Does Not Accumulate in PACE4-Negative Xenografts
To further validate the hypothesis that 64Cu-NOTA-ML would accumulate in PACE4 expressing prostate tumor, biodistribution studies were carried out on LNCaP xenografted Nu/Nu mice. After sacrifice 15 minutes post-injection, radioactivity levels in tumors and organs were measured. Figure 4B illustrates the peptide distribution and illustrates a very high tumor uptake, which was blockable by more than half of the full signal by co-injection with unlabeled peptide (13 ± 2% vs 6 ± 2%). Peptide distribution patterns in both mouse strains were similar with modest blocking yields (either significant reductions or trends) in the organs with low uptake such as pancreas, spleen, and lungs. These low blocking yields are most likely due to the fact that PACE4 levels are low in these organs so that those observed in these organs come mostly from unspecific peptide uptake. Again liver-specific uptake, which was the organ with the highest specific uptake in CD1 healthy mice (Figure 4A), was clearly discernable in the nude mice (11 ± 1% vs 7.0 ± 0.8%).
μPET imaging was further performed on Nu/Nu mice xenografted with LNCaP cells. Fig. 5, A and B show coronal, sagittal, and transaxial images where tumors as well as liver uptake can be appreciated with and without cold peptide co-injections. Kidney and bladder were the main background sources following radiolabeled peptide injection. As illustrated on the full mouse projections, tissues and organs displayed low uptake and tumor uptake was clearly discernable. Regions of interest were traced on PET images to quantify uptake in tumors and the easily discernable organs, which are presented in Figure 5C. These PET-derived biodistribution data (express as %ID/cm3) are consistent with biodistribution observations (Figure 4B) and present blockable uptake in liver, heart, and LNCaP xenografts but not in muscles and kidneys. The blockability was easier to observe on image quantification than in the in vivo biodistributions, most likely due to the limitation of image-specific zone quantification in contrast to whole organs. Differences between blocking yields (Figure 4B) are also inherent to quantification methodologies as biodistribution data come from saline washed organs, whereas image quantification comprises circulating compound with the tissues.
Figure 5.
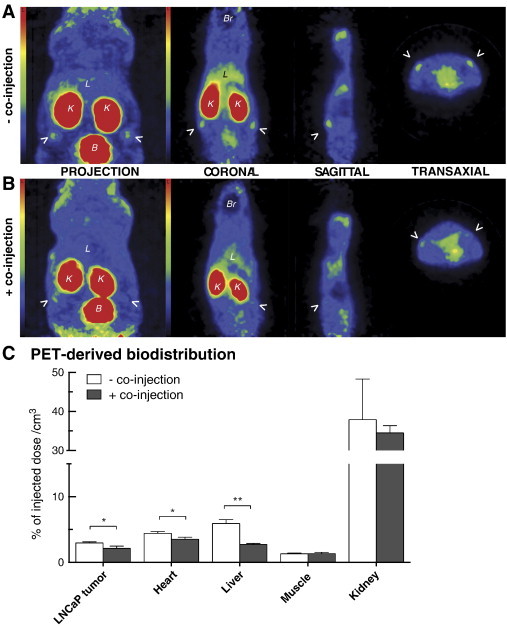
PET imaging of LNCaP tumor-bearing athymic nude mice. PET images and three-dimensional projection of 20-minute scans following injection of radiolabeled peptide (A) with or (B) without unlabeled peptide co-injection. Coronal, sagittal, and transaxial planes are shown for both conditions with their respective projections and scale bars. Arrows point out the LNCaP xenografts; L, liver; B, bladder; K, kidney; Br, brain. (C) PET-derived quantitative biodistribution with and without co-injection performed on a full 20-minute scan reconstruction. Pictures presented are taken from a representative mouse in both conditions. Data are mean ± SEM (n = 3). *P < .05, **P < .01.
To further demonstrate PACE4-specific uptake, PACE4-deficient PC3 cells were used to generate tumors. PET scan imaging of these PC3 xenografted mice with 64Cu-NOTA-ML showed that PC3 xenograft uptakes were remarkably low (more than four-fold lower as quantified on images), as shown in Figure 6. [18F]-FDG PET scans were performed to assess PC3 tumor uptake to validate proper vascularization and cell viability within the tumor, which might lead to data misinterpretation. As shown in Figure 6B, PC3 tumor [18F]-FDG uptake was faultless, confirming that tumor accessibility was not compromised by any means and that the peptide was able to access the tumor. This observation also supports the fact that the absence of signal is attributable to the lack of PACE4 in these tumor cells.
Figure 6.
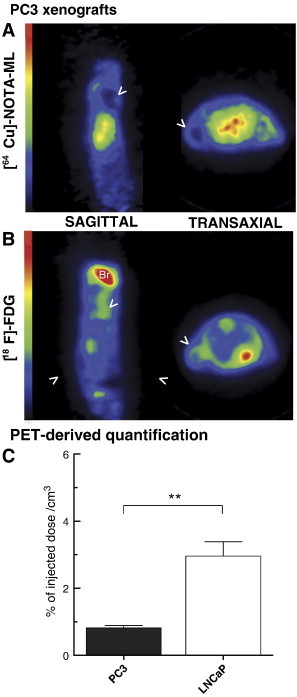
PACE4-lacking PC3 xenograft PET imaging. PET images from 20-minute scans of PC3 xenografted Nu/Nu mice following injection of (A) 64Cu-NOTA-ML or (B) [18F]-FDG. Sagittal and transaxial planes are shown for both compounds with their respective projections and scale bars. Arrows point out the PC3 xenografts; Br, brain. Pictures presented are taken from a representative mouse in both conditions. (C) PET-derived quantitative analyses of tumor uptake. Data are mean ± SEM (n = 3). **P < .01.
Discussion
Typical radiopeptides developed for PET imaging are cell surface receptor radioligands, which allow the binding of circulating molecules to receptor-expressing tumor sites and subsequent internalization [35]. Peptides present numerous advantages over antibodies as imaging tools due to their small size, ease of synthesis and radiolabeling, their rapid clearance, and the possibility to attain high specificity for their targets. Herein, we designed target-specific PET suitable imaging probes targeting the enzyme PACE4 by the addition of a NOTA chelating group on the N terminus of the already characterized ML peptide scaffold. Addition of such bifunctional chelate to a peptide significantly simplify its labeling using radiometal compared to labeling strategies using prosthetic group as single step labeling (10 minutes) is used to obtain high specific activity [30].
In this case, enzyme targeting was based on specific and tight interaction with PACE4 by the radiolabeled inhibitor. As previously described [22], NOTA moiety addition, just like other chemical modifications at the N terminus of the peptide, did not interfere with the C-terminal PC recognition pattern nor does it lead to a significant affinity variation toward PACE4, as assessed by enzyme kinetic assay where the nanomolar affinity constant (Ki = 22 nM) was preserved (Figure 1C). This is consistent with the well-characterized PC catalytic pocket known to accommodate substrates harboring the minimal motif RXXR. Thus, the RVKR motif within the ML peptide is essential for PC competitive inhibition, whereas the leucine extension is required for PACE4 selectivity [22]. When cell uptake was evaluated, cell entry of 64Cu-NOTA-ML was coherent with previous observation suggesting fast cell entry in a diffuse pattern [22]. However, this experiment enhanced the knowledge of ML peptide entry in cells by showing the relation between the compound retention and PACE4 levels (Figure 2).
As the ML peptide is composed of natural amino acids, its stability toward proteases and peptidases in biologic conditions was predictably compromised. In vivo stability studies (Figure 3) allowed us to establish that within 30 minutes of post-intravenous injection in mice, the circulating peptide was considerably degraded. However, at 15 minutes post-injection, the circulating molecule remained mostly uncleaved, indicating that further studies should be limited to no more than 20 minutes to prevent possible misinterpretation, such as obtaining signals that are due to metabolite accumulations. These in vivo stability data contrast considerably with those obtained in vitro with the ML peptide in plasma stability assay where the peptide half-life was 5 hours [34]. Consistent with the liver uptake, the main organ associated with first-pass metabolism, these results suggest that either liver or plasmatic proteases might be the main cause of 64Cu-NOTA-ML degradation in vivo. Moreover, the fact that the observed metabolites are still labeled with 64Cu (Figure 3) is coherent with the expected C-terminal degradation characterized in Kwiatkowska et al. [34].
Since PACE4 is overexpressed in prostate cancer [9], the rationale of using a specific high-affinity PACE4 inhibitor has great potential for the detection of prostate adenocarcinoma. LNCaP cell xenografts were used as they are the closest cell models to human clinical prostate adenocarcinoma [36] and because they have high PACE4 expression levels [21], [22]. As expected, radiolabeled peptide accumulation within LNCaP tumors was relatively high and was blocked by the co-injection of unlabeled peptide (Figures 4B and 5), indicating target-specific competition. Additionally, it is of interest to mention that since the competition was carried out with Cu-free peptide, the signal shift corresponding to specific binding in co-injection is directly related to the molecule. This discards the possibility of Cu-64Cu competition, especially since no free 64Cu was detected in circulating blood (Figure 3) as we previously observed in Fournier et al. [30]. Taken together, these data suggest that despite its short-term stability in blood circulation, the peptide is capable of specifically targeting prostate tumors, with high tumor uptake (14%; Figure 4B), and allows their visualization by PET imaging. This is probably enabled by the fast peptide uptake and retention by LNCaP cells, as observed in vitro (Figure 2A). This short-term stability could be a significant advantage for human studies, since a rapidly eliminated probe is a general requirement in most imaging studies for rapid accumulation at the target, not long-term period accumulation. Additionally, the fast kidney clearance can also be considered as an advantage since it is most likely the major contributor to the low image background, enhancing the contrast and hence tumor visualization, especially for distant metastasis. For further primary prostate tumor imaging in humans, combination with either Foley catheterization or diuretic administration should be envisioned in case of bladder signal interference.
Additionally, the PACE4-deficient PC3 xenografts were used to confirm peptide uptake specificity [22]. PET scans confirmed the very low uptake of 64Cu-NOTA-ML in these xenografts (Figure 6), lending support to the notion that this peptide could be used to determine tumor PACE4 expression status. This point is important to mention knowing that PACE4 expression is particularly widespread suggesting that molecular imaging of PACE4 alone might have some limitations. However, combined with other imaging modalities, getting information concerning tumor PACE4 status could be pertinent in clinical studies to distinguish prostate adenocarcinoma that are high PACE4 expressors as these patients may be better subjects for a PACE4-directed therapeutic intervention. PET-based approaches have previously been used to evaluate tumor status for estrogen receptor [37], [38], EGFR (epithelial growth factor receptor) [39], gastrin-releasing peptide receptor [30], [40], and integrins [41] using either radiolabeled antibodies or small molecule inhibitors and short peptides, to state some examples. The present proof of concept may be expanded since the ML peptide could be a potential radiotracer for other PACE4 overexpressing cancers such as breast, pancreas (as suggested by Oncomine databases), glioma [42], oral tongue [43], hepatocellular carcinoma [44], and ovarian cancer [45]. If true, each of these cancers could be further targeted for an anti-PACE4 therapy regimen [46].
The tumor-specific uptake of 64Cu-NOTA-ML indicates that systemic administration of the ML-based PACE4 targeting could be used as a therapeutic agent. The ML peptide inhibitor has already demonstrated important anti-proliferative properties on prostate cancer cells [22]. Furthermore, PACE4 gene silencing has dramatic effects on prostate cancer cells, leading to cell proliferation reduction and tumor progression blockade with attenuated angiogenesis in vivo [21]. While the specific uptake of the 64Cu-NOTA-ML probe in LNCaP xenografts is very promising for imaging applications, it remains to be seen if its therapeutic application can be equally effective, with the most important obstacles being an important kidney clearance and poor in vivo stability of the peptide. It is therefore clear that future improvements in pharmacokinetic properties, for example, through peptidomimetic modifications and/or substitution for unnatural amino acids, are required for in vivo pharmacological use. However, for imaging applications, such modifications may not necessarily be beneficial, as this may result in undesirable increased background levels.
In addition to proving itself useful for PACE4 PET imaging, the 64Cu-NOTA-ML probe also provided us with a biodistribution map of endogenous PACE4-expressing tissues. The images obtained are accurately mapped PACE4 biologic distribution [47]. The knowledge of PC distribution is mainly based on mRNA expression analyses (see Table 1). Mouse and human PACE4 proteins have a high homology (77%), which increase to 97% when comparing solely their catalytic domain, which is the binding domain of the ML peptide acting as an enzymatic inhibitor, which infers species specificity of the molecule. Aside from the pharmacokinetic limitations of the peptide and its rapid kidney clearance (Figure 4, Figure 5), the obtained biodistribution is consistent with reported expression studies, if we consider tracer accumulation versus blocking yields in each of the analyzed organs (Table 1). As for the brain, a PACE4-rich organ [48], [49], uptake was very low, most likely due to the inability of 64Cu-NOTA-ML probe to reach the brain (Figure 4), because of the blood-brain barrier impermeability to most peptides [50], [51].
Conclusion
The early detection of primary prostate cancer or its metastases, as with many other cancer types, remains an effective approach for effective management of this disease. PACE4 overexpression in prostate neoplastic cells represents a promising target for diagnosis and for therapy. We have customized a PACE4-selective peptide inhibitor for PET imaging and validated its target-specific uptake by human prostate cancer cell xenografts. We also determined that the radiopeptide organ distribution correlated well with endogenous PACE4 expression. These results strongly suggest that the described ML-based tracer is a useful tool to characterize PACE4-expressing tumors, which in turn could lead to specific therapies. The data also show that a pharmacological intervention with ML-based PACE4 inhibitors would likely distribute and accumulate in the tumor to exert their anti-proliferative effects.
The following are the supplementary data related to this article.
Physicochemical Properties of the Presented Peptide
Acknowledgments
The authors acknowledge the technical assistance of Marc-André Bonin (probe synthesis) and Jean-François Beaudoin (animal PET imaging). R.D. and B.G. are members of the Centre de Recherche Clinique Étienne-LeBel (Sherbrooke, Québec).
Footnotes
This work is funded by the Canadian Cancer Society (grant 701590) and the Fonds de Recherche du Québec-Santé (FRQ-S; Fond Innovant; to R.D. and B.G). This work is also supported by Prostate Cancer Canada and the Movember Foundation (to R.D.; grants 2012-951 and D2013-8). The Fondation Mon Étoile (http://fondationmonetoile.org) for cancer research also supports R.D. for this work. F.C. and C.L. hold PhD Student Scholarships from the FRQ-S. F.C. also holds a Graduate Studentship award from Prostate Cancer Canada (grant GS2014-02). Disclosure: There are no potential conflicts of interest with regard to this paper.
This article refers to supplementary material, which is designated by Table S1 and is available online at www.neoplasia.com.
Contributor Information
Robert Day, Email: robert.day@usherbrooke.ca.
Brigitte Guérin, Email: brigitte.guerin2@usherbrooke.ca.
References
- 1.American Cancer Society American cancer statistics. 2012. http://www.cancer.org
- 2.Chierichetti F., Pizzolato G. 18F-FDG-PET/CT. Q J Nucl Med Mol Imaging. 2012;56:138–150. [PubMed] [Google Scholar]
- 3.Effert P.J., Bares R., Handt S., Wolff J.M., Büll U., Jakse G. Metabolic imaging of untreated prostate cancer by positron emission tomography with 18fluorine-labeled deoxyglucose. J Urol. 1996;155:994–998. [PubMed] [Google Scholar]
- 4.Fletcher J.W., Djulbegovic B., Soares H.P., Siegel B.A., Lowe V.J., Lyman G.H., Coleman R.E., Wahl R., Paschold J.C., Avril N. Recommendations on the use of 18F-FDG PET in oncology. J Nucl Med. 2008;49:480–508. doi: 10.2967/jnumed.107.047787. [DOI] [PubMed] [Google Scholar]
- 5.Groves A.M., Win T., Haim S.B., Ell P.J. Non-[18F]FDG PET in clinical oncology. Lancet Oncol. 2007;8:822–830. doi: 10.1016/S1470-2045(07)70274-7. [DOI] [PubMed] [Google Scholar]
- 6.Salminen E., Hogg A., Binns D., Frydenberg M., Hicks R. Investigations with FDG-PET scanning in prostate cancer show limited value for clinical practice. Acta Oncol. 2002;41:425–429. doi: 10.1080/028418602320405005. [DOI] [PubMed] [Google Scholar]
- 7.Neves A.A., Brindle K.M. Assessing responses to cancer therapy using molecular imaging. Biochim Biophys Acta. 1766:242–261. doi: 10.1016/j.bbcan.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Kircher M.F., Hricak H., Larson S.M. Molecular imaging for personalized cancer care. Mol Oncol. 2012;6:182–195. doi: 10.1016/j.molonc.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Anjou F., Routhier S., Perreault J.P., Latil A., Bonnel D., Fournier I., Salzet M., Day R. Molecular validation of PACE4 as a target in prostate cancer. Transl Oncol. 2011;4:157–172. doi: 10.1593/tlo.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klee E.W., Bondar O.P., Goodmanson M.K., Dyer R.B., Erdogan S., Bergstralh E.J., Bergen H.R., III, Sebo T.J., Klee G.G. Candidate serum biomarkers for prostate adenocarcinoma identified by mRNA differences in prostate tissue and verified with protein measurements in tissue and blood. Clin Chem. 2012;58:599–609. doi: 10.1373/clinchem.2011.171637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S., Zhao Y., Hu K., Xu C., Wang L., Liu J., Yao A., Zhang H., Cao F. miR-124 exhibits antiproliferative and antiaggressive effects on prostate cancer cells through PACE4 pathway. Prostate. 2014;74(11):1095–2106. doi: 10.1002/pros.22822. [DOI] [PubMed] [Google Scholar]
- 12.Seidah N.G., Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 13.Couture F., D'Anjou F., Day R. On the cutting edge of proprotein convertase pharmacology: from molecular concepts to clinical applications. Biomol Concepts. 2011;2:421–438. doi: 10.1515/bmc.2011.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugère M., Day R. Cutting back on pro-protein convertases: the latest approaches to pharmacological inhibition. Trends Pharmacol Sci. 2005;26:294–301. doi: 10.1016/j.tips.2005.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatib A.M., Siegfried G., Chrétien M., Metrakos P., Seidah N.G. Proprotein convertases in tumor progression and malignancy: novel targets in cancer therapy. Am J Pathol. 2002;160:1921–1935. doi: 10.1016/S0002-9440(10)61140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassi D.E., Fu J., Lopez de Cicco R., Klein-Szanto A.J. Proprotein convertases: “master switches” in the regulation of tumor growth and progression. Mol Carcinog. 2005;44:151–161. doi: 10.1002/mc.20134. [DOI] [PubMed] [Google Scholar]
- 17.Tsuji A., Sakurai K., Kiyokage E., Yamazaki T., Koide S., Toida K., Ishimura K., Matsuda Y. Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim Biophys Acta. 2003;1645:95–104. doi: 10.1016/s1570-9639(02)00532-0. [DOI] [PubMed] [Google Scholar]
- 18.Fuller J.A., Brun-Zinkernagel A.M., Clark A.F., Wordinger R.J. Subtilisin-like proprotein convertase expression, localization, and activity in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2009;50:5759–5768. doi: 10.1167/iovs.08-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon V.M., Rehemtulla A., Leppla S.H. A role for PACE4 in the proteolytic activation of anthrax toxin protective antigen. Infect Immun. 1997;65:3370–3375. doi: 10.1128/iai.65.8.3370-3375.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nour N., Mayer G., Mort J.S., Salvas A., Mbikay M., Morrison C.J., Overall C.M., Seidah N.G. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol Biol Cell. 2005;16:5215–5226. doi: 10.1091/mbc.E05-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couture F., D'Anjou F., Desjardins R., Boudreau F., Day R. Role of proprotein convertases in prostate cancer progression. Neoplasia. 2012;14:1032–1042. doi: 10.1593/neo.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levesque C., Fugère M., Kwiatkowska A., Couture F., Desjardins R., Routhier S., Moussette P., Prahl A., Lammek B., Appel J.R. The multi-Leu peptide inhibitor discriminates between PACE4 and furin and exhibits antiproliferative effects on prostate cancer cells. J Med Chem. 2012;55(23):10501–10511. doi: 10.1021/jm3011178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reubi J.C., Maecke H.R. Peptide-based probes for cancer imaging. J Nucl Med. 2008;49:1735–1738. doi: 10.2967/jnumed.108.053041. [DOI] [PubMed] [Google Scholar]
- 24.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 25.Williams H.A., Robinson S., Julyan P., Zweit J., Hastings D. A comparison of PET imaging characteristics of various copper radioisotopes. Eur J Nucl Med Mol Imaging. 2005;32:1473–1480. doi: 10.1007/s00259-005-1906-9. [DOI] [PubMed] [Google Scholar]
- 26.Anderson C.J., Ferdani R. Copper-64 radiopharmaceuticals for PET imaging of cancer: advances in preclinical and clinical research. Cancer Biother Radiopharm. 2009;24:379–393. doi: 10.1089/cbr.2009.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guerin B., Ait-Mohand S., Tremblay M.C., Dumulon-Perreault V., Fournier P., Bénard F. Total solid-phase synthesis of NOTA-functionalized peptides for PET imaging. Org Lett. 2010;12:280–283. doi: 10.1021/ol902601x. [DOI] [PubMed] [Google Scholar]
- 28.Zeisler S.K., Pavan R.A., Orzechowski J., Langlois R., Rodrigue S., van Lier J.E. Production of 64Cu on the Sherbrooke TR-PET cyclotron. J Radioanal Nucl Chem. 2003;257:175–177. [Google Scholar]
- 29.McCarthy D.W., Shefer R.E., Klinkowstein R.E., Bass L.A., Margeneau W.H., Cutler C.S., Anderson C.J., Welch M.J. Efficient production of high specific activity 64Cu using a biomedical cyclotron. Nucl Med Biol. 1997;24:35–43. doi: 10.1016/s0969-8051(96)00157-6. [DOI] [PubMed] [Google Scholar]
- 30.Fournier P., Dumulon-Perreault V., Ait-Mohand S., Langlois R., Bénard F., Lecomte R., Guérin B. Comparative study of 64Cu/NOTA-[d-Tyr6, βAla11, Thi13, Nle14]BBN(6-14) monomer and dimers for prostate cancer PET imaging. EJNMMI Res. 2012;2:8. doi: 10.1186/2191-219X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamacher K., Coenen H.H., Stocklin G. Efficient stereospecific synthesis of no-carrier-added 2-[18F]-fluoro-2-deoxy-d-glucose using aminopolyether supported nucleophilic substitution. J Nucl Med. 1986;27:235–238. [PubMed] [Google Scholar]
- 32.Fugère M., Limperis P.C., Beaulieu-Audy V., Gagnon F., Lavigne P., Klarskov K., Leduc R., Day R. Inhibitory potency and specificity of subtilase-like pro-protein convertase (SPC) prodomains. J Biol Chem. 2002;277:7648–7656. doi: 10.1074/jbc.M107467200. [DOI] [PubMed] [Google Scholar]
- 33.Selivanov V.V., Picard Y., Cadorette J., Rodrigue S., Lecomte R. Detector response models for statistical iterative image reconstruction in high resolution PET. IEEE Trans Nucl Sci. 2000;47:1168–1175. [Google Scholar]
- 34.Kwiatkowska A., Couture F., Levesque C., Ly K., Desjardins R., Beauchemin S., Prahl A., Lammek B., Neugebauer W., Dory Y.L. Design, synthesis, and structure-activity relationship studies of a potent PACE4 inhibitor. J Med Chem. 2014;57(1):98–109. doi: 10.1021/jm401457n. [in press] [DOI] [PubMed] [Google Scholar]
- 35.Fani M., Maecke H.R., Okarvi S.M. Radiolabeled peptides: valuable tools for the detection and treatment of cancer. Theranostics. 2012;2:481–501. doi: 10.7150/thno.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai S., Sun Y., Squires J.M., Zhang H., Oh W.K., Liang C.Z., Huang J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate. 2011;71:1668–1679. doi: 10.1002/pros.21383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su H., Seimbille Y., Ferl G.Z., Bodenstein C., Fueger B., Kim K.J., Hsu Y.T., Dubinett S.M., Phelps M.E., Czernin J. Evaluation of [18F]gefitinib as a molecular imaging probe for the assessment of the epidermal growth factor receptor status in malignant tumors. Eur J Nucl Med Mol Imaging. 2008;35:1089–1099. doi: 10.1007/s00259-007-0636-6. [DOI] [PubMed] [Google Scholar]
- 38.Dehdashti F., Mortimer J.E., Siegel B.A., Griffeth L.K., Bonasera T.J., Fusselman M.J., Detert D.D., Cutler P.D., Katzenellenbogen J.A., Welch M.J. Positron tomographic assessment of estrogen receptors in breast cancer: comparison with FDG-PET and in vitro receptor assays. J Nucl Med. 1995;36:1766–1774. [PubMed] [Google Scholar]
- 39.Cai W., Chen K., He L., Cao Q., Koong A., Chen X. Quantitative PET of EGFR expression in xenograft-bearing mice using 64Cu-labeled cetuximab, a chimeric anti-EGFR monoclonal antibody. Eur J Nucl Med Mol Imaging. 2007;34:850–858. doi: 10.1007/s00259-006-0361-6. [DOI] [PubMed] [Google Scholar]
- 40.Fournier P., Dumulon-Perreault V., Ait-Mohand S., Tremblay S., Benard F., Lecomte R., Guerin B. Novel radiolabeled peptides for breast and prostate tumor PET imaging: 64Cu/and 68Ga/NOTA-PEG-[d-Tyr6, βAla11, Thi13, Nle14]BBN(6-14) Bioconjug Chem. 2012;23:1687–1693. doi: 10.1021/bc3002437. [DOI] [PubMed] [Google Scholar]
- 41.Haubner R., Weber W.A., Beer A.J., Vabuliene E., Reim D., Sarbia M., Becker K.F., Goebel M., Hein R., Wester H.J. Noninvasive visualization of the activated αvβ3 integrin in cancer patients by positron emission tomography and [18F]Galacto-RGD. PLoS Med. 2005;2:e70. doi: 10.1371/journal.pmed.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delic S., Lottmann N., Jetschke K., Reifenberger G., Riemenschneider M.J. Identification and functional validation of CDH11, PCSK6 and SH3GL3 as novel glioma invasion-associated candidate genes. Neuropathol Appl Neurobiol. 2012;38:201–212. doi: 10.1111/j.1365-2990.2011.01207.x. [DOI] [PubMed] [Google Scholar]
- 43.Estilo C.L., Oc P., Talbot S., Socci N.D., Carlson D.L., Ghossein R., Williams T., Yonekawa Y., Ramanathan Y., Boyle J.O. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurokawa Y., Matoba R., Nakamori S., Takemasa I., Nagano H., Dono K., Umeshita K., Sakon M., Monden M., Kato K. PCR-array gene expression profiling of hepatocellular carcinoma. J Exp Clin Cancer Res. 2004;23:135–141. [PubMed] [Google Scholar]
- 45.Longuespée R., Couture F., Levesque C., Kwiatkowska A., Desjardins R., Gagnon S., Vergara D., Maffia M., Fournier I., Salzet M. Implications of proprotein convertases in ovarian cancer cell proliferation and tumor progression: insights for PACE4 as a therapeutic target. Transl Oncol. 2014;7(3):410–419. doi: 10.1016/j.tranon.2014.04.008. [in press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehemtulla A. Cancer subclonal genetic architecture as a key to personalized medicine. Neoplasia. 2013;15:1410–1420. doi: 10.1593/neo.131972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seidah N.G., Chrétien M., Day R. The family of subtilisin/kexin like pro-protein and pro-hormone convertases: divergent or shared functions. Biochimie. 1994;76:197–209. doi: 10.1016/0300-9084(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 48.Beaubien G., Schäfer M.K., Weihe E., Dong W., Chrétien M., Seidah N.G., Day R. The distinct gene expression of the pro-hormone convertases in the rat heart suggests potential substrates. Cell Tissue Res. 1995;279:539–549. doi: 10.1007/BF00318166. [DOI] [PubMed] [Google Scholar]
- 49.Akamatsu T., Daikoku S., Nagamune H., Yoshida S., Mori K., Tsuji A., Matsuda Y. Developmental expression of a novel Kexin family protease, PACE4E, in the rat olfactory system. Histochem Cell Biol. 1997;108:95–103. doi: 10.1007/s004180050150. [DOI] [PubMed] [Google Scholar]
- 50.Egleton R.D., Davis T.P. Bioavailability and transport of peptides and peptide drugs into the brain. Peptides. 1997;18:1431–1439. doi: 10.1016/s0196-9781(97)00242-8. [DOI] [PubMed] [Google Scholar]
- 51.Witt K.A., Gillespie T.J., Huber J.D., Egleton R.D., Davis T.P. Peptide drug modifications to enhance bioavailability and blood-brain barrier permeability. Peptides. 2001;22:2329–2343. doi: 10.1016/s0196-9781(01)00537-x. [DOI] [PubMed] [Google Scholar]
- 52.Roebroek A.J., Taylor N.A., Louagie E., Pauli I., Smeijers L., Snellinx A., Lauwers A., Van de Ven W.J., Hartmann D., Creemers J.W. Limited redundancy of the proprotein convertase furin in mouse liver. J Biol Chem. 2004;279:53442–53450. doi: 10.1074/jbc.M407152200. [DOI] [PubMed] [Google Scholar]
- 53.Mori K., Imamaki A., Nagata K., Yonetomi Y., Kiyokage-Yoshimoto R., Martin T.J., Gillespie M.T., Nagahama M., Tsuji A., Matsuda Y. Subtilisin-like proprotein convertases, PACE4 and PC8, as well as furin, are endogenous proalbumin convertases in HepG2 cells. J Biochem. 1999;125:627–633. doi: 10.1093/oxfordjournals.jbchem.a022329. [DOI] [PubMed] [Google Scholar]
- 54.Kiefer M.C., Tucker J.E., Joh R., Landsberg K.E., Saltman D., Barr P.J. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991;10:757–769. doi: 10.1089/dna.1991.10.757. [DOI] [PubMed] [Google Scholar]
- 55.Constam D.B., Calfon M., Robertson E.J. SPC4, SPC6, and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at distinct sites during embryogenesis. J Cell Biol. 1996;134:181–191. doi: 10.1083/jcb.134.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuasa K., Futamatsu G., Kawano T., Muroshita M., Kageyama Y., Taichi H., Ishikawa H., Nagahama M., Matsuda Y., Tsuji A. Subtilisin-like proprotein convertase paired basic amino acid-cleaving enzyme 4 is required for chondrogenic differentiation in ATDC5 cells. FEBS J. 2012;279:3997–4009. doi: 10.1111/j.1742-4658.2012.08758.x. [DOI] [PubMed] [Google Scholar]
- 57.Yuasa K., Masuda T., Yoshikawa C., Nagahama M., Matsuda Y., Tsuji A. Subtilisin-like proprotein convertase PACE4 is required for skeletal muscle differentiation. J Biochem. 2009;146:407–415. doi: 10.1093/jb/mvp090. [DOI] [PubMed] [Google Scholar]
- 58.Croissandeau G., Basak A., Seidah N.G., Chretien M., Mbikay M. Proprotein convertases are important mediators of the adipocyte differentiation of mouse 3T3-L1 cells. J Cell Sci. 2002;115:1203–1211. doi: 10.1242/jcs.115.6.1203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Physicochemical Properties of the Presented Peptide


