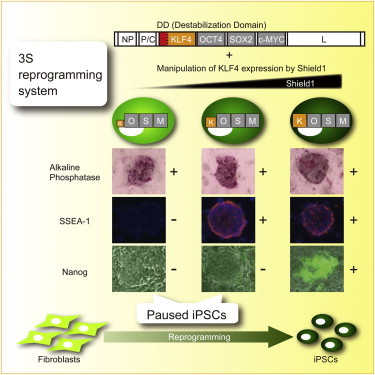
Summary
The detailed mechanism of reprogramming somatic cells into induced pluripotent stem cells (iPSCs) remains largely unknown. Partially reprogrammed iPSCs are informative and useful for understanding the mechanism of reprogramming but remain technically difficult to generate in a predictable and reproducible manner. Using replication-defective and persistent Sendai virus (SeVdp) vectors, we analyzed the effect of decreasing the expression levels of OCT4, SOX2, KLF4, and c-MYC and found that low KLF4 expression reproducibly gives rise to a homogeneous population of partially reprogrammed iPSCs. Upregulation of KLF4 allows these cells to resume reprogramming, indicating that they are paused iPSCs that remain on the path toward pluripotency. Paused iPSCs with different KLF4 expression levels remain at distinct intermediate stages of reprogramming. This SeVdp-based stage-specific reprogramming system (3S reprogramming system) is applicable for both mouse and human somatic cells and will facilitate the mechanistic analysis of reprogramming.
Graphical Abstract
Highlights
-
•
Reducing KLF4 expression generates partially reprogrammed cells
-
•
Different KLF4 levels produce iPSCs stably paused at distinct stages of reprogramming
-
•
Upregulation of KLF4 allows paused iPSCs to resume reprogramming
-
•
Homogenous populations of paused iPSCs are generated predictably and reproducibly
In this article, Hisatake, Nishimura, and colleagues report an SeVdp-based vector system that generates partially reprogrammed iPSCs by the precise control of the KLF4 expression level via an FKBP-derived degradation domain and Shield1. The system generates homogeneous populations of partially reprogrammed iPSCs stably paused at distinct stages of reprogramming and is applicable for both mouse and human somatic cells.
Introduction
Induced pluripotent stem cells (iPSCs) can be generated by introducing OCT4, SOX2, KLF4, and c-MYC (Takahashi et al., 2007; Takahashi and Yamanaka, 2006) or other combinations of reprogramming factors (Stadtfeld and Hochedlinger, 2010) into many types of mouse and human somatic cells. The understanding of its mechanism is crucial for generating high-quality iPSCs for cell therapy applications and also provides an insight into reprogramming and normal development. The reprogramming is an orderly process consisting of distinct stages, which can be distinguished by expression of specific markers, including stage-specific embryonic antigen-1 (SSEA-1), alkaline phosphatase (ALP), E-Cadherin (CDH1), and NANOG (Brambrink et al., 2008; Samavarchi-Tehrani et al., 2010; Stadtfeld et al., 2008). However, the precise molecular mechanisms behind iPSC generation remain to be elucidated.
Reprogramming mechanisms have been investigated by cell sorting a subpopulation of intermediate cells that express a specific combination of markers. These enriched cells are initially at a specific intermediate stage but continue reprogramming upon further cell culture, subsequently becoming a heterogeneous population of asynchronously reprogrammed cells (Hansson et al., 2012; O’Malley et al., 2013; Polo et al., 2012; Takahashi et al., 2014). Alternatively, more stable cell populations, isolated during iPSC generation, were also used for mechanistic analyses (Mikkelsen et al., 2008; Sridharan et al., 2009). These cells show phenotypes that are intermediate between somatic cells and iPSCs, thereby yielding mechanistic insights into reprogramming. Despite their usefulness, the stable cell populations are derived from rare by-products that appear sporadically during iPSC generation and difficult to obtain reproducibly in a predictable manner.
The efficiency of reprogramming and the characteristics of iPSCs are influenced by the expression levels and stoichiometry of reprogramming factors (Carey et al., 2011; Nagamatsu et al., 2012; Papapetrou et al., 2009; Sui et al., 2014; Tiemann et al., 2011). Detailed quantitative analyses show that merely increasing the expression levels of OCT4, SOX2, KLF4, and c-MYC does not necessarily lead to higher efficiency of iPSC generation; rather, the stoichiometry of the four factors plays a greater role in the efficiency of iPSC generation (Nagamatsu et al., 2012; Papapetrou et al., 2009; Tiemann et al., 2011). For example, high OCT4 and low SOX2 expression levels relative to other factors tend to generate iPSCs more efficiently than equal expression levels of the four factors. In addition, the reprogramming factor stoichiometry influences the gene expression pattern (Nagamatsu et al., 2012; Tiemann et al., 2011) and epigenetic status of iPSCs (Carey et al., 2011) as well as their ability to contribute to chimeric mice (Carey et al., 2011), suggesting that the stoichiometry of reprogramming factors may impact the reprogramming process as well.
We previously developed a unique gene transfer system, named SeVdp vectors, based on a mutant strain of Sendai virus, which remains persistently in the cytoplasm without integrating into the host genome (Nishimura et al., 2007). SeVdp vectors enable a long-term expression of multiple genes from a single vector with a constant stoichiometry and do not suffer from transcriptional silencing (Nishimura et al., 2011). These properties make SeVdp vectors an ideal tool for generating iPSCs. In fact, SeVdp vectors harboring four reprogramming factors (OCT4, SOX2, KLF4, and c-MYC) reprogram mouse and human somatic cells very efficiently (Nishimura et al., 2011; Nishimura et al., 2013; Takayama et al., 2010; Tateno et al., 2013; Wakao et al., 2013).
Here, we report the SeVdp-based stage-specific reprogramming system (3S reprogramming system) that generates paused iPSCs by controlling the KLF4 expression level with a destabilization domain (DD) and a small molecule, Shield1. We found that low KLF4 expression gives rise to partially reprogrammed cells, which are stably arrested at different stages of reprogramming depending on the level of KLF4 expression. Global gene expression analyses support that the partially reprogrammed iPSCs show intermediate gene expression profiles. Upregulation of KLF4 expression by Shield1 addition allows the partially reprogrammed cells to resume reprogramming toward pluripotency. The 3S reprogramming system establishes homogenous populations of paused iPSCs in a reproducible and predictable manner and is applicable for both mouse and human cells.
Results
Induction of Partially Reprogrammed Cells by SeVdp Vectors
In the course of developing SeVdp vectors for reprogramming, we created a vector named SeVdp(GKOSM) (Figure 1A), which is derived from SeVdp(KOSM) that induces fully reprogrammed iPSCs (Nishimura et al., 2011; Nishimura et al., 2013; Wakao et al., 2013). SeVdp(GKOSM) encodes EGFP in addition to KLF4, OCT4, SOX2, and c-MYC and enables monitoring of the SeVdp vector in living cells during reprogramming (Figure S1A available online). Mouse embryonic fibroblasts (MEFs) infected with SeVdp(GKOSM) or SeVdp(KOSM) gave rise to almost the same number of ALP-positive (ALP+) colonies (Table S1) consisting of small and rapidly growing cells (Figure 1B). Indeed, the colonies induced by SeVdp(GKOSM) were morphologically indistinguishable from SeVdp(KOSM)-induced ones, as exemplified by the cellular localization of β-CATENIN (Figures 1B and S1B). However, the colonies induced by SeVdp(GKOSM) were negative for SSEA-1 as opposed to the ones induced by SeVdp(KOSM), which were positive for SSEA-1 (Figure 1B). Expression of Thy1, Snail1, Snail2, and Tgfb3 was repressed 6 days after infection in the cells induced by SeVdp(GKOSM) and SeVdp(KOSM) (Figure 1C), consistent with the downregulation of somatic cell-specific genes at the early stage of reprogramming (Stadtfeld et al., 2008). On the other hand, at day 49, expression of mesenchymal-specific genes such as Cdh2, Zeb1, Zeb2, and Twist1 remained higher in SeVdp(GKOSM)-induced cells than in SeVdp(KOSM)-induced ones (Figure 1C), indicating that the SeVdp(GKOSM)-induced cells have not completed the mesenchymal-to-epithelial transition (MET), which normally occurs during iPSC generation (Li et al., 2010; Samavarchi-Tehrani et al., 2010). Moreover, these cells expressed only low levels of embryonic stem cell (ESC) marker genes including Cdh1, Fgf4, Rex1 (also called Zfp42), and Nanog even at day 49 (Figure 1C), showing that they persist as partially reprogrammed cells. Independent reprogramming by SeVdp(GKOSM) yielded cells with low expression levels of Cdh1, Fgf4, and Rex1 reproducibly (Figure 1D). Quantitative western blots (Figure S1C) showed that expression of all the four reprogramming factors, especially KLF4, were lower in SeVdp(GKOSM)-infected cells than in those infected by SeVdp(KOSM) (Figure 1E), presumably because Sendai virus vectors express genes located in its 3′ region at a higher level than those in the 5′ region (Tokusumi et al., 2002) (Figure 1A), and the presence of the EGFP gene in the 3′ region of SeVdp(GKOSM) is detrimental for the expression of the four factors. These findings raise the possibility that lowered expression of reprogramming factors induces partially reprogrammed cells.
Figure 1.
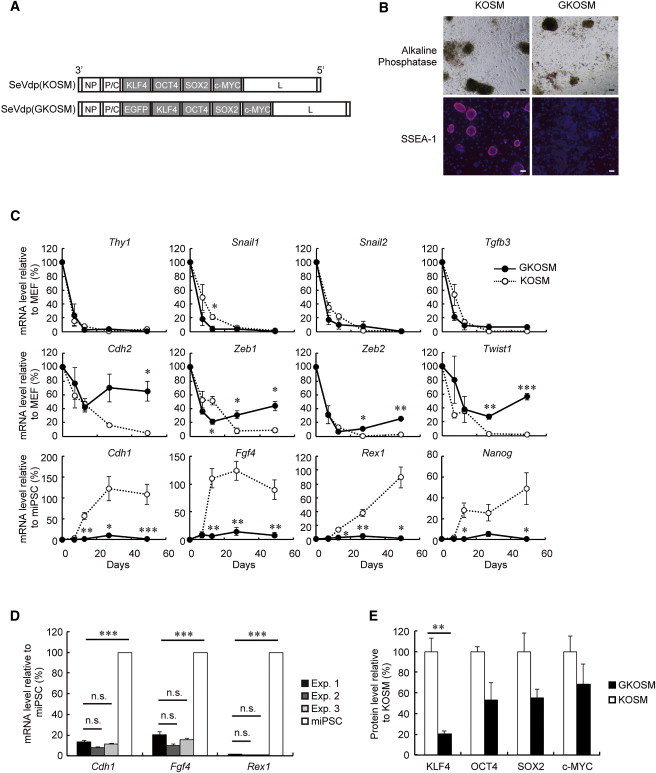
Generation of Partially Reprogrammed iPSCs by SeVdp Vectors
(A) Structures of SeVdp(KOSM) and SeVdp(GKOSM).
(B) Expression of ESC markers. Colonies induced by SeVdp(KOSM) or SeVdp(GKOSM) were stained for ALP or SSEA-1 at day 10 or 18, respectively. Scale bars, 100 μm.
(C) Time course of mRNA expression. Cells were collected at day 0, 6, 13, 25, and 49, and the mRNA levels of indicated genes were determined by quantitative RT-PCR and plotted relative to those in MEFs or mouse iPSCs (miPSC) (Nishimura et al., 2011). Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus SeVdp(KOSM)-infected cells.
(D) Reproducibility of reprogramming by SeVdp(GKOSM). Cells were collected at day 25 (Exp. 1) or day 23 (Exp. 2 and 3), and the mRNA levels of Cdh1, Fgf4, and Rex1 were determined by quantitative RT-PCR. Data represent means ± SEM of three independent PCR reactions. n.s., not significant; ∗∗∗p < 0.001 versus Exp. 1.
(E) Protein levels of reprogramming factors expressed from SeVdp vectors. SeVdp(KOSM) or SeVdp(GKOSM)-infected MEFs were collected 1 day after infection, and the protein level of each reprogramming factor was determined by western blotting as described in the Experimental Procedures. The protein level of each factor is plotted relative to that in SeVdp(KOSM)-infected cells. Data represent means ± SEM of three independent experiments. ∗∗p < 0.01.
Low KLF4 Expression Induces Partially Reprogrammed Cells
To identify which factor was responsible for partial reprogramming, we created four different vectors, SeVdp(fK-OSM), SeVdp(K-fOSM), SeVdp(KO-fSM), and SeVdp(KOS-fM). In each vector, one of the four factors was tagged N-terminally with a destabilization domain (DD), derived from a mutant FKBP12 (Banaszynski et al., 2006) (Figure 2A). This domain facilitates degradation of the tagged protein and decreases its expression level (Figure 2B). In addition, the DD-tagged protein is stabilized by a low molecular weight ligand, Shield1, in a precisely controlled manner (Figure 2B). When each vector was infected to MEFs in the absence of Shield1, the expression levels of DD-tagged KLF4, OCT4, SOX2, and c-MYC were 34.4% ± 6.0%, 11.5% ± 5.7%, 22.4% ± 15.8%, and 45.1% ± 14.5% of those in MEFs infected with SeVdp(KOSM), but the three other nontagged factors did not show similar decreases in expression (Figure 2C). Moreover, the expression levels of DD-tagged KLF4, OCT4, SOX2, and c-MYC in the absence of Shield1 were 34.9% ± 6.2%, 40.3% ± 20.7%, 40.5% ± 28.6%, and 46.0% ± 14.8% of those in the presence of 100 nM Shield1, but the three other nontagged factors showed lesser fluctuations in their expression levels by the addition of Shield1 (Figure 2D).
Figure 2.
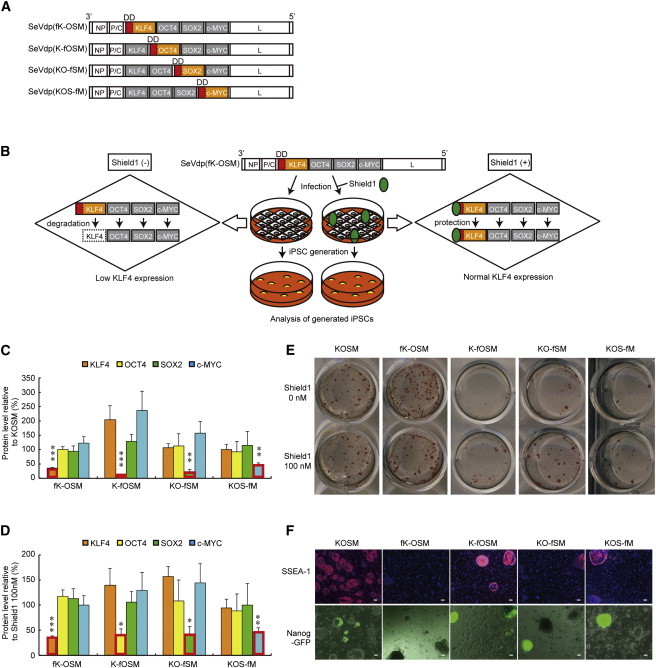
Effects of Downregulated KLF4, OCT4, SOX2, or c-MYC on Reprogramming
(A) Structures of SeVdp vectors in which one of the reprogramming factors is tagged N-terminally with DD.
(B) Stability of DD-tagged reprogramming factor during iPSC generation is regulated by the addition of Shield1.
(C and D) Protein levels of reprogramming factors expressed from SeVdp vectors. Cell extracts were prepared from MEFs infected with each indicated vector one day after infection in the presence or absence of 100 nM Shield1. The protein level of each reprogramming factor is determined by western blotting, and the level in the absence of 100 nM Shield1 is plotted relative to that in SeVdp(KOSM)-infected cells (C) or in the presence of 100 nM Shield1 (D). Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus SeVdp(KOSM) (C) or versus in the presence of 100 nM Shield1 (D).
(E) ALP+ colonies induced by each vector. MEFs (1.0 × 104) infected with the indicated vector were cultured in the presence or absence of 100 nM Shield1 and stained for ALP at day 13.
(F) Expression of SSEA-1 and NANOG. SSEA-1 was detected by immunofluorescence staining at day 18, and NANOG was detected by GFP fluorescence from the Nanog-GFP reporter transgene at day 22. Scale bars, 100 μm.
SeVdp(K-fOSM), SeVdp(KO-fSM), and SeVdp(KOS-fM) gave rise to smaller numbers of ALP+ colonies in the absence of Shield1 than in the presence of 100 nM Shield1 or by the infection with SeVdp(KOSM) (Figure 2E; Table S1). However, these colonies in the absence of Shield1 were positive for SSEA-1 and NANOG (Figure 2F) and far more advanced in reprogramming than the SeVdp(GKOSM)-induced colonies (Figure 1B), indicating that low levels of OCT4, SOX2, and c-MYC merely reduce the efficiency of generating fully reprogrammed iPSCs. By contrast, SeVdp(fK-OSM) gave rise to similar numbers of ALP+ colonies as compared with SeVdp(KOSM) (Figure 2E; Table S1). Moreover, the number of ALP+ colonies induced by SeVdp(fK-OSM) was essentially the same regardless of the presence of Shield1 (Figure 2E; Table S1), and the colonies induced in the absence of Shield1 were negative for SSEA-1 and NANOG (Figure 2F), displaying a phenotype common with the SeVdp(GKOSM)-induced ones (Figure 1B). Thus, low KLF4 expression in SeVdp(GKOSM)-infected cells is the main cause of generating partially reprogrammed iPSCs, and this phenomenon can be recapitulated by SeVdp(fK-OSM) in the absence of Shield1.
To further confirm that partially reprogrammed iPSCs were induced by low KLF4 expression, we prepared vectors that express KLF4 tagged C-terminally to the PEST or CL1 degradation signal (Gilon et al., 1998; Rogers et al., 1986) (Figure S2A). When KLF4 was tagged only with PEST [SeVdp(Kp-OSM)], KLF4 expression was decreased only moderately (48.0% ± 7.8%) (Figure S2B), and the induced cells were not only positive for ALP (Figure S2C) but also expressed ESC markers, Cdh1, Fgf4, and Rex1 (Figure S2D). Tagging KLF4 with both PEST and CL1 [SeVdp(Kcp-OSM)] decreased KLF4 expression slightly lower than that by SeVdp(fK-OSM) in the absence of Shield1 (23.8% ± 13.6%) (Figure S2B). Indeed, SeVdp(Kcp-OSM) induced ALP+ colonies (Figure S2C) that did not express ESC markers (Figure S2D), indicating that the induced cells are partially reprogrammed as in the case of SeVdp(fK-OSM) (Figures 2E and 2F). However, removal of KLF4 [SeVdp(OSM)] resulted in a drastically reduced number of colonies (Figure S2C). These results show that partially reprogrammed iPSCs are generated by low KLF4 expression and not by the untoward effect by the tagged DD.
Different Levels of KLF4 Expression Reprogram Cells to Different Degrees
We next examined if iPSCs generated by SeVdp(fK-OSM) acquire pluripotency when KLF4 expression is increased by Shield1. When SeVdp(fK-OSM) was infected to MEFs in the presence of 100 or 200 nM Shield1, the KLF4 expression level was restored almost to that by SeVdp(KOSM) (Figure 3A), which generates iPSCs that contribute to mouse chimeras (Nishimura et al., 2011). Expression analyses showed that cells induced by SeVdp(fK-OSM) in the presence of 100 nM Shield1 expressed the 12 marker genes in analogous time courses to those induced by SeVdp(KOSM) (Figure 3B). By contrast, the cells induced by SeVdp(fK-OSM) in the absence of Shield1 failed to completely downregulate MET-related genes and to upregulate ESC marker genes, displaying gene expression patterns similar to SeVdp(GKOSM)-induced cells (Figures 1C and 3B). As in the case of the colonies induced by SeVdp(GKOSM), the expression and localization of β-CATENIN in SeVdp(fK-OSM)-induced colonies were indistinguishable from those in SeVdp(KOSM)-induced ones (Figure S1B). Importantly, addition of Shield1 to the cells induced by SeVdp(Kcp-OSM) was not accompanied by the expression of the ESC-specific marker genes (Figure S2D), showing that Shield1 itself has no effect on induction of pluripotency.
Figure 3.
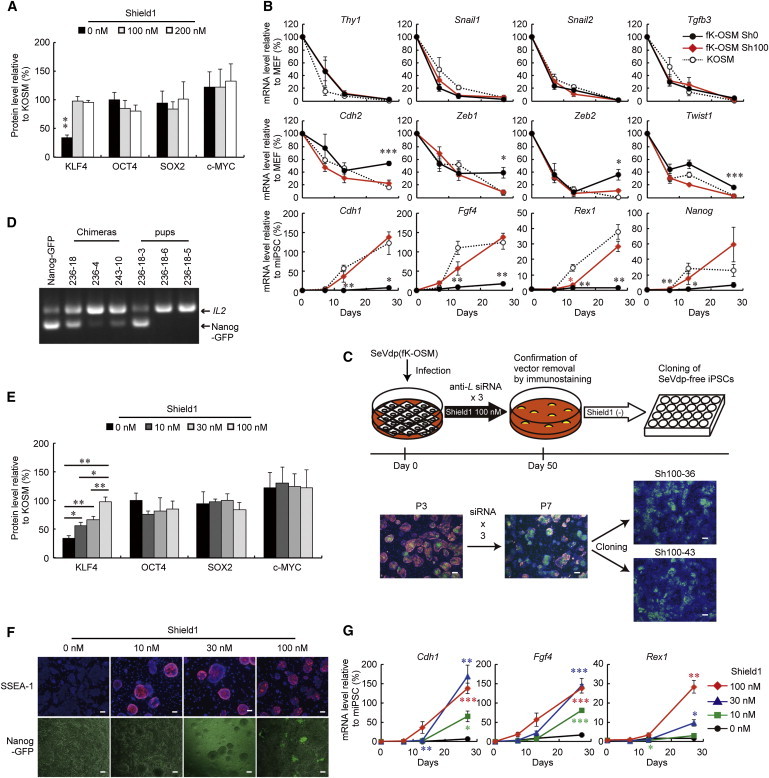
iPSC Generation by Manipulating the Protein Level of Exogenous KLF4
(A) Protein levels of reprogramming factors expressed from SeVdp(fK-OSM). Cell extracts were prepared from MEFs infected with SeVdp(fK-OSM) in the presence of the indicated concentration of Shield1 for one day. Protein levels are plotted relative to those in SeVdp(KOSM)-infected cells. Data represent means ± SEM of three independent experiments. ∗∗p < 0.01 versus SeVdp(KOSM).
(B) Time course of mRNA expression. SeVdp(fK-OSM)- or SeVdp(KOSM)-infected cells cultured in the presence (Sh100) or absence (Sh0) of 100 nM Shield1 were collected at days 7, 13, and 27 for quantitative RT-PCR. Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus SeVdp(KOSM)-infected cells.
(C) Vector-free iPSCs generated by SeVdp(fK-OSM). The cells induced by SeVdp(fK-OSM) in the presence of 100 nM of Shield1 (P3) were treated three times with L527 siRNA (P7), and the vector-free iPSCs were isolated at day 50 (Sh100-36 and Sh100-43). Green, Nanog-GFP. Red, NP of SeV. Scale bars, 100 μm.
(D) Production of chimeras from iPSCs established by SeVdp(fK-OSM) to show germline transmission. Genomic DNAs of the chimeric mice generated by the microinjection of clone Sh100-36 or Sh100-43, and their pups were analyzed by PCR to detect the Nanog-GFP transgene. The IL-2 gene was amplified as an internal control.
(E) Gradual increase of KLF4 expression by Shield1. Cell extracts were prepared as in (A). Protein levels are plotted relative to those in SeVdp(KOSM)-infected cells. Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01.
(F) Expression of SSEA-1 (day 27) and NANOG (day 29) in the cells cultured at indicated concentrations of Shield1. Scale bars, 100 μm.
(G) The Cdh1, Fgf4, and Rex1 mRNA levels were determined by quantitative RT-PCR at days 7, 13, and 27 for SeVdp(fK-OSM)-infected cells cultured at indicated concentrations of Shield1. Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus SeVdp(fK-OSM) in the absence of Shield1.
See also Figures S1 and S3 and Table S2.
These iPSCs, induced in the presence of 100 nM Shield1, were easily cleared of the SeVdp(fK-OSM) genome by an small interfering RNA (siRNA) against the L gene (Nishimura et al., 2011) that encodes the viral polymerase (Figures 2A and 3C), confirming that they have acquired a self-sustaining pluripotency state. When injected into mouse blastocysts, the vector-free iPSCs (Sh100-36) contributed to mouse chimeras, which subsequently gave rise to a live pup (236-18-3) carrying the Nanog-GFP transgene (Figure 3D). Thus, in the presence of 100 nM Shield1, DD-tagged KLF4 functions indistinguishably from nontagged KLF4 and permits SeVdp(fK-OSM) to reprogram MEFs to acquire pluripotency.
We then used SeVdp(fK-OSM) and different concentrations of Shield1 to analyze the dose-dependent effect of KLF4 on the reprogramming process. Infection of MEFs with SeVdp(fK-OSM) in the presence of 10, 30, or 100 nM Shield1 increased the KLF4 level in a dose-dependent manner while maintaining the expression of OCT4, SOX2, and c-MYC essentially unchanged (Figure 3E). In the presence of 10 or 30 nM Shield1, SeVdp(fK-OSM) gave rise to reprogrammed cells, most of which expressed SSEA-1, but not NANOG. The reprogramming of these cells is more advanced than ALP+, SSEA-1-negative (SSEA-1−), NANOG− cells induced by SeVdp(fK-OSM) in the absence of Shield1 but less advanced than ALP+, SSEA-1+, NANOG+ cells induced by SeVdp(fK-OSM) in the presence of 100 nM Shield1 (Figure 3F) or by SeVdp(KOSM) (Figure 2F). The intermediate degree of reprogramming is also supported by the expression levels of Cdh1, Fgf4, and Rex1 (Figure 3G).
It is of note that independently performed reprogramming by SeVdp(fK-OSM) yielded cells with similar expression profiles of Cdh1, Fgf4, and Rex1 (Figure S3A), showing that generation of partially reprogrammed iPSCs by SeVdp(fK-OSM) is, by and large, reproducible in independent experiments. Moreover, in the presence or absence of 10 nM Shield1, individual colonies isolated from the same plate showed similar gene expression profiles of Cdh1 and Fgf4 (Figure S3B), showing that SeVdp(fK-OSM)-induced colonies in the same plate are relatively homogeneous. Although Rex1 was expressed at somewhat varied levels, between 2% and 30% of the miPSC level, in the independent clones induced in the presence of 10 nM Shield1, this may reflect the intrinsically heterogeneous expression of Rex1 in mouse ESCs (Toyooka et al., 2008). Together, these results demonstrate that low KLF4 expression arrests the reprogramming process prematurely, and SeVdp(fK-OSM) yields partially reprogrammed iPSCs arrested at distinct stages depending on the amount of added Shield1. Given these unique properties, we named this as SeVdp-based stage-specific reprogramming system (3S reprogramming system).
Upregulation of KLF4 Expression Allows Partially Reprogrammed iPSCs to Resume Reprogramming
We addressed whether the partially reprogrammed cells have arrested but remained on a path toward pluripotency or have diverted from the path and become aberrantly reprogrammed. After MEFs were infected by SeVdp(fK-OSM) in the absence of Shield1 for 17 days, 100 nM Shield1 was added to the cell culture (Figure 4A). This increased the expression of DD-tagged KLF4 by more than 3-fold (Figure 4B), as revealed by anti-FKBP12 and anti-KLF4 antibodies. The upregulation of KLF4 expression at day 17 caused a gradual increase in the expression of ESC markers, NANOG (Figure 4C), Cdh1, Fgf4, and Rex1 (Figure 4D), indicating that the cells resumed reprogramming toward pluripotency. Similarly, partially reprogrammed cells, initially induced by SeVdp(fK-OSM) in the presence of 10 or 30 nM Shield1, also showed a gradual increase in the expression of NANOG upon addition of 100 nM Shield1 (Figure S4). Thus, the partially reprogrammed iPSCs generated by the 3S reprogramming system are competent to resume reprogramming upon the increased KLF4 level and hence should be regarded as “paused iPSCs.”
Figure 4.
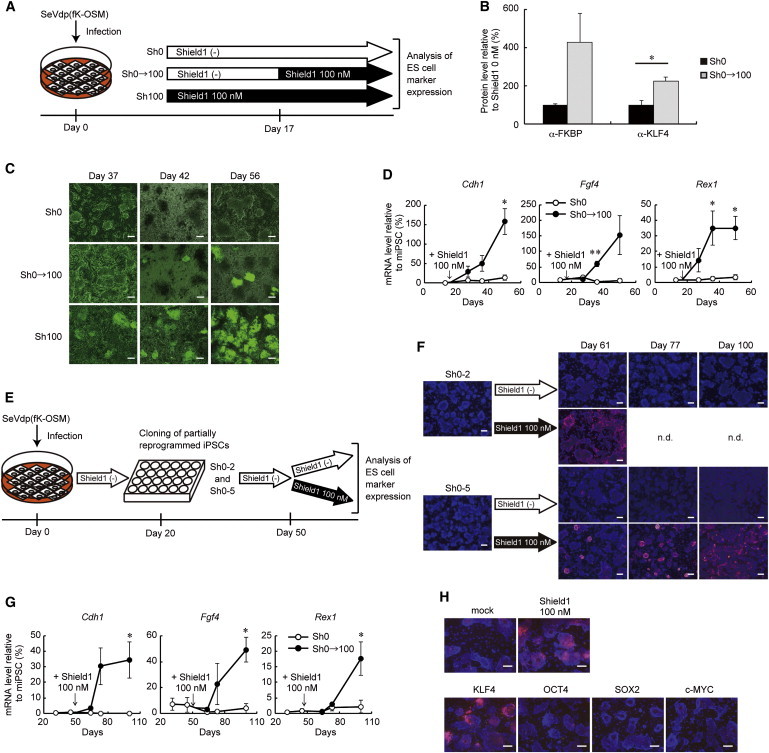
Upregulation of Exogenous KLF4 Resumes Reprogramming
(A) Outline of the experiments to restart reprogramming in partially reprogrammed iPSCs generated by SeVdp(fK-OSM) through the addition of Shield1 at day 17.
(B) Upregulation of the DD-tagged KLF4 protein level in partially reprogrammed iPSCs after 5 days of Shield1 addition. The protein levels of DD-tagged KLF4 or total KLF4 was determined by western blotting using an anti-FKBP or anti-KLF4 antibody, respectively, and were plotted relative to those in the cells cultured in the absence of Shield1. Data represent means ± SEM of three independent experiments. ∗p < 0.05.
(C) GFP fluorescence of the Nanog-GFP transgene was observed at indicated time points. Scale bars, 100 μm.
(D) The Cdh1, Fgf4, and Rex1 mRNA levels were determined by quantitative RT-PCR at day 16, 27, 36, and 49. 100 nM Shield1 was added at day 17. Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus no addition of Shield1.
(E) Partially reprogrammed iPSC clones were isolated at day 20 (Sh0-2 and Sh0-5) and cultured in the absence of Shield1 until day 50, when Shield1 was added.
(F) SSEA-1 expression was analyzed at indicated time points. Scale bars, 100 μm.
(G) Clone Sh0-5 was cultured in the absence of Shield1 until day 50, when Shield1 was added. The mRNA levels of Cdh1, Fgf4, and Rex1 were determined at day 31, 45, 64, 73, and 100. Data represent means ± SEM of three independent experiments. ∗p < 0.05 versus no addition of Shield1.
(H) SSEA-1 expression was analyzed in paused iPSCs (Sh0-5) 29 days after the addition of 100 nM Shield1 or infection with a retroviral vector expressing KLF4, OCT4, SOX2, or c-MYC. Scale bars, 100 μm.
Next, we asked if the paused iPSCs are stable and retain the ability to resume reprogramming after a long period of cell culture. We induced partially reprogrammed cells using SeVdp(fK-OSM) in the absence of Shield1, and individual clones were isolated at day 20 and cultured for additional 30 days under the same condition (Figure 4E). Even after additional culture for 30 days (between day 20 and 50), the cloned cells, Sh0-2 and Sh0-5, remained negative for SSEA-1 (Figure 4F) as well as for Cdh1, Fgf4, and Rex1 (Figure 4G). Moreover, addition of 100 nM Shield1 at day 50 allowed the cells to resume reprogramming and to express SSEA-1 (Figure 4F) as well as Cdh1, Fgf4, and Rex1 (Figure 4G), in similar time courses to those observed for the cells to which 100 nM Shield1 was added at day 17 (Figure 4D).
To further confirm that increased expression of KLF4, but not other Shield1-induced genes, allowed the partially reprogrammed cells to resume reprogramming, we used retrovirus-mediated gene transfer to introduce KLF4, OCT4, SOX2, or c-MYC into the paused iPSCs generated by SeVdp(fK-OSM) in the absence of Shield1. As shown in Figure 4H, additional expression of KLF4, but not other reprogramming factors, caused the cells to express SSEA-1 to a similar extent to the iPSCs generated by SeVdp(fK-OSM) in the presence of 100 nM Shield1, suggesting that the cells resume reprogramming even when KLF4 is provided by retrovirus-mediated gene transfer. Together, these results show that the paused iPSCs generated by the 3S reprogramming system are relatively stable and retain the ability to resume reprogramming for over a month.
Global Gene Expression Analyses of Paused iPSCs
To further characterize the paused iPSCs, we performed the analysis of their global gene expression. Hierarchical clustering of cells using differently expressed genes (DEGs) between MEFs and mouse iPSCs (miPSCs) (Nishimura et al., 2011) showed that SeVdp(fK-OSM)-induced cells are more similar to miPSCs than MEFs, and the cells are clustered together according to the amount of added Shield1 (Figure 5A). Global gene expression profiles also corroborate that the paused iPSCs generated in the absence of Shield1 are stable (Sh0 22d and Sh0 79d, Figure 5A) and resume reprogramming upon the addition of 100 nM Shield1 (Sh0→100 79d, Figure 5A). Principal component analysis indicates that SeVdp(fK-OSM)-induced cells have paused at intermediate stages between MEFs and miPSCs, and that the addition of a higher amount of Shield1 reprograms the cells closer to miPSCs (Figure S5A). The analysis also showed that the paused iPSCs generated by SeVdp(fK-OSM) in the absence of Shield1 resume reprogramming and advance toward pluripotency upon the addition of 100 nM Shield1 (Figure S5B). A recent study reported that mesendodermal genes are transiently activated during reprogramming and subsequently repressed again in iPSCs (Takahashi et al., 2014). Consistent with this, paused iPSCs express some of the mesendodermal genes (Eomes, Mixl1, and T), which are nonetheless repressed in fully reprogrammed iPSC produced by SeVdp(KOSM) or SeVdp(fK-OSM) in the presence of 100 nM Shield1 (Figures S5C–S5E). These gene expression profiles again confirm that paused iPSCs are intermediates during reprogramming.
Figure 5.
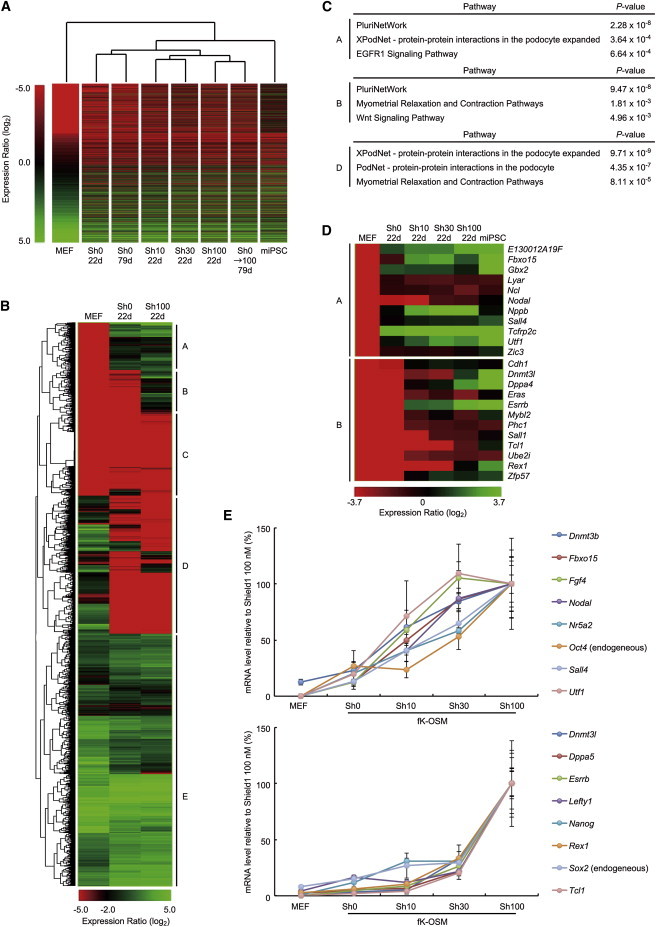
Global Gene Expression Profiles of Paused iPSCs
(A) Hierarchical clustering of paused iPSCs based on gene expression profiles of the differentially expressed genes (DEGs) between MEFs and miPSCs (p < 0.05). Analyzed samples include cells induced by SeVdp(fK-OSM) at the indicated concentrations (0, 10, 30, or 100 nM) of Shield1 for 22 or 79 days. Cells that resumed reprogramming (Sh0→100 79d) were first induced by SeVdp(fK-OSM) in the absence of Shield1 for 25 days and then cultured in the presence of 100 nM Shield1 before collection at day 79.
(B) Hierarchical clustering of DEGs. DEGs between MEFs and miPSCs (p < 0.05) were clustered based on gene expression profiles of the indicated cells and classified into five groups (group A–E).
(C) Enriched pathways in the genes of groups A, B, or D.
(D) Heatmap of the expression profiles of PluriNetWork genes clustered in group A or B.
(E) Two distinct expression profiles of pluripotency-related genes at increasing concentrations of Shield1. SeVdp(fK-OSM)-infected cells were cultured in the presence of indicated concentrations of Shield1, and the cells were collected at day 27. The mRNA levels of pluripotency-related genes were determined by quantitative RT-PCR. Data represent mean ± SEM of three independent experiments.
Hierarchical clustering of DEGs also revealed the presence of five distinct gene groups (Figure 5B). Group D includes a set of genes whose expression is already downregulated in paused iPSCs (Sh0 22d). The genes in groups A and B increase expression in a stepwise manner from MEFs to paused iPSCs (Sh0 22d) and from paused iPSCs (Sh0 22d) to iPSCs (Sh100 22d), respectively. Pathway analysis shows that Podocyte-related genes, which are involved in the epithelial-to-mesenchymal transition (EMT) of podocytes, are enriched in groups A and D (Figure 5C), indicating that the mesenchymal-to-epithelial transition (MET), a reverse process of EMT, separates paused iPSCs (Sh0 22d) from MEFs. This result is consistent with a previous report that showed the important role for KLF4 in MET during reprogramming (Li et al., 2010). Furthermore, PluriNetWork genes, which are associated with pluripotency, are enriched in groups A and B (Figure 5C), and heatmaps show that all the PluriNetWork genes in groups A and B increase their expression in a Shield1-dependent manner (Figure 5D), as if increasing the amount of KLF4 recapitulates the chronological progression of reprogramming. Independent quantitative RT-PCR analysis of selected pluripotency-related genes (Kim et al., 2008; O’Malley et al., 2013; Xu et al., 2010) identified two categories of genes that show distinct expression profiles in response to the Shield1 amount; genes that increase expression gradually as the amount of Shield1 is increased (upper panel, Figures 5E and S5F–S5M) and genes that increase expression markedly at 30 nM or higher concentrations of Shield1 (lower panel, Figures 5E and S5N–S5U). The former and the latter categories of genes correspond more or less to the genes in group A and group B, respectively (Figures 5D and 5E), demonstrating that pluripotency-related genes respond to the KLF4 level by at least two distinguishable modes. Together, the global gene expression analyses indicate that the iPSCs generated by the 3S reprogramming system are paused at critical stages of reprogramming and suggest that the KLF4-dependent transitions from one stage to the next is accompanied by changes in the expression of genes that are crucial for reprogramming.
Generation of Partially Reprogrammed Human iPSCs
Although molecular mechanisms of mouse iPSC generation have been investigated substantially, there have been relatively few mechanistic analyses of human iPSCs. Because our SeVdp vectors have been used successfully to generate human iPSCs from various somatic cells (Nishimura et al., 2013; Takayama et al., 2010; Tateno et al., 2013; Wakao et al., 2013), we tested if the 3S reprogramming system generates human iPSCs that have paused during reprogramming. When SeVdp(KOSM) and SeVdp(GKOSM) were infected to human fibroblasts, both vectors produced ALP+ colonies of small, rapidly growing cells in a similar manner to MEFs, although SeVdp(GKOSM) induced slightly more colonies than SeVdp(KOSM) (Figure 6A; Table S1). SeVdp(GKOSM)-induced human cells are similar to their mouse counterparts (Figure 1B) in that they are positive for ALP but negative for ESC markers, CDH1, FGF4, and REX1 (Figure 6B).
Figure 6.
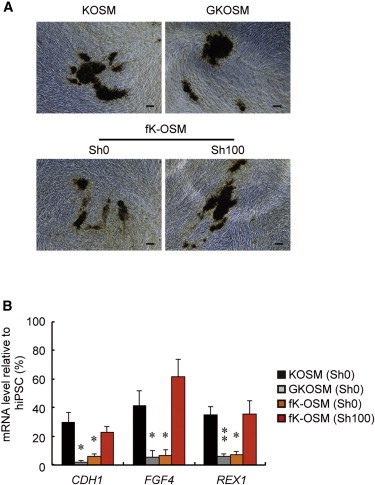
Generation of Partially Reprogrammed iPSCs from Human Somatic Cells
(A) Generation of ALP+ human iPSCs by SeVdp(fK-OSM). The indicated SeVdp vectors were used to generate human iPSCs, and the cells were stained for ALP at day 16. Scale bars, 100 μm.
(B) Expression of ESC markers in partially reprogrammed human iPSCs. The mRNA levels of CDH1, FGF4, and REX1 in the cells infected by the indicated vector in the presence or absence of 100 nM Shield1 were determined by quantitative RT-PCR at day 18. Data represent means ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.01 versus SeVdp(KOSM)-infected cells.
When human fibroblasts were infected with SeVdp(fK-OSM) in the presence or absence of 100 nM Shield1, similar numbers of ALP+ colonies were obtained regardless of the presence of Shield1 (Figure 6A; Table S1). However, induction in the absence of Shield1 generated cells that were negative for CDH1, FGF4, and REX1, whereas induction in the presence of 100 nM Shield1 generated cells that expressed these ESC marker genes (Figure 6B). Moreover, as in the case of mouse iPSCs (Figure 3C), hiPSCs generated by SeVdp(fK-OSM) in the presence of 100 nM Shield1 were cleared of the vector by the siRNA against the L gene to establish vector-free hiPSCs (Figure S6). These results indicate that SeVdp(fK-OSM) generates partially reprogrammed human iPSCs that are similar to the paused mouse iPSCs as well as fully reprogrammed iPSCs that have acquired a self-sustaining pluripotency state. Thus, the 3S reprogramming system can be utilized to generate partially reprogrammed human iPSCs, and these cells will be useful for analyzing the mechanism of human iPSC generation.
Discussion
Understanding the mechanisms of iPSC generation is important for application for cell therapy and also provides insights into the molecular mechanisms of normal developmental processes. The analysis, however, suffers from technical limits because the intermediate cells in the process of reprogramming are transient, asynchronous and heterogeneous. Cell sorting enriches intermediate cells, but they are transient and limited in amount (Hansson et al., 2012; O’Malley et al., 2013; Polo et al., 2012). For example, SSEA-1+ Oct4− reprogramming intermediates, initially isolated by fractionation using SSEA-1 antigen, diversify into a heterogeneous population of cells upon continued cell culture; some of them revert to lose SSEA-1 expression, whereas others advance to initiate Oct4 expression (Polo et al., 2012). Partially reprogrammed cell populations obtained during iPSC generation are relatively stable and expandable, but they are usually sporadic and unpredictable byproducts of reprogramming (Mikkelsen et al., 2008; Sridharan et al., 2009), sometimes generating aberrantly reprogrammed cells that do not transit into the pluripotent state (Mikkelsen et al., 2008).
Unlike these previous methods, the 3S reprogramming system generates partially reprogrammed iPSCs in a reproducible and predictable manner by regulating the amount of KLF4. These cells resume reprogramming upon increased KLF4 expression and therefore are not aberrantly reprogrammed but have paused during iPSC generation. Different KLF4 levels yield paused iPSCs at a series of stages during iPSC generation, enabling the molecular analysis of orderly events that proceed during reprogramming. Moreover, the paused iPSCs are relatively homogeneous and phenotypically stable, allowing expansion of cells for further analyses. Last, this system is based upon a defective Sendai virus, which replicates exclusively in the cytoplasm (Nishimura et al., 2011), and thus is free from both the positional effects and the silencing of transgenes that may arise in the systems based upon the retrovirus or lentivirus (Jaenisch et al., 1981). Therefore, our system to generate paused iPSCs is a significant advance over the sorted cells and partially reprogrammed cell populations previously used for mechanistic analyses of iPSC generation.
It remains to be determined how the low expression of KLF4, but not the other three factors, generates paused iPSCs. It is known that KLF4 represses somatic gene expression together with c-MYC at an early stage of iPSC generation and enhances pluripotency gene expression in cooperation with OCT4 and SOX2 at its late stage (Polo et al., 2012). Therefore, reduced levels of KLF4 may be sufficient for repressing somatic cell-specific genes, but not for activating pluripotency-related genes. A recent study also showed that KLF4 organizes long-range chromosomal interactions with the Oct4 locus by recruiting cohesion (Wei et al., 2013). These chromosomal interactions are observed not in SSEA-1− cells but only in SSEA-1+ cells during reprogramming, suggesting that KLF4 may mediate the chromosomal reorganization during the transition from the SSEA-1− to SSEA-1+ cells. Interestingly, this corresponds to a stage where SeVdp(fK-OSM) generates paused iPSCs in the absence of Shield1. It is thus possible that such dynamic chromosomal reorganization requires a high level of KLF4 and presents a roadblock for reprogramming with a low expression level of KLF4.
Recently, Sui et al. reported production of partially reprogrammed iPSCs using trimethoprim (TMP)-regulated destabilizing domain to control the expression of reprogramming factors from a piggyBac transposon vector (Sui et al., 2014). In contrast to our 3S reprogramming system, SOX2 rather than KLF4 plays a predominant role in regulating the progression of reprogramming. Several reasons may account for this apparent discrepancy. First, each system employed a different combination of the vector and reprogramming factors for iPSC generation. Sui et al. used only OCT4, SOX2, and KLF4 in piggyBac transposon vectors whereas our system used OCT4, SOX2, KLF4, and c-MYC in SeVdp vectors. Second, unlike the FKBP-based domain used in our system, the destabilizing domain used by Sui et al. seems to promote more complete protein degradation. Third, reprogramming may proceed via distinct pathways in each system and therefore the role for each reprogramming factor may not be identical between different systems. In fact, recent studies indicate the presence of multiple pathways by which reprogramming proceeds toward pluripotency (O’Malley et al., 2013; Parchem et al., 2014; Rais et al., 2013). Given this possibility, the availability of different systems that may generate iPSCs stalled at alternative intermediate states should be invaluable for more comprehensive understanding of the reprogramming process.
Regardless of the effect of the KLF4 expression level on the progression of reprogramming, the 3S reprogramming system described here provides a powerful tool for analyzing the mechanism of iPSC generation, particularly with respect to its ill-defined intermediate stages. One such example is MET, a reverse process of EMT that occurs during normal development. Consistent with the role for KLF4 in MET (Li et al., 2010), the paused iPSCs generated in the absence of Shield1 have stalled around MET and show gene expression profiles reminiscent of the ongoing MET. Another is the primitive streak-like mesendodermal (PSMN) state, where cells transiently display gene expression profiles resembling mesendoderm (Takahashi et al., 2014), which forms part of the primitive streak during early embryonic development. Paused iPSCs generated in the presence of 10–30 nM Shield1 express mesendoderm-related genes but have not reached the fully reprogrammed state. Because the addition of Shield1 permits the paused iPSCs to advance toward the fully reprogrammed state in a controlled manner, the 3S reprogramming system may be especially suitable for analyzing the MET stage, the mesendodermal stage as well as the late stage toward pluripotency. Finally, in addition to MEF-based iPSC generation, the current system also works for human somatic cells. Thus, the 3S reprogramming system will offer a unique opportunity to analyze transient and unstable stages of iPSC generation from mouse and human somatic cells.
Experimental Procedures
Cell Culture and Generation of iPSCs
Mouse iPSCs were generated from mouse embryonic fibroblasts (MEFs) isolated from a transgenic mouse carrying the Nanog-GFP-IRES-Puror reporter construct (Okita et al., 2007) (provided by the RIKEN BioResource Center) (Nanog-GFP MEFs). After Nanog-GFP MEFs were infected with each SeVdp vector at 32°C for 14 hr at an MOI of about 5, 2.0 × 104 of infected cells were seeded onto mitomycin C-treated MEFs in 6-well plates and cultured in KSR medium (Knockout-DMEM (Life Technologies) supplemented with 15% Knockout Serum Replacement (Life Technologies), 2 mM GlutaMAX (Life Technologies), 0.1 mM nonessential amino acids (Life Technologies), 55 μM 2-mercaptoethanol (Life Technologies), 100 U/ml penicillin-streptomycin (Wako), and 1,000 U/ml LIF [Wako]) for 7 days, and then in mES medium (DMEM; nacalai tesque) supplemented with 15% fetal bovine serum (Hyclone), 0.1 mM nonessential amino acids, 55 μM 2-mercaptoethanol, 100 U/ml penicillin-streptomycin and 1,000 U/ml LIF), in the presence or absence of Shield1 (Takara Bio). The presence of the vector in MEFs was confirmed 1 day after infection by immunostaining using an anti-SeV NP monoclonal antibody (Nishimura et al., 2011) as described in Supplemental Experimental Procedures. Reprogrammed cells were maintained on mitomycin C-treated SNL76/7 feeder cells (DS Pharma Biomedical) in the same mES medium, which was changed every 2 days. To remove SeVdp vectors from the reprogrammed cells, the cells were treated with 40 nM of L527 siRNA (Nishimura et al., 2011) mixed with Lipofectamine RNAi MAX Transfection Reagent (Life Technologies) in every passage after about day 20.
Human iPSCs were generated from human skin fibroblast NB1RGB cells (RIKEN BioResource Center). The cells were infected with SeVdp vectors at room temperature for 2 hr followed by 37°C for 14 hr at an MOI of about 5, and 2.0 × 104 of infected cells were seeded onto mitomycin C-treated MEFs in 6-well plates and cultured in Primate ESC culture medium (ReproCell) with 5 ng/ml basic FGF (Wako).
Retroviral vectors were infected to partially reprogrammed Sh0-5 cells cultured under a feeder-free condition at an MOI of about 10 for 2 days with 8 μg/ml of Hexadimethrine bromide (polybrene; Sigma). The cells were then passaged onto feeder cells and cultured in mES medium.
Determination of Protein Expression by Western Blotting
After SeVdp vectors were infected to MEFs, the cells were collected one day after infection and lysed on ice in SDS-PAGE gel loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 0.1 mg/ml Bromophenol blue, 5% 2-mercaptoethanol), followed by boiling for 5 min, and the whole-cell extracts were subjected to western blot analysis. Antibodies against SOX2, KLF4 or c-MYC were prepared by immunizing rabbits with each purified recombinant protein, as described previously (Fukuda et al., 2013). Each full-length recombinant protein was expressed in BL21(DE3)pLysS, separated by SDS-PAGE, and electroeluted. The purified recombinant proteins were used to immunize rabbits by Japan Bio Serum, and the obtained sera were affinity-purified by a corresponding antigen column, in which glutathione S-transferase (GST)-tagged proteins was crosslinked to glutathione Sepharose 4B (GE Healthcare). Anti-OCT4 (1:5,000, ab19857, Abcam) and anti-α-TUBULIN (1:10,000, ab7291, Abcam) were purchased commercially. Blots were probed with anti-rabbit or anti-mouse IgG-HRP secondary antibody (1:5,000, GE Healthcare) and visualized using Luminol, p-Coumaric acids, and H2O2 (Mruk and Cheng, 2011). Intensity of the chemiluminescence was quantified by LAS4000 (GE Healthcare) using ImageQuant TL software (GE Healthcare). We calculated the relative protein expression level of each reprogramming factor by normalizing on the basis of α-TUBULIN protein expression as described in Figure S1C. Determination of protein expression was carried out from biologically triplicate experiments.
Characterization of Mouse and Human iPSCs
Detailed methods for immunofluorescence, ALP staining, quantitative RT-PCR, microarray analysis, and generation of chimeric mice are described in Supplemental Experimental Procedures.
All animal experiments were performed according to the Guide for the Care and Use of Laboratory Animals at the University of Tsukuba.
Acknowledgments
We thank T. Fujiwara for providing pST-blue-T7-GFP-CL/PEST plasmid. We also thank T. Narita for help with antibody production, E. Noguchi for technical advice of DNA microarray analyses, and T. Nishimura for technical assistance. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) (to K.N., A.F., and K.H.), Takeda Science Foundation (to K.N.), and Program to Disseminate Tenure Tracking System by MEXT (to K.N.).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Ken Nishimura, Email: ken-nishimura@md.tsukuba.ac.jp.
Koji Hisatake, Email: kojihisa@md.tsukuba.ac.jp.
Accession Numbers
The NCBI Gene Expression Omnibus (GEO) accession number for the gene expression data sets reported in this paper is GSE56406.
Supplemental Information
References
- Banaszynski L.A., Chen L.C., Maynard-Smith L.A., Ooi A.G., Wandless T.J. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Fukuda A., Shimada M., Nakadai T., Nishimura K., Hisatake K. Heterogeneous nuclear ribonucleoprotein R cooperates with mediator to facilitate transcription reinitiation on the c-Fos gene. PLoS ONE. 2013;8:e72496. doi: 10.1371/journal.pone.0072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilon T., Chomsky O., Kulka R.G. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J., Rafiee M.R., Reiland S., Polo J.M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Reports. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Jähner D., Nobis P., Simon I., Löhler J., Harbers K., Grotkopp D. Chromosomal position and activation of retroviral genomes inserted into the germ line of mice. Cell. 1981;24:519–529. doi: 10.1016/0092-8674(81)90343-3. [DOI] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk D.D., Cheng C.Y. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis. 2011;1:121–122. doi: 10.4161/spmg.1.2.16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu G., Saito S., Kosaka T., Takubo K., Kinoshita T., Oya M., Horimoto K., Suda T. Optimal ratio of transcription factors for somatic cell reprogramming. J. Biol. Chem. 2012;287:36273–36282. doi: 10.1074/jbc.M112.380683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Segawa H., Goto T., Morishita M., Masago A., Takahashi H., Ohmiya Y., Sakaguchi T., Asada M., Imamura T. Persistent and stable gene expression by a cytoplasmic RNA replicon based on a noncytopathic variant Sendai virus. J. Biol. Chem. 2007;282:27383–27391. doi: 10.1074/jbc.M702028200. [DOI] [PubMed] [Google Scholar]
- Nishimura K., Sano M., Ohtaka M., Furuta B., Umemura Y., Nakajima Y., Ikehara Y., Kobayashi T., Segawa H., Takayasu S. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Kaneko S., Kawana-Tachikawa A., Tajima Y., Goto H., Zhu D., Nakayama-Hosoya K., Iriguchi S., Uemura Y., Shimizu T. Generation of rejuvenated antigen-specific T cells by reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:114–126. doi: 10.1016/j.stem.2012.11.002. [DOI] [PubMed] [Google Scholar]
- O’Malley J., Skylaki S., Iwabuchi K.A., Chantzoura E., Ruetz T., Johnsson A., Tomlinson S.R., Linnarsson S., Kaji K. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013;499:88–91. doi: 10.1038/nature12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Papapetrou E.P., Tomishima M.J., Chambers S.M., Mica Y., Reed E., Menon J., Tabar V., Mo Q., Studer L., Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14:617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y., Zviran A., Geula S., Gafni O., Chomsky E., Viukov S., Mansour A.A., Caspi I., Krupalnik V., Zerbib M. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui D., Sun Z., Xu C., Wu Y., Capecchi M.R., Wu S., Li N. Fine-tuning of iPSC derivation by an inducible reprogramming system at the protein level. Stem Cell Rep. 2014;2:721–733. doi: 10.1016/j.stemcr.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Sasaki A., Yamamoto M., Nakamura M., Sutou K., Osafune K., Yamanaka S. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat. Commun. 2014;5:3678. doi: 10.1038/ncomms4678. [DOI] [PubMed] [Google Scholar]
- Takayama N., Nishimura S., Nakamura S., Shimizu T., Ohnishi R., Endo H., Yamaguchi T., Otsu M., Nishimura K., Nakanishi M. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J. Exp. Med. 2010;207:2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno H., Matsushima A., Hiemori K., Onuma Y., Ito Y., Hasehira K., Nishimura K., Ohtaka M., Takayasu S., Nakanishi M. Podocalyxin is a glycoprotein ligand of the human pluripotent stem cell-specific probe rBC2LCN. Stem Cells Transl. Med. 2013;2:265–273. doi: 10.5966/sctm.2012-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann U., Sgodda M., Warlich E., Ballmaier M., Schöler H.R., Schambach A., Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–435. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- Tokusumi T., Iida A., Hirata T., Kato A., Nagai Y., Hasegawa M. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus Res. 2002;86:33–38. doi: 10.1016/s0168-1702(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wakao H., Yoshikiyo K., Koshimizu U., Furukawa T., Enomoto K., Matsunaga T., Tanaka T., Yasutomi Y., Yamada T., Minakami H. Expansion of functional human mucosal-associated invariant T cells via reprogramming to pluripotency and redifferentiation. Cell Stem Cell. 2013;12:546–558. doi: 10.1016/j.stem.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Wei Z., Gao F., Kim S., Yang H., Lyu J., An W., Wang K., Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Xu H., Lemischka I.R., Ma’ayan A. SVM classifier to predict genes important for self-renewal and pluripotency of mouse embryonic stem cells. BMC Syst. Biol. 2010;4:173. doi: 10.1186/1752-0509-4-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


