Summary
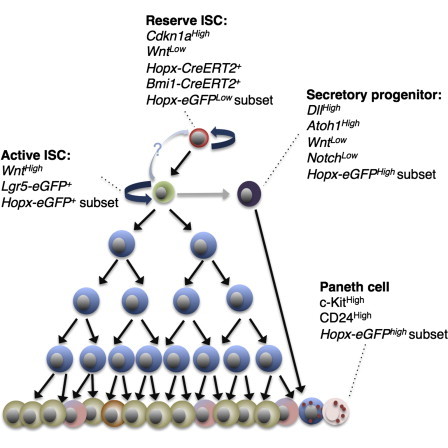
The recent development of targeted murine reporter alleles as proxies for intestinal stem cell activity has led to significant advances in our understanding of somatic stem cell hierarchies and dynamics. Analysis of these reporters has led to a model in which an indispensable reserve stem cell at the top of the hierarchy (marked by Bmi1 and Hopx reporters) gives rise to active intestinal stem cells (marked by an Lgr5 reporter). Despite these advances, controversy exists regarding the specificity and fidelity with which these alleles distinguish intestinal stem cell populations. Here, we undertake a comprehensive comparison of widely used proxy reporters including both CreERT2 and EGFP cassettes targeted to the Lgr5, Bmi1, and Hopx loci. Single-cell transcriptional profiling of these populations and their progeny reveals that reserve and active intestinal stem cells are molecularly and functionally distinct, supporting a two-stem-cell model for intestinal self-renewal.
Graphical Abstract
Highlights
-
•
Proxy intestinal stem cell reporter alleles often mark heterogeneous populations
-
•
Discrepancies exist between proxy reporter activity and endogenous transcripts
-
•
Reserve and active intestinal stem cells are molecularly distinct
-
•
Reserve intestinal stem cells give rise to active stem cells during homeostasis
In this study, Lengner and colleagues utilize single-cell profiling to provide a comparative analysis of epithelial cells marked by a number of putative intestinal stem cell proxy reporter alleles. They confirm the existence of two molecularly distinct stem cell populations governing homeostasis, with a long-term reserve stem cell giving rise to a short-term active stem cell.
Introduction
The intestinal epithelium provides a paradigmatic model for understanding stem cell organization and dynamics in highly proliferative tissues. The past decade has seen numerous breakthroughs in our understanding of intestinal stem cells (ISCs). Prior to 2007, the existence of ISCs at the base of small intestinal crypts was a subject of speculation. Undifferentiated, radiosensitive label-retaining cells (LRCs) around the +4 position from the crypt base had long been postulated to be ISCs (Potten et al., 2002); however, no functional data verifying the developmental capacity of these cells existed.
Beginning in 2007, a series of landmark studies identified several loci that marked functional intestinal stem cells upon insertion of an inducible Cre recombinase (CreERT2). The first locus identified was Lgr5, a canonical Wnt/β-catenin target gene that encodes an R-spondin receptor whose activity, in turn, potentiates canonical Wnt signaling (de Lau et al., 2011). An EGFP-IRES-CreERT2 reporter at the Lgr5 transcriptional start site marks actively cycling crypt base columnar cells (CBCs) that self-renew and give rise to all the differentiated progeny of the small intestine (Barker et al., 2007). Lgr5+ CBCs are capable of in vitro intestinal organoid formation and contribute to the colonic epithelium upon transplantation (Sato et al., 2009; Yui et al., 2012). These findings were surprising in light of the longstanding belief that LRCs represented the ISC population.
Shortly after the identification of Lgr5+ CBCs, the Capecchi group inserted an IRES-CreERT2 cassette into the Bmi1 locus following findings that this polycomb complex component played a critical role in hematopoietic and neural stem cell self-renewal (Molofsky et al., 2003; Park et al., 2003). Remarkably, the Bmi1-CreERT2 reporter marked relatively rare cells residing at the +4 position, on average, from the intestinal crypt base (Sangiorgi and Capecchi, 2008). As with Lgr5, Bmi1-CreERT2-marked cells continually gave rise to all functional cell types of the intestinal epithelium over long periods of time, clearly demonstrating their functional capacity as ISCs (Sangiorgi and Capecchi, 2008).
The functional hierarchy of these stem cells was elucidated through the use of diphtheria toxin (DT)-mediated cell ablation. Bmi1-CreERT2 mice containing a lox-stop-lox-DT transgene enabled the ablation of Bmi1-expressing ISCs. This resulted in loss of intestinal crypts and tissue integrity (Sangiorgi and Capecchi, 2008). Remarkably, an analogous experiment performed with a diphtheria toxin receptor inserted into the endogenous Lgr5 locus (Lgr5-DTR) followed by DT treatment efficiently ablated all CBCs with no functional consequences for the homeostatic epithelium (Tian et al., 2011). In this model, an increase in the frequency of cells marked with a Bmi1-EGFP+ knockin reporter was observed upon Lgr5+ CBC ablation, and lineage tracing with Bmi1-CreERT2 demonstrated that these cells give rise to Lgr5+ CBCs. Interestingly, Bmi1-CreERT2-marked cells were found to be insensitive to stimulation and antagonism of the canonical Wnt pathway that drives self-renewal of CBCs (Yan et al., 2012). These findings support a model in which Bmi1-CreERT2 cells represent a reserve ISC that gives rise to an active, Lgr5+ CBC stem cell that bears the proliferative burden necessary to maintain homeostasis.
Insight into the benefits of such a two-stem-cell system (Li and Clevers, 2010) came from studying the response of the epithelium to acute injury. High-dose (12–14 Gy) γ-irradiation (γ-IR) quantitatively ablates the vast majority if not all Lgr5+ CBCs (Yan et al., 2012), as well as LRCs (Potten et al., 2002). Reserve ISCs are resistant to high-dose radiation and become activated to generate new Lgr5+ CBCs in order to repopulate the epithelium (Tian et al., 2011; Yan et al., 2012). In this context, Lgr5+ cells are indispensable, possibly due to the tremendous proliferative output required to regenerate the entire tissue and/or activation of the Lgr5-DTR allele in reserve ISCs as they convert to CBCs (Metcalfe et al., 2014).
Further support for the hierarchical two-stem-cell model came with the discovery of an additional reserve ISC marker locus, Hopx, encoding an atypical homeodomain protein with functions in early heart development (Chen et al., 2002). An IRES-CreERT2 cassette inserted into the endogenous Hopx locus revealed that, like Bmi1-CreERT2 cells, Hopx-CreERT2 cells are capable of giving rise to Lgr5+ cells (Takeda et al., 2011). Thus, reserve ISCs give rise to progeny including active Lgr5+ CBCs that become dependent on canonical Wnt activity. The precise relationship between Hopx-CreERT2- and Bmi1-CreERT2-marked cells remains unexamined.
Despite the elegant genetic evidence supporting the existence of a two-stem-cell system (active CBCs and reserve ISCs), considerable controversy exists regarding the identity of these stem cells and their relationship to one another. Specifically, the messenger RNAs emanating from the endogenous loci used in generation of marker alleles including Bmi1, Tert, and Hopx exist at higher levels in the Lgr5-EGFP-high population in comparison to the Lgr5-EGFP-low population (Muñoz et al., 2012), and the endogenous Bmi1 and Tert transcripts can be detected throughout almost all cells of the crypt below the transit-amplifying (T/A) zone (Itzkovitz et al., 2012). These findings led to suggestions that the marked stem cells may represent a single population or that they exist in a continuum, not discernible as distinct populations. Many of these discrepancies could be accounted for if, in fact, these reporter alleles mark heterogeneous populations that are mistakenly assumed to be homogenous in population-based analyses and/or if the presence of endogenous mRNAs does not correlate with reporter activity emanating from a single locus.
Further complexities in our understanding of ISC biology arose in recent reports describing the existence of secretory precursor cells of the intestine. One report described these secretory precursors as long-lived LRCs that express high levels of Lgr5 and resist intermediate doses of γ-IR (6 Gy; Buczacki et al., 2013). This finding was particularly curious in light of classic studies describing the intestinal LRC as being exquisitely radiosensitive (undergoing apoptosis in response to as little as 1 Gy γ-IR; Potten et al., 2002), and studies using highly sensitive multi-isotope imaging mass spectrometry suggest that there are no LRCs in the intestinal epithelium (Steinhauser et al., 2012). Thus, the existence and identity of LRCs of the intestinal epithelium remains controversial, and how these cells relate to the reserve ISCs marked by Bmi1- or Hopx-CreERT2 activity is entirely unknown.
In a separate study, another group identified secretory precursor cells as a proliferative population marked by expression of the Notch ligand Dll1 (van Es et al., 2012). These cells have a very specific gene-expression pattern with high expression of Notch ligands (Dll), low levels of Notch receptors and target genes (Hes), and high levels of Atoh1 (Math1), which is suppressed by Notch signaling and promotes differentiation into the secretory lineage (van Es et al., 2012). In contrast to the secretory LRCs, the Dll1+ secretory precursors do not express Wnt target genes including Lgr5 (van Es et al., 2012). Interestingly, both the Dll1+ and label-retaining secretory precursor cells exhibited broad stem cell activity (generating not only secretory lineages) in response to epithelial damage, although these were rare events (Buczacki et al., 2013; van Es et al., 2012).
In an attempt to reconcile conflicting reports in the literature and provide a foundation for understanding intestinal stem cell dynamics and hierarchy moving forward, we undertook a comprehensive comparative analysis of established ISC knockin reporter alleles including Lgr5-EGFP-IRES-CreERT2, Bmi1-EGFP, Bmi1-CreERT2, Hopx-CreERT2, and Hopx-EGFP. We apply single-cell analyses to address the heterogeneity inherent in these populations, whether they exist as molecularly distinct stem cell pools, and how they differ in their proliferative output. Through the analysis of Wnt, Notch, proliferation, differentiation, and stem-cell-related transcripts along with lineage tracing and cell cycle analyses, we place these marked populations in a model hierarchy. Our findings begin to reconcile the contrasting literature regarding the identity of ISC populations marked by proxy reporter alleles and support the existence of a two-stem-cell model.
Results
Comparative Analysis of Reporter Activity in Reserve and Active ISC Populations
To directly compare CBC ISCs, reserve ISCs, and putative reserve ISCs, we examined the spatial distribution of cells marked by Lgr5-EGFP-IRES-CreERT2 (CBC marker; Barker et al., 2007), Bmi1-CreERT2 (reserve ISC; Sangiorgi and Capecchi, 2008; Tian et al., 2011; Yan et al., 2012), Hopx-CreERT2 (reserve ISC; Takeda et al., 2011), Bmi1-EGFP (putative reserve ISC; Tian et al., 2011), and Hopx-EGFP (putative reserve ISC; Takeda et al., 2013) in the proximal jejunum of mice that were maintained on a C57Bl/6 background and cohoused (Figure S1 available online).
CBCs marked with Lgr5-EGFP-IRES-CreERT2 exhibited a robust, crypt-localized signal that decreased rapidly into the early T/A zone, although not every crypt contained Lgr5-EGFP+ cells, consistent with the known mosaic activity of this allele (Figure 1A). Activation of a ROSA26-lox-stop-lox-tdTomato reporter (referred to as LSL-Tomato) in Lgr5-EGFP-IRES-CreERT2 mice by a single injection of tamoxifen (Tam) marked cells in a position and with a frequency consistent with that of Lgr5-EGFP+ cells, indicating that, in this model, enhanced GFP (EGFP) correlates well with CreER activity (Figure 1B). This notion is supported by flow cytometric analysis of EGFP+ and Tomato+ populations. Lgr5-EGFP+ cells comprised approximately 1.7% of the epithelial preparation, whereas the Lgr5-LSLTomato+ population comprised 0.32%, consistent with some inefficiency in CreERT2 nuclear translocation and genomic recombination in response to Tam (Figures 1C and 1D). All Lgr5-Tomato+ cells were also Lgr5-EGFP+, confirming the correlation between these reporters and indicating that insufficient cell division occurs in the 18 hr lineage trace for Tomato+ cells to generate daughters that have lost EGFP.
Figure 1.
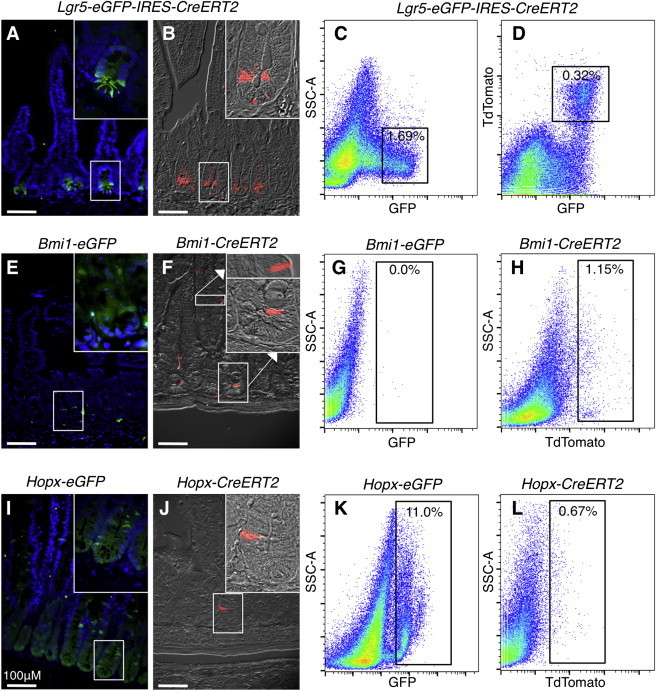
Histological and Flow Cytometric Analysis of Intestinal Stem Cell Proxy Reporter Alleles
(A and B) Immunofluorescence detection of EGFP from the Lgr5-EGFP-IRES-CreERT2 locus (A) or tdTomato from the Lgr5-EGFP-IRES-CreERT2::LSL-tdTomato reporter (B).
(C and D) Flow cytometric analysis of Lgr5-EGFP (C) or EGFP versus tdTomato in Lgr5-EGFP-IRES-CreERT2::LSL-tdTomato.
(E and F) Immunofluorescence detection of Bmi1-EGFP (E) or Bmi1-CreERT2::LSL-tdTomato (F).
(G and H) Flow cytometric analysis of Bmi1-EGFP (G) or Bmi1-CreERT2::LSL-tdTomato (H).
(I and J) Immunofluorescence detection of Hopx-EGFP (I) or Hopx-CreERT2::LSL-tdTomato (J).
(K and L) Flow cytometric analysis of Hopx-EGFP (K) or Hopx-CreERT2::LSL-tdTomato (L).
All tdTomato immunofluorescence was performed 24 hr after a single dose of Tam. All tdTomato flow cytometry was performed 18 hr after a single dose of Tam (n = 3 independent experiments per allele). See also Figure S1.
Putative reserve ISCs marked with Bmi1-EGFP were not reliably detectable by immunostaining; it was difficult to determine whether cells in the crypt were Bmi1-EGFP+ over background (Figure 1E). In contrast, functional reserve ISCs marked by Bmi1-CreERT2-LSL-Tomato were observed as single cells within the crypt; however, this allele also frequently marked cells within the villi (Figure 1F). Approximately 1% of epithelial cells are marked by Bmi1-CreERT2, whereas nearly zero cells were detectable in Bmi1-EGFP small intestine (Figures 1G and 1H). This finding highlights a discrepancy between Bmi1-EGFP- and Bmi1-CreERT2-marked populations.
We next examined the activity of Hopx knockin alleles. Consistent with published reports, Hopx-CreERT2-LSL-Tomato predominantly marked single cells above the crypt base, similar in location and frequency to those marked by Bmi1-CreERT2 (Figure 1J). Unlike Bmi1-CreERT2, Hopx-CreERT2 did not frequently mark cells outside of the crypt. In stark contrast to Hopx-CreERT2, analysis of Hopx-EGFP+ revealed activity of this reporter throughout the crypt in the CBC, Paneth cell, and +4 zones (Figure 1I). In epithelial cells, 0.7% was marked by Hopx-CreERT2 and 11% were marked by Hopx-EGFP+ (Figures 1K and 1L). This finding again highlights discrepancies between the functionally validated CreERT2 marker alleles and the EGFP reporters. These discrepancies may be due to the differing sites of integration of these reporters (Figure S1), the perdurance of EGFP in rapidly cycling cells, differences due to efficiency of CreERT2 target allele excision, or a combination of these factors. Our findings thus far call into question the validity of using the EGFP reporter alleles as proxies for functional reserve ISC identity defined by Hopx- and Bmi1-CreERT2 activity.
Cell-Cycling Dynamics in Active and Reserve Intestinal Stem Cells
CBCs cells are actively cycling, with estimates that they divide approximately daily (Snippert et al., 2010). In contrast, reserve ISCs are often referred to as being quiescent; however, published evidence for their quiescence does not exist. Given the recent study defining LRCs as secretory progenitor cells (Buczacki et al., 2013), we compared the dynamics of DNA synthesis between Lgr5+ CBCs and the reserve ISCs. We observed that, during the course of a 2 hr pulse labeling with 5-ethynyl-2′-deoxyuridine (EdU), approximately 40% of Lgr5+ cells underwent DNA synthesis (Figures 2A, 2B, and S2A). In contrast, both Bmi1- and Hopx-CreERT2-marked populations had approximately 20% of cells synthesize DNA during the same period, with EdU+ reserve ISCs observed as single cells near the crypt base (Figures 2A, 2B, and S2A). This result supports the rapidly cycling nature of CBCs but also indicates that reserve ISCs undergo DNA synthesis relatively frequently, likely too frequently to be label-retaining cells if they represent homogenous populations.
Figure 2.
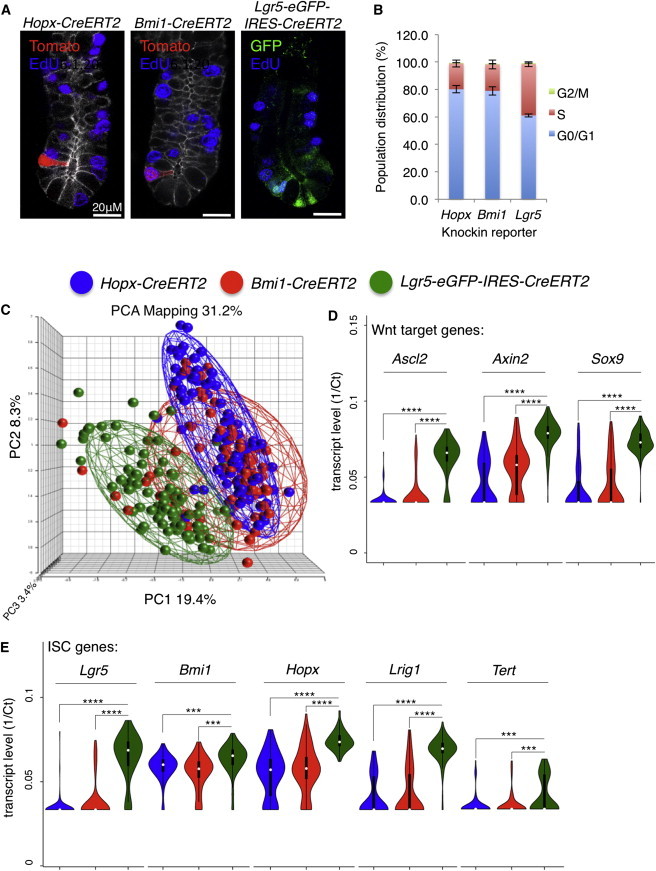
Single-Cell Analysis of ISC Proliferation and Gene Expression
(A) Immunofluorescence costaining for proliferative cells marked by a 2 hr pulse of EdU along with Tomato in Bmi1- and Hopx-CreERT2 crypts or EGFP in Lgr5-EGFP crypts.
(B) Quantification of cell-cycle distribution in the ISC marker models shown in (A) (n = 3 independent experiments; ±SD).
(C) Principal component analysis (PCA) of single-cell mRNA profiles generated from Bmi1-CreER, Hopx-CreER, and Lgr5-EGFP cells isolated from small intestinal crypts.
(D) Violin plots showing transcript levels of canonical Wnt target genes in individual cells plotted in (C).
(E) Violin plots showing transcript levels of putative ISC-marker genes in individual cells plotted in (C) (n = 93, Bmi1-CreERT2; n = 93, Lgr5-EGFP; n = 91, Hopx-CreERT2). Two-sided t test; ∗∗∗∗p < 1 × 10−10; ∗∗∗p < 1 × 10−4.
See also Figures S2 and S3.
Single-Cell Transcriptional Profiling Reveals Heterogeneity in Reporter-Marked ISC Populations
We next sought to understand the heterogeneity and molecular identity of ISCs both within and across the marked populations. We initially compared cells marked by reporter alleles that were functionally shown to mark ISCs. Single cells marked by Tomato expression (for Bmi1- and Hopx-CreERT2) or by EGFP (for Lgr5) were isolated from small intestinal crypt preparations (after removal of villi) by fluorescence-activated cell sorting (FACS) 18 hr after Tam induction and subjected to profiling on Fluidigm Biomark HD dynamic arrays. The transcript levels of 48 genes representing both components and targets of the Wnt and Notch pathways, ISC-related genes, and proliferation- and metabolism-related genes were analyzed in single cells with two distinct primer pairs per gene (Figure S2B; Tables S1, S2, and S3).
Principal component analysis (PCA) of single cells within these populations revealed clear differences in the molecular identity of Lgr5+ CBCs in comparison to the reserve ISCs (Figure 2C). PCA plots of these same populations annotated by experiment number rather than reporter identity revealed no experimental bias across three distinct epithelial preparations that were independently isolated, sorted, amplified, and analyzed (Figure S2C). Interestingly, analysis of housekeeping genes Gapdh and Gusb (Wang et al., 2010) revealed significant differences in their expression, both between individual cells within a group and between groups, indicating that normalization of RNA quantification to expression of these genes can introduce bias (Figure S2D).
A number of Bmi1-CreERT2+ cells exhibited a “CBC-like” identity in the PCA, possibly reflecting the transition of these cells from the reserve state to an active CBC state (Figure 2C). This was observed less frequently in Hopx-CreERT2+ cells (Figure 2C). Calculations of the coefficient of variation (CV) in gene expression across individual cells in these two populations further reveals that the Hopx-CreERT2+ cells represent a more homogenous population than Bmi1-CreERT2+ (mean CV% = 13.61 and 14.84, respectively). Consistent with this notion, fewer Hopx-CreERT2+ cells expressed Wnt target genes whose expression characterizes the CBC state in comparison to Bmi1-CreERT2+ cells (including Ascl2, Axin2, Sox9, and Lgr5; Figures 2C and 2D). It is important to highlight here that the Hopx- and Bmi1-CreERT2+ cells that we profiled were derived from crypt preparations, and thus, the Bmi1-CreERT2+-marked cells within the villi (Figure 1F) were excluded in the analysis.
Examination of ISC-related genes including Lgr5, Bmi1, Hopx, Lrig1, and Tert revealed that all of these genes are expressed at higher levels in Lgr5-EGFP+ cells than in the reserve ISC populations. Whereas the endogenous putative reserve ISC transcripts Bmi1 and Hopx were highly expressed relative to Lgr5 and other Wnt signature genes in Hopx- and Bmi1-CreERT2+ cells, their expression was higher still in Lgr5-EGFP+ cells (Figures 2D and 2E). This finding supports prior bulk transcriptome profiling of Lgr5-EGFP+ cells and in situ hybridization studies (Itzkovitz et al., 2012; Muñoz et al., 2012) and highlights the discrepancy between Hopx- and Bmi1-CreERT2+ reporter activity and the levels of endogenous Hopx and Bmi1 transcripts. This also demonstrates that the presence of high levels of Hopx or Bmi1 mRNA is not only a poor indicator of reserve ISC identity but in fact is more indicative of the CBC state. Further, this result reconciles contradictions in the literature suggesting that the CBC and reserve ISCs represent the same population based on expression of Bmi1 and Hopx in Lgr5+ cells. We conclude that the use of endogenous Bmi1 or Hopx mRNA as evidence for reserve intestinal stem cell identity is invalid.
Similar to canonical Wnt target genes, Notch target genes such as Hes1 and Olfm4 were also preferentially expressed in Lgr5-EGFP+ cells in comparison to reserve ISCs (Figure S3A). Further, the master secretory cell fate determinant Atoh1 (Math1) appeared consistent across the three ISC populations, further suggesting that the secretory progenitor population may be a distinct pool of cells not profiled in this assay.
We next asked which differentially expressed genes were the best predictors of reserve ISC identity (Hopx-CreERT2+ or Bmi1-CreERT2+) versus CBC identity (Lgr5-EGFP+). We found that low expression of Wnt and cell-cycle-related genes Ccnd1, Myc, Myb, and Lgr5 along with high expression of Cdkn1a (encoding p21) and Cubn (encoding a vitamin B12 receptor) best predict reserve stem cell versus active CBC stem cell identity (Figure S3B; not shown). This finding is not surprising given the dependence of CBCs on high canonical Wnt pathway activity and their rapidly cycling state relative to the reserve ISCs.
Fidelity of EGFP Reporters Relative to CreERT2 Reporters
Having established the heterogeneity and identity of cells within the Hopx- and Bmi1-CreERT2+ populations versus Lgr5+ CBCs, we next compared these populations to their counterparts marked by EGFP insertions. This is an important comparison, as functional studies such as lineage tracing and diphtheria-toxin-based cell ablation have only been carried out using CreERT2 alleles, yet the activity of their EGFP counterparts is often used as proxy evidence for stem cell identity. We initially examined the profiles of cells marked by EGFP versus Tomato in Lgr5-EGFP-IRES-CreERT2::LSL-Tomato mice and found that these populations were largely overlapping in PCA plots (Figure S3C). This is consistent with our flow cytometric profiling, demonstrating that the Tomato+ population marked a subset of EGFP+ cells (Figure 1D). Interestingly, there was less variation in the Tomato+ population than the EGFP+ population, likely due to the perdurance of EGFP into the previously described “Lgr5-mid/low” state in which CBCs begin committing to differentiation (Merlos-Suárez et al., 2011). Thus, there is good correlation between the activity of the CreER and EGFP reporters in the Lgr5 locus.
Detection of Bmi1-EGFP+ in the proximal small intestine was difficult; however, we attempted to collect enough Bmi1-EGFP+ cells to perform single-cell profiling. Profiling of Bmi1-EGFP+ revealed that many of the sorted cells were either debris or gave highly inconsistent signatures, with no clear cellular identity (Figure S3D). There are several potential explanations for this result, including the possibility that the few Bmi1-EGFP+ cells we were able to collect were not bona fide reserve ISCs or that the excessive sort time required to collect these cells resulted in RNA degradation and sorting errors. Thus, histological, flow cytometric, and single-cell gene-expression profiling indicate that there are discrepancies between the activity of the Bmi1-CreER and the Bmi1-EGFP alleles, and such Bmi1-EGFP+ cells may not represent bona fide reserve intestinal stem cells.
In contrast, the Hopx-EGFP allele was active throughout the crypt (Figures 1I and 1K). To better understand the populations marked by Hopx-EGFP, we separated Hopx-EGFP+ cells into three distinct gates for sorting and profiling. These include an EGFPHigh population, an EGFPLow population, and an EGFPLow population with a low degree of side scatter (low complexity; designated Hopx-EGFPSmall; Figure 3A). PCA indicated that these populations were largely distinct, with some overlap between the Hopx-EGFPHigh cells and the Lgr5-EGFP+ population and between the Hopx-EGFPLow cells and the Hopx-CreERT2+ population (Figures 3B and S4A). Overall, the Hopx-EGFP+ populations had varying expression of Hopx, low expression of Lgr5 and Wnt pathway genes, and formed intestinal organoids from single cells in vitro with an efficiency significantly lower than Lgr5-EGFPHigh cells but comparable to Lgr5-EGFPMid cells, demonstrating that stem cell potential exists within this population (Figures 3C and 3D; not shown).
Figure 3.
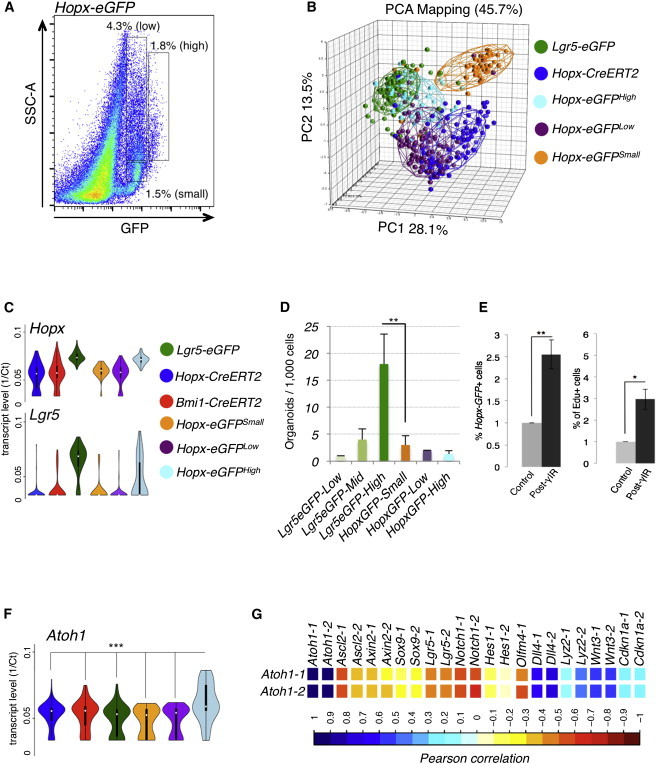
Characterization of Hopx-EGFP-Marked Cells
(A) FACS gating strategy separating the Hopx-EGFP+ population into Hopx-EGFPHigh (n = 48), Low (n = 94), and Small (low side scatter; n = 47) subgroups.
(B) PCA plots of populations shown in (A) in comparison to Lgr5-EGFP- and Hopx-CreERT2-marked cells.
(C) Hopx and Lgr5 expression across the Hopx-EGFP populations in comparison to Lgr5-EGFP-, Bmi1-CreERT2-, and Hopx-CreERT2-marked cells (as shown in Figure 2E and provided here as reference).
(D) In vitro intestinal organoid formation efficiencies from single cells from Hopx-EGFP populations in comparison to Lgr5-EGFP-High, Medium, and Low populations.
(E) Flow cytometric quantification of total Hopx-EGFP+ cell frequency (left panel) and frequency of actively proliferating Hopx-EGFP+ cells in control mice and mice treated with 12 Gy γ-IR and quantified 48 hr later. For (D) and (E), n = 3 independent experiments, ±SD, ∗p < 0.1, and ∗∗p < 0.01.
(F) Atoh1 transcript levels in single cells across the populations shown in (B). ∗∗∗p < 0.0005 for difference in mean expression of Atoh1 in Hopx-EGFPHigh cells versus any other population, Two-sided t test.
(G) Pearson correlation matrix between expression of Atoh1 and transcripts characteristic of secretory progenitor cells.
See also Figure S4.
We also examined how Hopx-EGFP+ cells respond to injury after irradiation. A 12 Gy dose of γ-IR is sufficient to quantitatively ablate Lgr5+ CBCs (Yan et al., 2012). Within 2–4 days after γ-IR, surviving reserve ISCs begin actively proliferating, with Lgr5+ cells arising shortly thereafter (Yan et al., 2012). We administered 12 Gy γ-IR to Hopx-EGFP+ mice and observed a significant increase in both the frequency and proliferative rate of Hopx-EGFP+ cells 2 days after γ-IR relative to nonirradiated controls (Figure 3E), providing evidence that a subset of the Hopx-EGFP+ population contains a radioresistant cell capable of surviving and proliferating in response to high-dose γ-IR, properties consistent with a reserve ISC.
Given that Hopx-EGFP+ cells labeled nearly all cells at the base of crypts (Figure 1I), we reasoned that Paneth cells should also be contained in one of the Hopx-EGFP+ subpopulations. Indeed, c-Kit+, CD24+ Paneth cells were observed in the Hopx-EGFP+ population, and these cells constituted a large fraction of the Hopx-EGFPHigh population (Figure S4B). These findings provide clear evidence that Hopx-CreER and Hopx-EGFP mark different cell populations and thus do not support the use of Hopx-EGFP as a surrogate marker for reserve ISCs.
Identification of the Secretory Precursor Cell Molecular Signature
Several recent studies have identified secretory precursor cells with seemingly disparate properties. One study identified LRCs with very high Lgr5 expression as precursors of the intestinal secretory lineages (Buczacki et al., 2013). Because these cells were found to be slow cycling, it was posited that the reserve ISCs marked by Bmi1-CreERT2 may in fact represent these secretory precursors rather than a distinct, general reserve ISC. Our cell-cycle analysis suggests that Bmi1-CreERT2 likely cycle too frequently to be LRCs, and our single-cell profiling of the Bmi1-CreERT2 cells revealed that they express little to no Lgr5, indicating that they are not label-retaining secretory precursors. Further, numerous published studies have demonstrated that reserve ISCs (marked either by Bmi1- or Hopx-CreERT2) act as stem cells during homeostasis and generate all cell types of the intestinal epithelium, not only secretory lineages. Thus, our study and numerous published reports provide compelling evidence that the reserve ISCs are both functionally and molecularly distinct from label-retaining secretory precursor cells.
Another independent study identified a distinct population of secretory precursors through the use of Dll knockin reporter alleles (van Es et al., 2012). In contrast to the LRCs, the Dll+ secretory precursors exhibited a very specific gene-expression pattern with high expression of genes encoding Notch ligands (Dll), low levels of Notch receptors and Notch pathway target genes (Hes), and very high levels of Atoh1 (Math1; van Es et al., 2012). Unlike the label-retaining precursors, these cells also exhibit very low levels of canonical Wnt target genes including Lgr5. We therefore searched for cells with patterns of anticorrelation between Atoh1 and the Notch/Wnt pathway (Atoh1High, NotchLow, WntLow, and DllHigh) within the various populations. Correlation matrices between genes across all of the populations (Table S4) revealed the expected anticorrelation only within the Hopx-EGFPHigh group. This group had the largest population of AtohHigh cells relative to all other groups (Figure 3F), and the Hopx-EGFPHigh cells exhibited significant anticorrelation between Atoh1 expression and that of Wnt target genes including Ascl2 and Lgr5, as well as Notch pathway genes Notch1, Hes1, and Olfm4 (Figures 3G and S4C). Further, there was a significant positive correlation between Atoh1 and the Notch ligand Dll4 in this population (Figure 3G), consistent with the lateral inhibition model of Notch signaling and the previously established secretory precursor cell molecular signature. These molecular correlations were not significantly observed in the functional reserve ISC populations, in the Lgr5-EGFP+ population, or other Hopx-EGFP populations (Table S4). Thus, our findings strongly suggest that the Dll+ secretory progenitor cells exist as a subpopulation within the Hopx-EGFPHigh population and are distinct from the functionally validated Bmi1- and Hopx-CreERT2-marked reserve ISCs.
Examining the Functional Capacity of Active and Reserve Intestinal Stem Cells
Tam-induced lineage tracing initiated by Lgr5-, Hopx-, or Bmi1-CreERT2 demonstrated that these populations are all capable of producing all of the functional cell types of the intestine; however, the frequency and dynamics with which they self-renew produce other ISC types, produce non-stem cell progeny, or undergo exhaustion is poorly understood. We therefore set out to compare the proliferative output of CBCs and reserve ISCs during intestinal homeostasis. To study the reserve ISC behavior, we chose to use the Hopx-CreERT2 allele rather than the Bmi1-CreERT2 because Hopx-CreERT2 did not label differentiated cells in the villi and the Hopx-CreERT2+ population was more homogenous than the Bmi1-CreERT2+ population based on its lower coefficient of variation (Figure S5A).
Our experimental strategy was to initiate tracing from Hopx-CreERT2 or Lgr5-CreERT2 with a single Tam dose on the LSL-Tomato background and then collect and profile progeny produced 4 and 7 days later (Figure S5B). We initiated activation of the Tomato reporter in Hopx-CreERT2+ mice and assessed clonal expansion. Figure 1J shows the first histologically detectable Tomato+ cells 24 hr after Tam treatment as single cells within crypts. Four days after Tam, Hopx-CreERT2+ progeny consisted primarily of small clusters of one to four cells in the crypts or around the crypt-villus junction, and 7 days later, some of these clusters had gone on to label a ribbon of differentiated cells in the villi (Figures 4A and S5C). In contrast, the proliferative output from Lgr5-CreERT2 was more rapid and robust. Four days after initiating labeling, Lgr5 progeny had already encompassed the entire crypt-villus axis (Figure 4B).
Figure 4.
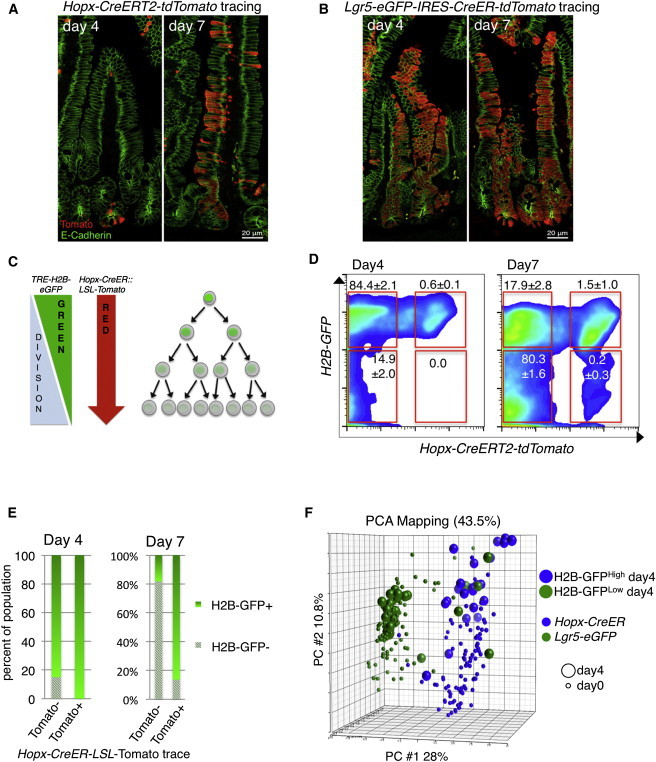
Defining the Proliferative Output of Reserve ISCs
(A and B) Lineage tracing from Hopx-CreERT2 (A) and Lgr5-CreERT2 (B) 4 and 7 days after activation of the LSL-tdTomato reporter.
(C) Lineage-tracing strategy from Hopx-CreERT2 while simultaneously monitoring cell division through loss of the H2B-GFP label.
(D) Flow cytometric analysis of proliferation in the intestinal epithelium (through loss of H2B-GFP label) relative to HopxCre-tdTomato lineage tracing at 4 and 7 days post-Tomato activation and H2B-GFP chase (n = 3 independent experiments; ±SD).
(E) Quantification of H2B-GFP loss in progeny of Hopx-CreERT2 cells (Tomato+) versus bulk epithelium (Tomato−) as in (D).
(F) PCA plot of single cells derived from Hopx-CreERT2 reserve ISCs (Tomato+) that have either not undergone cell division (H2B-GFPHigh; n = 48) or have divided (H2B-GFPLow; n = 48). Parental Hopx-CreERT2 cells and Lgr5-EGFP cells are those shown in Figure 2 and included in the plot for reference.
See also Figure S5.
Whereas the proliferative dynamics of Lgr5+ CBCs have been extensively characterized (Snippert et al., 2010), analogous studies on reserve ISC populations are lacking. To begin understanding these dynamics, we crossed a doxycycline (dox)-inducible H2B-GFP allele (TRE-H2BGFP) into the Hopx-CreERT2-LSL-Tomato reporter mice and maintained these mice on dox for 6 weeks, starting at postnatal day 14 in order to fully label nuclei with GFP. We then withdrew dox and simultaneously initiated Tomato tracing (Figures 4C and S5B), enabling us to gauge the amount of cell division that has occurred in daughter cells emanating from the Hopx-CreERT2 reserve ISC by monitoring loss of H2B-GFP. Four days after initiating tracing, we observed that the Hopx-CreERT2 progeny had undergone limited cell division compared to the bulk (Tomato−) population and could be divided into two populations of H2B-GFP intensity (Figures 4D and S5D). One week after initiation of the lineage trace, a subpopulation of Tomato+ cells had undergone sufficient divisions to lose their H2B-GFP label; however, the progeny of the Hopx-CreERT2 ISC still retained more H2B-GFP than the Tomato− population, consistent with these cells cycling slower than CBCs (Figures 4D and 4E).
To understand what cell-fate decisions the reserve ISC makes in its initial divisions, we separated Hopx-CreERT2 progeny into two populations 4 days after initiation of lineage tracing: an H2B-GFPHigh and H2B-GFPLow population (Figure S5D). These were then subjected to single-cell profiling. Remarkably, PCA shows a clear division of cellular identity between these two groups: the H2B-GFPHigh population retained a reserve ISC identity, whereas the cells that had divided (H2B-GFPLow) primarily exhibited an Lgr5+ CBC identity (Figures 4F and S5E). This indicates that, in their earliest cell-fate decision, reserve ISCs give rise to Lgr5+ CBCs, providing molecular evidence to support prior histological observations that cells traced from Bmi1- or Hopx-CreERT2 give rise to Lgr5+ cells (Li and Clevers, 2010; Takeda et al., 2011; Tian et al., 2011).
We next compared the proliferative output of reserve ISCs to actively cycling CBCs 7 days after initiation of tracing. Because of the complexity of this population, we developed a mathematical model of stem cell identity based on the initial HopxCreERT2-Tomato+ and Lgr5-EGFP+ populations discussed above. We used these populations as references to train an algorithm to build an idealized reserve versus CBC ISC molecular identity and then retrospectively asked how many of the cells in these reference populations fit the mathematically idealized identities. The vast majority of Hopx-CreERT2+ cells at day 0 exhibited a reserve ISC identity, with only a few cells being categorized as CBC-like or “other” (i.e., differentiated or T/A cells; Figures 5A and 5E). Similarly, the vast majority of Lgr5-EGFP+ cells at day 0 were categorized as CBC-like, with only two cells in this group being identified as reserve ISCs (Figures 5B and 5E). To further examine the fidelity of the algorithm, we interrogated Bmi1-CreERT2+ cells at day 0 and found that this population contained more CBC-like cells than the Hopx-CreERT2+ population but fewer than the Lgr5-EGFP+ population, as well as a slight increase in the number of cells that fall into neither category (Figure S6A). This is exactly what would be expected given the physical distribution of Bmi1-CreERT2+ cells, their PCA clustering, and the frequency of cells expressing the canonical Wnt/CBC signature genes in the Bmi1-CreERT2+ population relative to the other populations.
Figure 5.
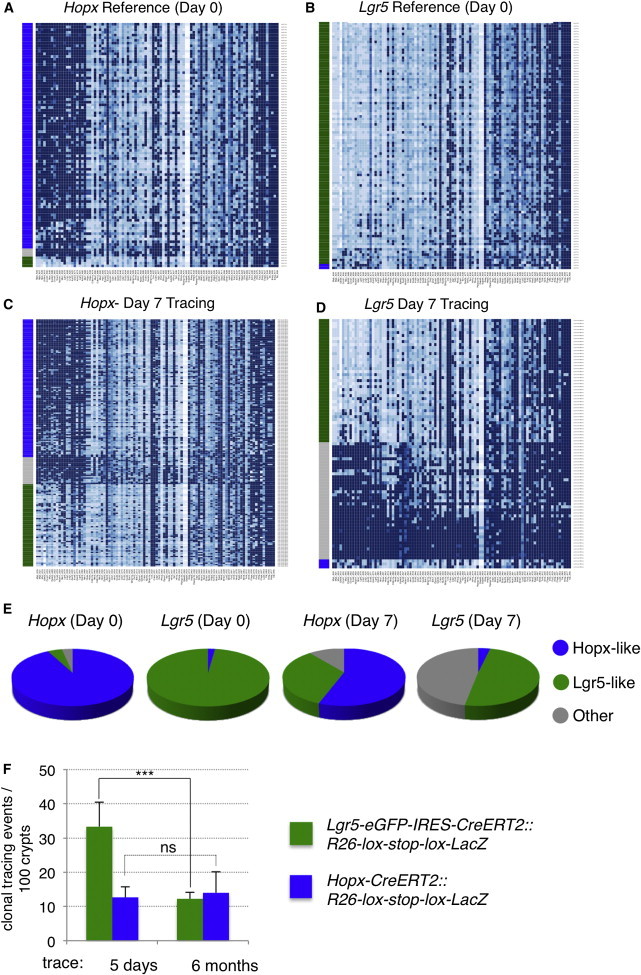
Molecular Characterization of Reserve ISC versus Active CBC Stem Cell Progeny
(A–D) Heatmaps of gene expression of 48 transcripts across populations of single Hopx-CreERT2 cells in the reference population (18 hr after Tomato activation; A), in the Lgr5-EGFP reference population (B), in the progeny of Hopx-CreERT2 cells after 7 days of lineage tracing (C; n = 188), and in the progeny of Lgr5-EGFP-IRES-CreERT2 cells after 7 days of lineage tracing (D; n = 95).
(E) Distribution of cellular identity defined by the algorithm in the four populations pictured in (A)–(D).
(F) Frequency of clonal-tracing events initiated by a single dose of Tam in Lgr5-CreERT2::LSL-Lacz or Hopx-CreERT2::LSL-Lacz mice (n = 15 independent experiments; ±SD); ∗∗∗p < 1 × 10−4.
See also Figure S6.
One week after initiation of the lineage trace, Hopx-CreERT2 progeny generated primarily more reserve ISCs as well as CBC-like cells and a small percentage of differentiated other cells (Figures 5C and 5E). In contrast, Lgr5-CreERT2 gave rise primarily to differentiated progeny (other), followed by CBC cells, with no discernable increase in the fraction of reserve ISCs (Figures 5D and 5E). Taken together, these findings provide compelling evidence that, under homeostatic conditions, reserve ISCs either self-renew or generate CBCs upon dividing. In contrast, CBCs self-renew and generate the differentiated progeny necessary for tissue function. We find no evidence that CBCs give rise to reserve ISCs with any appreciable frequency under homeostatic conditions, although we cannot rule out that such conversion occurs at low frequency or in response to tissue damage.
Given our observations that Hopx-CreERT2 cells give rise to Lgr5+ CBCs in their initial divisions and that Hopx-CreERT2 clones are long-lived, generating all cell types of the epithelium for up to 1 year (Takeda et al., 2011), one would predict that this would either result in a net accumulation of Lgr5+ CBCs over time if these cells are equally long-lived or that Lgr5+ CBCs undergo loss over time necessitating replacement. To address this, we analyzed extinction of lineage-tracing events in clones derived from either Lgr5-CreERT2::R26-LSL-Lacz mice or Hopx-CreERT2::R26-LSL-Lacz mice shortly after initiation of tracing with a single Tam dose (5 days) and after a 6-month chase period (Takeda et al., 2011). Interestingly, Lgr5-CreER-derived clones exhibited significant clonal extinction, with approximately a 75% reduction in the number of clones persisting over the 6-month chase period (Figures 5F and S6B). In contrast, the frequency of Hopx-CreER-derived clones remained constant over the same time course. This finding highlights differences in proliferative output of these two ISC types, with reserve ISC clones maintaining clonal proliferative output significantly longer than active ISCs.
Hierarchical Model of Intestinal Cells Marked by ISC Proxy Knockin Marker Alleles
In order to compare populations marked by the knockin proxy reporter alleles shown in Figure S1, we performed unsupervised hierarchical clustering of all cells profiled in this study (Figure 6A). As expected, the bulk of Lgr5+ CBCs comprise one end of the hierarchy, and the reserve Hopx-CreERT2-marked ISCs comprise the other end of the spectrum. Bmi1-CreERT2-marked reserve ISCs tend to cluster with Hopx-CreERT2 cells, with the exception of the few cells that have a CBC identity. Hopx-EGFPLow cells cluster near the reserve ISCs, and Hopx-EGFPHigh and Hopx-EGFPSmall cells generally fall into distinct clusters between the two extremes of the spectrum. Interestingly, a small subset of cells from all of these groups falls into the CBC cluster, indicative of the heterogeneity within these populations (Figure 6A).
Figure 6.
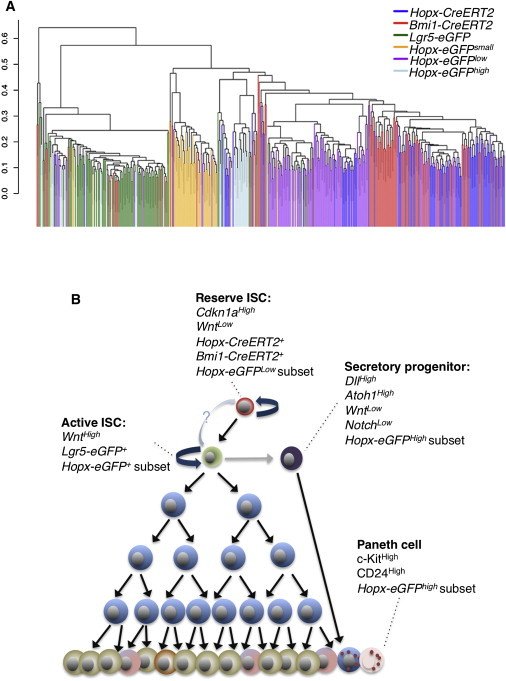
Hierarchical Model of Cellular Identity
(A) Unsupervised hierarchical clustering of single cells from proxy reporter allele-marked groups.
(B) Working model of intestinal stem cell hierarchy.
Discussion
The characterization of ISC populations by histological and bulk molecular profiling approaches has resulted in a number of seemingly contradictory findings in the published literature. Specifically, indispensable reserve ISCs marked by Bmi1- and Hopx-CreERT2 alleles have been shown to give rise to dispensable Lgr5-EGFP+ CBCs (Sangiorgi and Capecchi, 2008; Tian et al., 2011), indicating that two functionally distinct ISC populations exist. The broad acceptance of these findings has, however, been precluded by observations that endogenous Bmi1 and Hopx transcripts are readily detectable in Lgr5+ cells, a finding that has led to speculation that these ISC populations may mark cells along a continuum rather than two distinct populations. Our findings clearly demonstrate that the Bmi1-CreER and Hopx-CreER-marked populations contain primarily reserve ISCs that are molecularly distinct from the Lgr5-EGFP+ CBCs, providing molecular support for the cell ablation studies demonstrating their functional dissimilarity.
The interpretation of ISC data is further confounded by the use of EGFP proxy reporters inserted into the reserve ISC loci. Whereas the Hopx-EGFP allele marks some cells with active CBC stem cell identity, as well as some reserve ISCs based on expression profiles and γ-IR studies, we conclude that this reporter acts as a nonspecific marker of intestinal crypt cells. This includes the presence of secretory precursor cells within the Hopx-EGFP+ population based on their gene-expression signature characterized by high Atoh1 to Wnt/Notch pathway expression ratio (van Es et al., 2012).
Conversely, we also provide evidence that the Bmi1-EGFP allele does not recapitulate the expression pattern of its CreERT2 counterpart, and in fact, we are unable to reliably detect any Bmi1-EGFP+ cells despite procuring these mice directly form Jackson Laboratories stocks. Thus, our data provide evidence that their use as proxies for reserve ISC activity is unreliable, at best, and can be misleading if presented as evidence for stem cell identity (as only the CreER reporters in the Bmi1 and Hopx loci have been functionally demonstrated to mark reserve stem cells).
In contrast, the Lgr5-EGFP-CreER allele exhibits good correlation between the EGFP and CreER reporters. The Lgr5 reporter is bicistronic, with EGFP and CreER emanating from a single transcript, whereas the Bmi1 and Hopx reporters are derived from distinct targeting events with differing sites of integration. It is tempting to speculate that the alternative usage of untranslated regions governing mRNA stability accounts for the observed discrepancies, although no direct evidence for this hypothesis exists.
We also resolve the controversy surrounding the identity of reserve versus active ISCs resulting from the observation that endogenous Bmi1 and Hopx transcripts are readily detectable in Lgr5+ cells. Whereas we observe endogenous Bmi1 and Hopx transcripts in Bmi1-CreER and Hopx-CreER-marked ISCs, the highest levels of these endogenous transcripts are actually present in Lgr5-EGFP+ cells. This demonstrates a clear disjunction between the activity of the reporter cassettes and the endogenous transcripts, which could be a result of message stability, CreER activity, or other unknown causes. Regardless of the underlying cause, our findings indicate that, in the case of reserve ISCs, the only alleles that reliably mark these cells are Bmi1- and Hopx-CreER and that the presence of the Bmi1 and Hopx transcripts cannot act as proxies for reserve ISC activity.
Finally, we provide functional evidence supporting the previously proposed two-intestinal-stem-cell system (Li and Clevers, 2010). Through lineage tracing and single-cell profiling of daughter cells derived from either active (Lgr5-CreER+) or reserve (Hopx-CreER+) ISCs over time, we observe striking differences in the proliferative output of these two cell types. In their initial divisions, reserve ISCs self-renew and generate active Lgr5+-CBCs. In contrast, active Lgr5+-CBCs self-renew and generate differentiated progeny (Figure 6B).
These findings predict that Lgr5+-CBCs must have a significantly shorter lifespan than the reserve ISCs; otherwise, an age-related accumulation of Lgr5+-CBCs would result from the constant generation of CBCs from the reserve ISCs. Indeed, this finite lifespan of Lgr5+-CBCs can be observed in prior studies where the random activation of any one of four fluorophores in the Lgr5-CreER::R26R-Confetti mouse model results in multiple unique labeling events in any given crypt, and over time, clonal extinction is observed as a drift to monoclonality (Snippert et al., 2010).
In the current study, we directly compare the extinction of Lgr5-CreER- and Hopx-CreER-marked clones and demonstrate that, whereas approximately two thirds of Lgr5 clones are lost over a 6-month period, there is no loss of Hopx-CreER-derived clones. This result provides additional evidence that the reserve ISC is capable of longer proliferative output than the Lgr5+ CBC. Because CBCs have a shorter lifespan than the reserve Hopx-CreER ISCs, it is tempting to speculate that lost CBCs are replaced by new CBCs generated de novo from reserve ISCs. Although there is clear evidence that Lgr5 cells compete with one another for crypt dominance through neutral drift (existing Lgr5 cells replacing lost Lgr5 cells through symmetric division), there is also clear evidence that reserve ISCs generate Lgr5 cells, and thus, further studies are necessary to determine which mechanism of Lgr5 cell replacement occurs during homeostasis and with what frequency.
Taken together, our findings provide support for the hierarchical two-stem-cell hypothesis previously proposed, where active and reserve stem cells exists in functionally and molecularly distinct states and argue against the notion that these two stem cell populations simply represent different stages of a single stem cell continuum.
Experimental Procedures
Mouse Strains
Lgr5−EGFP-IRES-CreERT2 (JAX mice strain 008875), Bmi1-CreERT2 (JAX strain 010531), and Bmi1-EGFP (JAX strain 017351) mice were obtained from the Jackson Laboratory. Hopx-CreERT2 (JAX strain 017606) and Hopx-EGFP mice were generated at the University of Pennsylvania. Mice were maintained on a C57/BL6N background (or C57BL/Ka for Bmi1-EGFP). TRE-H2BGFP mice were obtained from Jackson Laboratory (Jax strain 016836). For all Cre induction experiments, mice received a single intraperitoneal injection of 100 μl Tam (10 mg/ml in corn oil; Sigma; T5648). All mouse protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania under protocol 803415 to Dr. Lengner.
H2B-GFP Labeling
TRE-H2BGFP::Hopx-CreERT2::lox-stop-lox-tdTomato mice were maintained on dox for 6 weeks starting at postnatal day 14 in order to fully label all nuclei with GFP. Dox was withdrawn when mice reached 8 weeks of age and tdTomato lineage tracing was simultaneously initiated one dose of Tam. Mice were sacrificed 4–7 days after dox withdrawal and initiation of tracing.
Two-Step Single-Cell Gene Expression
The two-step single-cell gene-expression protocol (advanced development protocol 33) from Fluidigm was adopted for this study. Briefly, 5 μl of RT Mix Solution which includes 1.2 μl 5× VILO Reaction Mix (Life Technologies; 11754-250), 0.3 μl SUPERase-In (Life Technologies; AM2696), and 0.25 μl 10% NP40 (Thermo Scientific; 28324) was dispensed into each well of 96-well plate. Single cells were sorted into the well directly. The plate was vortexed and immediately frozen on dry ice. For room temperature cycling, the plate was thawed on ice and RNA denatured by incubating at 65°C for 90 s and then chilled on ice for 5 min. Each well was supplemented with 1 μl mixture of 10× SuperScript Enzyme Mix (Life Technologies; 11754-250) and T4 Gene 32 Protein (New England BioLabs; PN M0300S). mRNA was reverse transcribed into cDNA following the thermal cycling conditions below: 25°C, 5 min/50°C, 30 min/55°C, 25 min/60°C, 5 min/70°C, and 10 min. Resulting cDNA was preamplified with 50 nM primer mix for 23 PCR cycles (96°C for 5 s and 60°C for 4 min) and then treated with ExoI for 30 min to remove unincorporated primers. The final product was diluted 1:3 with Tris-EDTA (TE) buffer. For each chip sample inlet, 2.25 μl diluted cDNA, 2.5 μl 2× Sso Fast EvaGreen supermix with low ROX, and 0.25 μl of Fluidigm sample loading agent were added. Individual gene-specific DELTAgene assays were diluted at 1:10 ratios with TE buffer. Two and a half microliters of each primer was then mixed with 2.5 μl assay loading agent inserted into chip “assay” inlets. Chip loading and PCR was performed according to the manufacturer’s protocol. The data were analyzed by Fluidigm Gene Expression Analysis Package.
Genes analyzed include: Areg, Ascl2, Atoh1, Axin2, Bmi1, Bmpr1a, Ccnd1, Cdkn1a, Cdx1, Chga, Cubn, Dll4, Dvl2, Efnb1, Epas1, Ephb2, Ereg, Fut2, Gapdh, Gsk3b, Gusb, H6pd, Hes1, Hes5, Hif1a, Hopx, Jag1, Lgr5, Lrig1, Lyz2, Msi1, Msi2, Myb, Myc, Notch1, Numb, Olfm4, Pcna, Ppargc1b, Rhoa, Saa2, Sirt3, Sox9, Tat, Tcf4, Tert, Wnt3, and Wnt6. Details regarding the generation and training of the ISC identity-calling algorithm can be found in the Supplemental Information.
Histological Scoring of Hopx-CreER and Lgr5-CreER-Derived R26R-Lacz Clones
Archived tissue from Hopx-CreER::R26-LSL-Lacz and Lgr5-EGFP-IRES-CreER::R26-LSL-Lacz small intestines were provided by and described in Takeda et al. (2011). The frequency of clonal tracing events in crypts (Lacz+ clones) from 5-day and 6-month lineage traces was scored in freshly cut paraffin sections. For Hopx-CreER, a total of 641 crypts were scored at day 5 and 739 at 6 months. For Lgr5-CreER, a total of 809 crypts were scored at day 5 and 1,048 at 6 months.
Acknowledgments
We thank Drs. Anil Rustgi and John Lynch in the Division of Gastroenterology, Department of Medicine, University of Pennsylvania for reagents and fruitful discussions. C.J.L. is funded by R01 CA168654 from the National Cancer Institute. This work was supported in part by a pilot award from the University of Pennsylvania Institute for Regenerative Medicine and the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306) and its core facilities (molecular pathology and imaging, molecular biology/gene expression, cell culture, and mouse), and C.J.L. was supported by the center’s pilot and feasibility grant program.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Chen F., Kook H., Milewski R., Gitler A.D., Lu M.M., Li J., Nazarian R., Schnepp R., Jen K., Biben C. Hop is an unusual homeobox gene that modulates cardiac development. Cell. 2002;110:713–723. doi: 10.1016/s0092-8674(02)00932-7. [DOI] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Itzkovitz S., Lyubimova A., Blat I.C., Maynard M., van Es J., Lees J., Jacks T., Clevers H., van Oudenaarden A. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Metcalfe C., Kljavin N.M., Ybarra R., de Sauvage F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I.K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Potten C.S., Owen G., Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Steinhauser M.L., Bailey A.P., Senyo S.E., Guillermier C., Perlstein T.S., Gould A.P., Lee R.T., Lechene C.P. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., Leboeuf M.R., Padmanabhan A., Wang Q., Li L., Lu M.M., Millar S.E., Epstein J.A. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es J.H., Sato T., van de Wetering M., Lyubimova A., Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang J., Liu D., Su Y. Normalizing genes for real-time polymerase chain reaction in epithelial and nonepithelial cells of mouse small intestine. Anal. Biochem. 2010;399:211–217. doi: 10.1016/j.ab.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


