Summary
It is clear that neural differentiation from human pluripotent stem cells generates cells that are developmentally immature. Here, we show that the let-7 plays a functional role in the developmental decision making of human neural progenitors, controlling whether these cells make neurons or glia. Through gain- and loss-of-function studies on both tissue and pluripotent derived cells, our data show that let-7 specifically regulates decision making in this context by regulation of a key chromatin-associated protein, HMGA2. Furthermore, we provide evidence that the let-7/HMGA2 circuit acts on HES5, a NOTCH effector and well-established node that regulates fate decisions in the nervous system. These data link the let-7 circuit to NOTCH signaling and suggest that this interaction serves to regulate human developmental progression.
Graphical Abstract
Highlights
-
•
let-7 miRNAs influence developmental maturity of neural progenitors
-
•
let-7 miRNAs act through HMGA2 and NOTCH to regulate gliogenesis
-
•
HMGA2 expression regulates access of NICD to HES5 promoter
-
•
Induction of let-7 miRNAs can accelerate oligodendrogenesis
Lowry and colleagues investigate the role of let-7 miRNAs in human neural progenitor cell developmental maturation and fate decisions. They identify HMGA2, a let-7 target gene, and HES5, a Notch effector, as critical nodes in a pathway regulating a transition from neurogenesis to gliogenesis, and demonstrate a regulatory mechanism by which HMGA2 allows access to the HES5 promoter.
Introduction
We previously determined that, by both gene expression and functional analyses, the derivatives of human pluripotent stem cells (hPSCs) more closely resembled cell types found prior to 6 weeks of gestation than later time points (Patterson et al., 2012). In fact, this appears to be an emerging theme in hPSC differentiation (Chang et al., 2011; Mariani et al., 2012; Zambidis et al., 2005). This suggests that hPSC derivatives are developmentally immature, which could stem from either inadequate culturing methods or could suggest that developmental timing is somewhat conserved in vitro.
Among the most differentially expressed genes in all PSC derivatives are LIN28A and LIN28B, RNA binding proteins known to regulate the let-7 family of miRNAs (Patterson et al., 2012). LIN28B seems to function primarily in the nucleus by sequestering pri-let-7s to prevent maturation by Microprocessor, whereas LIN28A functions in the cytoplasm by recruiting uridylyl transferase to polyuridylate the pre-let-7s and prevent their further processing by Dicer (Graf et al., 2013; Hagan et al., 2009; Kim and Nam, 2006; Lee et al., 2014; Piskounova et al., 2011). In lower organisms, Lin28A expression is strongly correlated with the differentiation status and self-renewing capacity of cells throughout development. Although there is less evidence for the role of this pathway in human development specifically, many groups have demonstrated that LIN28A is reexpressed in a variety of human cancers and is highly correlated with prognosis and disease progression (Viswanathan and Daley, 2010; West et al., 2009). Furthermore, LIN28A has also been used to reprogram somatic cells back to the pluripotent state (Yu et al., 2007). All of these known roles are linked to developmental progression and make LIN28A/B-let-7 an attractive candidate for manipulating the maturity of hPSC-derived cells.
Previous work by other groups in lower organisms has argued that Lin28A plays a role in maturation of the nervous system (Balzer et al., 2010), and some have shown that overexpression of LIN28B in human adult hematopoietic stem/progenitor cells can reverse their developmental progression to a fetal-like state (Yuan et al., 2012). Downstream of LIN28A/B, however, a role for let-7 in human gestational maturation in the nervous system has not been established. In fact, one study of a murine model has suggested that the role of Lin28A in developmental progression was let-7 independent (Balzer et al., 2010). Recent work has also suggested that LIN28/let-7 regulates neurogenesis by controlling the proliferation of progenitors (Cimadamore et al., 2013; Nishino et al., 2013). Here, we explore the role of the LIN28/let-7 pathway in the developmental progression of human neural progenitor cells (NPCs). We demonstrate that LIN28B plays a clear role in gestational progression of the developing human nervous system through regulation of let-7 miRNAs. These miRNAs then go on to regulate HMGA2, which has also been implicated in developmental progression (Sanosaka et al., 2008). We also show here that HMGA2 appears to regulate cell-fate decisions in neural progenitors (NPCs) in this context through HES5, a key node in the NOTCH pathway and previously implicated in neurogenesis.
Results
PSC-NPCs Are Functionally and Transcriptionally Distinct from Tissue-Derived Counterparts
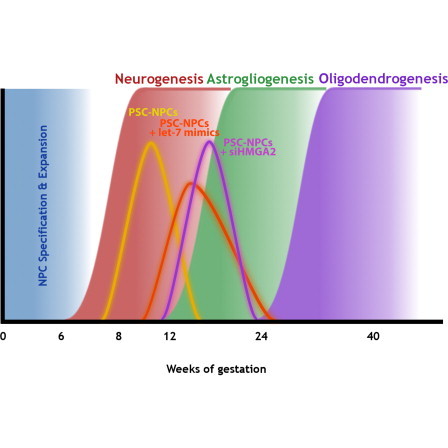
During mammalian development, NPCs progress through several phases: an early expansion phase, characterized by symmetric divisions; a neurogenic phase, characterized by asymmetric divisions and resulting in new neurons; a gliogenic phase where astrocytes are primarily produced; and finally a phase where oligodendrocytes are generated (Figure 1). The result is that neurogenesis precedes gliogenesis on a developmental timescale.
Figure 1.
let-7 Activity Correlates with Human Gliogenesis
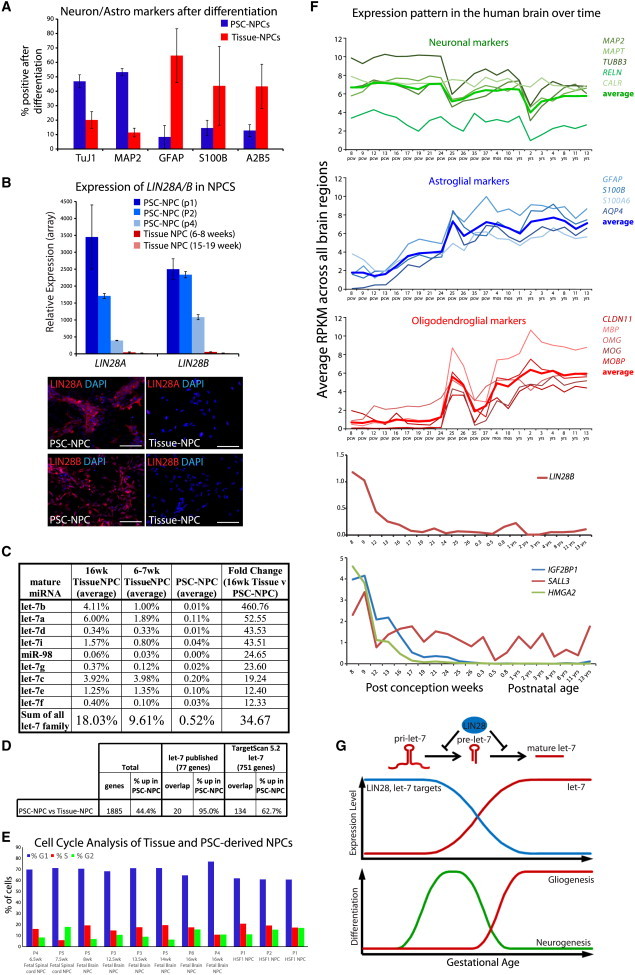
(A) Differentiation of human NPCs by growth factor withdrawal drives the generation of neurons and astrocytes. Percentage of positive PSC-NPCs and Tissue-NPCs (7–19 weeks of gestation) undergoing neuronal (TUJ1, MAP2) versus glial (GFAP, S100B, A2B5) differentiation. Error bars represent standard error of the mean (SEM) over three at least three biological replicates.
(B) Average expression of LIN28A and LIN28B probe sets from (Patterson et al., 2012). PSC-NPCs p1 (n = 7), PSC-NPCs p2 (n = 2), PSC-NPCs p4 (n = 2), 6–8 week tissue-NPCs (n = 6), 15–19 week fetal NPCs (n = 5). Error bars represent SEM over biological replicate lines. Scale bars, 100 μm.
(C) Percentage of contribution of let-7 miRNA family members to all miRNA present in cells derived from 16 week Tissue-NPCs (n = 2), 6–7 week Tissue-NPCs (n = 2), and PSC-NPCs (n = 3) by miRNA-seq. Fold change of normalized expression between 16 week Tissue-NPCs and PSC-NPCs for each family member is shown at right. Data for all miRNAs can be found in Table S1.
(D) Genes differentially expressed between PSC-NPCs (n = 7) and Tissue-NPCs (n = 5) as identified (Patterson et al., 2012) and the number of let-7 targets represented in the data. let-7 target lists were generated from published articles and TargetScan 5.2.
(E) Despite having significantly different levels of let-7, PSC- and Tissue-NPCs show similar proliferative potential as measured by cell-cycle analysis based on propidium iodide staining and flow cytometry.
(F) RNA-seq data from the Allen Institute’s Brainspan developmental transcriptome database displayed as log-scale reads per kilobase measured (log2 RPKM) across the developing human brain. Top, transcription of neuronal, glial and oligodendrocyte marker genes (the average of all markers indicated by boldface line); middle, transcription of LIN28B; bottom, transcription of selected let-7 target genes.
(G) Top, schematic of LIN28’s role in the progression of let-7 microRNAs and the pathway’s temporal correlation with development. Bottom, differentiation propensity of NPCs correlated with gestational age.
NPCs were derived from either hPSCs or from fetal tissue sources and were validated by immunostaining and judged to be relatively homogenous (Figure S1A; Patterson et al., 2012). We determined that PSC-NPCs across all passages had a higher propensity to differentiate into MAP2/TUJ1+ neurons (∼50%) over GFAP/S100/A2B5+ glia (<10%) (Figure 1A). Meanwhile, tissue-derived NPCs isolated from fetal brain or spinal cord samples at 12–19 weeks of gestation (Tissue-NPCs) were more apt to differentiate into glia (∼70%) over neurons (<20%) (Figure 1A; Patterson et al., 2012). These data suggest that PSC-NPCs were functionally less mature than tissue-derived counterparts (neurogenesis precedes gliogenesis). Furthermore, we have previously shown that upon subsequent passage of PSC-NPCs the propensity for gliogenesis increased but still did not approach that of tissue-derived cells (Patterson et al., 2012).
To understand the molecular basis for this observed functional discrepancy, gene expression profiling was performed on PSC derivatives and tissue-derived counterparts (Patterson et al., 2012). Among the most differentially expressed genes were LIN28A and LIN28B (Figures 1B, top, and S1C), and this was confirmed at the protein level by immunostaining (Figure 1B, bottom). Although continued passaging reduces the levels of LIN28A and LIN28B in PSC-NPCs, their expression is not decreased to a level found in the Tissue-NPCs within the time points utilized for this study (Figure 1B; Patterson et al., 2012).
LIN28 homologs are known to negatively regulate the highly conserved let-7 family of miRNAs. Our previous microarray analyses demonstrated a significantly higher expression of some let-7 miRNAs in Tissue-NPCs (Figure S1C), and this result was confirmed by direct sequencing of mature miRNA (Figure 1C; Table S1). The latter analysis demonstrated that not only were all let-7 family members significantly higher in Tissue-NPCs, as we had previously shown by RT-PCR (Patterson et al., 2012), but all nine family members were found among the top 30 differentially expressed miRNAs between Tissue-NPCs and PSC-NPCs (Table S1). Furthermore, the let-7 family as a whole was the most abundantly expressed miRNA family in 16 week Tissue-NPCs, representing almost 18% of the total miRNA in these cells. In addition, let-7 family members were expressed at an intermediate level in 6–7 week Tissue-NPCs.
To determine whether let-7 target genes were among the differentially expressed mRNA distinguishing PSC-NPCs from their tissue-derived counterparts, two lists of let-7 targets were generated (Figure 1D): one with published let-7 targets that have been experimentally confirmed (77 genes) and one with predicted let-7 targets generated by TargetScan 5.2 (751 genes). Of the 77 published let-7 targets, 20 were differentially expressed between PSC-NPCs and Tissue-NPCs, and 134 of the 751 TargetScan predicted let-7 targets were differentially expressed. Notably, 95% and 63%, respectively, were specifically lower in Tissue-NPCs, consistent with a negative regulatory activity of let-7 in tissue-NPCs.
Recent work in mice has suggested that during neurogenesis, the induction of let-7 leads to exit of the cell cycle and differentiation toward neurons (Cimadamore et al., 2013; Nishino et al., 2013). Analysis of eight tissue-derived NPCs and three PSC-derived NPCs failed to find consistent differences in the percentage of cells in S or G2/M phase of the cell cycle (Figure 1E), suggesting that despite having orders of magnitude more let-7 (Figure 1C), Tissue-NPCs can still maintain a proliferative state.
To determine when the transition from neurogenesis to gliogenesis occurs endogenously, we probed developmental transcriptome data for glial hallmark genes from the Allen Brain Atlas. GFAP, AQP4, and S100B were all expressed at a low level in all brain regions prior to 12 weeks but surged thereafter suggesting that gliogenesis begins in the human brain at roughly 12 weeks of gestation. A similar examination of oligodendrocyte markers suggests that oligodendrogenesis begins at 24 weeks (Figure 1F, middle). These gene expression data are consistent with pathological data on fetal tissue showing oligodendrocyte progenitors prevalent at 20 weeks of gestation and mature oligodendrocytes at 30 weeks (Craig et al., 2003; Dean et al., 2011; Jakovcevski et al., 2009). Notably, the expression of LIN28B in human brain was only detectable in weeks 8–9 of gestation and dropped off significantly thereafter (Figure 1F, middle), whereas LIN28A was undetectable in these analyses. let-7 miRNAs were also detectable in these data sets, and this family showed a striking induction across the human brain in the 9–12 postconception week (PCW) time frame (Figure S1B). Taken together, these data suggested that there is a strong correlation between the transition from highLIN28/lowlet-7 to lowLIN28/highlet7 state and the switch from neuro- to gliogenesis. Therefore, we hypothesized that the let-7 plays a functional role in the decision to make either neurons or glia by neural progenitors (Figure 1G).
let-7 Expression and Processing across Development
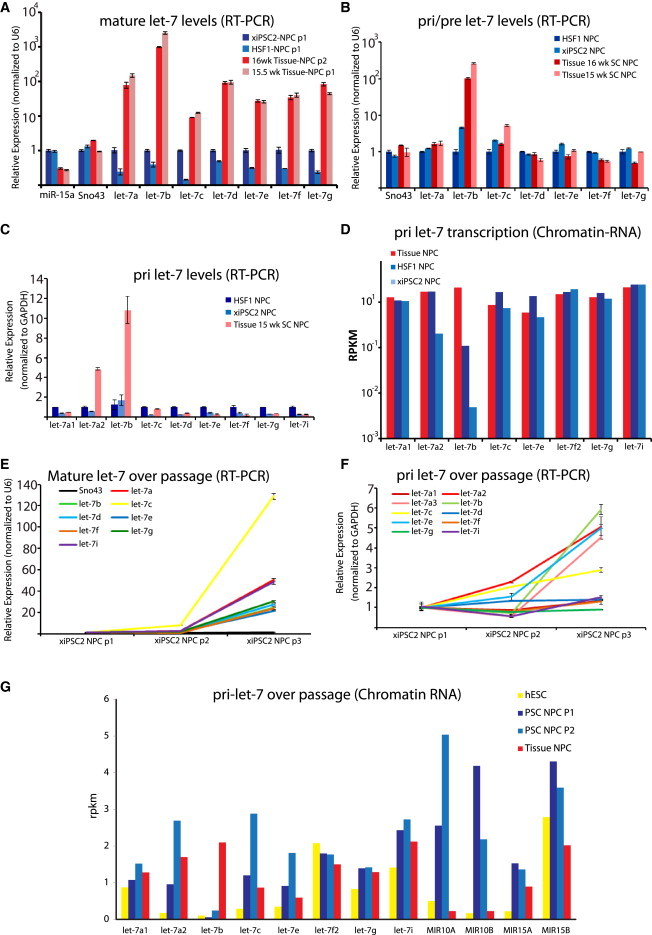
LIN28A/B controls the maturation step to generate mature let-7 (Hagan et al., 2009; Piskounova et al., 2008, 2011; Viswanathan et al., 2008), and LIN28A and B are also let-7 targets. A priori, we expected that levels of let-7 rose during development as a result of LIN28A/B downregulation over time. To understand the dynamics of let-7 processing during development, we assessed the levels of pri, pre, and mature let-7 miRNAs (Figure 2), we used RT-PCR to detect specifically mature let-7 family members in NPCs and found that, similar to what was found by miRNA sequencing (miRNA-seq) profiling (Figure 1D), all let-7 family members were dramatically higher in Tissue-NPCs compared to PSC-NPCs (Figure 2A; Patterson et al., 2012). Conversely, by using a miRNA RT-PCR approach that strictly measures primary let-7 transcripts, we determined that pri-let-7b and pri-let-7a2 transcript levels are significantly higher in Tissue-NPCs, whereas all the other tested members of the let-7 family were essentially unchanged (Figure 2C). Similarly, a separate approach that can distinguish pri- and pre-let-7 messages from mature let-7 miRNAs also showed that pre-let-7b was the only let-7 family member assayed that was differentially expressed in Tissue- versus PSC-NPCs (Figure 2B). Interestingly, let-7b and let-7a3 are known to be expressed from the same locus (Wang et al., 2012), whereas let-7a1 and let-7a2 are transcribed from different loci.
Figure 2.
Dynamics of let-7 Expression and Processing between PSC-NPCs and Tissue-NPCs
(A) RT-PCR for mature let-7 family members normalized against small nucleolar RNA U6 in two PSC-NPCs and two Tissue-NPCs. miR-15a and Sno43 were assayed as controls.
(B) Real-time RT-PCR for pre and pri let-7 family members normalized against small nucleolar RNA U6.
(C) Real-time RT-PCR for pri-let-7 miRNAs normalized against the relative levels of GAPDH in each cell type.
(D) miRNA-seq of chromatin-associated primary let-7 family miRNA transcripts between PSC-NPCs and Tissue-NPCs displayed as reads per kilobase measured (RPKM).
(E) Real-time RT-PCR for mature let-7 family miRNAs in PSC-NPCs over three passages normalized against U6. Sno43 was run as a negative control and was unchanged over passage.
(F) Real-time RT-PCR for primary let-7 family miRNA transcripts in PSC-NPCs over three passages normalized against U6.
(G) miRNA-seq of chromatin-associated primary let-7 family miRNA transcripts in PSC-NPCs over two passages (n = 2 for each group) displayed as reads per kilobase measured (RPKM). Data for miRs-10a, 10b, 15a, and 15b are shown to indicate that let-7 transcriptional regulation is unique in PSC versus Tissue-NPCs, even over passage, where let-7a1, a2, b, c, and e were induced. Error bars in all RT-PCR graphs represent SEM over three to four technical replicates, and results shown are representative of at least three independent experiments.
Passaging of PSC-NPCs results in a decrease of both LIN28A and LIN28B (Figure 1B; Patterson et al., 2012) and increase of mature let-7s (Figure 2E; Patterson et al., 2012), suggesting a functional link between LIN28 expression and levels of mature let-7 as expected (Hagan et al., 2009; Heo et al., 2008). Interestingly, when assaying for pri-miRNA transcripts, many of the pri-let-7s were also induced during passaging as detected by both RT-PCR (Figure 2F) and Chromatin-RNA-seq (discussed more below; Figure 2G), which presumably occurred independently of any direct regulation by LIN28 protein, because they are not thought to play any role in regulation of transcription of pri-let-7s. These results point toward the existence of undescribed regulatory mechanisms governing let-7 transcription independent of LIN28 during human gestation.
To further verify the apparent transcriptional regulation of the let-7 family, we employed an approach whereby chromatin-associated RNA is captured and sequenced (Bhatt et al., 2012). This approach generates data on relative amounts of nascent transcript, and therefore a bona fide measure of transcription as opposed to steady-state levels of RNA. Consistent with results from RT-PCR, transcription at the let-7a3/let-7b locus was significantly different between PSC-NPCs and Tissue-NPCs, demonstrating a LIN28A/B independent regulation of let-7 (Figures 2D and 2G). These data also suggest that the let-7 family of miRNAs is each subject to unique modes of transcriptional regulation that occurs prior to the actions of LIN28A/B on let-7 maturation. Also shown is the expression of several unrelated miRNAs (miR-10, 15A, 15B) to demonstrate that the let-7 effect is specific to this family and not indicative of imbalanced analysis (Figure 2G). This method allowed for a complete annotation of human let-7 transcriptional loci based on analysis of transcribed RNA, as opposed to in silico prediction, and showed a large degree of polycistronic expression of groups of let-7 family members (Table S2).
Manipulation of LIN28A/B Shows a Modest Effect on Developmental Maturity of NPCs
To determine what role LIN28 had on the developmental maturity of NPCs, we utilized two strategies. First, we used a small interfering RNA (siRNA) approach to knockdown both homologs. When siLIN28A and siLIN28B were introduced by transfection, mRNA for these genes was suppressed by 70%–75% as measured by RT-PCR (Figure S3A). This was also confirmed by western blot (Figure S3B). As a result of this knockdown, mature let-7 family miRNAs accumulated, as measured by RT-PCR, relative to transfection of a nontarget (siNT) control (Figure S3C). Also of note, siRNAs against both homologs were necessary to observe an induction of the let-7 family members, demonstrating their known semiredundant roles in let-7 maturation (Nam et al., 2011).
Microarray profiling identified many genes differentially expressed as a result of siLIN28A/B dual knockdown. However, these differentially expressed genes did not significantly overlap with the original list of probe sets differentially expressed between PSC-NPCs and their tissue-derived counterparts (Figure S3H; Patterson et al., 2012), nor were let-7 downstream targets, besides LIN28A and LIN28B, selectively knocked down (Figure S3F). In addition, the dual knockdown had no effect on the propensity to differentiate into neurons or glia (Figure S3D). These findings suggested that RNAi based knockdowns can lower LIN28 expression and induce let-7 maturation, but the resulting subtle let-7 induction exerted little effect functional effect. The second strategy to assess the functional relevance of LIN28B in NPC maturation utilized re-expression in Tissue-NPCs. When LIN28B was overexpressed in Tissue-NPCs by viral infection, mature let-7 levels decreased (Figure S3E), a portion of the differentially expressed genes between Tissue-NPCs and PSC-NPCs were corrected (Figure S3H), including some let-7 target genes (Figure S3F). Importantly, neurogenesis was promoted at the expense of gliogenesis upon induction of LIN28B in Tissue-NPCs (Figure S3G).
Direct Manipulation of let-7 Levels Alters Cell Fate in Neural Progenitors
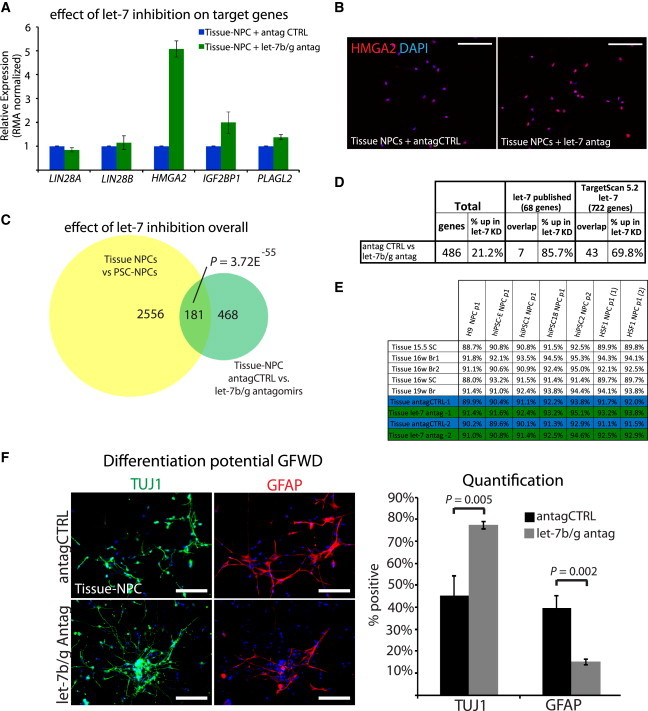
Previous studies either did not directly address the role of let-7 in LIN28-mediated developmental progression or ruled it out altogether (Balzer et al., 2010; Yuan et al., 2012). To assay whether let-7 miRNAs play a role in Tissue-NPCs, we introduced antagomirs against let-7b and let-7g. Microarray analysis on two independent experiments demonstrated that HMGA2 specifically was strongly induced by antagomirs compared to nonspecific controls at both the RNA and protein levels (Figures 3A and 3B). Looking at global gene expression, 649 probe sets were differentially expressed between let-7b/g antagomir-treated and nontarget controls (antag CTRL). Of these probe sets, 181 overlapped with previous comparisons between PSC- and Tissue-NPCs (p = 3.72 × 10−55, Figure 3C), and let-7 targets were specifically increased in these cells as expected (Figure 3D). Furthermore, Tissue-NPCs transfected with let-7b/g antagomirs were significantly more similar to PSC-NPCs than antag-CTRL-transfected cells on a global transcriptome level as measured by Pearson correlation (Figure 3E). Together, these data indicated that levels of let-7 in PSC- and Tissue-NPCs play a role in the transcriptional disparity between these sources of NPCs, and that manipulation of let-7 levels can shrink the dissimilarity between them. Finally, when antagomirs against let-7b/g were introduced into Tissue-NPCs, they became significantly more neurogenic and less gliogenic (Figure 3F; p < 0.005).
Figure 3.
A Role for LIN28/let-7 in the Developmental Progression of Human Tissue-NPCs
(A) Averaged robust multiarray average (RMA) normalized expression of selected let-7 target genes in Tissue-NPCs transfected with let-7b/g antagomiRs or nontargeted control antagomiRs by gene expression microarray. Error bars represent SEM across two biological replicates.
(B) Immunostaining for HMGA2 in Tissue-NPCs showed that let-7 antagomirs induce HMGA2 at the protein level.
(C) Venn diagram demonstrating the original differences identified by Patterson et al. (2012) (yellow) and the overlap with gene expression differences (>1.54-fold change) between let-7b/g antagomirs versus nonspecific control (antagCTRL) in Tissue-NPCs (green; n = 2).
(D) Overlap of published and predicted let-7 targets with genes changed after transfection of Tissue-NPCs with let-7b/g antagomiRs or nontargeted control antagomiRs measured by gene expression microarray.
(E) Pearson correlations of global gene expression similarity between Tissue-NPCs, Tissue-NPCs transfected with let-7b/g antagomiRs or nontargeted control antagomiRs, and PSC-NPCs.
(F) Immunofluorescence (LEFT) and quantification (RIGHT) for TUJ1 (neurons) and GFAP (glia) on Tissue-NPCs differentiated for 3 weeks in growth factor withdrawal following transfection with let-7b/g antagomirs or antag CTRL. p value was calculated with Student’s t test for at least 1,200 cells across eight to ten fields of view. Error bars represent SEM over fields of view, and results shown are representative of at least three independent experiments. Scale bar, 100 μm.
To determine whether gliogenesis in PSC-NPCs could be regulated by let-7, mature oligonucleotides (mimics) for let-7b and let-7g were transfected into PSC-NPCs. Several let-7 targets (LIN28A, LIN28B, HMGA2, IGF2BP1, and PLAGL2) were suppressed as measured by microarray profiling (Figure 4A). Global expression analysis of the of let-7 mimic-treated NPCs over two independent experiments identified 331 probe sets differentially expressed between let-7b/g-transfected PSC-NPCs over respective nonspecific mimic control (miCTRL). Furthermore, among the transcriptional differences observed between these two conditions a significant number overlapped with the original list of genes differentially expressed between PSC- and Tissue-NPCs (p = 3.76 × 10−46, Figure 4B). Focusing on direct let-7 downstream targets, expression profiling determined that not only were let-7 target genes overrepresented in the data, but they were specifically downregulated in let-7b/g-transfected NPCs over miCTRLs (Figure 4C). Furthermore, let-7b/g-transfected PSC-NPCs were significantly more similar to 16–19 week tissue-NPCs than miCTRL-transfected PSC-NPCs on a transcriptome level as measured by Pearson correlation (Figure 4D). Following let-7 induction, the PSC-NPCs were differentiated for 3 weeks in growth factor withdrawal media. miCTRL-transfected NPCs overwhelmingly produced TUJ1+ neurons, whereas only rare GFAP+ cells were found. When let-7b/g mimics were transfected prior to differentiation, the PSC-NPCs were slightly less neurogenic and significantly more gliogenic (Figure 4E; p = 3.58E−05).
Figure 4.
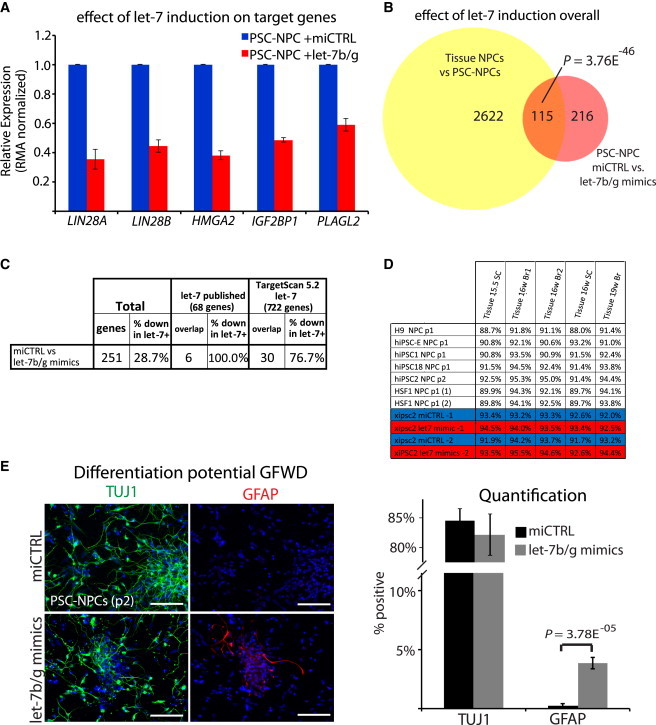
Direct Introduction of let-7 miRNAs Affects Developmental Progression of PSC-NPCs
(A) Averaged RMA normalized expression of selected let-7 target genes in PSC-NPCs transfected with let-7b/g mimics or nontargeted control miRNAs by gene expression microarray. Error bars represent SEM across two biological replicates.
(B) Venn diagram demonstrating the original differences identified by Patterson et al. (2012) (yellow), and the overlap with gene expression differences (>1.54-fold change) between let-7b/g mimics versus nonspecific control (miCTRL) in PSC-NPCs (red; n = 2).
(C) Overlap of published and predicted let-7 targets with genes changed after transfection of PSC-NPCs with let-7b/g mimics or nontargeted control miRNAs measured by gene expression microarray.
(D) Pearson correlations of global gene expression similarity between PSC-NPCs, PSC-NPCs transfected with let-7b/g mimics or nontargeted control miRNAs, and Tissue-NPCs.
(E) Immunofluorescence (left) and quantification (right) for TUJ1 (neurons) and GFAP (glia) on PSC-NPCs differentiated for 3 weeks in growth factor withdrawal following transfection with let-7b/g mimics or miCTRL. p value indicated reflects Student’s t test for at least 800 cells in multiple wells and across six fields of view. Error bars represent SEM over fields of view, and results shown are representative of at least three independent experiments. Scale bar, 100 μm.
Taken together, experiments that employed direct regulation of let-7 levels showed significant effects in both PSC- and Tissue-NPCs at both the molecular and functional levels, whereas attempts to manipulate LIN28A/B yielded more subtle effects. This observation is summarized in Figure S3, where manipulation of LIN28A/B only showed modest effects on both gene expression and differentiation in NPCs (Figures S3G and S3H). Instead, direct manipulation of let-7 clearly shows that this miRNA family can control the fate of NPCs (Figures 3 and 4), in contrast to what was shown in a murine model (Balzer et al., 2010).
let-7 Acts through HMGA2 to Control Cell Fate
We hypothesized that the key let-7 target genes responsible for cell fate in our model should show differences in gene expression levels in experimental settings presented here where NPC fate was affected (Figures 1A, 4E, and 5F). Taking the genes changed in every experimental model we produced that correlated with changes in neurogenesis versus gliogenesis, we narrowed the list of candidate genes. We furthered narrowed the list by taking gene expression data from the Allen Brain Atlas to identify genes changed from the pregliogenic state to the gliogenic stage (8–9 weeks PCW versus 16–18 weeks postconception) (Figure 6A). Just six genes appeared to have expression patterns that correlate with the switch from neurogenesis to gliogenesis in all these settings (Figure 5A). Of these six genes, two were predicted let-7 target genes (HMGA2 and GABBR2), but only HMGA2 has been implicated in neurogenesis (Sanosaka et al., 2008). HMGA2 was shown to affect cell fate in the murine brain (Nishino et al., 2008; Sanosaka et al., 2008) as well as various tissues (Monzen et al., 2008; Yu et al., 2013) and is thought to be mostly expressed prenatally (Ayoubi et al., 1999; Gattas et al., 1999). We found that HMGA2 was among the genes whose expression was limited to fetal brain prior to 10 weeks of gestation, just prior to the surge of gliogenesis predicted by the Allen Database (Figure 1F).
Figure 5.
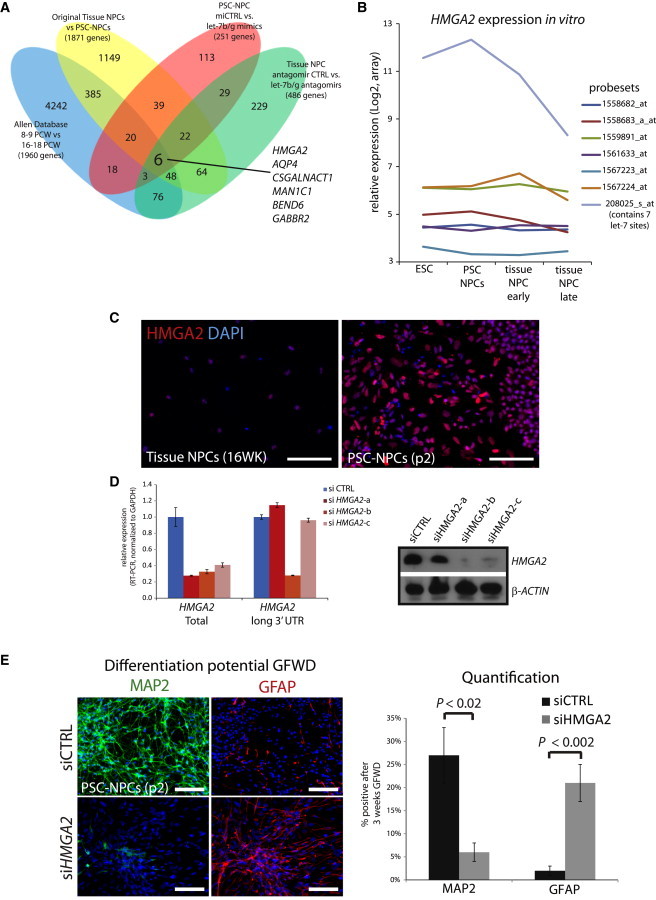
HMGA2 Is a Critical Target of let-7 in Developmental Progression
(A) Venn diagram demonstrating the overlap of genes changed in comparisons between neurogenic and gliogenic NPCs in the indicated contexts. Original list (yellow): n = 7 PSC-NPC lines and n = 5 Tissue-NPC lines; mimic experiment (red): n = 2 for both mimic- and control-treated lines; antagomir experiment (green): n = 2 for both antag- and control-treated lines.
(B) Normalized expression of all probe sets recognizing HMGA2 in cell types representing developing NPCs. Probe set 208025_s_at is the only one that recognizes the HMGA2 isoform containing multiple let-7 target sites.
(C) Immunostaining for HMGA2 in PSC and Tissue-derived NPCs.
(D) Real-time RT-PCR and western blot for HMGA2 expression in PSC-NPCs transfected with either control siRNA or each of three different siRNAs against HMGA2.
(E) Percentage of positive PSC-NPCs transfected with either control siRNA or siRNA against HMGA2 undergoing neuronal (MAP2) versus glial (GFAP) differentiation following 3 weeks growth factor withdrawal. Images and quantification are representative of three independent experiments. At least 1,900 cells were analyzed across at least eight fields of view. Error bars represent SEM over fields of view, and these data are representative of at least three independent experiments. Scale bar, 100 μm.
Figure 6.
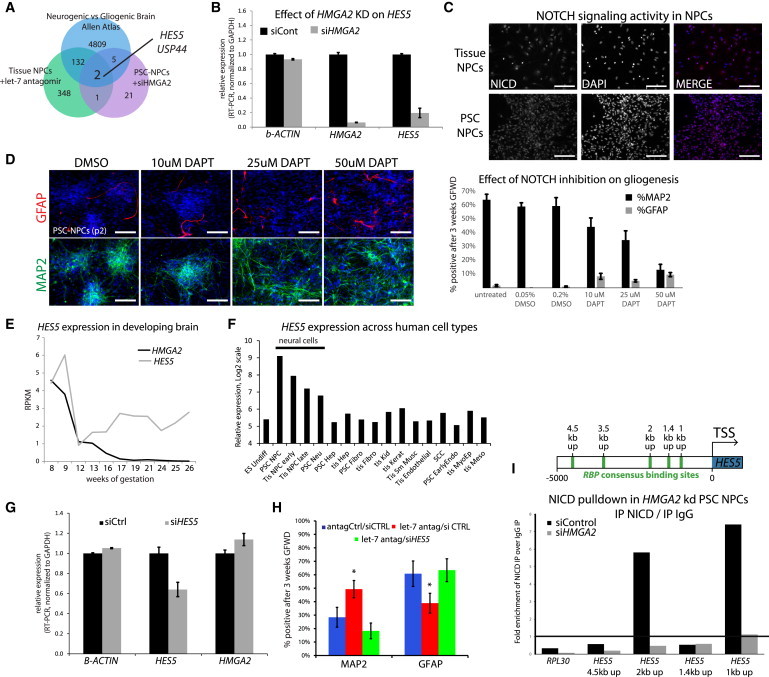
let-7/HMGA2 Regulates Notch Sensitivity through HES5
(A) Venn diagram demonstrating overlap of genes changed in comparisons between neurogenic and gliogenic NPCs in the indicated contexts.
(B) RT-PCR for relative expression after siRNA transfection to suppress HMGA2 expression.
(C) PSC-NPCs and Tissue-NPCs show similar levels of nuclear NICD accumulation suggesting that both have similar activation of the NOTCH pathway. Scale bar, 100 μm.
(D) Using a gamma-secretase inhibitor to block NOTCH activation decreases neuronal differentiation and induces gliogenesis as measured by MAP2 and GFAP staining and quantification. Scale bar, 100 μm.
(E) Transcription of HMGA2 and HES5 quantified by RNA-seq across the developing human brain.
(F) Gene expression data taken from an in-house database of a variety of human cell types suggests that HES5 is mostly expressed in the nervous system, whereas all the other family members are scattered throughout various types of cells from different organs (Log2 scale, RMA transformation).
(G) siRNA silencing of HES5 shows a 40% knockdown of message compared to scramble control.
(H) As shown previously in Figure 5E, inhibition of let-7 with antagomirs increased neurogenesis at the expense of gliogenesis. Silencing HES5 blocks this effect demonstrating a clear link between let-7 and HES5 activity in neurogenesis. ∗Student’s t test p < 0.05 for n > 450 cells in multiple wells. Paired t tests were performed for each manipulation, and only those with significant changes were indicated by ∗. Error bars represent SEM over technical replicates. Results shown are representative of at least three independent experiments.
(I) Chromatin immunoprecipitation with antibody against the active NOTCH1 product, NICD, shows strong enrichment for NICD binding in at two predicted RBPj binding sites in the HES5 upstream region (2 kp up and 1 kb up, see schematic). This enrichment was lost in cells with siHMGA2. Binding is represented as enrichment of pull-down over input and calculated as a function of enrichment of IgG over input. RPL30 was employed as a negative control locus for a constitutively expressed gene not sensitive to NOTCH1 signaling. Shown is the average of two experiments.
A closer look at HMGA2 transcription shows that at least eight transcripts are produced from this locus. One of these transcripts contains a large 3′ UTR that contains six let-7 binding sites, whereas the other transcripts are predicted to be much less sensitive to suppression by let-7. Reanalysis of expression analysis by array with various probe sets that each recognize distinct transcripts showed that the dominant transcript expressed contains multiple let-7 sites (identified by 208025_s_at), and that this is the only one affected by let-7 manipulation (data not shown; Figure 5B). Furthermore, this particular transcript is the only one differentially expressed between PSC- and Tissue- NPCs (Figure 5B). The difference in expression of HMGA2 between Tissue- and PSC- NPCs was also obvious at the protein level (Figure 5C). Taken together, these data suggested that suppression of HMGA2 levels by induction of let-7 correlated with the switch from neurogenesis to gliogenesis in the nervous system. To directly test the role of HMGA2 in the developmental progression of PSC-NPCs, we silenced this gene by RNAi prior to terminal differentiation. Three distinct siRNAs were used (a–c), and just one of them (siHMGA2b) was predicted to target the long isoform with let-7 sites. HMGA2 message was suppressed by this method at both the RNA (Figure 5D, left) and protein levels (Figure 5D, right). As with induction of let-7 mimics, suppression of HMGA2 significantly induced gliogenesis in PSC-NPCs (Figure 5E).
The Notch Pathway Is a Key Effector of let-7 in Gliogenesis
Profiling RNA expression after HMGA2 KD uncovered a modest number of genes changed. Intersection of these genes with those that were changed during human neurogenesis (Allen Database) or in Tissue-NPCs where let-7 was inhibited by antagomirs found just two genes consistently altered (HES5 and USP44)(Figure 6A). Significantly, HES5 is a key target gene and effector of the NOTCH pathway, and perhaps the best known regulator of gliogenesis in mice (Hojo et al., 2000; Kageyama and Ohtsuka, 1999; Kageyama et al., 2008; Ohtsuka et al., 1999, 2001). RT-PCR of PSC-NPCs with knockdown of HMGA2 clearly showed decreased HES5 levels, suggesting a functional correlation between the two (Figure 6B). This correlation did not appear to be due to a difference in the activity of NOTCH, as both PSC and Tissue-NPCs showed similar activity (Figure 6C). Although NOTCH signaling has been shown to either promote or inhibit gliogenesis depending on the context, in our human model abrogation of all NOTCH signaling with gamma-secretase inhibitor (DAPT) stimulated gliogenesis and suppressed neuronal differentiation (Figure 6D).
Using the Allen database, it is clear that HES5 is coexpressed with HMGA2 at 8–11 weeks of gestation just prior onset of gliogenesis, and expression of both genes drop significantly thereafter (Figure 6E). On the other hand, most other NOTCH effectors did not significantly change over the same time (Figure S4A). Expression patterns for HES5 across nine different human cell types representing derivatives of all three germ layers suggested that HES5 is restricted to the nervous system, whereas the other HES/HEY family members had a more widespread distribution throughout various organs (Figures 6F and S4B). This analysis also showed that PSC-NPCs expressed the highest levels of HES5, followed by lower expression in Tissue-NPCs, which were more gliogenic (Figure 1A; Patterson et al., 2012).
In order to functionally probe for a link between HES5 and let-7, this miRNA was experimentally abrogated in Tissue-NPCs with antagomirs while suppressing HES5 induction with siRNA. As shown above (Figure 3F), suppression of let-7s with antagomirs increased neurogenesis and decreased gliogenesis in Tissue-NPCs, but when HES5 induction was simultaneously blocked by siRNA (Figure 6G), this effect was lost (Figure 6H). This result was consistent with the described role for HES5 in murine neural development in the absence of LIF, whereas, in the presence of LIF, HES5 has a progliogenic role (Chambers et al., 2001; Chenn, 2009; Hirabayashi and Gotoh, 2005).
As an AT-hook binding protein, HMGA2 is well known to be broadly associated with chromatin. To begin to understand why HMGA2 expression strongly correlated with HES5 levels in NPCs, we probed whether HMGA2 can regulate access of NICD/RBPj transcriptional complexes to the HES5 promoter in response to NOTCH activation. Chromatin immunoprecipitation (ChIP) for NICD uncovered significant binding to two RBP binding sites within the HES5 regulator region (Figure 6I), as expected in cells with strong NOTCH activation (Figure 6C). On the other hand, when HMGA2 expression was abrogated by siRNA, NICD did not appear to significantly associate with these same sites, consistent with the notion that HMGA2 plays a role in regulating access of NICD to the HES5 promoter.
The let-7 Effect on Gliogenesis Extends to Oligodendrocyte Formation
The fact that induction of let-7 drove PSC-NPCs toward a more astrocytic fate suggested that let-7 could play an important role in developmental switches. In the same vein, established protocols to generate oligodendrocytes with PSC-NPCs normally take at least 17 weeks (Stacpoole et al., 2013; Wang et al., 2013), which is akin to the time it takes to produce them in utero (Figure 1F). Using one of these protocols, we observed oligodendrocyte progenitors (OPCs) and mature oligodendrocytes after just 6–7 weeks of culture, but only in PSC-NPCs cultures following let-7 induction at the NPC stage (Figure 7). These cells were judged to be bona fide oligodendrocytes on the basis of coexpression of O4 and Myelin Basic Protein (MBP) in confocal imaging. This result again suggested that let-7 can play a role in the developmental maturity of PSC-NPCs and provides an approach to speed the generation of astrocytes and oligodendrocytes from human PSCs.
Figure 7.
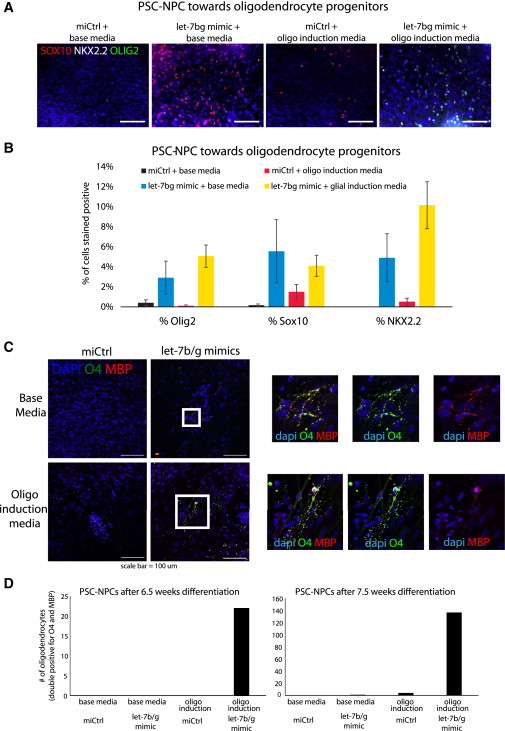
Generation of Oligodendrocytes Is Facilitated by Induction of let-7
(A) At 6 weeks of differentiation toward the oligodendrocyte lineage, OPC markers were identified by immunostaining and quantified in (B).
(B) Quantification of results presented in (A).
(C) Following at least 6 weeks of differentiation in either base or glial induction medium, mature oligodendrocytes were identified by immunostaining and quantified in (D). The bottom panels followed a glial induction protocol (Wang et al., 2013) and produced oligodendrocytes after just 6.5 weeks, again, only with let-7 induction at the NPC stage prior to directed differentiation. At 7.5 weeks, the number of oligodendrocytes produced was considerably larger. Scale bar, 100 mm.
(D) A quantification of oligodendrocytes in each condition at two time points in each well. Cells were judged as bona fide oligodendrocytes if positive for both O4 and MBP.
Discussion
The critical advance of this study is definitively showing that let-7 levels not only correlate with developmental progression in the human nervous system, but also serve to functionally drive the transition. Furthermore, whereas the let-7 target gene HMGA2 had previously been shown to promote neurogenesis in murine NPCs, we showed here that this is also the case for humans as well. Finally, we provide evidence that the mechanism by which let-7 miRNAs drives progression in the nervous system is by altering expression of a key NOTCH effector gene HES5. This NOTCH effector, and NOTCH signaling in general, has been shown to be a promoter of gliogenesis in murine models, whereas our data suggest a proneurogenic role. This discrepancy could be due to several obvious differences in context: murine versus human models, the presence or absence of LIF signaling, and/or in vitro versus in vivo settings. Nevertheless, the effect in this human setting was clear: inhibiting NOTCH activity generated NPCs that went from negligible numbers to a state where over 10% of differentiated cells were astrocytes. We did not try the converse experiment, inducing NOTCH activity, because it appeared as though all NPCs tested, whether neurogenic or gliogenic, showed considerable NOTCH activity. Interestingly, data presented here suggest that no other NOTCH effectors were affected by let-7 or HMGA2 manipulation, only HES5. These data suggest that the Notch pathway can be regulated by HES/HEY effectors by fine-tuning access to the binding sites of its effectors.
However, this study provides only a partial understanding of the relation between HMGA2 and regulation of Notch signaling. This is due in part to a poorly defined binding pattern of HMGA2 throughout the genome. It is known to be abundant when expressed during development and, as an AT-binding protein, can in theory bind nearly the entire genome. Thus, whereas we show here that the expression of HMGA2 appears to affect access of NICD to the HES5 promoter, it is unclear whether this is due to direct binding at this locus, or a general effect on chromatin compaction.
This study also highlights a role for transcriptional control of let-7 miRNAs in development. It was thought that primary let-7 transcription occurred at a constant level, and that levels of mature let-7 were strictly a matter of LIN28 activity. However, data provided here clearly demonstrate that let-7b is regulated at the transcriptional level even in cells that do not express LIN28A/B. Our data suggest that, developmentally, one let-7 family member (pri-let-7b) is induced at the transcriptional level prior to the loss of LIN28A/B protein and may be a key driver of LIN28A/B suppression as gliogenesis is initiated. It will be important to determine whether the methods developed here to facilitate developmental progression also apply to cells that have been transplanted into tissue.
Experimental Procedures
Cell Culture
Undifferentiated hPSC lines HSF1, H9, and xiPSC2 were maintained as previously described (Lowry et al., 2008) and in accordance with UCLA ESCRO. Neural progenitors were derived through formation of neural rosettes and maintained and further differentiated as described (Karumbayaram et al., 2009). NPCs were derived from fetal tissue (either brain or spinal cord) as described. All NPCs were judged to be pure by immunostaining as described (Patterson et al., 2012) (Figure S1).
Transfection
siRNAs (Thermo Dharmacon), let-7 mimics (Thermo Dharmacon), and let-7 antagomirs (Thermo Dharmacon “inhibitors”) were transfected with Lipofectamine RNAiMAX (Invitrogen) at a ratio of 5 μl lipofectamine:20 nM siRNA or 5 μl lipofectamine:40 nM mimic or antagomirs for each well of a 6-well plate. A reverse transfection method was used. Briefly, Lipofectamine and oligos were premixed in 1 ml of OptiMEM (Gibco) for 25 min in the precoated receiving plate. Cells were then passaged with TrypLE (Gibco), resuspended in 1 ml of NPC media without antibiotics, and plated on top of transfection media. Transfections were incubated overnight at 37°C after which time media was replaced with standard NPC culturing media with antibiotics for the indicated lengths of time. Lentivirus for CMV-LIN28B-SV40-mCherry (Genecopoeia) or UBC-mCherry (Kohn lab) was made by the UCLA vector core. NPCs were reverse infected overnight in NPC media without antibiotic.
Immunostaining
PSC-NPCs or Tissue-NPCs differentiated for 3 weeks were passaged with TrypLE (GIBCO) to glass coverslips 2–7 days before fixing. Coverslips were fixed and stained using standard protocols as described (Patterson et al., 2012). Primary antibodies included rabbit × TUJ1 (1:2,000; Covance), rabbit × GFAP (1:1,000; Dako), rabbit × S100 (1:1,000; Dako), chicken × GFAP (1:2,000, Abcam), mouse × MAP2 (1:500; Abcam), rabbit × Cleaved Notch1 (Val1744) (1:500; Cell Signaling Technologies), rabbit × LIN28A (1:300; Cell Signal), rabbit × LIN28B (1:300; Cell Signal), rabbit × HMGI-C (HMGA2) (1:200, Santa Cruz Biotechnology), mouse × O4 (1:300; R&D Systems), rat × MBP (1:50; Abcam), mouse × A2B5 (1:1,000; Abcam). All images were captured on a Zeiss microscope using Axiovision software (Zeiss) for image capture. GFAP or TUJ1 positive cells were counted using ImageJ software as a percentage of the total DAPI-labeled nuclei or as a percentage of the mCherry+ cells within the field. Average percentage positive cells and SEM was calculated over at least six fields of view. Western blot analysis was performed using standard procedures as described (Lowry et al., 2005). Primary western blot antibodies include rabbit × HMGI-C (1:500; Santa Cruz), rabbit × LIN28A (1:500; Cell Signal), rabbit × LIN28B (1:500; Cell Signal), and mouse × actin (1:1,000).
Additional materials and methods can be found in the Supplemental Information.
Author Contributions
M.P. and X.G.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; K.L., M.E., J.C., Y.X., J.L., S.A., S.M.D.: collection and assembly of data; S.S. and M.P.: data analysis and interpretation; W.E.L.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript, and financial support.
Acknowledgments
We would like to thank the core facilities at UCLA (Flow Cytometry Core, EEBRCM; Clinical Genomics, Pathology). We would also like to thank Wen Gu for help with cell-cycle analysis and Gerry Weinmaster for useful suggestions on the Notch pathway. M.P. and Y.X. were supported by a CIRM Training Grant (TG2-01169). X.G. was supported by the MSTP program, David Geffen School of Medicine at UCLA. W.E.L. was supported by the Maria Rowena Ross Chair in Cell Biology and Biochemistry, and this work was supported by grants from CIRM (RB3-05207), NIH (P01GM99134-NIGMS), and an Eli & Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA Innovation Award.
Published: October 2, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2014.08.015.
Supplemental Information
References
- Ayoubi T.A., Jansen E., Meulemans S.M., Van de Ven W.J. Regulation of HMGIC expression: an architectural transcription factor involved in growth control and development. Oncogene. 1999;18:5076–5087. doi: 10.1038/sj.onc.1202881. [DOI] [PubMed] [Google Scholar]
- Balzer E., Heine C., Jiang Q., Lee V.M., Moss E.G. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G., Black D.L., Smale S.T. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers C.B., Peng Y., Nguyen H., Gaiano N., Fishell G., Nye J.S. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development. 2001;128:689–702. doi: 10.1242/dev.128.5.689. [DOI] [PubMed] [Google Scholar]
- Chang C.J., Mitra K., Koya M., Velho M., Desprat R., Lenz J., Bouhassira E.E. Production of embryonic and fetal-like red blood cells from human induced pluripotent stem cells. PLoS ONE. 2011;6:e25761. doi: 10.1371/journal.pone.0025761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A. A top-NOTCH way to make astrocytes. Dev. Cell. 2009;16:158–159. doi: 10.1016/j.devcel.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Cimadamore F., Amador-Arjona A., Chen C., Huang C.T., Terskikh A.V. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc. Natl. Acad. Sci. USA. 2013;110:E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A., Ling Luo N., Beardsley D.J., Wingate-Pearse N., Walker D.W., Hohimer A.R., Back S.A. Quantitative analysis of perinatal rodent oligodendrocyte lineage progression and its correlation with human. Exp. Neurol. 2003;181:231–240. doi: 10.1016/s0014-4886(03)00032-3. [DOI] [PubMed] [Google Scholar]
- Dean J.M., Moravec M.D., Grafe M., Abend N., Ren J., Gong X., Volpe J.J., Jensen F.E., Hohimer A.R., Back S.A. Strain-specific differences in perinatal rodent oligodendrocyte lineage progression and its correlation with human. Dev. Neurosci. 2011;33:251–260. doi: 10.1159/000327242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattas G.J., Quade B.J., Nowak R.A., Morton C.C. HMGIC expression in human adult and fetal tissues and in uterine leiomyomata. Genes Chromosomes Cancer. 1999;25:316–322. [PubMed] [Google Scholar]
- Graf R., Munschauer M., Mastrobuoni G., Mayr F., Heinemann U., Kempa S., Rajewsky N., Landthaler M. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 2013;10:1146–1159. doi: 10.4161/rna.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan J.P., Piskounova E., Gregory R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y., Gotoh Y. Stage-dependent fate determination of neural precursor cells in mouse forebrain. Neurosci. Res. 2005;51:331–336. doi: 10.1016/j.neures.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Hojo M., Ohtsuka T., Hashimoto N., Gradwohl G., Guillemot F., Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Jakovcevski I., Filipovic R., Mo Z., Rakic S., Zecevic N. Oligodendrocyte development and the onset of myelination in the human fetal brain. Front Neuroanat. 2009;3:5. doi: 10.3389/neuro.05.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res. 1999;9:179–188. doi: 10.1038/sj.cr.7290016. [DOI] [PubMed] [Google Scholar]
- Kageyama R., Ohtsuka T., Kobayashi T. Roles of Hes genes in neural development. Dev. Growth Differ. 2008;50(Suppl 1):S97–S103. doi: 10.1111/j.1440-169X.2008.00993.x. [DOI] [PubMed] [Google Scholar]
- Karumbayaram S., Novitch B.G., Patterson M., Umbach J.A., Richter L., Lindgren A., Conway A.E., Clark A.T., Goldman S.A., Plath K. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N., Nam J.W. Genomics of microRNA. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Cho S., Sun Kim M., Choi K., Cho J.Y., Gwak H.S., Kim Y.J., Yoo H., Lee S.H., Park J.B., Kim J.H. The ubiquitin ligase human TRIM71 regulates let-7 microRNA biogenesis via modulation of Lin28B protein. Biochim. Biophys. Acta. 2014;1839:374–386. doi: 10.1016/j.bbagrm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- Lowry W.E., Blanpain C., Nowak J.A., Guasch G., Lewis L., Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W.E., Richter L., Yachechko R., Pyle A.D., Tchieu J., Sridharan R., Clark A.T., Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Simonini M.V., Palejev D., Tomasini L., Coppola G., Szekely A.M., Horvath T.L., Vaccarino F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzen K., Ito Y., Naito A.T., Kasai H., Hiroi Y., Hayashi D., Shiojima I., Yamazaki T., Miyazono K., Asashima M. A crucial role of a high mobility group protein HMGA2 in cardiogenesis. Nat. Cell Biol. 2008;10:567–574. doi: 10.1038/ncb1719. [DOI] [PubMed] [Google Scholar]
- Nam Y., Chen C., Gregory R.I., Chou J.J., Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim I., Chada K., Morrison S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino J., Kim S., Zhu Y., Zhu H., Morrison S.J. A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. eLife. 2013;2:e00924. doi: 10.7554/eLife.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Ishibashi M., Gradwohl G., Nakanishi S., Guillemot F., Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Sakamoto M., Guillemot F., Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- Patterson M., Chan D.N., Ha I., Case D., Cui Y., Van Handel B., Mikkola H.K., Lowry W.E. Defining the nature of human pluripotent stem cell progeny. Cell Res. 2012;22:178–193. doi: 10.1038/cr.2011.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Viswanathan S.R., Janas M., LaPierre R.J., Daley G.Q., Sliz P., Gregory R.I. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Piskounova E., Polytarchou C., Thornton J.E., LaPierre R.J., Pothoulakis C., Hagan J.P., Iliopoulos D., Gregory R.I. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanosaka T., Namihira M., Asano H., Kohyama J., Aisaki K., Igarashi K., Kanno J., Nakashima K. Identification of genes that restrict astrocyte differentiation of midgestational neural precursor cells. Neuroscience. 2008;155:780–788. doi: 10.1016/j.neuroscience.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Stacpoole S.R., Spitzer S., Bilican B., Compston A., Karadottir R., Chandran S., Franklin R.J. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Reports. 2013;1:437–450. doi: 10.1016/j.stemcr.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q. Lin28: a microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.J., Legesse-Miller A., Johnson E.L., Coller H.A. Regulation of the let-7a-3 promoter by NF-κB. PLoS ONE. 2012;7:e31240. doi: 10.1371/journal.pone.0031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Bates J., Li X., Schanz S., Chandler-Militello D., Levine C., Maherali N., Studer L., Hochedlinger K., Windrem M., Goldman S.A. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Viswanathan S.R., Yabuuchi A., Cunniff K., Takeuchi A., Park I.H., Sero J.E., Zhu H., Perez-Atayde A., Frazier A.L. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu K.R., Park S.B., Jung J.W., Seo M.S., Hong I.S., Kim H.S., Seo Y., Kang T.W., Lee J.Y., Kurtz A., Kang K.S. HMGA2 regulates the in vitro aging and proliferation of human umbilical cord blood-derived stromal cells through the mTOR/p70S6K signaling pathway. Stem Cell Res. 2013;10:156–165. doi: 10.1016/j.scr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Yuan J. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambidis E.T., Peault B., Park T.S., Bunz F., Civin C.I. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–870. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


