ABSTRACT
Bacterial ribosomes frequently translate to the 3′ end of an mRNA without terminating at a stop codon. Almost all bacteria use the transfer-messenger RNA (tmRNA)-based trans-translation pathway to release these “nonstop” ribosomes and maintain protein synthesis capacity. trans-translation is essential in some species, but in others, such as Caulobacter crescentus, trans-translation can be inactivated. To determine why trans-translation is dispensable in C. crescentus, a Tn-seq screen was used to identify genes that specifically alter growth in cells lacking ssrA, the gene encoding tmRNA. One of these genes, CC1214, was essential in ΔssrA cells. Purified CC1214 protein could release nonstop ribosomes in vitro. CC1214 is a homolog of the Escherichia coli ArfB protein, and using the CC1214 sequence, ArfB homologs were identified in the majority of bacterial phyla. Most species in which ssrA has been deleted contain an ArfB homolog, suggesting that release of nonstop ribosomes may be essential in most or all bacteria.
IMPORTANCE
Genes that are conserved across large phylogenetic distances are expected to confer a selective advantage. The genes required for trans-translation, ssrA and smpB, have been found in >99% of sequenced bacterial genomes, suggesting that they are broadly important. However, these genes can be deleted in some species without loss of viability. The identification and characterization of C. crescentus ArfB reveals why trans-translation is not essential in C. crescentus and suggests that many other bacteria are likely to use ArfB to survive when trans-translation is compromised.
INTRODUCTION
The efficient translation cycle of initiation, elongation, termination, and recycling allows rapid protein synthesis and cellular growth. Most ribosomes pass through this cycle many times every cell generation, so breaks in the cycle can lead to rapid depletion of protein synthesis capacity. A major source of translation stress in bacteria is truncated mRNA. Because bacterial ribosomes do not require any information from the 3′ end of an mRNA for initiation, they can start translating mRNAs that are incompletely transcribed or damaged (1). If the mRNA does not have a stop codon or if the ribosome reads through the stop codon and reaches the 3′ end of the mRNA without terminating, the translation cycle will be broken. A stop codon in the decoding center is required for release factors RF1 or RF2 to hydrolyze the peptidyl-tRNA and allow the ribosome to be recycled (2). When the ribosome is at the 3′ end of the mRNA but there is no stop codon in the decoding center, trans-translation is used to resolve the “nonstop” translation complex and allow the translation cycle to resume (3). During trans-translation, transfer-messenger RNA (tmRNA), encoded by the ssrA gene, and SmpB, a small protein, recognize a nonstop translation complex and divert the ribosome onto a reading frame within tmRNA. A stop codon at the end of the tmRNA reading frame allows translation termination and ribosome recycling. trans-translation also targets the nascent polypeptide and problematic mRNA for rapid degradation, clearing the cell of all components of the nonstop translation complex (4, 5).
Genes encoding tmRNA and SmpB are found in >99% of sequenced bacterial genomes (6), suggesting that trans-translation confers a selective advantage under most environmental conditions. In culture, trans-translation is essential in some species. However, ssrA can be deleted in some bacteria without causing a severe phenotype (7).
The apparent contradiction between the universal conservation of trans-translation and the mild phenotype caused by removing this system in Escherichia coli was explained by the discovery of ArfA (alternative rescue factor A). The arfA gene was identified in a screen for mutations that were synthetically lethal with an ssrA deletion (8). ArfA is a small protein that allows RF2 to hydrolyze peptidyl-tRNA on nonstop translation complexes (9, 10). ArfA is a true backup system for trans-translation, because ArfA itself is made from a truncated mRNA (11, 12). When trans-translation is functional, ArfA is tagged by trans-translation and degraded. If trans-translation is disabled or overwhelmed, untagged ArfA is produced to release the accumulating nonstop translation complexes. ArfA activity allows E. coli to live without trans-translation, but in ΔarfA cells, ssrA is essential (8). Likewise, ssrA is essential in Shigella flexneri, which does not have arfA, but ssrA can be deleted in S. flexneri cells expressing the E. coli arfA gene (13). arfA genes have been identified in many Gammaproteobacteria and some Betaproteobacteria, and this may explain why ssrA can be deleted in species such as Salmonella enterica and Yersinia pestis (7, 8, 11). Other bacteria, such as Caulobacter crescentus, do not have an arfA homolog but still do not require trans-translation for viability.
This paper describes the results of a screen using transposon mutagenesis followed by sequencing (Tn-Seq) to identify genetic interactions with ssrA in C. crescentus. One gene, CC1214, was shown to be essential in ΔssrA cells but not in wild-type cells. The CC1214 protein could hydrolyze peptidyl-tRNA in nonstop translation complexes in vitro and had homology to E. coli ArfB. The phylogenetic distribution of CC1214 homologs raises the possibility that all or most bacteria require at least one system to resolve nonstop translation complexes.
RESULTS
Identification of genetic interactions with ssrA.
Genetic interactions with ssrA were probed using Tn-seq. Wild-type and ΔssrA cultures were individually mutagenized with the himar1 transposon and plated on selective medium to allow the growth of cells with an integrated transposon. Colonies were pooled and sequenced to identify the location of transposon insertions. In this experiment, the number of times each sequence is recovered depends on the frequency of transposon insertions at that site and the amount of colonies grown from the mutant cells. Because the frequency of transposition at each site is a function of the DNA sequence and chromosomal context, which are the same in both strains, the ratio of the number of times a sequence is recovered in ΔssrA and wild-type cultures provides a measure of the relative fitness of the mutant in each strain background. Mutants with a low ratio are relatively more fit in the wild-type background, whereas mutants with a high ratio are relatively more fit in ΔssrA cells. Sequence data were pooled to generate a ratio for each gene (Table 1; see also Table S1 in the supplemental material).
TABLE 1 .
Selected genes identified by Tn-seq
| Gene | No. of insertions in indicated backgrounda |
Log10 ratiob | |
|---|---|---|---|
| ΔssrA | Wild type | ||
| CC1214 | 1 | 564 | −2.45 |
| tRNAArg(CCU) | 0 | 214 | −2.33 |
| rluD | 39 | 2,930 | −1.87 |
| lepA | 90 | 5,478 | −1.78 |
| Efp | 3 | 139 | −1.54 |
| pth | 0 | 24 | −1.39 |
| CC1215 | 85,851 | 1,011 | 1.93 |
| smpB | 1,644 | 38 | 1.62 |
Number of insertions normalized with respect to total number of insertions in ΔssrA background.
Ratio as calculated after adding 1 to the number of insertions.
Genes with a low ratio include rluD, lepA, efp, pth, and tRNAArg(CCU), which have genetic or functional interactions with trans-translation in E. coli. Deletion of rluD increases the frequency of stop codon readthrough and increases the tagging of E. coli proteins (14). LepA (EF4) is a translation elongation factor, and overexpression of lepA inhibits trans-translation (15). EF-P promotes translation through polyproline sequences and other sequences that stall translation elongation (16). Deletion of rluD, lepA, or efp is likely to generate more substrates for trans-translation, so it is not unexpected that mutations in these genes would have a synthetic phenotype in the ΔssrA background. Multiple genetic interactions between pth and ssrA have been reported in E. coli, including exacerbation of pth mutant phenotypes in ΔssrA cells, so it is not surprising that mutation of pth in C. crescentus ΔssrA cells results in relatively lower fitness (17). tRNAArg(CCU) is a rare tRNA, and the amount of trans-translation of genes with an AGG codon in E. coli is negatively correlated with the abundance of tRNAArg(CCU) (18).
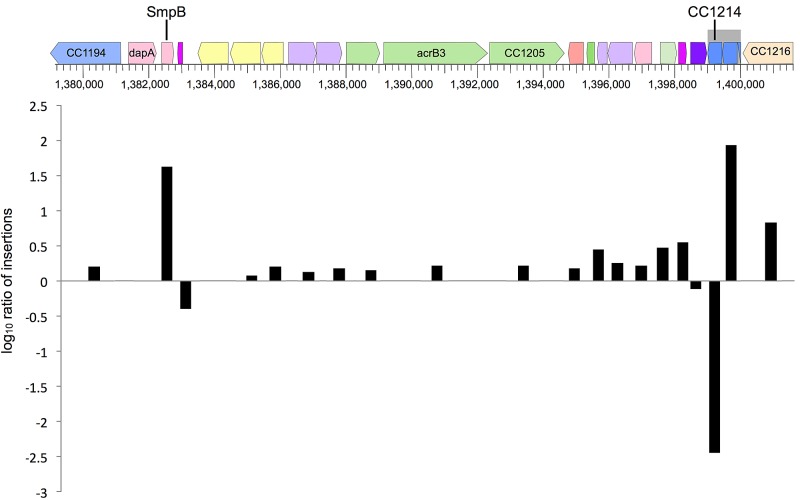
CC1214, annotated as a peptidyl-tRNA hydrolase family protein, had the lowest ratio of any gene (Fig. 1, Table 1). Sequence homology searches suggested that CC1214 encoded a homolog of E. coli ArfB, a gene identified as a multicopy suppressor of lethality in E. coli cells lacking both ssrA and arfA (19). E. coli arfB cannot support viability from its chromosomal context (8). Nevertheless, the sequence similarity to ArfB made CC1214 a candidate for releasing nonstop complexes in the absence of trans-translation in C. crescentus.
FIG 1 .
Tn-seq identification of genes interacting with ssrA. After Tn-seq, the log10 ratio of the normalized number of transposon insertions in ΔssrA cells to the number in wild-type cells was calculated and plotted for a portion of the C. crescentus chromosome. The organization of genes and the direction of transcription are represented with colored arrows.
Genes with a high ratio include smpB and CC1215 (Table 1). Deletion of smpB in wild-type cells causes slow growth, but deletion of smpB in ΔssrA cells does not cause any additional growth defect, resulting in a relatively greater fitness in the ΔssrA background. Because a large number of insertions in CC1215 were recovered in both strains, the high ratio for CC1215 is likely due to increased fitness in ΔssrA cells. Because CC1214 and CC1215 are in an operon, the interactions of both these genes with ssrA were investigated.
CC1214 deletion is synthetically lethal with ssrA deletion.
The Tn-seq data suggest that cells lacking both trans-translation and CC1214 are at a severe competitive disadvantage. To characterize the phenotype of cells lacking CC1214, the gene was replaced with an Ω cassette encoding spectinomycin resistance, using 2-step recombination (20). The CC1214::Ω strain grew at the same rate as the wild type (Table 2) and showed no morphological defects. In contrast, CC1214 could not be deleted in ΔssrA cells using the same procedure, suggesting that CC1214 is essential in the absence of trans-translation.
TABLE 2 .
Doubling times of mutant strains
| Strain description | Doubling time ± SD (min) |
|---|---|
| Wild type | 103 ± 1 |
| ΔssrA | 127 ± 1 |
| CC1214::Ω | 103 ± 1 |
| CC1215::cat (sense) | 101 ± 1 |
| CC1215::cat (antisense) | 99 ± 5 |
| ΔssrA CC1215::cat (sense) | 122 ± 1 |
| ΔssrA CC1215::cat (antisense) | 114 ± 4 |
A cotransduction experiment was used to confirm that loss of CC1214 is lethal in the ΔssrA background. A gene conferring kanamycin resistance was inserted in the chromosome of C. crescentus CC1214::Ω ~22 kb from the CC1214 locus to make strain KCK428. Based on their separation on the chromosome, the kanamycin and spectinomycin resistance markers would be expected to cotransduce using phage φCR30 at a frequency of 45%. Consistent with this prediction, when a φCR30 lysate was prepared from KCK428 cells and used to infect wild-type C. crescentus, 52% of kanamycin-resistant transductants were also spectinomycin resistant (Table 3). In contrast, when the φCR30 lysate was used to infect ΔssrA cells, none of the 150 kanamycin-resistant colonies were spectinomycin resistant. When ΔssrA cells containing a plasmid-borne copy of ssrA were used as the recipients, the cotransduction frequency was restored to 51%. Likewise, when ΔssrA cells containing a plasmid-borne copy of CC1214 were used as the recipients, the cotransduction frequency was 55%. These results indicate that C. crescentus cells lacking both ssrA and CC1214 cannot grow under typical culture conditions.
TABLE 3 .
Cotransduction of CC1214 alleles with Kanra
| Description of recipient strain | % of transductants with CC1214::Ω | % of transductants with CC1214 |
|---|---|---|
| Wild type | 52 | 48 |
| ΔssrA | 0 | 100 |
| ΔssrA pssrA | 51 | 49 |
| ΔssrA pCC1214 | 55 | 45 |
| ΔssrA pCC1214G30A | 0 | 100 |
Kanr, kanamycin resistance.
CC1215 interactions with ssrA are due to polar effects on CC1214.
The Tn-seq data indicated that himar1 insertions in CC1215 confer a competitive advantage in ΔssrA cells. CC1215 could be replaced with the cat gene, encoding chloramphenicol resistance, in both wild-type and ΔssrA cells. Insertions of cat in the sense and antisense orientation relative to CC1215 were recovered in both strains. Deletion of CC1215 did not alter the growth rate of strains in liquid culture or on plates (Table 2), indicating that the increased recovery of himar1 insertions in ΔssrA cells in the Tn-seq experiment was not due to loss of the CC1215 gene product. One likely explanation for the observed interaction between CC1215 and ssrA is that himar1 insertion resulted in increased expression of CC1214, which is immediately downstream from CC1214, making the ΔssrA cells more fit. Consistent with that hypothesis, CC1215 could not be replaced with an Ω cassette in ΔssrA cells, but in wild-type cells, the CC1215::Ω mutation was viable. Because the Ω cassette contains transcriptional terminators flanking the antibiotic resistance gene, it produces strong polar effects (21).
CC1214 hydrolyzes peptidyl-tRNA on nonstop translation complexes.
CC1214 was annotated as a peptidyl-tRNA hydrolase, but it has greater sequence similarity to E. coli arfB. Because purified E. coli ArfB can release peptidyl-tRNA from nonstop complexes in vitro, the CC1214 protein was purified and tested for nonstop ribosome release activity. In vitro transcription/translation assays were programmed with a dihydrofolate reductase (DHFR) gene lacking a stop codon (DHFR-ns) and resolved on bis-Tris-polyacrylamide gels that preserve the peptidyl-tRNA bond. Ribosomes translating DHFR-ns reach the 3′ end of the mRNA and cannot be released by RF1 or RF2, so the percentage of DHFR protein in the peptidyl-tRNA band is a measure of the ribosome release activity. For untreated reaction mixtures, 52% of the DHFR protein was in the peptidyl-tRNA band (Fig. 2). When puromycin, a drug that hydrolyzes peptidyl-tRNA in the ribosome, was added to the reaction mixtures prior to electrophoresis, 16% of the DHFR protein remained as peptidyl-tRNA. When purified CC1214 was added to the reaction mixtures, 11% of the DHFR was in the peptidyl-tRNA, indicating that CC1214 can hydrolyze ~80% of peptidyl-tRNA from nonstop complexes under these conditions. This amount of release activity was similar to the release activity observed for RF1 and RF2 when the DHFR template contained a TAA stop codon (not shown).
FIG 2 .
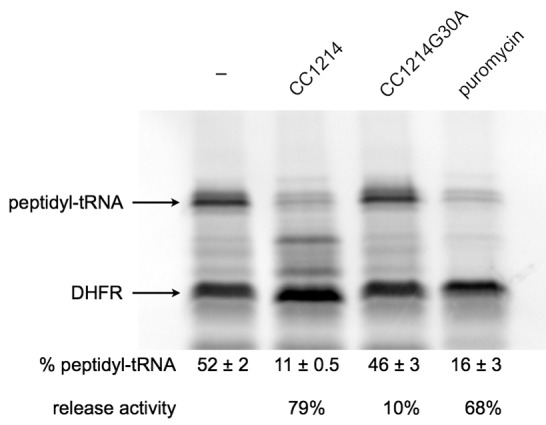
CC1214 hydrolyzes peptidyl-tRNA on nonstop translation complexes. In vitro transcription/translation reactions were programmed with a DHFR gene lacking a stop codon, treated with CC1214, CC1214G30A, or puromycin, and resolved on a bis-Tris gel. The positions of free DHFR protein and DHFR peptidyl-tRNA are indicated. The average (± standard deviation) percentages of DHFR in the peptidyl-tRNA band from 3 experiments and the percent release activities with respect to the release activity in the untreated control are shown.
Bacterial factors that hydrolyze peptidyl-tRNA on the ribosome, including RF1, RF2, and ArfB, have a GGQ motif that is important for activity (8, 22), but this motif is not found in Pth enzymes that hydrolyze free peptidyl-tRNA. A GGQ motif is conserved in CC1214. A variant of CC1214 in which the GGQ motif was replaced with GAQ (CC1214G30A) was purified and tested for nonstop ribosome release activity. In these reactions, 48% of the DHFR remained in the peptidyl-tRNA band, indicating that the GGQ motif is important for CC1214 activity. Consistent with that hypothesis, a plasmid-borne copy of CC1214G30A would not allow transduction of CC1214::Ω into ΔssrA cells (Table 3).
Multicopy expression of CC1214 suppresses the ΔssrA growth defect.
C. crescentus strains lacking ssrA or smpB grow slowly (23). To determine whether overexpression of CC1214 could suppress this phenotype, CC1214 was cloned under the control of a xylose-inducible promoter on a multicopy plasmid. During exponential growth phase in liquid culture, wild-type cells overexpressing CC1214 had a doubling time of 94 ± 3 min (mean ± standard deviation) (Fig. 3). Wild-type cells with an empty vector had a doubling time of 98 ± 5 min, indicating that overexpression of CC1214 does not have a large effect on growth rate when trans-translation is functional. Consistent with previously published data (23), the doubling time in ΔssrA cells with an empty vector was 125 ± 3 min. However, when CC1214 was overexpressed, the doubling time decreased to 108 ± 3 min. This partial suppression of the slow growth phenotype in ΔssrA cells suggests that release of nonstop complexes by CC1214 can compensate for the absence of trans-translation.
FIG 3 .
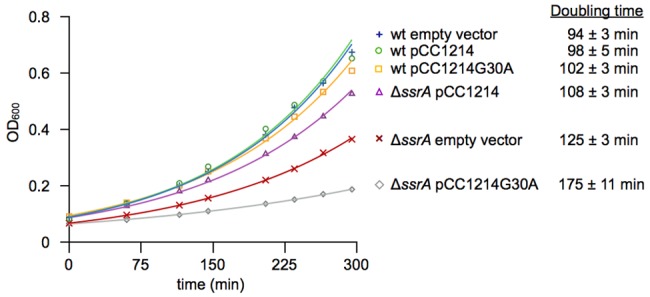
Overexpression of CC1214 suppresses the slow growth phenotype in ΔssrA cells. The growth of C. crescentus strains was monitored by measuring the optical density at 600 nm and plotted versus time. The doubling time (± standard deviation) of each strain is indicated. wt, wild type.
Suppression by CC1214 required nonstop ribosome release, because overproduction of the inactive CC1214G30A variant resulted in a decrease in growth rate instead of an increase. In wild-type cells overexpressing CC1214G30A, the doubling time was 102 ± 3 min, and in ΔssrA cells overexpressing CC1214G30A, the doubling time was 175 ± 11 min. These results suggest that the inactive mutant can compete with wild-type CC1214 for binding to nonstop ribosomes and inhibit nonstop ribosome release in the absence of trans-translation. Consistent with this interpretation, mutants of E. coli ArfB, RF1, and RF2 with mutations that alter the GGQ motif can bind nonstop ribosomes (19).
Genes encoding ArfB are widely distributed in bacteria.
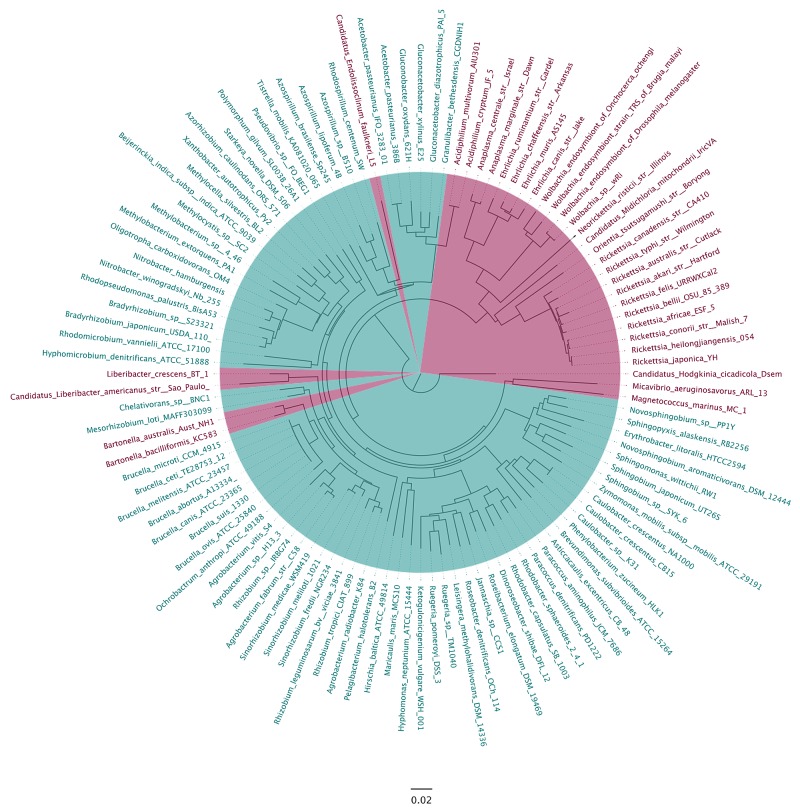
Because CC1214 has significant sequence similarity to the E. coli ArfB gene, including the catalytic GGQ motif and positively charged residues in the C-terminal tail that are required for release of nonstop ribosomes (Fig. 4), and because CC1214 can release nonstop ribosomes in vitro and can support the viability of strains in which ssrA is deleted, we propose that CC1214 and its homologs should be renamed ArfB. To determine how widely distributed genes encoding ArfB are within the Alphaproteobacteria, 236 completed alphaproteobacterial genomes were searched using BLAST (24) with the C. crescentus ArfB sequence. No homologs were identified in the rickettsial branch (including Rickettsia, Ehrlichia, Wolbachia, Candidatus, and Anaplasma species), but of the remaining 171 species, 80% encoded ArfB (Fig. 5). To determine whether ArfB is abundant throughout other lineages of bacteria, BLAST searches were performed on the NCBI set of representative genomes for the 21 different bacterial phyla. Homologs were found in 11 phyla (Table 4). More than 60% of Planctomycetes, Chlorobi, and Betaproteobacteria species encoded homologs, and >40% of Cyanobacteria, Bacteroidetes, Deltaproteobacteria, and Actinobacteria species had recognizable homologs.
FIG 4 .

ArfB homologs are found across the bacterial kingdom. ArfB sequences were aligned with Clustal Omega. Asterisks indicate conserved residues, colons indicate residues with strongly similar properties, and dots indicate residues with weakly similar properties. Residues which have been shown experimentally to be important for hydrolysis activity in E. coli are highlighted in green. Species abbreviations are as follows: Ccr, Caulobacter crescentus; Eco, Escherichia coli; Bja, Bradyrhizobium japonicum; Mxa, Myxcoccus xanthus; Sco, Streptomyces coelicolor; S6803, Synechocystis sp. strain PCC6803; SL21, Spirochaeta sp. strain L21-RPul-D2; SNBC, Sulfurovum sp. strain NBC37-1; Bth, Burkholderia thailandensis; and Dto, Desulfobacula toluolica strain Tol2.
FIG 5 .
ArfB homolog distribution in Alphaproteobacteria. A tree of alphaproteobacterial species generated using 16S rRNA gene sequences is shown, with species encoding ArfB homologs highlighted in turquoise and species that do not encode an ArfB homolog highlighted in magenta.
TABLE 4 .
Phylogenetic distribution of ArfB homologs
| Phylum | No. of representative genomes | No. with arfB | % with arfB |
|---|---|---|---|
| Actinobacteria | 174 | 83 | 48 |
| Aquificae | 9 | 0 | 0 |
| Cyanobacteria | 55 | 25 | 45 |
| Chlorobi/Bacteroidetes | 96 | 48 | 50 |
| Firmicutes | 268 | 0 | 0 |
| Alphaproteobacteria | 178 | 102 | 57 |
| Betaproteobacteria | 102 | 75 | 74 |
| Epsilonproteobacteria | 29 | 3 | 10 |
| Gammaproteobacteria | 271 | 101 | 37 |
| Deltaproteobacteria | 54 | 32 | 59 |
| Fusobacteria | 7 | 0 | 0 |
| Spirochaetes | 41 | 7 | 17 |
| Chlamydiae/Verrucomicrobia | 19 | 0 | 0 |
| Planctomycetes | 7 | 6 | 86 |
| Tenericutes | 48 | 0 | 0 |
| Synergistetes | 7 | 0 | 0 |
| Thermotogae | 17 | 0 | 0 |
| Deinococcus | 7 | 0 | 0 |
| Chloroflexi | 16 | 1 | 6 |
| Armatimonadetes | 1 | 0 | 0 |
| Caldiserica | 1 | 0 | 0 |
| Chrysiogenetes | 1 | 0 | 0 |
| Deferribacteres | 4 | 0 | 0 |
| Dictyoglomi | 2 | 0 | 0 |
| Elusimicrobia | 2 | 0 | 0 |
| Fibrobacteres/Acidobacteria | 9 | 0 | 0 |
| Gemmatimonadetes | 1 | 1 | 100 |
| Nitrospirae | 3 | 1 | 33 |
| Thermodesulfobacteria | 2 | 0 | 0 |
| Total | 1,431 | 383 | 34 |
DISCUSSION
The results described here show that C. crescentus can survive without trans-translation only because it has ArfB to release nonstop translation complexes. ArfB can hydrolyze peptidyl-tRNA on nonstop translation complexes in vitro, and C. crescentus cells require either trans-translation or ArfB activity for viability. Although E. coli ArfB was identified in a multicopy suppressor screen as a gene that would allow both ssrA and arfA to be deleted, E. coli ArfB will not support viability as the only nonstop complex release activity from its chromosomal locus (19). In contrast, the C. crescentus ArfB in its wild-type locus can act as a backup system for trans-translation during growth in culture. E. coli and C. crescentus ArfB have similar release activities in vitro, so it is likely that E. coli ArfB does not act as a backup system for trans-translation solely because it is expressed at low levels.
Homology searches using the C. crescentus ArfB sequence revealed that several other species that do not require trans-translation encode ArfB, including Bradyrhizobium japonicum and Streptomyces coelicolor (25, 26). With the addition of ArfB homologs similar to the C. crescentus protein, all species in which ssrA or smpB have been deleted encode either ArfB or ArfA, with the exception of Francisella tularensis and Bacillus subtilis. Tn-seq experiments or other synthetic-lethal screens should reveal whether F. tularensis and B. subtilis have a different backup system for trans-translation or whether resolution of nonstop complexes is not essential in these species.
C. crescentus cells lacking trans-translation grow slowly and have a cell cycle defect, and this phenotype is not complemented by ssrA-DD, a variant that releases nonstop complexes but does not target the nascent polypeptide for rapid degradation (23). These results were interpreted to indicate that degradation of one or more tagged proteins was required for normal growth in C. crescentus. However, this hypothesis is not consistent with the observation that overexpression of arfB in ΔssrA cells restored the wild-type growth rate. Instead, it appears that the slow growth phenotype in cells lacking trans-translation is caused by failure to release nonstop translation complexes, similar to phenotypes observed for ssrA deletions in other species.
Does C. crescentus ArfB perform unique functions for the cell, or is its only role to fill in when trans-translation activity is overwhelmed? This question is particularly interesting in C. crescentus, because the levels of tmRNA and SmpB fluctuate as a function of the cell cycle, with both molecules dropping to very low levels in early S phase (27, 28). Nevertheless, ArfB does not play a critical role in the cell cycle, because strains in which arfB is deleted grow at the same rate as the wild type. Moreover, transcriptome sequencing (RNA-seq) and ribosome profiling experiments indicated that arfB is not highly transcribed or translated during growth in culture (21), and experiments on synchronized cultures indicated that arfB transcription is not cell cycle regulated (29). Any role for ArfB other than acting as an emergency backup for trans-translation remains to be identified.
Mitochondria contain a protein, ICT1, that has 33% sequence identity with C. crescentus ArfB, and ICT1 has been shown to hydrolyze peptidyl-tRNA on nonstop translation complexes, as well as at stop codons (30). ICT1 is clearly important for mitochondria, because it is conserved in all mitochondrial genomes and is essential in HeLa cells (30, 31). Mitochondria are thought to derive from Alphaproteobacteria, likely from the rickettsial lineage (32), and the distribution of tmRNA and ArfB in current Alphaproteobacteria suggests that early Alphaproteobacteria had both trans-translation and ArfB. In fact, mitochondria from some single-celled eukaryotes retain tmRNA and SmpB in addition to ArfB (6). Why did the rickettsial lineage keep trans-translation and lose ArfB, while mitochondria in higher eukaryotes kept the ArfB homolog and lost trans-translation? Presumably, bacteria retain trans-translation because it targets the nascent polypeptide and nonstop mRNA for degradation in addition to releasing the ribosome. Mitochondria contain homologs of bacterial ClpXP, the protease that degrades most tmRNA-tagged proteins, so proteolysis of tagged proteins should have occurred in early mitochondria. It is possible that mitochondria have a different pathway to remove incomplete proteins, limiting the advantage of trans-translation. Alternatively, the simplicity of the single-chain ArfB might have been advantageous during genome reduction.
MATERIALS AND METHODS
Bacterial growth.
Strains are described in Table 5. C. crescentus strains were grown at 30°C in peptone-yeast extract (PYE) medium, supplemented with tetracycline (2 µg/ml), streptomycin (50 µg/ml), spectinomycin (100 µg/ml), or kanamycin (20 µg/ml) where appropriate. Culture growth was monitored by measuring the optical density at 600 nm.
TABLE 5 .
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| CB15N | Wild-type C. crescentus | 38 |
| CB15N ΔssrA | In-frame deletion of ssrA | 27 |
| CB15N CC1214::Ω | CC1214 replaced by omega cassette | This study |
| KCK426 | E. coli BL21(DE3) carrying pET28CC1214 | This study |
| KCK428 | CB15N CC1214::Ω with Kanr marker 22.4 kb downstream | This study |
| KCK429 | CB15N CC1215 replaced by cat gene in antisense orientation | This study |
| KCK430 | CB15N CC1215 replaced by cat gene in sense orientation | This study |
| KCK431 | CB15N ΔssrA CC1215 replaced by cat gene in antisense orientation | This study |
| KCK432 | CB15N ΔssrA CC1215 replaced by cat gene in sense orientation | This study |
| Plasmids | ||
| pML81 | pJS14-derived plasmid with xylose promoter and NdeI site | M. T. Laub, unpublished |
| pML81-His6-CC1214 | Expresses CC1214 from xylose promoter | This study |
| pBBR1-tet | pBBR1 plasmid with tetA cloned into NotI site | Gift from Camille Lochte, Pasteur Institute of Lille |
| pBBR-PBN | pBBR1-tet-derived plasmid with PstI, BamHI, and NdeI cloning sites. | This study |
| pCC1214 | Expresses CC1214 from xylose promoter | This study |
| pACC1214G30A | Expresses CC1214 active-site mutant (GAQ) from xylose promoter | This study |
| pSsrA | Expresses ssrA from its native promoter | 23 |
| pBGS18T(1349674–1350629) | pBGS18T containing targeting fragment from position 1349674 to 1350629 | 34 |
| pET28CC1214 | Expresses CC1214 from T7 promoter | This study |
| pDHFR | Expresses DHFR from T7 promoter | New England Biolabs |
| pMT425 | Plasmid with vanillate-inducible promoter, cat gene | 39 |
Plasmid construction.
Oligonucleotide sequences are shown in Table S2 in the supplemental material. Plasmid pBBR-PBN was constructed by amplifying plasmid pBBR1-tet by PCR using primers BBRtetPBN1 and BBRtetPBN2, digesting the product with BamHI, and ligating to circularize the plasmid. Plasmid pCC1214 was constructed by amplifying the CC1214 gene by PCR using primers 1214_His6F and 1214_Rev, digesting the resulting DNA with NdeI and HindIII, and ligating the product into pML81 cut with the same enzymes to create plasmid pML81-His6-1214. The xylose promoter and CC1214 were then amplified from pML81-His6-1214 by PCR using primers PstXyl224F and 1214PstIR and ligated into plasmid pBBR-PBN, cut with PstI, to produce pCC1214. pCC1214G30A was made by primer extension mutagenesis. pML81-His6-1214 was used as the template in two separate PCRs using primers PstXyl224F and G30A_R2 or primers G30A_F2 and 1214PstIR. The products of these reactions were used in a second PCR with primers PstXyl224F and 1214PstIR. The resulting product was cloned into the PstI site of pBN and sequenced to verify the mutation. pET28CC1214 was constructed by amplifying CC1214 with primers 1214_Fwd and 1214_Rev, digesting with NdeI and HindIII, and ligating into pET28b that had also been digested with NdeI and HindIII.
Tn-seq.
Overnight cultures of wild-type and ΔssrA C. crescentus strains were grown and mutagenized with a himar1 transposon. More than 100,000 colonies were pooled, and chromosomal DNA was extracted and sequenced using an Illumina HiSeq 2000 as previously described (33). Data sets were analyzed in SeqMonk version 0.27.0 by normalizing to the total number of reads in the largest data set, adding 1 to eliminate infinite and zero ratios, and calculating the ratio of insertions in ΔssrA cultures to insertions in wild-type cultures (33).
Genetic deletions.
Regions of ~1.5 kb flanking CC1214 were amplified by PCR using primers 1214_UF and 1214_UR or 1214_DF and 1214_DR. The products were cut with SpeI and EcoRI or NheI and EcoRI, respectively, and ligated into the SpeI and NheI sites of integrating plasmid pNPTS138. The omega cassette from pHP45Ω was inserted at the EcoRI site between the flanking regions. The resulting plasmid was used to construct CB15NCC1214::Ω using the two-step recombination method (20). Mutants were selected on spectinomycin, and the CC1214::Ω mutation was confirmed by PCR with primers 1214_ScrnF and 1214_ScrnR. CB15NCC1215::cat was constructed similarly, using primers 1215_UF and 1215_UR to amplify a region ~1.5 kb upstream from position 1215. A downstream flanking region was amplified using primers 1215_DF and 1215_DR. The PCR fragments were digested with NheI and EcoRI or BamHI and EcoRI and ligated into pNPTS138 digested with NheI and BamHI. The cat gene from pMT425 was amplified with primers cat220F and EcoCatR, cut with EcoRI, and ligated into the EcoRI site between the two flanking regions. The direction of the cat insertion was determined by PCR, and two-step recombination was used to construct CB15NCC1215::cat with the cat cassette facing in either the sense (KCK429) or antisense (KCK430) direction with respect to CC1215. The presence of the mutation in selected clones was verified by PCR using primers 1215_ScrnF and 1215_ScrnR. To construct strains KCK431 and KCK432, φCR30 phage lysate was prepared from strain KCK429 and KCK430, respectively, and used to infect CB15N ΔssrA. The presence of the mutation was verified using primers 1215_ScrnF and 1215_ScrnR.
Phage transduction.
A kanamycin resistance marker was inserted 22.4 kb from CC1214::Ω to make strain KCK428 by transforming CB15N CC1214::Ω with the nonreplicating plasmid pBGS18T(1349674–1350629) (34). φCR30 phage lysate was prepared from KCK428 and used to infect CB15N or CB15N ΔssrA (35). Transductants were selected on plates containing kanamycin and patched onto plates containing streptomycin and spectinomycin to determine the cotransduction frequency. The expected frequency was predicted by the equation log(distance) = −1.56(ctf) + 5.06, where distance represents the number of bases between the two markers and ctf represents cotransduction frequency (34).
Purification of CC1214.
KCK426 was grown to an optical density at 600 nm of ≈0.8, and the expression of CC1214 was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 1 mM. Cells were harvested by centrifugation, resuspended in guanidinium lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate [pH 7.8], 400 mM NaCl), and sonicated. The lysate was cleared by centrifugation at 11,000 × g for 10 min. A column was loaded with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) that had been equilibrated with denaturing binding buffer (8 M urea, 20 mM sodium phosphate [pH 7.8], 500 mM NaCl). The column was loaded with cleared lysate and washed by adding 10 volumes of denaturing binding buffer and rocking for 15 min. The column was then washed with 10 volumes of denaturing wash buffer (8 M urea, 20 mM sodium phosphate [pH 6.0], and 500 mM NaCl) and 10 volumes of denaturing wash buffer equilibrated to pH 5.3. Bound protein was eluted with denaturing elution buffer (8 M urea, 20 mM sodium phosphate [pH 4.0], and 500 mM NaCl) and visualized by SDS-PAGE. Fractions containing 6×His-ArfB were dialyzed against buffer H (10 mM HEPES [pH 7.5], 150 mM NaCl).
In vitro translation and peptidyl hydrolysis assays.
CC1214 was assayed for peptidyl hydrolysis activity using the PURExpress system (New England Biolabs) similarly to previous assays (36), with some modifications. Nonstop DHFR was amplified using primers HAF_T7 and UTR_DHFR_FL and used as the template for the reaction. After 1 h of incubation at 37°C, CC1214 or CC1214G30A was added to a final concentration of 200 nM. A control sample was treated with 70 µg/ml puromycin. After 1 h of incubation at 37°C, total protein was precipitated by the addition of acetone, resuspended in sample loading buffer (5 mM sodium bisulfite, 50 mM MOPS [morpholinepropanesulfonic acid], 50 mM Tris base, 1 µM EDTA, 0.1% SDS, 5% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue), and resolved on a bis-Tris gel using MOPS running buffer.
Sequence homology searches.
The protein sequence of CC1214 was used in a tblastn search (24) of the June 2014 NCBI database of representative microbial genomes. An E value cutoff of 10−7 and a molecular-mass cutoff of 20 kDa were used to identify homologs. In cases where a homolog was identified for a given phylum, that homolog was then used in a tblastn search of the remaining representative genomes in that phylum. If a homolog was not found, tblastn searches were performed with homologs from closely related phyla. A tree of Alphaproteobacteria species was constructed by collecting 16S rRNA gene sequences from PATRIC (37) and aligning them with BLAST. The tree was viewed and colored using Fig tree software (version 1.4.2 [http://tree.bio.ed.ac.uk/software/figtree/]).
SUPPLEMENTAL MATERIAL
Tn-Seq data. The gene names and coordinates are in the first five columns, followed by the normalized number of insertions in the ΔssrA and wild-type strains. The last column has the log10 ratio of ΔssrA/wild-type insertions.
DNA oligonucleotides used in this study.
ACKNOWLEDGMENT
This work was supported by NIH grant GM68720 to K.C.K.
Footnotes
Citation Feaga HA, Viollier PH, Keiler KC. 2014. Release of nonstop ribosomes is essential. mBio 5(6):e01916-14. doi:10.1128/mBio.01916-14.
REFERENCES
- 1. Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. 2005. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69:101–123. 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schmeing TM, Ramakrishnan V. 2009. What recent ribosome structures have revealed about the mechanism of translation. Nature 461:1234–1242. 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 3. Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 4. Keiler KC. 2008. Biology of trans-translation. Annu. Rev. Microbiol. 62:133–151. 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- 5. Moore SD, Sauer RT. 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76:101–124. 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 6. Hudson CM, Lau BY, Williams KP. 2014. Ends of the line for tmRNA-SmpB. Front. Microbiol. 5:421. 10.3389/fmicb.2014.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keiler KC, Feaga HA. 2014. Resolving nonstop translation complexes is a matter of life or death. J. Bacteriol. 196:2123–2130. 10.1128/JB.01490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chadani Y, Ono K, Ozawa S, Takahashi Y, Takai K, Nanamiya H, Tozawa Y, Kutsukake K, Abo T. 2010. Ribosome rescue by Escherichia coli ArfA (YhdL) in the absence of trans-translation system. Mol. Microbiol. 78:796–808. 10.1111/j.1365-2958.2010.07375.x. [DOI] [PubMed] [Google Scholar]
- 9. Chadani Y, Ito K, Kutsukake K, Abo T. 2012. ArfA recruits release factor 2 to rescue stalled ribosomes by peptidyl-tRNA hydrolysis in Escherichia coli. Mol. Microbiol. 86:37–50. 10.1111/j.1365-2958.2012.08190.x. [DOI] [PubMed] [Google Scholar]
- 10. Shimizu Y. 2012. ArfA recruits RF2 into stalled ribosomes. J. Mol. Biol. 423:624–631. 10.1016/j.jmb.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 11. Schaub RE, Poole SJ, Garza-Sánchez F, Benbow S, Hayes CS. 2012. Proteobacterial ArfA peptides are synthesized from non-stop messenger RNAs. J. Biol. Chem. 287:29765–29775. 10.1074/jbc.M112.374074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garza-Sánchez F, Schaub RE, Janssen BD, Hayes CS. 2011. tmRNA regulates synthesis of the ArfA ribosome rescue factor. Mol. Microbiol. 80:1204–1219. 10.1111/j.1365-2958.2011.07638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramadoss NS, Zhou X, Keiler KC. 2013. tmRNA is essential in Shigella flexneri. PLoS ONE 8:e57537. 10.1371/journal.pone.0057537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schaub RE, Hayes CS. 2011. Deletion of the RluD pseudouridine synthase promotes SsrA peptide tagging of ribosomal protein S7. Mol. Microbiol. 79:331–341. 10.1111/j.1365-2958.2010.07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shoji S, Janssen BD, Hayes CS, Fredrick K. 2010. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie 92:157–163. 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85. 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 17. Singh NS, Varshney U. 2004. A physiological connection between tmRNA and peptidyl-tRNA hydrolase functions in Escherichia coli. Nucleic Acids Res. 32:6028–6037. 10.1093/nar/gkh924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hayes CS, Bose B, Sauer RT. 2002. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:3440–3445. 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chadani Y, Ono K, Kutsukake K, Abo T. 2011. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol. Microbiol. 80:772–785. 10.1111/j.1365-2958.2011.07607.x. [DOI] [PubMed] [Google Scholar]
- 20. Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schrader JM, Zhou B, Li G-W, Lasker K, Childers WS, Williams B, Long T, Crosson S, McAdams HH, Weissman JS, Shapiro L. 2014. The coding and noncoding architecture of the Caulobacter crescentus genome. PLoS Genet. 10:e1004463. 10.1371/journal.pgen.1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mora L, Heurgué-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH. 2003. The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol. Microbiol. 47:267–275. 10.1046/j.1365-2958.2003.03301.x. [DOI] [PubMed] [Google Scholar]
- 23. Keiler KC, Shapiro L. 2003. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185:573–580. 10.1128/JB.185.2.573-580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25. Ebeling S, Kündig C, Hennecke H. 1991. Discovery of a rhizobial RNA that is essential for symbiotic root nodule development. J. Bacteriol. 173:6373–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang C, Glover JR. 2009. The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS One 4:e4459. 10.1371/journal.pone.0004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keiler KC, Shapiro L. 2003. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 185:1825–1830. 10.1128/JB.185.6.1825-1830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hong SJ, Tran QA, Keiler KC. 2005. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 57:565–575. 10.1111/j.1365-2958.2005.04709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fang G, Passalacqua KD, Hocking J, Llopis PM, Gerstein M, Bergman NH, Jacobs-Wagner C. 2013. Transcriptomic and phylogenetic analysis of a bacterial cell cycle reveals strong associations between gene co-expression and evolution. BMC Genomics 14:450. 10.1186/1471-2164-14-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. 2010. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 29:1116–1125. 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Handa Y, Inaho N, Nameki N. 2011. YaeJ is a novel ribosome-associated protein in Escherichia coli that can hydrolyze peptidyl-tRNA on stalled ribosomes. Nucleic Acids Res. 39:1739–1748. 10.1093/nar/gkq1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams KP, Sobral BW, Dickerman AW. 2007. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 189:4578–4586. 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murray SM, Panis G, Fumeaux C, Viollier PH, Howard M. 2013. Computational and genetic reduction of a cell cycle to its simplest, primordial components. PLoS Biol. 11:e1001749. 10.1371/journal.pbio.1001749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. West L, Yang D, Stephens C. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155-2166. 10.1128/JB.184.8.2155-2166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ely B, Johnson RC. 1977. Generalized transduction in Caulobacter crescentus. Genetics 87:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramadoss NS, Alumasa JN, Cheng L, Wang Y, Li S, Chambers BS, Chang H, Chatterjee AK, Brinker A, Engels IH, Keiler KC. 2013. Small molecule inhibitors of trans-translation have broad-spectrum antibiotic activity. Proc. Natl. Acad. Sci. U. S. A. 110:10282–10287. 10.1073/pnas.1302816110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJ, Yoo HS, Zhang C, Zhang Y, Sobral BW. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42:D581–D591. 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evinger M, Agabian N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tn-Seq data. The gene names and coordinates are in the first five columns, followed by the normalized number of insertions in the ΔssrA and wild-type strains. The last column has the log10 ratio of ΔssrA/wild-type insertions.
DNA oligonucleotides used in this study.