Abstract
The search for new natural products is leading to the isolation of novel actinomycete species, many of which will ultimately require genetic analysis. Some of these isolates will likely exhibit low intrinsic frequencies of homologous recombination and fail to sporulate under laboratory conditions, exacerbating the construction of targeted gene deletions and replacements in genetically uncharacterised strains. To facilitate the genetic manipulation of such species, we have developed an efficient method to generate gene or gene cluster deletions in actinomycetes by homologous recombination that does not introduce any other changes to the targeted organism's genome. We have synthesised a codon optimised I-SceI gene for expression in actinomycetes that results in the production of the yeast I-SceI homing endonuclease which produces double strand breaks at a unique introduced 18 base pair recognition sequence. Only those genomes that undergo homologous recombination survive, providing a powerful selection for recombinants, approximately half of which possess the desired mutant genotype. To demonstrate the efficacy and efficiency of the system, we deleted part of the gene cluster for the red-pigmented undecylprodiginine complex of compounds in Streptomyces coelicolor M1141. We believe that the system we have developed will be broadly applicable across a wide range of actinomycetes.
Actinomycetes are Gram-positive mycelial bacteria with a high genomic GC content that produce a wide variety of industrially and medically relevant compounds including chemotherapeutics, fungicides, herbicides, immunosuppressants and many clinically used antibiotics. The search for new pharmacologically active and industrially relevant natural products has led to the recent isolation of many novel actinomycete strains, many of which are recalcitrant to genetic manipulation.
Homologous recombination is a highly conserved process present in all forms of life, from bacteria to eukaryotes1. It is used regularly to target gene replacement and deletion in a wide range of organisms including bacteria2, although the efficiency of this process is very much dependent on the propensity of the targeted organism to carry out homologous recombination.
Gene deletion, replacement or disruption in bacteria generally involves the use of a non-replicative vector carrying the targeted DNA, with selection for antibiotic resistance resulting in integration of the construct into the host's genome by homologous recombination (yielding the first cross-over recombinant). The resulting strain is then screened for loss of the resistance marker carried on the vector, and the sensitive isolates obtained screened phenotypically and/or by PCR to identify the required second cross-over recombinant. In organisms with a low intrinsic level of homologous recombination, screening for loss of antibiotic resistance can be a very tedious process, and in mycelial actinomycetes that do not sporulate, sometimes extremely difficult if not impossible to achieve. To expedite this process and to select for this second recombination event, it is possible to induce a DNA double-strand break adjacent to a targeted locus by using a meganuclease such as I-SceI. I-SceI is a monomeric 235 amino acid homing endonuclease encoded in the mitochondrial DNA of Saccharomyces cerevisiae3 which is able to cut double stranded DNA at an 18 bp recognition sequence: TAGGGATAACAGGGTAAT. The length of this sequence ensures that it is absent from all actinomycete genome sequences determined thus far. Chromosome repair and viability can only result through homologous recombination between duplicated regions of the genome, thus providing a selection for the second cross-over event which should yield a mixture of both the original genotype and the required mutant.
Lu et al.4 previously reported the successful exploitation of the I-SceI meganuclease in Streptomyces coelicolor to delete the gene cluster for the blue-pigmented polyketide antibiotic actinorhodin using a two plasmid system in which the vector delivering the meganuclease, an integrative pSET derivative5, remained, with its antibiotic resistance gene, in the genome of the targeted organism once the required second cross-over recombinant had been obtained. This precludes the subsequent use in the mutant of any vector that relies on the widely-used integration system of the Streptomyces phage ΦC31, and eliminates the further use of the thiostrepton resistance marker carried by the pSET derivative. Moreover, integration at the ΦC31 attB site can considerably reduce the level of production of some specialised metabolites, including antibiotics6.
Here we report a I-SceI-based mutagenesis strategy that enables the construction of marker-less deletions of genes and gene clusters that is applicable to many actinomycete species, and that should be particularly useful for strains that are difficult to manipulate genetically.
Results
Validating the system with a marker-less deletion in S. coelicolor
To validate the system and assess its efficiency, we chose to delete an essential region of the undecylprodiginine (Red) biosynthetic gene cluster of S. coelicolor that contained the cluster-situated regulatory gene redD and the polyketide synthase gene redX (Fig. 17;). Red is a red-pigmented tripyrrole compound that remains associated with the mycelium, thus providing a facile visual screen for loss of Red production and a quantitative estimation of deletion frequency.
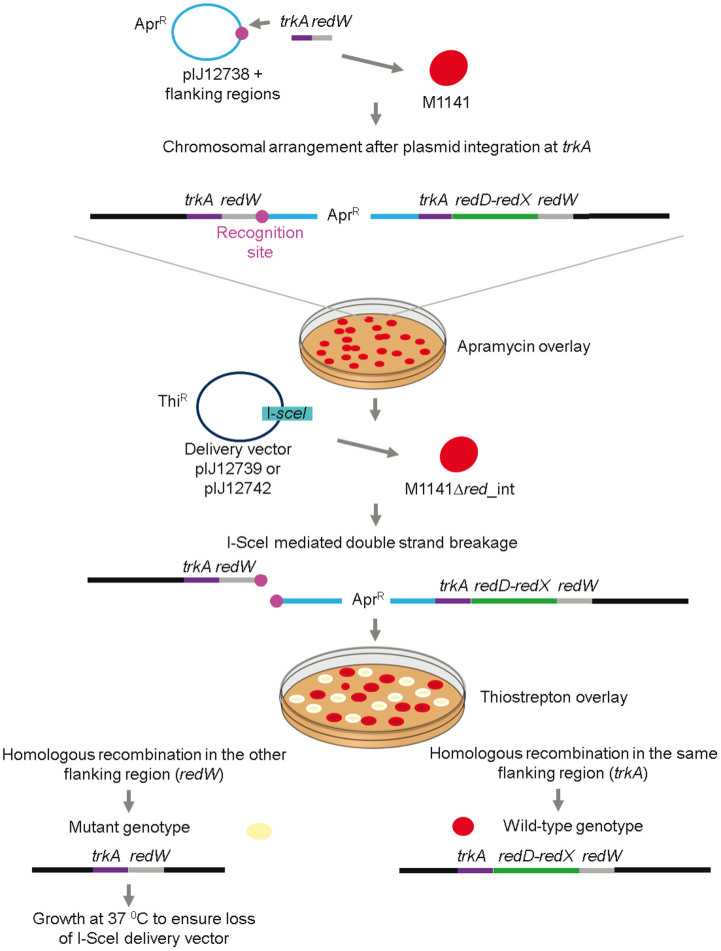
Figure 1. Overview of the protocol developed to obtain marker-less gene deletions in actinomycetes.
pIJ12738 containing the I-SceI recognition sequence (shown by a pink circle) and the flanking regions (in this example, trkA and redW, corresponding to the left and right flanking regions of redD and redX, respectively) of the chromosomal region to be deleted was conjugated into M1141. Apramycin resistant exconjugants were analysed by PCR to confirm integration of the plasmid (in this schematic, integration occurred by homologous recombination at trkA). A single exconjugant (M1141Δred_int) was selected as recipient of the delivery vectors pIJ12739 and pIJ12742 expressing the I-SceI meganuclease gene. After conjugation, the plate was overlaid with thiostrepton to select for exconjugants. I-SceI creates double strand breaks at its introduced recognition sequence, and the only genomes to survive are those that undergo homologous recombination to reconstitute an intact chromosome. If recombination occurs in the same flanking region that was used in the first cross-over event (in this example, trkA), then the exconjugant will revert to a wild type genotype. In contrast, if recombination takes place in the other flanking region (in our example, redW), the exconjugant will have a mutant genotype. Exconjugants were analysed by PCR to confirm the required mutant genotype. To ensure loss of the I-SceI delivery vector, the mutant strain was grown at 37°C, and loss of the vector confirmed by PCR.
First cross-over in one of the homologous regions flanking redDX
Streptomyces coelicolor M11418 is a derivative of S. coelicolor M1459 in which the gene cluster for production of the blue-pigmented polyketide antibiotic actinorhodin has been deleted. Thus colonies of M1141 are red in appearance, while derivatives that are unable to produce Red would be cream in colour.
pIJ12740, containing sequences flanking redD (trkA) and redX (redW) and the adjacent I-SceI recognition site, was introduced into S. coelicolor M1141 from Escherichia coli by conjugation. Apramycin-resistant exconjugants, all pigmented due to their ability to synthesise Red, were isolated and the presence of the non-replicative plasmid confirmed by PCR. Integration by homologous recombination through trkA was confirmed for some of the exconjugants using primers corresponding to the chromosomal sequence to the left of trkA (as shown in Fig. 1) and to a sequence internal to redW (data not shown). One of these exconjugants was designated M1141Δred_int (for Δred integrant).
Delivery and expression of I-SceI to select for the second homologous recombination event
To select for second cross-over recombinants, pIJ12739 (Fig. 2), containing the codon optimised I-SceI gene under the control of the thiostrepton inducible tipA promoter, was introduced into M1141Δred_int by conjugation from E. coli and exconjugants selected by overlaying the agar plate with thiostrepton. Spores were collected directly from the conjugation plate, diluted and plated on R5 agar containing nalidixic acid (to prevent growth of the E. coli donor strain) and the number of red and cream colonies on each plate counted (Fig. 3). After examining over 2000 exconjugants from each of four independent matings, the frequency of colonies displaying a Red− phenotype after the second crossover event varied between 29 and 52 percent. Ten cream and ten red colonies were analysed by PCR to confirm deletion of redDX in the cream isolates (M1141Δred), reversion to the wild type genotype in the red colonies (Fig. 3), and loss of the pIJ12740 backbone by homologous recombination in all of them (data not shown).
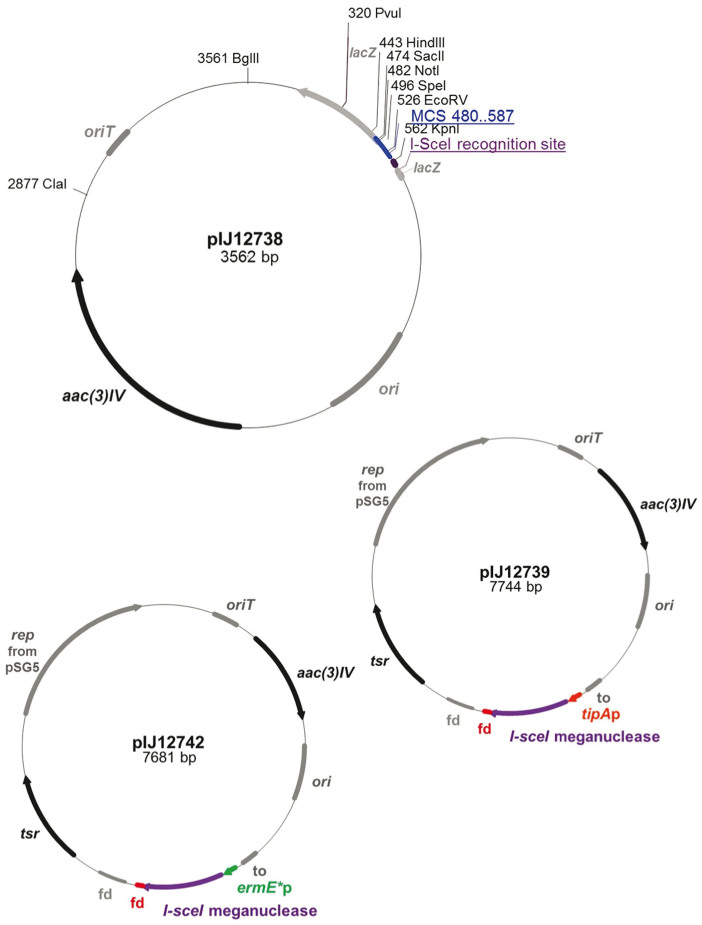
Figure 2. Restriction maps of the plasmids constructed in this study.
aac(3)IV, apramycin resistance gene; ori, E. coli origin of replication; oriT, origin of transfer; MCS, multiple cloning site; lacZ, α fragment of lacZ; rep, origin of replication from pSG5; tsr, thiostrepton resistance gene; fd, transcriptional terminator from phage fd; to, transcriptional terminator; tipAp, thiostrepton inducible tipA promoter; ermE*p, mutated constitutive promoter from ermE.
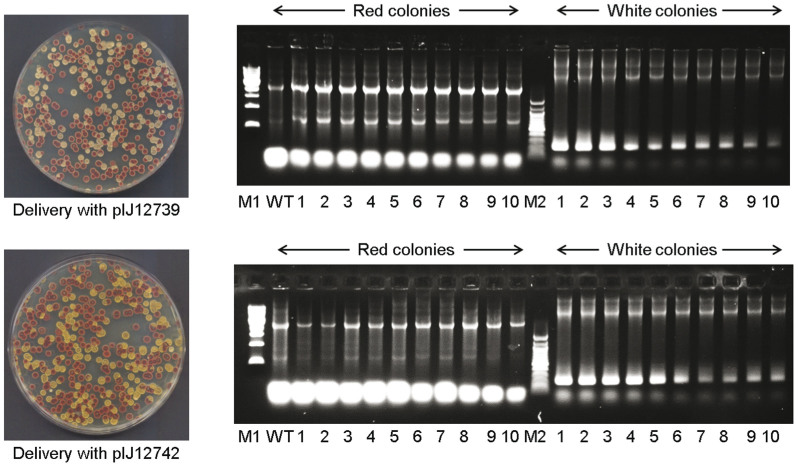
Figure 3. Phenotype and genotype verification of wild type and mutant exconjugants of M1141Δred_int.
R5 agar plates containing exconjugants resulting from conjugation of either pIJ12739 (top) or pIJ12742 (bottom). Colony PCR (to the right of each agar plate) carried out on ten red (M1141 with intact red gene cluster, expected fragment size 2697 bp), and ten cream (M1141Δred, expected fragment size 160 bp) randomly selected exconjugants confirmed that the phenotypes corresponded to the presence or absence of redDX, respectively. M1, 1 kb ladder (New England Biolabs); M2, 100 bp ladder (New England Biolabs). WT, genomic DNA from M1141 used as a control.
Induction of the tipA promoter by thiostrepton requires the presence of the tipA gene10, which is not present in all actinomycetes. As an alternative strategy, and to overcome this potential limitation, pIJ12742 (Fig. 2), containing the codon optimised I-SceI gene under the control of the strong constitutive ermE* promoter, was delivered to the first cross-over recombinant following exactly the same protocol as described above for pIJ12739. After screening over 2000 colonies from each of three independent conjugations, the frequency of colonies displaying a Red− mutant phenotype varied between 27 and 49 percent, similar to the results obtained with pIJ12739. Again, ten of the cream and ten of the red colonies were analysed by PCR to verify that the phenotype of the colony corresponded to the predicted genotype (Fig. 3) and that the pIJ12740 backbone had been lost by a second homologous recombination event.
Loss of the temperature-sensitive I-SceI delivery vectors pIJ12739 and pIJ12742
Both pIJ12739 and pIJ12742 are derivatives of pGM1190, an intermediate copy number, conjugative plasmid containing the temperature-sensitive replication origin of pSG511. When exconjugants carrying either pIJ12739 or pIJ12742 are grown above 34°C, the plasmids are unable to replicate and are lost. After plasmid curing, the exconjugants can be grown at their lower optimal temperature. Growth of ten red and ten cream colonies containing each of the I-SceI delivery plasmids on SFM agar at 37°C resulted in complete loss of each of the pGM1190 derivatives, confirmed by their sensitivity to thiostrepton and PCR analysis (data not shown). Thus, after growth at the non-permissive temperature, all of the M1141Δred exconjugants with a marker-less deletion of redDX had also lost the nuclease delivery vector, leaving a mutant with no introduced DNA or antibiotic resistance markers.
Interestingly, when S. coelicolor M1141Δred derivatives carrying either pIJ12739 or pIJ12742 were passaged through two rounds of sporulation at 30°C in the absence of thiostrepton, 60 percent of the resulting colonies were sensitive to the antibiotic suggesting loss of the plasmids, which was confirmed by PCR analysis (data not shown). This inherent instability in the absence of antibiotic selection could be useful when working with any strains that may not be able to grow at higher temperatures.
Discussion
In bacteria, double stranded breakage of the chromosome is a lethal event unless the lesion is repaired by homologous recombination. We have taken advantage of the mitochondrial homing endonuclease I-SceI to select for second cross-over recombinants, with 27–52 percent yielding the required mutational event. In this study, we have demonstrated the utility of this approach by constructing a two plasmid system to create a marker-less deletion in S. coelicolor. This system has also been used to efficiently delete, in an iterative manner, entire gene clusters from the genome of Streptomyces venezuelae (Neil Holmes, personal communication). In this study we used conjugation from E. coli for plasmid transfer (conjugation is the most widely-used procedure for introducing DNA into actinomycetes), but in principle protoplast transformation could be used instead with targeted strains for which conjugation protocols do not yet exist. In addition to being readily applicable to well-studied academic and industrial strains (where there is clear potential for use in the construction of engineered strains with improved expression of heterologous genes and gene clusters), we believe that the system described here will be particularly useful when attempting to make mutants in novel and genetically uncharacterised actinomycetes, especially if they exhibit low levels of homologous recombination and/or fail to sporulate, which generally provides a very useful means of segregating wild type and mutant genomes in a multi-nucleoid mycelial organism. If dealing with a non-sporulating strain, mycelia from each of the conjugation plates could be harvested, fragmented (for example, by using a Potter homogeniser) and then subjected to the same protocol.
Methods
Bacterial strains, plasmids and general methods
The strains and plasmids used and generated in this study are listed in Table 1. Plasmid isolation, PCR amplification and E. coli growth and manipulation were carried out following standard methods12,13,14. S. coelicolor strains were grown on SFM and R5 agar as described previously9.
Table 1. Strains and plasmids used in this study.
| Strain/Plasmid | Genotype/Description | Reference |
|---|---|---|
| Escherichia coli DH5α | F− ϕ80 lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk−, mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen™ |
| E. coli ET12567 | dam-13::Tn9 dcm-6 hsdM CmlR, carrying helper plasmid pUZ8002 | 22 |
| S. coelicolor M1141 | M145 derivative Δact | 8 |
| S. coelicolor M1141Δred_int | M1141 containing pIJ12740 | This study |
| S. coelicolor M1141Δred | M1141 ΔredDX | This study |
| pUC57-Simple_SceI | pUC57-Simple with tcp830p-I-SceI gene-fd terminator-MCS-I-SceI site | GeneScript (Piscataway, USA) |
| pKC1132 | Conjugative vector, non-replicative in Streptomyces, AprR | 5 |
| pIJ12738 | pKC1132 with MCS and I-SceI site from pUC57-Simple_SceI | This study |
| pGM1190 | tsr (ThiR), aac(3)IV (AprR), oriT, to terminator, tipAp, RBS, fd terminator | 11 |
| pIJ12739 | pGM1190 with tipAp-I-SceI gene, ThiR | This study |
| pIJ12551 | Conjugative and ϕC31-integrative vector, AprR | 19 |
| pIJ12590 | pIJ12551 with I-SceI gene | This study |
| pIJ12742 | pGM1190 with ermE*p-I-SceI gene, ThiR | This study |
| pIJ12740 | pIJ12738 with redXW left and right flanking sequences | This study |
Plasmid construction
Oligonucleotides generated and used in this study are described in Table 2. DNA manipulation and cloning were carried out according to standard protocols12. Plasmid constructs were confirmed by sequencing. A fragment containing the tcp830 promoter15, followed by the synthetic I-SceI gene codon optimised for actinomycetes, the Streptomyces lividans multi-copy plasmid pIJ101 terminator region16, a multiple cloning site (MCS) containing the following unique sites: SacII, NotI, XbaI, SpeI, PstI, EcoRI and EcoRV and the I-SceI 18 bp recognition sequence was synthesised by GeneScript (Piscataway, USA) and flanked by HindIII and MfeI restriction sites. The fragment was provided on the plasmid pUC57-Simple_SceI. This plasmid was treated with XbaI and SspI, and the 117 bp fragment containing the MCS followed by the I-SceI recognition site was cloned into pKC11325 cut with XbaI and EcoRV to generate pIJ12738 (Fig. 2).
Table 2. Oligonucleotides used in this study.
| Oligonucleotide | Sequence | Notes |
|---|---|---|
| ermESceI_F_SnaBI | AATACGTAGGATCCAGCCCGACCCGAGC | Amplify ermE*p-I-SceI gene fusion from pIJ12590 |
| ermESceI_R_EcoRI | ACGAATTCGATATCGCGCGCGG | Amplify ermE*p-I-SceI gene fusion from pIJ12590 |
| redD_left_flank_XbaI_F | AATCTAGAGGACTCGTTGAAGAGCCACT | Amplify the region 5′ to redDW |
| redD_left_flank_BamHI_R | TTGGATCCGTTCTCCGCACTCCCATGA | Amplify the region 5′ to redDW |
| redX_right_flank_BamHI_F | TTGGATCCCACCTGTTGATCGAGGAAGG | Amplify the region 3′ to redDW |
| redX_right_flank_KpnI_R | ATAGGTACCCACCGAGATGCTCTCGAAGT | Amplify the region 3′ to redDW |
| KC1132_flank_SacII_check_F | GTTTTCCCAGTCACGACGTT | Confirm presence of pIJ12740 in M1141 |
| KC1132_flank_EcoRV_check_R | TGTGGAATTGTGAGCGGATA | Confirm presence of pIJ12740 in M1141 |
| Red_deletion_check_F | CTGTACAACTTCGGGGGATG | Confirm redXW deletion in exconjugants |
| Red_deletion_check_R | GCGTCGAAGTCGAAGTTCAT | Confirm redXW deletion in exconjugants |
| SceI_check_F | CATGCAGTTCGAGTGGAAGA | Confirm presence of pIJ12739 or pIJ12742 |
| SceI_check_R | CGGGATGGTCTTCTTGTTGT | Confirm presence of pIJ12739 or pIJ12742 |
To construct the plasmid carrying the codon optimised I-SceI gene, pUC57-Simple_SceI was cut with NdeI and EcoRI to generate a 853 bp fragment that was cloned into pGM119011 cleaved with the same enzymes. This generated pIJ12739 (Fig. 2) in which the I-SceI gene is under the control of the inducible tipA promoter17.
To express the I-SceI gene from the strong constitutive ermE* promoter18, a 806 bp NdeI-SacII fragment containing the gene was excised from pIJ12739 and cloned into pIJ1255119 digested with the same enzymes to generate pIJ12590. A 1055 bp fragment containing ermE*p-I-SceI gene was amplified by PCR using oligonucleotides containing SnaBII or EcoRI restriction sites (Table 2). This fragment was cloned into pGM1190 digested with the same enzymes to generate pIJ12742 (Fig. 2).
Construction of deletion mutants
Chromosomal regions flanking the adjacent cluster-situated regulatory gene redD and the polyketide synthase gene redX of the undecylprodiginine (Red) biosynthetic gene cluster were amplified by PCR and cloned into pIJ12738. Both genes are essential for Red biosynthesis20,21. The flanking region 5′ of redD was amplified to generate a 1516 bp fragment with terminal XbaI and BamHI sites and the region 3′ of redX was amplified to generate a 1419 bp fragment with terminal BamHI and KpnI sites. These two flanking fragments were cloned into pIJ12738 digested with XbaI and KpnI to generate pIJ12740 with the I-SceI site adjacent to the introduced red gene fragments. pIJ12740 was then introduced into E. coli ET12567/pUZ800222 by transformation. Conjugation between E. coli ET12567/pUZ8002 carrying the oriT-containing plasmid and Streptomyces coelicolor M1141 was carried out as previously described9. Chromosomal integration of pIJ12740 generated M1141Δred_int which was still capable of Red production. pIJ12739 or pIJ12742 carrying the I-SceI gene was then conjugated into M1141Δred_int to induce a double strand break at the introduced I-SceI site. Ten individual red and cream colonies isolated from each of the conjugations were streaked on SFM agar and grown at 37°C until sporulation. Spores were harvested and plated on SFM agar with or without thiostrepton. No colonies grew in the presence of thiostrepton, indicative of plasmid loss, which was confirmed by PCR analysis (data not shown).
Author Contributions
L.T.F.-M. and M.J.B. conceived and designed the project, and wrote the manuscript; L.T.F.-M. performed the experiments.
Acknowledgments
This work was supported financially by the Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant “Understanding and Exploiting Plant and Microbial Secondary Metabolism” (BB/J004561/1). We thank Günther Muth for kindly providing pGM1190.
References
- West S. C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 (2003). [DOI] [PubMed] [Google Scholar]
- Smithies O. Forty years with homologous recombination. Nat. Med. 7, 1083–1086 (2001). [DOI] [PubMed] [Google Scholar]
- Jacquier A. & Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell 41, 383–394 (1985). [DOI] [PubMed] [Google Scholar]
- Lu Z., Xie P. & Qin Z. Promotion of markerless deletion of the actinorhodin biosynthetic gene cluster in Streptomyces coelicolor. Acta Biochim. Biophys. Sin. (Shanghai). 42, 717–721 (2010). [DOI] [PubMed] [Google Scholar]
- Bierman M. et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992). [DOI] [PubMed] [Google Scholar]
- Baltz R. H. Genetic manipulation of antibiotic-producing Streptomyces. Trends Microbiol. 6, 76–83 (1998). [DOI] [PubMed] [Google Scholar]
- Cerdeño A. M., Bibb M. J. & Challis G. L. Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 8, 817–829 (2001). [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P. & Bibb M. J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F. & Hopwood D. A. Practical Streptomyces genetics. (John Innes Foundation, Norwich, U.K., 2000). [Google Scholar]
- Chiu M. L., Viollier P. H., Katoh T., Ramsden J. J. & Thompson C. J. Ligand-induced changes in the Streptomyces lividans TipAL protein imply an alternative mechanism of transcriptional activation for MerR-like proteins. Biochemistry 40, 12950–12958 (2001). [DOI] [PubMed] [Google Scholar]
- Muth G., Nuβbaumer B., Wohlleben W. & Pühler A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in Streptomycetes. Mol. Gen. Genet. 219, 341–348 (1989). [Google Scholar]
- Sambrook J., Fritsch E. F. & Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. (Cold Spring Harbor, NY, 1989). [Google Scholar]
- Gust B., O'Rourke S., Bird N., Kieser T. & Chater K. F. Recombineering in Streptomyces coelicolor. (John Innes Foundation, Norwich, U.K., 2003). [Google Scholar]
- Gust B. et al. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54, 107–128 (2004). [DOI] [PubMed] [Google Scholar]
- Rodríguez-García A., Combes P., Pérez-Redondo R., Smith M. C. A. & Smith M. C. M. Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces. Nucleic Acids Res. 33, e87 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T. & Hopwood D. A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 15, 2665–2675 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Holt T. G. & Thompson C. J. Thiostrepton-induced gene expression in Streptomyces lividans. J. Bacteriol. 171, 1459–1466 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R. & Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38, 215–226 (1985). [DOI] [PubMed] [Google Scholar]
- Sherwood E. J., Hesketh A. R. & Bibb M. J. Cloning and analysis of the planosporicin lantibiotic biosynthetic gene cluster of Planomonospora alba. J. Bacteriol. 195, 2309–2321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson J. S., Malpartida F. & Hopwood D. A. Genetic and biochemical characterization of the red gene cluster of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 131, 2431–2441 (1985). [DOI] [PubMed] [Google Scholar]
- Takano E. et al. Transcriptional regulation of the redD transcriptional activator gene accounts for growth-phase-dependent production of the antibiotic undecylprodigiosin in Streptomyces coelicolor A3(2). Mol. Microbiol. 6, 2797–2804 (1992). [DOI] [PubMed] [Google Scholar]
- MacNeil D. J. et al. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 115, 119–125 (1992). [DOI] [PubMed] [Google Scholar]