Abstract
Nonenveloped viruses are generally released from the cell by the timely lysis of host cell membranes. SV40 has been used as a model virus for the study of the lytic nonenveloped virus life cycle. The expression of SV40 VP4 at later times during infection is concomitant with cell lysis. To investigate the role of VP4 in viral release and its mechanism of action, VP4 was expressed and purified from bacteria as a fusion protein for use in membrane disruption assays. Purified VP4 perforated membranes as demonstrated by the release of fluorescent markers encapsulated within large unilamellar vesicles or liposomes. Dynamic light scattering results found that VP4 treatment did not cause membrane lysis or change the size of the liposomes. Liposomes encapsulated with bodipy-labeled streptavidin were used to show that VP4 formed stable pores in membranes. These VP4 pores had an inner diameter of between 1 and 5 nm. Asymmetrical liposomes containing pyrene-labeled lipids in the outer monolayer were employed to monitor transbilayer lipid diffusion. Consistent with VP4 forming toroidal pore structures in membranes, VP4 induced transbilayer lipid diffusion or lipid flip-flop. Altogether, these studies support a central role for VP4 acting as a viroporin in the disruption of cellular membranes to trigger SV40 viral release by forming toroidal pores that unite the outer and inner leaflets of membrane bilayers.
Newly assembled viral particles are released from the infected host cell to efficiently propagate the infection process. Enveloped viruses generally exit the host cell by a budding or membrane fission event (1, 2). In contrast, nonenveloped viruses are frequently released by the timely execution of the death of the host cell by a poorly defined cytolytic process (3, 4). Enveloped and nonenveloped viruses utilize viral encoded proteins termed viroporins to mediate membrane disruption during various stages of the viral life cycle including viral penetration and release (5). In general, viroporins contain one or two hydrophobic transmembrane domains and a basic amino acid cluster that supports their interaction with host cell membranes. They are commonly small hydrophobic proteins that oligomerize to form pores in host cell membranes. Examples of well studied viroporins include influenza virus M2 protein that acts as a proton conducting channel in the viral envelope that supports the acidification of the viral particle within endosomes to trigger viral penetration (6, 7), as well as assisting in the membrane fission process involved in viral release (8). The nonenveloped reovirus µ1N protein forms pores in endosomal membranes for release of the subviral particle into the cytoplasm (9). The adenovirus protein VI and poliovirus VP4 protein also disrupt endosomal membranes for viral penetration (10, 11). Nonenveloped viruses are generally released from their host cell through a lytic mechanism triggered by viroporins so that viruses are free of membranes as is the case with the blue tongue non-structural viral protein 3 (NS3) that increases membrane permeability of mammalian cells and is associated with release of viral particles (4).
Simian Vacuolating virus 40 (SV40) is a well-characterized polyomavirus that has been utilized as a paradigm for understanding the viral life cycle of nonenveloped viruses. SV40 appears to initiate cell lysis by expressing the late protein VP4 during the later stages of viral infection to support virus release (12–14). VP4 is a 125 amino acid protein expressed from a downstream Met codon in the VP2/3 transcript, therefore its sequence overlaps with the C-termini of both VP2 and VP3 (14). It possesses a central hydrophobic domain and a C-terminal nuclear localization sequence (NLS). VP4 is not found in the virus but rather acts directly on the host cell where it traffics to the nuclear envelope (12). In support of the role of SV40 VP4 as a viroporin, bacterially expressed and purified VP4 was shown to form pores in biological membranes (13). However, little is known about the mechanisms of membrane disruption utilized by viroporins.
Some eukaryotic cells secrete antimicrobial lytic peptides as a defense against microbial attack. Studies using these antimicrobial peptides have shown that they can disrupt bacterial membranes using three possible mechanisms (15). First, the barrel-stave model describes the formation of aqueous pores created by amphipathic alpha-helices integrated into the lipid bilayer. Second, the carpet model states that peptides accumulate on the membrane surface through electrostatic forces where positively charged amino acids bind anionic lipid head groups. At high concentrations, it is hypothesized that these peptides disrupt the membrane in a detergent-like manner resulting in the formation of peptide-lipid micelles. Finally, a toroidal pore model proposes that protein segments insert into the membrane and by interacting with the lipid head groups force curvatures in the interacting lipids, resulting in the fusion of the inner and outer leaflets at the lipid-protein interaction site to form a toroidal pore structure.
In this study, we investigated the mechanism SV40 VP4 employs to bind and disrupt lipid membranes. For this purpose, a tagged version of VP4 was constructed and its properties of membrane disruption were analyzed using model membranes. These studies showed that VP4 efficiently formed stable pores that perforated lipid membrane bilayers. Interestingly, VP4 was found to induce transbilayer lipid diffusion or lipid flip-flop consistent with VP4 acting as a viroporin by forming toroidal pores in membranes to initiate the dissemination of viral particles.
EXPERIMENTAL PROCEDURES
Reagents
The glutathione-S-transferase (GST)-Tag (12G8) mouse monoclonal antibody was purchased from Abmart (Arlington, MA). A pyrene-labeled lipid, 1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine (pyPC), was purchased from Life Technologies (Grand Island, NY). Additional phospholipids were obtained from Avanti Polar Lipids (Alabaster, AL). All other reagents were purchased from Sigma (St. Louis, MO).
DNA constructs
The pGEX-6P-1 plasmid (Amersham Bioscience; Piscataway, NJ) was modified to include a tobacco etch virus (TEV) protease site and a C-terminal 6xHis epitope upstream and downstream of the multiple cloning site, respectively, to create pGEX-6P-1-TEV-His. The GST-tagged version of full length VP4 (GST-TEV-VP4-His) was created by PCR cloning into the bacterial expression plasmid pGEX-6P-1-TEV-His using standard techniques (13). GST-tagged VP2 and VP3 were created by the same method as described for VP4. Phusion site-directed mutagenesis (New England BioLabs, Ipswich, MA) was used according to the manufacturer’s recommendations to generate hydrophobic domain (HD) point mutants L71K/L75E (VP4 K/E), L71D/L75D (VP4 D/D), and P70A. Site-directed mutagenesis of the nuclear localization sequence (NLS, KKKRK) to QAQGE was used to create VP4ΔNLS. The NLS sequence KKKRK was introduced at residues 51 to 55 or 113 to 117 in the VP4ΔNLS backbone to create VP4-NLS 51–55 or VP4-NLS 113–117, respectively. Mutagenesis was confirmed by sequencing.
Expression and purification of proteins
The BL21 E. coli Rosetta strain (DE3: pLysS) (Merck KGaA, Darmstadt, Germany) was transformed with GST-TEV-VP4-His and grown at 37 °C to an OD of 0.4 at 600 nm. NaCl (300 mM) was added to increase the osmolality of the nutrient medium. Simultaneous with the osmotic increase, the medium was supplied exogenously with 20 mM Pro and the culture was induced with 1 mM IPTG for 4 hr at 30 °C. Cells were centrifuged, suspended in phosphate buffered saline (PBS) (pH 7.4)/10 mM dithiothreitol (DTT) with 200 µg/ml lysozyme and protease inhibitors (1 mM phenylmethanesulfonylfluoride, PMSF), 1 µg/ml leupeptin, 10 µM pepstatin), and rotated for 30 min at 37 °C. Triton X-100 (1%) was added to the cells, which were then sonicated and the insoluble debris was sedimented by centrifugation for 20 min at 12,000 × g. The clarified supernatant was then added to glutathione sepharose 4B (GE healthcare) matrix pre-equilibrated with PBS (pH 7.4)/10 mM DTT/ 1% Triton X-100. The matrix was washed three times with PBS (pH 7.4), and the protein was eluted with freshly prepared 10 mM reduced glutathione (in 50 mM Tris-HCl, pH 8.0). The eluate from glutathione sepharose resin was further purified by addition to Ni-NTA His Bind resin (Merck KGaA) pre-equilibrated with PBS (pH 7.4)/10 mM imidazole and additional 150 mM NaCl. The matrix was washed three times with PBS (pH 7.4)/50 mM imidazole and the protein was eluted with 250 mM imidazole/PBS (pH 7.5). Protein purity was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and protein concentration was determined using a Bradford assay using BSA as a reference protein (Bio-Rad, Hercules, CA). The expression and purification of VP4 mutants and GST-TEV-His were performed similarly. Expression and purification of GST-TEV-VP2-His and GST-TEV-VP3-His was performed as described above with minor modifications. For expression of VP2 protein, expression was induced with 1 mM IPTG in the presence of 400 mM NaCl and 20 mM Pro for 3 h at 30 °C. Expression of VP3 was carried out at 16 °C for 12 h after inducing with 1 mM IPTG.
Liposome preparation
Large unilamellar vesicles (LUVs) or liposomes were generated using a Liposofast extruder (Avestin Inc., Ottawa, Canada)(16). Chloroform solutions of lipids PC (phosphatidylcholine) and PG (phosphoglycerol) were mixed in the respective ratios and chloroform was evaporated at 37 °C with a mild N2 flow. The lipid film was kept under vacuum for at least 3 hr to eliminate traces of the organic solvent. To hydrate the lipid mixture, 0.25 ml of Hepes buffer saline (HBS, pH 7.5) was added to the dried phospholipid mixture (final total lipid concentration 30 mM) and the samples were incubated for 30 min at 37 °C. The lipids were then suspended by vortexing. The suspended lipid mixtures were frozen in liquid N2 and thawed at 37 °C a total of five times to reduce the number of multilamellar liposomes and to enhance the trapped volumes of the vesicles (16). The samples were passed at room temperature (20–23 °C) through the extruder equipped with a 100 nm pore size polycarbonate filter a total of 21 times. The resulting liposomes were stored at 4 °C and used within 2 weeks of production. Liposomes containing the fluorophore, Terbium-Dipicolinic acid [Tb(DPA)33−], were prepared as above, except that HBS buffer included 3 mM TbCl3 (Alfa Aesar, Ward Hill, MA), and 9 mM 2,6-pyridinedicarboxylic acid (DPA, neutralized to pH 7.0). The resulting liposomes were separated from non-encapsulated [Tb(DPA)33−] by size-exclusion chromatography (sepharose CL-6B-200, 0.7 cm inner diameter × 50 cm) in HBS buffer. Liposomes encapsulating streptavidinBodipy were prepared in a similar manner with HBS buffer including 5 µM streptavidinBodipy conjugate.
Liposome permeabilization assay
Liposomes (100 µM total lipids) were suspended in 300 µl of buffer A (PBS, pH 7.4) containing 5 mM EDTA. The net initial emission intensity (F0) was determined after equilibration of the sample at 25 °C for 5 min. Protein was added to samples at 116 nM final concentration and incubated for 30 min at 37 °C. After re-equilibration at 25 °C, the final net emission intensity (Ff) of the sample was determined (i.e., after blank subtraction and dilution correction) and the fraction of marker quenched was estimated using (F0-Ff)/(F0-FT), where FT is the net emission intensity obtained when the same liposomes are treated with 3 mM Triton X-100 (i.e., under conditions of maximal release of the fluorophore).
Steady state fluorescence spectroscopy
Intensity measurements were performed using a Fluorolog 3–21 photon-counting spectrofluorimeter equipped with a double monochromator in the excitation light path, a single emission monochromator, cooled photo multiplier tube housing, a 450 W xenon lamp and temperature controlled sample holder. For pore formation activity assays employing [Tb(DPA)33−] liposomes, excitation/emission wavelengths were set to 278/544 nm and a 385 nm long pass filter was placed in the emission channel in order to block second-order harmonic light from passing through the emission monochromator. The band pass was typically 2 nm for excitation and 4 nm for emission. Measurements were done in 4 × 4 mm quartz microcells stirred with a 2 × 2 mm magnetic bar as described previously (16). For experiments using streptavidin-Bodipy samples were excited at 492 nm and the emission intensity was measured at 510 nm. Emission scans of pyrene containing samples were carried out at 1 nm intervals between 355 nm and 520 nm, excitation wavelength was 344 nm.
Dynamic light scattering
The average size of the liposomes, before and after incubation with VP4, was determined by dynamic light scattering (DLS). A Malvern Zetasizer Nano ZS instrument was used to measure hydrodynamic radius of LUVs at 25 °C. VP4 and liposome concentrations were the same as that employed for the liposome permeabilization assays.
Pore stability
The presence of stable pores formed by VP4 was assessed by measuring the ability of biocytin (~1 nm diameter) or biotin-labeled β-amylase (~5 nm diameter) to diffuse through the pores formed by VP4. Liposomes encapsulating streptavidinBodipy (~5 nm diameter) were treated with VP4, and diffusion of the biotin markers through the pores was detected as an increase in streptavidinBodipy fluorescence as described previously (15). Briefly, liposomes loaded with streptavidinBodipy (100 µM total lipids) were suspended in HBS buffer (pH 7.5) containing 1 µM biocytin or 100 nM biotin-labeled β-amylase. The net initial emission intensity (F0) was determined after equilibration of the sample at 25 °C for 5 min. VP4 was added at a final concentration of 232 nM, and samples were incubated at 37 °C for 30 min. After re-equilibration at 25 °C, the final net emission intensity (F) of the sample was determined (i.e., after blank subtraction and dilution correction) and the enhancement of streptavidinBodipy fluorescence emission was estimated using F/F0, which is proportional to the amount of biotinylated marker that can diffuse through membrane pores and bind to streptavidinBodipy. As a control, samples containing biotin-labeled β-amylase were treated with Triton X-100 after F had been recorded to disrupt membranes and determine the maximum binding between biotin-labeled β-amylase and streptavidinBodipy (not shown).
Measurement of transbilayer lipid diffusion (lipid flip-flop assay) using asymmetric LUVs
Transbilayer lipid diffusion or flip-flop was measured by using a method based on monitoring the leaflet dilution of a pyrene labeled PC analog (pyPC) as a result of transbilayer diffusion. Since the spectral properties of pyPC depend on the probe concentration at a membrane leaflet, transbilayer diffusion in the case of asymmetrically labeled liposomes results in detectable changes in the fluorescence emission (17). The fluorescence intensity emitted by the probe in the 397 nm spectral region (Im) is dominated by its monomeric form, while the fluorescence emission in the 479 nm region (Ie) is dominated by the excimeric (excited-state dimer) form. Since the excimer/monomer ratio is dependent on the concentration of the probe, a decrease in the ratio Ie/Im with respect to control is indicative of transbilayer probe diffusion. Asymmetrically and symmetrically pyrene-labeled LUVs were prepared using a method described by Muller et al. (18). To prepare asymmetrically pyrene-labeled LUVs, dried pyPC was dissolved in 100 µl of ethanol, and then further diluted up to 2 ml with HBS (pH 7.5)(5% final ethanol concentration) to form a pyPC micelle solution. The pyPC micelle solution (3% of the final product) and unlabeled LUVs were mixed at 37 °C with agitation for a few min, and the mixture was incubated for 6–8 h at 22 °C. This results in the probe partitioning into the outer monolayer at a concentration of approximately 5 mol % of the total lipid or 10 mol % of the outer monolayer. VP4-mediated lipid flip-flop was detected by acquiring fluorescence emission spectra in the 355 nm to 520 nm region with excitation at 344 nm. The emission intensity of pyrene for the excimer (Ie) and for the monomer fluorescence (Im) was recorded before and 10 min after addition of VP4 at two different wavelengths, 479 nm and 397 nm, respectively. The fluorescence spectra of symmetrically and asymmetrically pyrene-labeled LUVs served as positive and negative controls, respectively. When lipid flip-flop occurs in the presence of VP4, there is a reduction in the excimer-to-monomer ratio because of the dilution of the probe from one monolayer to two. The extent of flip-flop was estimated from the spectrum Ie/Im ratio. Values of the ratio Ie/Im were normalized to that of the LUVs alone. The symmetrically labeled LUVs were prepared by extrusion as described above, with the pyrene-labeled lipid mixed with the other lipids in organic solvent prior to preparing the LUVs.
Hemolysis assay
Bovine red blood cells (RBCs) were washed repeatedly in cold PBS immediately before use. Reactions (700 µl) were incubated at 37 °C in hemolysis buffer (PBS, pH 7.4, 1 mM DTT and 200 ng bovine serum albumen (BSA)) with 0.5% RBCs, without or with 232 nM or 464 nM minor viral protein or GST, unless otherwise noted. Samples were removed after 30 min for end-point assays. Reactions were centrifuged at 6,000 × g for 5 min at 4 °C to pellet unlysed cells. The absorbance of hemoglobin in the supernatant was measured at 414 nm. The percentage of hemolysis was calculated as [(A414 (sample) - A414 (blank))/(A414 (water) - A414 (blank))] × 100. The blank reaction contained all components except protein. As a 100% release control, RBCs were hypotonically lysed by adding 50% water.
RBC binding and integration
For determining RBC surface binding, RBCs were incubated with the proteins as described above at 37 °C for 30 min. The cells were then centrifuged at 6,000 x g for 5 min and the supernatant and pellet fractions were separated. Alkaline extractions were performed by suspending the cell pellet in 700 µl of ice-cold 0.1 M Na2CO3 (pH 11.5), followed by incubation on ice for 30 min. The solution was layered on top of a 100 µl sucrose cushion, and the membrane-bound fraction was isolated by ultracentrifugation for 20 min at 65,000 × g at 4 °C. The membrane pellet was suspended in sample buffer, and the supernatant containing the peripherally associated proteins was TCA precipitated, washed with cold acetone, and suspended in sample buffer for resolution by SDS-PAGE (12% acrylamide) and immunoblotting.
RESULTS
VP4 permeabilizes liposomal membranes
VP4 was expressed and purified from E. coli with GST and His tags added to its N and C termini, respectively, to optimize its solubility and facilitate purification (Figure S1 of the Supporting Information). Removal of the GST-tag by TEV proteolysis resulted in aggregation of VP4 (data not shown)(13), therefore the studies were carried out using the GST-tagged construct. A fluorescence-based membrane perforation assay was employed to determine if VP4 disrupted liposomes (Figure 1). The membrane perforation assay detected the release of liposome-encapsulated fluorophores when membranes were adequately disrupted. Liposomes were suspended in buffer containing quenchers of the encapsulated fluorophore. A decrease in the fluorescence intensity was observed when the fluorophore was released from the liposomes or the quencher was allowed to enter the liposome as a result of membrane disruption permitting contact between the quencher and the fluorophore (Figure 1A). In contrast, quenching was not observed if the membrane remained intact and the quencher and the fluorophore were unable to cross the membrane bilayer. This experimental system provides a highly tractable approach to characterize the membrane disruption properties of VP4.
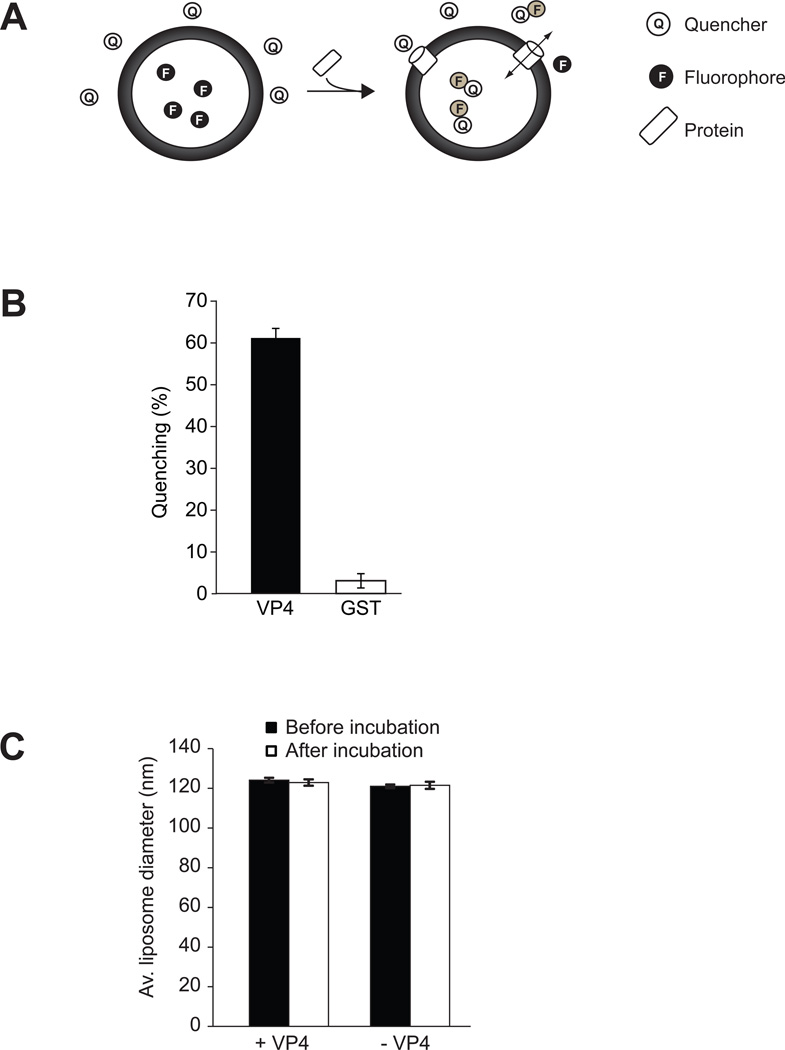
Figure 1. VP4 disrupts liposomal membranes.
(A) Scheme showing the liposome disruption assay employed. Membrane disruption was examined by encapsulating [Tb(DPA) 33−] fluorophore into LUVs. When these LUVs were suspended in a solution containing EDTA (quencher), protein mediated membrane disruption was monitored by the quenching of [Tb(DPA) 33−] emission as the encapsulated molecules were released, and terbium ions were chelated by EDTA. (B) VP4 disrupts LUVs. LUVs were prepared to examine the membrane disruption activity of VP4. Liposome disruption was evaluated using LUVs and the percentage of fluorophore quenched is indicated. Each data point shows the average of at least two independent measurements with error bars representing standard deviations. (C) Average diameter of LUVs before and after 30 min incubation with VP4 as determined by DLS. Mock LUVs were incubated in absence of any protein. Each data point shows the average of at least two independent measurements with error bars representing standard deviations.
Liposomes comprised of PC and PG were prepared as previously described (18–20). Mammalian membranes are highly enriched in PC and the cytoplasmic leaflet also contains negatively charged lipids such as PG, which are important for protein-lipid interactions. The ability of VP4 to disrupt these liposomes was tested. VP4 efficiently permeabilized liposomes comprised largely of PC (PC-PG 90:10) as 61% of the encapsulated fluorescence was quenched upon VP4 addition (Figure 1B). GST alone displayed no significant membrane disruption. The size of the liposomes was unaffected by VP4 treatment as dynamic light scattering found that the liposomes were ~123 nm in diameter before and after exposure to VP4 (Figure 1C). These results demonstrated that VP4 efficiently disrupted liposome membranes by forming pores in the membranes.
VP4 forms stable size-selective membrane pores
Liposome disruption by VP4 led to the question of whether VP4 formed transient or stable pores in membranes. To examine this issue closer, liposomes encapsulating a streptavidinBodipy fluorescent marker (~5 nm diameter for the monomer) were prepared. Pore stability was analyzed by employing a fluorescence-based spectroscopic assay, which detected the binding of streptavidinBodipy with the fluorescence enhancers biocytin (biotin moiety covalently attached to the amino acid Lys, ~1 nm diameter) or biotin attached to the protein β-amylase (~5 nm diameter). Binding of the biotin-labeled molecules (enhancers) to streptavidinBodipy produced a 2- or 3-fold enhancement in the fluorescence intensity of the streptavidinBodipy marker (21–23). No change in fluorescence was observed when the enhancers were exposed to the intact streptavidinBodipy encapsulated liposomes (data not shown). When the biotin-labeled molecules are added externally to the liposomes following incubation with VP4 for different time periods, fluorescence enhancement will be detected only if VP4 formed stable pores thereby allowing interaction of streptavidinBodipy and the biotin markers (Figure 2A). The size of the pore will dictate which molecules can cross the membrane. On the other hand, if VP4 does not form stable pores and closes after a brief period of time then the fluorescence will increase only when the enhancer is present before the addition of VP4, and not when added after the incubation.
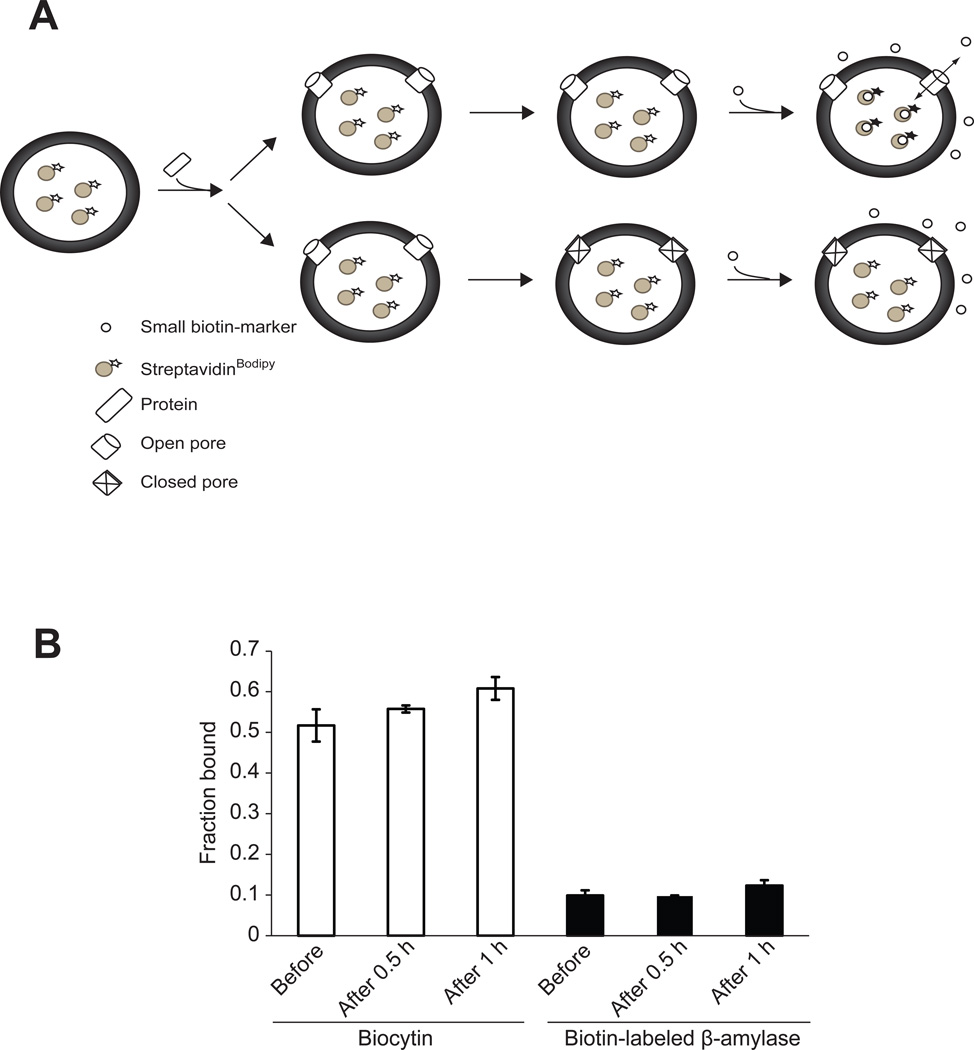
Figure 2. VP4 forms stable and size selective membrane pores.
(A) Scheme showing the pore stability assay employed. Pore stability was analyzed by employing a fluorescence-based spectroscopic assay, which detected the binding of streptavidinBodipy with fluorescence enhancers after the addition of the enhancer after incubation with VP4. The passage of biocytin (~1 nm diameter), biotin-labeled β-amylase (~5 nm diameter), or streptavidinBodipy (~5 nm diameter) through the pores formed by GST-VP4 was measured as detailed in Experimental Procedures. When the liposomes encapsulating streptavidinBodipy are used and the biotin-labeled molecules are externally added following incubation with VP4, fluorescence enhancement will be detected only if VP4 formed stable pores thereby allowing interaction of streptavidinBodipy and the biotin markers (top scenario). On the other hand, if VP4 does not form stable pores and closes after a brief period of time then the fluorescence will not increase when the enhancer is added after incubation with GST-VP4 (bottom scenario). The size of the pore will dictate which molecules can cross the membrane. (B) The stability of the pores was examined by measuring the increase in the fluorescence intensity of encapsulated streptavidinBodipy when the enhancers biocytin or biotin-labeled β-amylase were present in the external buffer solution either before the addition of VP4, added after incubation for 0.5 hr or 1 hr with VP4. Only biocytin was able to diffuse through the formed pores. The pores formed by VP4 remained open after incubation for 1 hr. The total lipid concentration was 100 µM, and the concentration of the protein was 232 nM. Each data point shows the average of at least two independent measurements with error bars representing standard deviations.
The addition of VP4 resulted in an increase in fluorescence intensity only for the biocytin samples, and not for the biotin-labeled β-amylase samples (Figure 2B). Therefore, pores formed by VP4 had a diameter larger than 1 nm, but were of insufficient size to support the translocation of the biotin-labeled β-amylase or streptavidinBodipy across the liposomal membrane. These results are consistent with our earlier observation that VP4 formed ~3 nm diameter pores in biological membranes (13). The VP4 dependent fluorescence intensity increased when biocytin was present before the addition of VP4 or when it was added after the addition of the protein (0.5 hr and 1 hr, Figure 2B). This indicated that the VP4 pores were stable and remained open for at least 1 hr.
VP4 induces transbilayer lipid diffusion
Since VP4 forms discrete size pores in liposomes that did not alter the diameter of the liposomes, the carpet model of membrane disruption could be ruled out as the mechanism for membrane disruption by VP4. To delineate between the barrel-stave and toroidal pore mechanisms of membrane disruption, a fluorescence-based lipid flip-flop assay was utilized. This method is based on the dilution of the pyrene-lipid probe (pyPC) as a result of transbilayer diffusion. Lipid flip-flop is detected by determining if asymmetrically pyrene-labeled large unilamellar vesicles become symmetrical after VP4 treatment due to transbilayer diffusion. If VP4 behaves as a barrel-stave pore, the addition of VP4 to asymmetric LUVs will result in no significant movement of pyrene probe between the two leaflets of LUVs (Figure 3A, top scheme). However, formation of toroidal pores by VP4 will result in transbilayer pyrene-lipid probe movement from outer leaflet to the inner leaflet of the membrane bilayer leading to overall dilution of the fluorescence probe (Figure 3A, lower scheme).
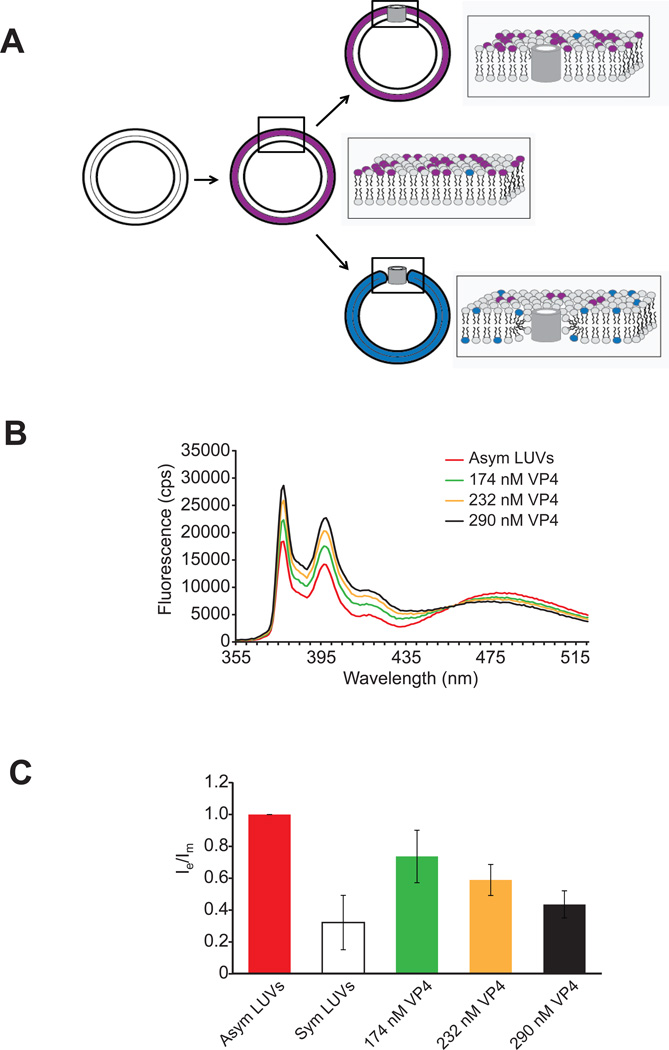
Figure 3. VP4 induces lipid flip-flop.
(A) Scheme showing the lipid flip-flop assay employed. The method is based on the dilution of the pyrene probe (pyPC) as a result of transbilayer diffusion. Lipid flip-flop was detected by using symmetrically and asymmetrically pyrene-labeled large unilamellar vesicles (LUVs). Addition of VP4 to asymmetric LUVs will result in no significant dilution of the pyrene probe from one leaflet to the other leaflet of LUVs if barrel-stave pores are formed (top). However, formation of toroidal pores by VP4 will result in transbilayer lipid movement resulting in dilution of the pyrene probe from outer leaflet to the inner leaflet of the membrane bilayer (bottom). The fluorescence spectrum of pyrene is characterized by two signals, one arising from monomer molecules (shown in blue) and the other from excited dimer (excimer) molecules (shown in purple). After incorporation of pyrene probe into the outer leaflet of membrane, the redistribution of pyrene to the inner leaflet is accompanied by a change of the analogue concentration in each leaflet and, thus, by a change of the intensity ratio between the excimer and the monomer signals (Ie/Im). (B) Fluorescence spectra of pyrene-labeled LUVs (PC-PG-pyPC, 87:10:3). The lipid concentration of the pyrene-labeled LUVs was 100 µM. (C) The ratio of transbilayer diffusion of pyrene probe (pyPC) is shown. A decrease in the ratio of Ie to Im reflects transbilayer diffusion of pyPC. The ratio Ie/Im has been normalized to a value of 1 with asymmetric LUVs alone. The extent of the transbilayer diffusion was negligible for LUVs in the absence of protein. The error bars correspond to standard deviations of measurements carried out in three different batches of liposomes.
The fluorescence spectral features of pyrene are sensitive to its microenvironment and oligomeric state. The pyrene probe exhibits a signature monomer fluorescence emission peak (397 nm) and the intensity of this peak reports on the overall number of monomeric probes present. An additional spectral band for pyrene excimers (excited state dimers) is also observed at a longer wavelength (479 nm)(24). Dilution of pyrene by its movement from one leaflet to the other will result in an increase in monomers and a decrease in excimers. Transbilayer lipid movement can be monitored by measuring the decrease and increase in fluorescence intensities of excimer (Ie) and monomer (Im) populations, respectively, by calculating the ratios of their intensities.
Lower Im and higher Ie values were observed in the spectrum of the asymmetrically pyrene-labeled LUVs when compared with the spectrum of samples treated with VP4 (Figure 3B and C). For symmetrically pyrene-labeled LUVs, a high fluorescence intensity of the monomer (Im) at 397 nm and a low fluorescence intensity of the excimer (Ie) at 479 nm were observed when compared with the spectrum of samples treated with VP4. Incubation with increasing concentrations of VP4 led to the gradual enhancement of Im and a reduction of Ie in the asymmetrically pyrene-labeled LUVs. These changes in pyrene fluorescence indicate that lipid flip-flop was induced by VP4 in a concentration dependent manner consistent with VP4 forming a toroidal pore in the LUV membranes.
VP4 mutations abolish transbilayer lipid diffusion
VP4 possesses two domains that control its subcellular localization: a central hydrophobic domain (HD) and a C-terminal NLS (12). To explore the requirement of these domains for transbilayer lipid diffusion by VP4, purified VP4 with mutations in these regions were used to determine their ability to perforate membranes and support lipid flip-flop. Purified VP4 mutants showed a single band on SDS-PAGE confirming the absence of any visible contaminants from E. coli (Figure S1 of the Supporting Information).
Charged residues were incorporated within the HD by altering hydrophobic amino acids and the mutant proteins were tested for their membrane perforation activity using LUVs encapsulating [Tb(DPA)33−]. The addition of two charged residues [L71D/L75D (VP4 D/D) or L71K/L75E (VP4 K/E)] abolished membrane disruption activity, suggesting that the overall hydrophobicity of the domain was important for the perforation activity of VP4 (Figure 4A).
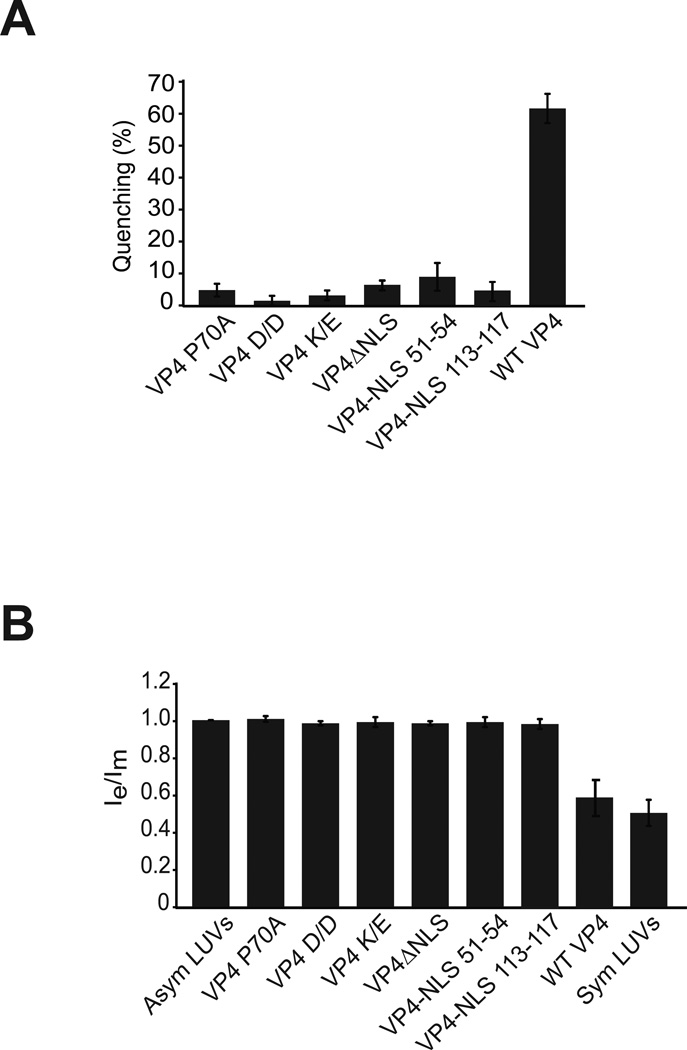
Figure 4. Mutating VP4 abolishes transbilayer lipid diffusion.
(A) VP4 mutants are deficient in disruption of LUVs. LUVs were prepared to examine the membrane disruption activity of VP4 and its mutants as in Figure 1. Liposome disruption was evaluated and the percentage of fluorophore quenched is indicated. Each data point shows the average of at least two independent measurements with error bars representing standard deviations. (B) The ratio of transbilayer diffusion of pyrene probe (pyPC). The lipid concentration of the pyrene-labeled LUVs was 100 µM. The ratio Ie/Im has been normalized to a value of 1 with asymmetric LUVs alone. The extent of the transbilayer diffusion was negligible for LUVs in the absence of protein. The error bars correspond to standard deviations of measurements carried out with three different batches of liposomes.
Sequence alignment of the minor structural protein VP2 from the Orthopolyomavirus genus showed high conservation of Pro in the last hydrophobic domain (shared by VP4) suggesting that these residues might be critical for its cellular activity (12). To test the role of the Pro for membrane disruption activity of VP4, the second Pro in the hydrophobic domain (Pro 70) was substituted with Ala (P70A). The fluorescence quenching was found to be 5% in the case of P70A mutant compared to about ~60% for the wild type VP4 (Figure 4A). This indicated that the central HD Pro was required for the membrane perforation activity of VP4.
Mutating amino acids KKKRK of the NLS to QAQGE (ΔNLS) drastically reduced the membrane disruptive activity of VP4 (Figure 4A). The proximal spacing of the basic cluster of amino acids that comprise the NLS was largely conserved in Orthopolyomaviruses (12). This led us to question whether this orientation and spacing of the NLS and HD were crucial for membrane perforation. The NLS was introduced eight residues upstream of the HD (NLS 51–55) or 29 residues carboxyl to the HD (NLS 113–117). Purified proteins were tested for membrane disruption of [Tb(DPA)33−] encapsulated LUVs. Interestingly, neither mutant rescued the membrane disruption activity of VP4 (Figure 4A). This implied that the basic cluster of residues in mutants NLS 51–55 and NLS 113–117 could not form a functional structure capable of perforating membranes. Similar observations were found using different lipid compositions of LUVs suggesting that VP4 NLS position mutants were deficient in membrane perforation activity (Figure S2 of the Supporting Information).
Consistent with the membrane perforation assay using [Tb(DPA)33−] encapsulated LUVs, the VP4 mutants did not show any lipid flip-flop activity (Figure 4B). The VP4 mutations adding charged residues to the HD, mutating the HD Pro residue or the NLS deletion and positioning mutants were all deficient in supporting lipid flip-flop as observed by excimer and monomer fluorescence intensity ratios. Together these results indicated that the hydrophobicity of the HD along with the spacing of the basic charged NLS and the central Pro were essential for membrane disruption and supporting lipid flip-flop between membrane leaflets of the LUVs.
VP2 and VP3 do not possess lipid flip-flop activity
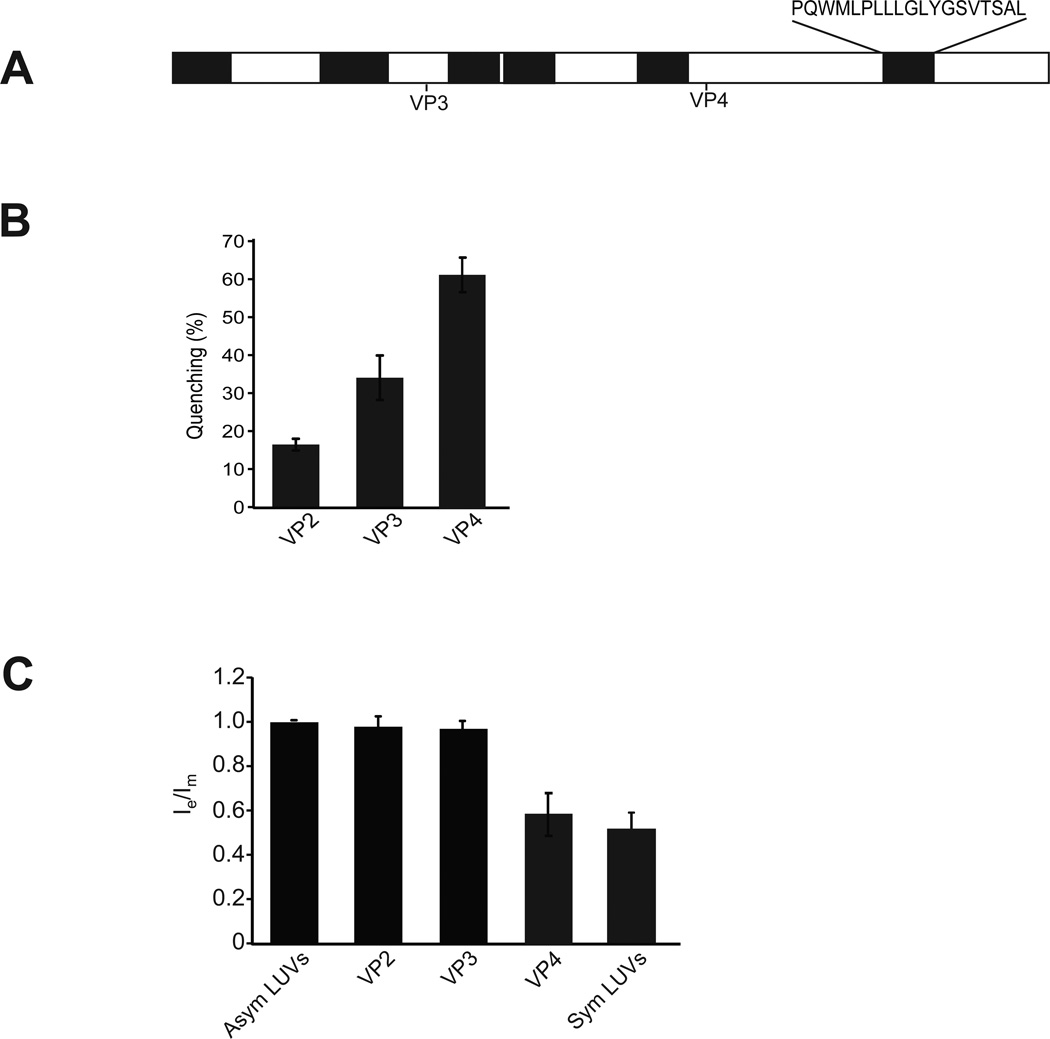
The viral minor structural proteins VP2 and VP3 serve critical roles in the SV40 viral life cycle, and these roles involve their ability to interact with host cell membranes (25–27). The mechanism of action of VP2 and VP3 is, however, poorly defined. The VP4 sequence is entirely contained within VP2 and VP3 (Figure 5A). To investigate if VP2 or VP3 possess liposome disruption activity, each protein was expressed and purified from E. coli. The isolated proteins were probed for their ability to disrupt LUVs encapsulating [Tb(DPA)33−]. Incubation of LUVs with purified VP2 resulted in about 15% of fluorescence quenching (Figure 5B) while VP3 displayed double the activity (Figure 5B). This suggested that VP2 and VP3 both have the ability to disrupt membranes but at diminished levels compared to VP4. To determine if VP2 and VP3 have lipid flip-flop activity, asymmetrical pyrene-labeled LUVs were incubated with the proteins. Neither VP2 nor VP3 caused a redistribution of the pyrene-labeled lipids when added to asymmetric labeled vesicles, suggesting that membrane disruption by these proteins occurred by a different mechanism than the one described for VP4 (Figure 5C). Altogether, this indicated that while VP2 and VP3 can perforate the LUV membranes, they appear to disrupt membranes using a different mechanism than VP4, as lipid flip-flop is not observed.
Figure 5. VP2 and VP3 do not possess lipid flip-flop activity.
(A) Schematic representation of the SV40 protein VP2 displaying predicted hydrophobic domains (27). These hydrophobic domains, which were identified by using a variety of algorithms, may act as transmembrane domains. Start sites for VP3 and VP4 are indicated. The amino acid sequence of the last HD that is shared by VP2, VP3 and VP4 is indicated. (B) VP2 and VP3 show membrane disruptive activity. The membrane disruption activity of VP2, VP3 and VP4 was examined as in Figure 1. The percentage of fluorophore quenched is indicated. Each data point shows the average of at least two independent measurements with error bars representing standard deviations. (C) The ratio of transbilayer diffusion of pyrene probe (pyPC) was determined as in Figure 3. The lipid concentration of the pyrene-labeled LUVs was 100 µM. The ratio Ie/Im has been normalized to a value of 1 with asymmetric LUVs alone. The extent of the transbilayer diffusion was negligible for LUVs in the absence of protein. The error bars correspond to standard deviations of measurements carried out with three different batches of liposomes.
VP2 and VP3 integrate into the lipid bilayer unlike VP4
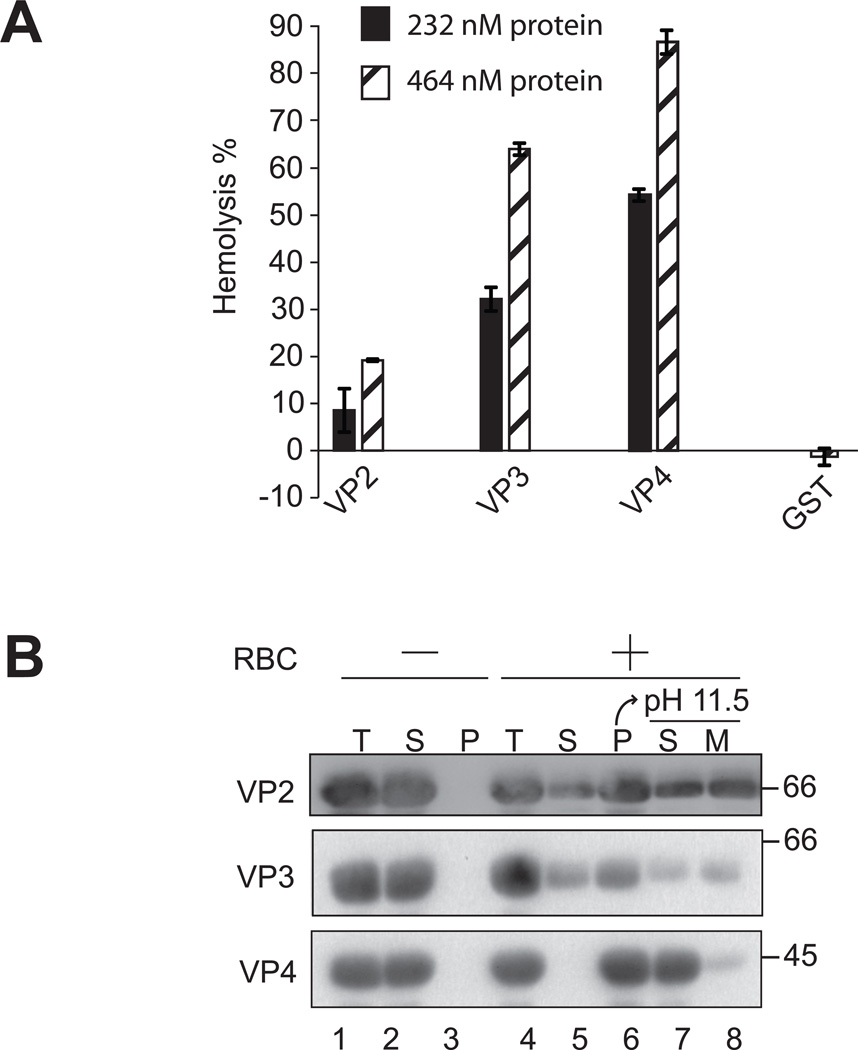
To investigate the differences in VP2, VP3 and VP4 membrane binding and disruption, a hemolysis assay was employed using bovine red blood cells (RBCs). RBCs constitute a simple and efficient model system to study protein-membrane interactions and lysis. RBCs were treated with purified VP2, VP3 or VP4 at 37 °C for 30 min. The level of hemolysis was measured by determining the fraction of hemoglobin released into the supernatant after centrifugation. GST alone did not display membrane disruptive activity. VP2 showed ~20% hemolysis compared to about ~60% hemolysis using VP3 (Figure 6A). VP4 possessed the most efficient hemolysis activity with ~90% hemolysis observed using equimolar protein concentration.
Figure 6. VP2 and VP3 integrate into the lipid bilayer unlike VP4.
(A) VP2, VP3, and VP4 possess hemolytic activity. VP2, VP3, and VP4 were incubated with bovine RBCs for 30 min at 37 °C. Released hemoglobin was measured by the A414 of the supernatant after centrifugation and the removal of unlysed cells. GST was used as a control to rule out its contribution to the hemolytic activity of minor viral proteins. The error bars represent the standard deviation from three independent experiments. (B) Hemolysis reaction mixtures (lane 4, Total) containing bovine RBCs and viral proteins were incubated at 37 °C for 30 min. RBC bound (lane 6, Pellet) and unbound (lane 5, Supernatant) proteins were separated by centrifugation. Membrane fractions (lane 6) were alkaline extracted with 0.1 M Na2CO3, pH 11.5 and ultracentrifuged to separate the soluble (S) and membrane (M) fractions (lane 7 and 8). Samples resolved by reducing SDS-PAGE were immunoblotted with antibody against GST. Separate reactions were performed in the absence of RBCs (lanes 1–3).
A cell-binding assay was employed to determine if the late viral proteins interact with cell membranes. VP2, VP3 and VP4 were separately incubated without and with RBCs for 30 min at 37 °C, and bound and unbound fractions were separated by centrifugation. Cell binding was determined by the amount of protein that sedimented with the cells. In the absence of RBCs, all of the proteins tested remained soluble and were therefore found in the supernatant (Figure 6B, lanes 2 and 3). However, in the presence of RBCs, ~85% of VP2, 60% of VP3 and 100% of VP4 were localized to the red blood cell pellets indicative of the binding of proteins to RBCs (Figure 6B, lane 6).
To determine if the bound protein was integrated into the lipid bilayer of the cells, the bound fractions were alkaline extracted with membrane and soluble fractions separated by ultracentrifugation (12, 13). Interestingly, ~65% of the membrane associated VP2 was found in the membrane pellet after alkaline extraction and centrifugation (Figure 6B, lane 8). This suggested that more than half of VP2 was integrated into the lipid bilayer. Similarly, ~60% of the membrane associated VP3 was found in the membrane pellet. Unlike VP2 and VP3, the majority of the membrane associated VP4 (~90%) was found in the supernatant after alkaline extraction and centrifugation (Figure 6B, lane 7 compared to lane 8). This suggested that VP4 was not fully integrated into the lipid bilayer. Together, these results suggested that VP2 and VP3 have different mechanism of membrane binding and disruption when compared to VP4.
DISCUSSION
Viroporins are a rapidly growing class of viral-encoded proteins that interact with host cell membranes to create pores, however relatively little is known about how these proteins form pores and assist the viral life cycle (5). We found that the SV40 late protein VP4, which is expressed in the host cell but is not a part of the mature virus particle (14), forms stable and size selective pores of 1–5 nm that disrupt model membranes. As VP4 membrane binding supports transbilayer lipid diffusion or lipid flip-flop, VP4 appears to form toroidal pores within the membrane bilayer of liposomes. The toroidal pore and membrane perforation activity of VP4 require an extended HD inclusive of a central Pro residue, as well as a C-terminally positioned positive charged NLS.
Based on several lines of evidence, our examination of purified VP4 demonstrates that VP4 acts as a viroporin. First, VP4 perforates liposomes as demonstrated by the ability of liposome-encapsulated fluorophores to be quenched by externally added reagents. Second, it forms stable pores that are present for at least an hr after membranes are exposed to VP4. Third, the VP4 pores are larger than 1 nm but smaller than 5 nm as they support the translocation of biocytin (1 nm diameter) but not biotin-labeled β-amylase or streptavidinBodipy (both have diameters of ~5 nm). Fourth, VP4 possess several characteristics that are representative of viroporins including its small size, central HD and positively charged cluster of amino acids. And finally, VP4 supports transbilayer lipid diffusion or lipid flip-flopping. Altogether, these results support the conclusion that VP4 is a viroporin that produces a toroidal pore structure in membranes.
There are three proposed models for the disruption of membranes by peptides: the carpet; barrel-stave; and toroidal pore models. The carpet model could be excluded as VP4 formed size selective pores in liposomes and the overall size of the liposomes were unaffected by VP4 treatment. Our results were also inconsistent with VP4 forming a barrel-stave pore structure in the membrane as VP4 was extracted from membranes upon alkaline treatment, suggesting that it was not fully integrated into the membrane bilayer as a traditional alpha-helical hydrophobic transmembrane segment. Rather, the loose association of VP4 with membranes that supported its alkaline extraction from membranes and its ability to initiate lipid transbilayer diffusion are both supportive of VP4 formation of toroidal pore structures. A toroidal structure that unites the lipid bilayer requires positive lipid curvature. Our earlier studies found that the lytic activity of VP4 was inhibited by phosphatidylethanolamine, a conical lipid that induces negative lipid curvature (13).
Alternatively, VP4 might be present in membranes in equilibrium of multimeric and monomeric species. The oligomeric structure that supports pore formation might be involved in solute leakage, while monomers might induce lipid transport from one leaflet to another. Proteinaceous lipid translocators have been postulated as biological membranes are generally asymetrical and phospholipid transbilayer movement is slow in lipid only bilayers; however, the identification of specific translocases has been elusive. From studies using membrane proteins and model peptides in lipid bilayers, some results indicate that monomeric single membrane-spanning proteins are most effective at inducing lipid flip-flop while others suggest that polytopic membrane proteins are able to flip lipids with more specificity (28–31). Proteins that appear to have flippase activity include leader peptidase, the potassium channel KcsA, opsin from photoreceptor discs and some SNARE proteins. The possibility that a monomeric form of VP4 could have lipid flippase activity will require further exploration.
Some antimicrobial peptides are known to kill microbes by forming toroidal pores in their membranes. The antimicrobial peptide magainin 2 is proposed to form toroidal pores in membranes that connect the outer and inner leaflets of the membrane through peptide-mediated bending of lipid monolayers (32). Colicin E1 is cytotoxic toward E. coli as it forms lipidic pores that depolarize the cytoplasmic membrane (33). Lipids having negative spontaneous curvature inhibit the formation of pores by colicin E1. This inhibition of pore formation by negative curvature inducing lipid was also observed with VP4 (13). Colicin E1 induces lipid flip-flop and the decrease in anion selectivity of the channel in the presence of negatively charged lipids, which implies a significant contribution of lipid to the structure of the channel. Bax protein is a pro-apoptotic Bcl-2 family member that activates pore formation in the outer mitochondria membrane to release cytochrome c. It has been shown to perturb membranes in a manner similar to that observed for antimicrobial peptides. The insertion of Bax into a membrane results in increased membrane conductivity and release of varying size particles due to the formation of lipidic or toroidal pores (20, 34). The association of the Bax protein with the membrane has also been described as an equilibrium between soluble and membrane-bound Bax (35), however this is unlikely the case for VP4 as VP4 was found predominantly in membrane fractions using a sedimentation assay (Figure 6B).
During the viral life cycle, VP4 is expressed 60 hr post infection (14). Its NLS supports its trafficking to the nucleus, where its positive charges combined with the hydrophobic domain initiate VP4 interaction with the nuclear membrane (12). The presence of a 1–5 nm pore in the inner nuclear membrane would provide a conduit that connects the nucleoplasm with nuclear membrane space and the contiguous endoplasmic reticulum lumen. Further studies will be required to determine how these membrane pores affect the nucleoplasm and lead to nuclear and cell membrane lysis. For instance, since the endoplasmic reticulum lumen is oxidizing and possesses high levels of free calcium; do the pores support elevations in the nucleoplasm calcium levels and its oxidation? This is of special interest as SV40 particles assemble in the nucleus and are stabilized by calcium and interpentameric disulfide bonds (36–38).
The entire sequence of VP4 is found in VP2 and VP3. VP2 and VP3 are also lytic proteins; however they did not support transbilayer diffusion of the labeled lipid probe when added to asymmetrical liposomes. If VP4 forms toroidal pores, why don’t VP2 or VP3? VP2 and VP3 were alkaline extractable from membranes to a much lesser extent than VP4, suggesting that the additional N-terminal HDs found in these proteins might interact with membranes as transmembrane segments integrated across the lipid bilayer. The membrane integration of N-terminal HDs could inhibit the ability of the C-terminal HD to form a toroidal structure observed in the context of VP4. Alternatively, a hybrid structure could be formed that involves both barrel stave and toroidal structures, however the barrel stave domains would inhibit the transbilayer diffusion of labeled lipids monitored by this assay. Structural approaches such as solid state NMR will be required to provide a better understanding of the structures that these late viral proteins form in membranes.
The small genome size for viruses such as polyomaviruses requires the gene products generated to be versatile to support the molecular and cellular gymnastics required to infect a cell. In the case of SV40, a single transcript encodes for VP2, VP3 and VP4 by initiating translation from three consecutive in-frame Met residues resulting in the production of three proteins with identical C-termini. In the context of VP4, the C-terminal HD supports transbilayer diffusion, which is consistent with the formation of a toroidal pore. A toroidal pore structure is previously associated with bacterial lysis by antimicrobial peptides (32, 39). Future studies will be required to increase our knowledge on the mechanism of action of viroporins, a rapidly expanding class of viral proteins. As viroporins play essential roles in the viral life cycle that require membrane perforation or disruption, they provide intriguing targets for viral therapies. In addition, directing their lytic effects towards aberrant cells may be used in oncolytic approaches to kill cancerous cells since these viroporins have evolved strategies to disrupt mammalian cells.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge members of the Hebert and Heuck labs for helpful discussions. We would also like to thank the Baldwin lab (UMass, Amherst, MA) for supplying the bovine RBCs.
FUNDING SOURCE STATEMENT
This work was supported by US Public Health grant (AI078142 and GM086874 to DNH) and an American Lung Association grant (RG-71111-N to APH). KMG was partially supported by National Science Foundation, Integrative Graduate Education and Research Traineeship (IGERT), Institute for Cellular Engineering (DGE-0654128), and both KMG and FBR were partially supported by National Institutes of Health Chemistry-Biology Interface training grant (T32GM00815).
ABBREVIATIONS
- Bodipy
4,4-difluoro-5,7-dimethyl-4-bora-3a,4a–diaza-3-indacene
- DTT
dithiothreitol
- DLS
dynamic light scattering
- DPA
2,6-pyridinedicarboxylic acid or dipicolinic acid
- EDTA
ethylenedinitrilotetraacetic acid
- GST
glutathione-S-transferase
- HBS
hepes buffer saline
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid
- Ie/Im
excimer:monomer emission intensity ratio
- IPTG
isopropyl α-D-thiogalactopyranoside
- LB
Luria-Bertani
- LUVs
large unilamellar vesicles
- NLS
nuclear localization sequence
- PC
phosphatidylcholine
- PG
phosphoglycerol
- PMSF
phenylmethanesulfonylfluoride
- pyPC
1-hexadecanoyl-2-(1-pyrenedecanoyl)-sn-glycero-3-phosphocholine
- RBC
red blood cell
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SV40
Simian Vacuolating virus 40
Footnotes
SUPPORTING INFORMATION
Figures S1 showing SDS-PAGE analysis of two-step affinity purified GST tagged SV40 VP4 and its mutants. Figure S2 showing liposome permeabilization and lipid flip-flop activity of VP4 and its mutants. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 2.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldabe R, Barco A, Carrasco L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem. 1996;271:23134–23137. doi: 10.1074/jbc.271.38.23134. [DOI] [PubMed] [Google Scholar]
- 4.Han Z, Harty RN. The NS3 protein of bluetongue virus exhibits viroporin-like properties. J Biol Chem. 2004;279:43092–43097. doi: 10.1074/jbc.M403663200. [DOI] [PubMed] [Google Scholar]
- 5.Nieva JL, Madan V, Carrasco L. Viroporins: structure and biological functions. Nat Rev Microbiol. 2012;10:563–574. doi: 10.1038/nrmicro2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:1–20. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 7.Pielak RM, Chou JJ. Influenza M2 proton channels. Biochim Biophys Acta. 2011;1808:522–529. doi: 10.1016/j.bbamem.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agosto MA, Ivanovic T, Nibert ML. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc Natl Acad Sci U S A. 2006;103:16496–16501. doi: 10.1073/pnas.0605835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danthi P, Tosteson M, Li QH, Chow M. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J Virol. 2003;77:5266–5274. doi: 10.1128/JVI.77.9.5266-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giorda KM, Raghava S, Hebert DN. The Simian Virus 40 Late Viral Protein VP4 Disrupts the Nuclear Envelope for Viral Release. J Virol. 2012;86:3180–3192. doi: 10.1128/JVI.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raghava S, Giorda KM, Romano FB, Heuck AP, Hebert DN. The SV40 Late Protein VP4 Is a Viroporin that Forms Pores to Disrupt Membranes for Viral Release. PLoS Pathog. 2011;7:e1002116. doi: 10.1371/journal.ppat.1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels R, Sadowicz D, Hebert DN. A Very Late Viral Protein Triggers the Lytic Release of SV40. PLoS Pathog. 2007;3:e98. doi: 10.1371/journal.ppat.0030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 16.Heuck AP, Tweten RK, Johnson AE. Assembly and topography of the prepore complex in cholesterol-dependent cytolysins. J Biol Chem. 2003;278:31218–31225. doi: 10.1074/jbc.M303151200. [DOI] [PubMed] [Google Scholar]
- 17.Vanderkooi JM, Callis JB. Pyrene. A probe of lateral diffusion in the hydrophobic region of membranes. Biochemistry. 1974;13:4000–4006. doi: 10.1021/bi00716a028. [DOI] [PubMed] [Google Scholar]
- 18.Muller P, Schiller S, Wieprecht T, Dathe M, Herrmann A. Continuous measurement of rapid transbilayer movement of a pyrene-labeled phospholipid analogue. Chem Phys Lipids. 2000;106:89–99. doi: 10.1016/s0009-3084(00)00134-1. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama F, Imura Y, Ohno K, Zendo T, Nakayama J, Matsuzaki K, Sonomoto K. Peptide-lipid huge toroidal pore, a new antimicrobial mechanism mediated by a lactococcal bacteriocin, lacticin Q. Antimicrob Agents Chemother. 2009;53:3211–3217. doi: 10.1128/AAC.00209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epand RF, Martinou JC, Montessuit S, Epand RM. Transbilayer lipid diffusion promoted by Bax: implications for apoptosis. Biochemistry. 2003;42:14576–14582. doi: 10.1021/bi035348w. [DOI] [PubMed] [Google Scholar]
- 21.Nicol F, Nir S, Szoka FC., Jr Orientation of the pore-forming peptide GALA in POPC vesicles determined by a BODIPY-avidin/biotin binding assay. Biophys J. 1999;76:2121–2141. doi: 10.1016/S0006-3495(99)77368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosconi MP, Zhao G, London E. Analyzing topography of membrane-inserted diphtheria toxin T domain using BODIPY-streptavidin: at low pH, helices 8 and 9 form a transmembrane hairpin but helices 5–7 form stable nonclassical inserted segments on the cis side of the bilayer. Biochemistry. 2004;43:9127–9139. doi: 10.1021/bi049354j. [DOI] [PubMed] [Google Scholar]
- 23.Romano FB, Rossi KC, Savva CG, Holzenburg A, Clerico EM, Heuck AP. Efficient isolation of Pseudomonas aeruginosa type III secretion translocators and assembly of heteromeric transmembrane pores in model membranes. Biochemistry. 2011;50:7117–7131. doi: 10.1021/bi200905x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bains G, Patel AB, Narayanaswami V. Pyrene: a probe to study protein conformation and conformational changes. Molecules. 2011;16:7909–7935. doi: 10.3390/molecules16097909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daniels R, Rusan NM, Wadsworth P, Hebert DN. SV40 VP2 and VP3 Insertion into ER Membranes Is Controlled by the Capsid Protein VP1: Implications for DNA Translocation out of the ER. Mol Cell. 2006;24:955–966. doi: 10.1016/j.molcel.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 26.Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol. 2011;13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- 27.Giorda KM, Raghava S, Zhang MW, Hebert DN. The viroporin activity of the minor structural proteins VP2 and VP3 is required for SV40 propagation. submitted. 2012 doi: 10.1074/jbc.M112.428425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kol MA, de Kroon AI, Killian JA, de Kruijff B. Transbilayer movement of phospholipids in biogenic membranes. Biochemistry. 2004;43:2673–2681. doi: 10.1021/bi036200f. [DOI] [PubMed] [Google Scholar]
- 29.Langer M, Sah R, Veser A, Gutlich M, Langosch D. Structural properties of model phosphatidylcholine flippases. Chem Biol. 2013;20:63–72. doi: 10.1016/j.chembiol.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Sanyal S, Menon AK. Flipping lipids: why an' what's the reason for? ACS Chem Biol. 2009;4:895–909. doi: 10.1021/cb900163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon I, Huber T, Sanyal S, Banerjee S, Barre P, Canis S, Warren JD, Hwa J, Sakmar TP, Menon AK. Opsin is a phospholipid flippase. Curr Biol. 2011;21:149–153. doi: 10.1016/j.cub.2010.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee MT, Hung WC, Chen FY, Huang HW. Many-body effect of antimicrobial peptides: on the correlation between lipid's spontaneous curvature and pore formation. Biophys J. 2005;89:4006–4016. doi: 10.1529/biophysj.105.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobko AA, Kotova EA, Antonenko YN, Zakharov SD, Cramer WA. Lipid dependence of the channel properties of a colicin E1-lipid toroidal pore. J Biol Chem. 2006;281:14408–14416. doi: 10.1074/jbc.M513634200. [DOI] [PubMed] [Google Scholar]
- 34.Terrones O, Antonsson B, Yamaguchi H, Wang HG, Liu J, Lee RM, Herrmann A, Basanez G. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J Biol Chem. 2004;279:30081–30091. doi: 10.1074/jbc.M313420200. [DOI] [PubMed] [Google Scholar]
- 35.Satsoura D, Kucerka N, Shivakumar S, Pencer J, Griffiths C, Leber B, Andrews DW, Katsaras J, Fradin C. Interaction of the full-length Bax protein with biomimetic mitochondrial liposomes: a small-angle neutron scattering and fluorescence study. Biochim Biophys Acta. 2012;1818:384–401. doi: 10.1016/j.bbamem.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Greber UFaKH. Nuclear targeting of adenovirus and simian virus SV40. Trends Cell Biol. 1996;6:189–195. doi: 10.1016/0962-8924(96)10016-7. [DOI] [PubMed] [Google Scholar]
- 37.Li PP, Nguyen AP, Qu Q, Jafri QH, Aungsumart S, Cheng RH, Kasamatsu H. Importance of calcium-binding site 2 in simian virus 40 infection. J Virol. 2007;81:6099–6105. doi: 10.1128/JVI.02195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzaki K, Sugishita K, Ishibe N, Ueha M, Nakata S, Miyajima K, Epand RM. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.