Abstract
Cross-linked polymer dots with intense and narrow yellow emission were designed using boron-dipyrromethene (BODIPY) polymer as the acceptor and poly[9,9-dioctylfluorenyl-2,7-diyl-co-1,4-benzo-{2,1′-3}-thiadiazole] (PFBT) polymer as the donor. The emission fwhm’s of the polymer dots (Pdots) were 37 nm. CL-BODIPY 565 Pdots were about 5 times brighter than commercial quantum dots (Qdots) 565 under identical experimental conditions. Specific cellular targeting indicated that the small, bright, and narrow emissive CL-BODIPY 565 Pdots are promising probes for biological applications.
Over the past two decades, semiconducting polymers have been the subject of great research interest because of their uses in a wide range of optoelectronic applications, such as polymer light emitting diodes (PLEDs),1 nonlinear optical materials (NLO),2 field effect transistors,3 sensors,4 information storage,5 electrochromic devices,6 and solar cells.7 Recently, research into semiconducting polymers as probes and biosensors has emerged as a new area of investigation because of their outstanding fluorescence characteristics.8 Semiconducting polymer dots (Pdots) with their high brightness and high photostability have attracted considerable attention. Besides green fluorescent proteins (GFPs),9,10 quantum dots (Qdots),11,12 carbon nanodots (Cdots),13 nanodiamonds,14 and fluorescent dyes, Pdots are one of the more promising nanosized probes that have practical applications in biomedical research.15 Pdots have been demonstrated in a number of applications, such as fluorescence bioimaging and biosensing.16 Pdots can form ultrabright probes17−19 and can be conjugated to biological molecules,20directly functionalized,21,22 and made to have narrow emission spectra.23−25
However, many scientific issues still await further investigation for Pdots to be used in biological applications. Research on smaller, brighter, and narrower emissive Pdots in the visible and near-infrared (NIR) region remain a challenge. Multicolor imaging is a powerful approach to study dynamic processes at cellular and subcellular levels. Though we have successfully developed green, orange, deep red, and NIR Pdots with narrow emission for bioimaging,23,25 there is a need to develop Pdots with other colors. Yellow Pdots, in particular, are needed to fill up the spectral gap between the green and orange Pdots.
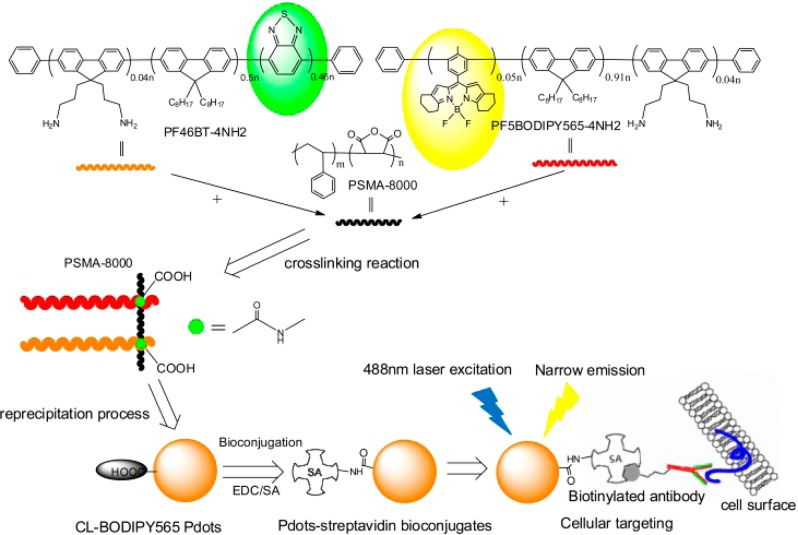
We have designed new yellow Pdots that consist of donor and acceptor semiconducting polymers that are cross-linked to an amphiphilic functional polymer. A narrow emissive fluorescent dye, boron-dipyrromethene (BODIPY), was copolymerized into the acceptor semiconducting polymer, while poly[9,9-dioctylfluorenyl-2,7-diyl-co-1,4-benzo-{2,1′-3}-thiadiazole] (PFBT) was used as the donor molecule in the donor semiconducting polymer. The emission spectrum of the donor overlapped well with the absorption of the acceptor to enable efficient Förster resonant energy transfer (FRET) between them. We then developed a cross-linking strategy to covalently bind these fluorescent semiconducting polymers together with the amphiphilic functional polymer, poly(styrene-co-maleic anhydride) (PSMA, with a Mw as 8000). This step generated cross-linked polymers with the donor and the acceptor semiconducting polymers. These polymers were then nanoprecipitated to form small and stable Pdots. The Pdots had high fluorescence brightness and narrow emission by the FRET mechanism (Scheme 1). Finally, we performed biomolecular conjugation and successfully used those bioconjugated Pdots for fluorescence bioimaging and flow cytometry.
Scheme 1. Schematic Illustration of the Preparation of Small, Bright, and Narrow Emissive Cross-Linked Semiconducting Polymers and Pdot Bioconjugates for Specific Cellular Targeting.
On the basis of the cross-linking strategy, we first synthesized the PFBT polymer with a 4% molar ratio of the amino functional group as the donor polymer (PF46BT-4NH2, Scheme S2, Supporting Information (SI)). Pdots made from PFBT have been widely studied because of their high absorption cross-section, single-particle brightness, and excellent photostability. PFBT also has an absorption peak of 450–460 nm, which is very close to the 488 nm excitation wavelength laser generally used in biological applications.
We then designed a new BODIPY semiconducting polymer as the acceptor polymer by modifying the structure of the BODIPY core (monomer 1, Scheme S1, SI) to tune its color so that its absorption spectrum has a good overlap with the donor polymer emission. The absorption spectrum of the BODIPY-monomer 1 (peaks at 546 nm) had good overlap with the PFBT emission (peaks at ∼540 nm) (Figure S1, SI), which increased the FRET efficiency to obtain bright, fluorescent Pdots. Finally, we copolymerized 5% molar ratio of monomer 1 with the fluorene monomer and 4% molar ratio of the amino-functional group to obtain the acceptor polymer (PF5BODIPY565-4NH2, Scheme S1, SI).
To determine the optimal ratio between the donor polymer (PF46BT-4NH2) and the acceptor polymer (PF5BODIPY565-4NH2) to get the highest FRET efficiency, we first blended the donor and acceptor polymers together to form Pdots with different ratios of the donor and acceptor polymers before we carried out the covalent cross-linking of the semiconducting polymers. Because the donor and acceptor polymers were closely packed into single Pdots, efficient FRET resulted in the complete quenching of the PFBT donor and intense fluorescence from the acceptor polymer (Figure S2, SI). The optimal ratio of 0.54 (weight ratio between PF5BODIPY565-4NH2 and PF46BT-4NH2) was determined for the blended Pdots, which exhibited an efficient yellow emission peak at 564 nm and a high fluorescence quantum yield of 0.32 (Figure S2b, SI). Once we obtained the optimal donor-to-acceptor ratio, we ran the cross-linking reaction to chemically bond the donor and acceptor polymer together via condensation reaction between amino and maleic anhydride groups. The cross-linked BODIPY 565 Pdots (CL-BODIPY 565 Pdots) showed narrow emission centered at 564 nm with a fwhm as narrow as 37 nm (Figure 1a). To the best of our knowledge, this is the narrowest emission bandwidth reported so far for any type of Pdot.
Figure 1.
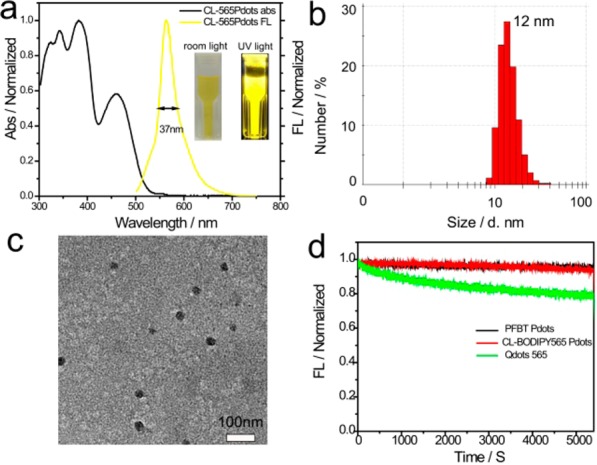
(a) Absorption and fluorescence spectra of CL-BODIPY 565 Pdots in water, (b) the histograms of the distribution of the sizes of CL-BODIPY 565 Pdots, measured by DLS (the mean size is 12 nm), (c) TEM images of CL-BODIPY 565 Pdots, (d) photobleaching curves of PFBT Pdots (black line), CL-BODIPY 565 Pdots (red line), and Qdots 565 (green line).
To evaluate its photophysical properties, CL-BODIPY 565 Pdots were prepared by the nanoprecipitation method with an average particle size of 12 nm, as characterized by dynamic light scattering (DLS) and transmission electron microscopy (TEM) (Figure 1b and 1c). For comparison, commercially available Qdots 565 (Invitrogen, Eugene, OR, USA) were dispersed in Milli-Q water and measured to have a comparable particle size of ∼13 nm (measured by DLS in the same conditions). As expected, unlike the larger particle size of Pdots we reported previously (∼18 nm, the average hydrodynamic diameter prepared under comparable conditions),23 CL-BODIPY 565 Pdots exhibit a significantly smaller particle size and a narrower size distribution. This can be attibuted to the amphiphilic polymer PSMA (Mw = 8000), which acted as a successful cross-linker to connect the donor and acceptor polymers tightly during the Pdot formation.
CL-BODIPY 565 Pdots exhibited remarkable photostability as seen with the PFBT Pdots under identical conditions (measured on the fluorimeter equipped with 450 W xenon lamp, and the excitation wavelength was centered at 488 nm). The photobleaching process was carried out over 1.5 h, which did not result in an observable decrease in the fluorescence intensity of CL-BODIPY 565 Pdots (Figure 1d). In contrast, Qdots 565 exhibited an obvious single-exponential photobleaching decay. The comparable high photostability of CL-BODIPY 565 Pdots can be attributed to the incorporation of the PFBT donor polymer into the CL-BODIPY 565 Pdots since our previous results have proved that PFBT is a photostable polymer.20,23,26−28
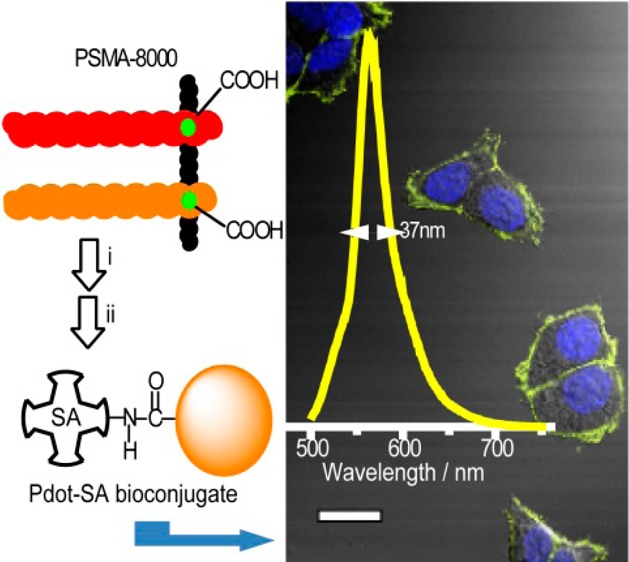
To evaluate the performance of the CL-BODIPY 565 Pdots for biological applications, we used them to label breast cancer MCF-7 cells and analyze these Pdot-labeled cells by flow cytometry and cellular imaging. CL-BODIPY 565 Pdots were conjugated with streptavidin via the 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide hydrochloride (EDC) catalyzed coupling reaction. These Pdot bioconjugates were used to label MCF-7 cells which were first incubated with biotinylated anti-EpCAM antibodies (Figure 2). There was excellent separation between cells labeled with Pdot-streptavidin and cells that were incubated with Pdot-streptavidin bioconjugates but in the absence of the biotinylated primary antibody. MCF-7 cells labeled with CL-BODIPY 565 Pdot-streptavidin bioconjugates (CL-BODIPY 565 Pdot-SA) exhibited approximately 5 times higher intensity than those labeled with Qdot 565-streptavidin bioconjugates under identical experimental conditions. The specific cellular labeling of the CL-BODIPY 565 Pdot-SA probes was further confirmed by confocal fluorescence imaging (Figure 3). CL-BODIPY 565 Pdot-SA probes were effectively labeled with biotin anti-EpCAM receptors on the MCF-7 cell surface; however, fluorescence was not detected in the negative control experiment which was carried out in the absence of the biotinylated primary antibody. Therefore, the result proved that CL-BODIPY 565 Pdot-SA could be used as probes for specific cellular labeling without nonspecific binding.
Figure 2.
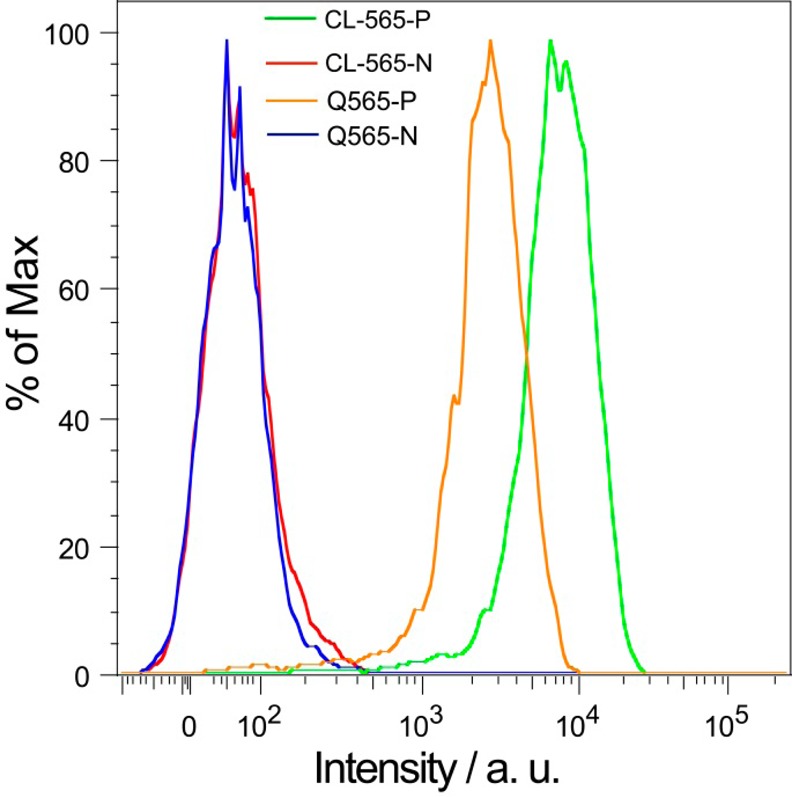
Flow-cytometry measurements of the intensity distributions of MCF-7 breast cancer cells labeled via nonspecific binding (N: negative control) and specific targeting (P: positive control), using Qdots 565 (Q565-N: Qdot negative control; Q565-P: Qdot positive control), CL-BODIPY 565 Pdots (CL-565-N: CL-BODIPY 565 negative control; CL-565-P: CL-BODIPY 565 positive control). All Qdots and Pdots were conjugated with streptavidin.
Figure 3.
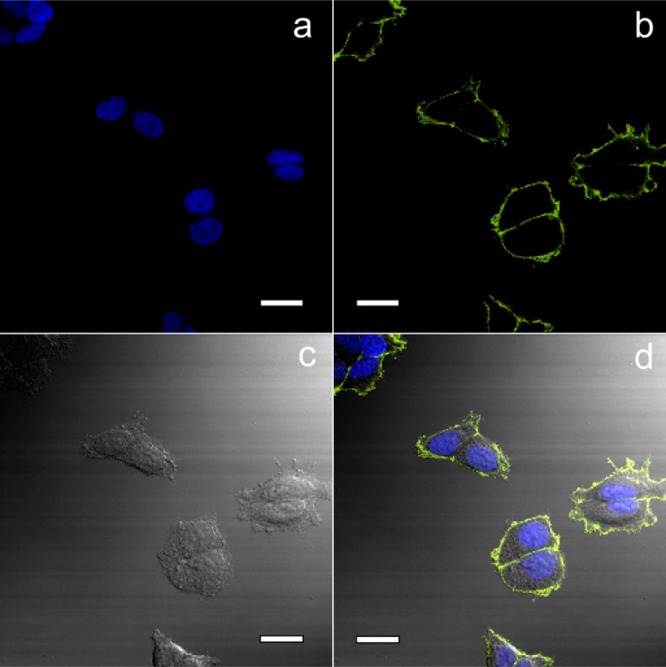
Confocal fluorescence microscopy images of MCF-7 cells labeled with CL-BODIPY 565 Pdot-SA probes: (a) blue fluorescence from the nuclear stain Hoechst 34580, (b) yellow fluorescence from CL-BODIPY 565 Pdot-SA, (c) Nomarski (differential interference contrast; DIC) microscopy, and (d) combined DIC and fluorescence images. Scale bars: 20 μm.
Acknowledgments
D.T.C. gratefully acknowledges support for this research from the National Institutes of Health (NS052637 and GM085485), the Life Sciences Discovery Fund, the Department of Defense CDMRP program (BC100510), and the University of Washington.
Supporting Information Available
Synthesis scheme of BODIPY monomers and polymers. Spectroscopic properties of BODIPY monomers and Pdots, and NMR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): D.T.C. and J.Y. have financial interest in Lamprogen, which has licensed the described technology from the University of Washington.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kraft A.; Grimsdale A. C.; Holmes A. B. Angew. Chem., Int. Ed. 1998, 37, 402–428. [DOI] [PubMed] [Google Scholar]
- Yesodha S. K.; Sadashiva Pillai C. K.; Tsutsumi N. Prog. Polym. Sci. 2004, 29, 45–74. [Google Scholar]
- Boudreault P. T.; Wakim S.; Blouin N.; Simard M.; Tessier C.; Tao Y.; Leclerc M. J. Am. Chem. Soc. 2007, 129, 9125–9136. [DOI] [PubMed] [Google Scholar]
- McQuade D. T.; Pullen A. E.; Swager T. M. Chem. Rev. 2000, 100, 2537–2574. [DOI] [PubMed] [Google Scholar]
- Möller S.; Perlov C.; Jackson W.; Taussig C.; Forrest S. R. Nature 2003, 426, 166–169. [DOI] [PubMed] [Google Scholar]
- Beaujuge P. M.; Reynolds J. R. Chem. Rev. 2010, 110, 268–320. [DOI] [PubMed] [Google Scholar]
- a Sun Y.; Welch G. C.; Leong W. L.; Takacs C. J.; Bazan G. C.; Heeger A. J. Nat. Mater. 2012, 11, 44–48. [DOI] [PubMed] [Google Scholar]; b Li G.; Zhu R.; Yang Y. Nat. Photonics 2012, 6, 153–161. [Google Scholar]
- Zhu C.; Liu L.; Yang Q.; Lv F.; Wang S. Chem. Rev. 2012, 112, 4687–4735. [DOI] [PubMed] [Google Scholar]
- Yu J.; Xiao J.; Ren X. J.; Lao K. Q.; Xie X. S. Science 2006, 311, 1600–1603. [DOI] [PubMed] [Google Scholar]
- Shaner N. C.; Steinbach P. A.; Tsien R. Y. Nat. Methods 2005, 2, 905. [DOI] [PubMed] [Google Scholar]
- Han M.; Gao X.; Su J. Z.; Nie S. Nat. Biotechnol. 2001, 19, 631–635. [DOI] [PubMed] [Google Scholar]
- Chan W. C. W.; Nie S. Science 1998, 281, 2016–2018. [DOI] [PubMed] [Google Scholar]
- Baker S. N.; Baker G. A. Angew. Chem., Int. Ed. 2010, 49, 6726–6744. [DOI] [PubMed] [Google Scholar]
- Vaijayanthimala V.; Chang H.-C. Nanomedicine 2009, 4, 47–55. [DOI] [PubMed] [Google Scholar]
- Baker M. Nat. Methods 2010, 12, 957–962. [Google Scholar]
- Wu C.; Chiu D. T. Angew. Chem., Int. Ed. 2013, 52, 3086–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Jin Y.; Schneider T.; Burnham D. R.; Smith P. B.; Chiu D. T. Angew. Chem., Int. Ed. 2010, 49, 9436–9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Hansen S.; Hou Q.; Yu J.; Zeigler M.; Jin Y.; Burnham D.; McNeill J.; Olson J.; Chiu D. T. Angew. Chem., Int. Ed. 2011, 50, 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F.; Wu C.; Jin Y.; Wang M.; Chan Y.-H.; Yu J.; Sun W.; Hayden S.; Chiu D. T. Chem. Commun. 2012, 48, 1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Schneider T.; Zeigler M.; Yu J.; Schiro P.; Burnham D.; McNeill J. D.; Chiu D. T. J. Am. Chem. Soc. 2010, 132, 15410–15417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Yu J.; Wu C.; Jin Y.; Rong Y.; Ye F.; Chiu D. T. ACS Nano 2012, 6, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Wu C.; Zhang X.; Ye F.; Gallina M. E.; Rong Y.; Wu I.; Sun W.; Chan Y.; Chiu D. T. Adv. Mater. 2012, 24, 3498–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.; Wu C.; Yu J.; Zhang X.; Ye F.; Zeigler M.; Gallina M. E.; Wu I.; Zhang Y.; Chan Y.; Sun W.; Uvdal K.; Chiu D. T. ACS Nano 2013, 7, 376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Yu J.; Deng R.; Rong Y.; Fujimoto B.; Wu C.; Zhang H.; Chiu D. T. Angew. Chem., Int. Ed. 2013, 52, 11294–11297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Yu J.; Rong Y.; Ye F.; Chiu D. T.; Uvdal K. Chem. Sci. 2013, 4, 2143–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Bull B.; Szymanski C.; Christensen K.; McNeill J. ACS Nano 2008, 2, 2415–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.; Wu C.; Sahu S.; Fernando L.; Szymanski C.; McNeill J. J. Am. Chem. Soc. 2009, 131, 18410–18414. [DOI] [PubMed] [Google Scholar]
- Yu J.; Wu C.; Tian Z.; McNeill J. Nano Lett. 2012, 12, 1300–1306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.