Abstract
Streptococcus pneumoniae colonizes the highly diverse polymicrobial community of the nasopharynx where it must compete with resident organisms. We have shown that bacterially produced antimicrobial peptides (bacteriocins) dictate the outcome of these competitive interactions. All fully-sequenced pneumococcal strains harbor a bacteriocin-like peptide (blp) locus. The blp locus encodes for a range of diverse bacteriocins and all of the highly conserved components needed for their regulation, processing, and secretion. The diversity of the bacteriocins found in the bacteriocin immunity region (BIR) of the locus is a major contributor of pneumococcal competition. Along with the bacteriocins, immunity genes are found in the BIR and are needed to protect the producer cell from the effects of its own bacteriocin. The overlay assay is a quick method for examining a large number of strains for competitive interactions mediated by bacteriocins. The overlay assay also allows for the characterization of bacteriocin-specific immunity, and detection of secreted quorum sensing peptides. The assay is performed by pre-inoculating an agar plate with a strain to be tested for bacteriocin production followed by application of a soft agar overlay containing a strain to be tested for bacteriocin sensitivity. A zone of clearance surrounding the stab indicates that the overlay strain is sensitive to the bacteriocins produced by the pre-inoculated strain. If no zone of clearance is observed, either the overlay strain is immune to the bacteriocins being produced or the pre-inoculated strain does not produce bacteriocins. To determine if the blp locus is functional in a given strain, the overlay assay can be adapted to evaluate for peptide pheromone secretion by the pre-inoculated strain. In this case, a series of four lacZ-reporter strains with different pheromone specificity are used in the overlay.
Keywords: Infectious Diseases, Issue 91, bacteriocins, antimicrobial peptides, blp locus, bacterial competition, Streptococcus pneumoniae, overlay assay
Introduction
Streptococcus pneumoniae, a common colonizer of the polymicrobial community of the nasopharynx, is able to compete with other pneumococci and closely related species through the production of bacterially produced antimicrobial peptides (bacteriocins). Every fully-sequenced pneumococcal strain examined to date contains a version of the bacteriocin-like peptide locus, blp. Bacteriocin production by pneumococcus has been demonstrated to be important in the outcome of both in vitro and in vivo competition1-4. Competitive dynamics are influenced by the expression of diverse bacteriocins along with cognate immunity proteins in response to a specific peptide pheromone. Induction of the blp locus is controlled by a quorum sensing system in which a peptide pheromone, BlpC, binds to the sensor kinase, BlpH, and initiates a signaling cascade resulting in the transcription of the blp locus5,6. There are four allelic variations of BlpC that each bind to its cognate BlpH6. For the cell to survive the effects of its own bacteriocin, the genes that encode cognate immunity proteins are transcribed on the same operon as each of the bacteriocin genes. Killing occurs if a competitor strain is not able to upregulate the blp locus in response to exogenous pheromone or, if the strain lacks the specific immunity gene required for protection. The transporter complex, BlpAB is required for the secretion and processing of the peptide pheromone and the bacteriocin peptides2,4. Recently, it has been shown that a significant proportion of pneumococcal strains are “cheaters” meaning they have a conserved mutation in the blpA gene which renders them unable to secrete pheromones and bacteriocins1,2. These strains are able to respond to exogenous BlpC2 secreted by neighboring strains allowing for the production of immunity proteins.
Although, crude preparations of bacteriocins from supernatants can be prepared from producer strains using biochemical methods steps to achieve purity, this approach is not useful for screening large collections of isolates and if the level of bacteriocin expression is low in broth grown cultures2,7-9. The overlay assay is used as a rapid way to investigate the competitive dynamics between strains that may exist in vitro. To do so, a pneumococcal strain is pre-inoculated onto an agar plate and allowed to grow to achieve local bacterial concentrations sufficient for induction of the blp locus. A second strain is then inoculated into a molten soft agar solution which is then applied over the pre-inoculated strain to test for sensitivity. To evaluate for activity of the blp locus (independent of inhibitory activity), strains can be overlaid with a series of three previously described BlpC reporter strains. These strains were constructed to contain one of three different blpH alleles that respond to the three most common BlpC pheromones secreted by the pneumococci. The reporter strains have an integrated lacZ gene that is under the control of a BlpH dependent promoter and carry a deletion in the blpC gene that prevents self-stimulation10,11.
Protocol
1. Preparation of Producer Strain
Streak a pneumococcal strain to be tested for bacteriocin production on a 5% sheep blood tryptic soy agar plate from -80 °C freezer stocks. NOTE: The use of blood plates for initial growth of the strain to be tested for inhibition greatly increases the reproducibility of the assay. Additionally, other non-pneumococcal strains that produce bacteriocins can be used although growth conditions may need to be modified for a specific organism. We have used Streptococcus mitis and Lactococcus lactis in the overlay assay successfully.
Incubate the plate overnight (O/N) at 37 °C with 5% CO2.
After incubation of the producer strain, prepare the tryptic soy agar (TSA) plate by pipetting 3,000 to 4,000 units of catalase onto plates containing 25 ml of TSA and spread the catalase solution using sterile glass beads. Let the plate dry for about 10 min under a biological safety cabinet.
- Collect a visible quantity of bacteria onto a pipette tip. Stab the pipette tip into the dried TSA plate. NOTE: More than one producer strain can be stabbed into a single TSA plate. It is important to space out the stabs so that resulting halos do not overlap.
- If possible, include known bacteriocin producer as a positive control and the strain to be used in the overlay as a negative control. NOTE: Positive and negative controls are have been previously published and are available upon request10.
Incubate TSA plate containing multiple stabs at 37 °C with 5% CO2 for 6 hr.
2. Making Glycerol Stocks of the Overlay Strain
Prepare glycerol stocks (cultures grown to mid-exponential phase and then stored at -80 °C after the addition of glycerol) prior to the day of the overlay assay. Inoculate 5-10 colonies of pneumococcal strain to be tested for sensitivity or pheromone secretion in Todd-Hewitt medium with 0.5% yeast extract (THY). NOTE: Construction of BlpC inducible reporters has been previously described2 and strains are available upon request.
Incubate 5-10 colonies in glass tubes with 5 ml of THY with tightly closed lids in a 37 °C water bath without shaking.
- Incubate cultures at 37 °C until they reach an OD620 of 0.3 to 0.5. NOTE: This will take approximately 4-5 hr depending on the strain used.
- Gently invert the tube a few times prior to measuring the OD.
If the experiment cannot be performed the same day or the strain will be used multiple times in the future, make glycerol stocks and resume the experiment at a later date. For glycerol stocks, add glycerol to cultures to a final concentration of 20%. Aliquot cultures into 1.5 ml microcentrifuge tubes and store at -80 °C. NOTE: Samples can be stored for months at -80 °C. If not making glycerol stocks, use samples directly for overlay assay.
3. Overlay Assay
Just prior to overlay application, mix the overlay components. In a 15 ml conical tube, add 5 ml of THY, 3,000-4,000 U of catalase, and 200 µl of the overlay strain broth culture that was either grown that day or a glycerol stock of the culture that was thawed at room temperature. If using the reporter strain, add 50 µl of X-gal (40 mg/ml) to the overlay mixture. Use one conical tube per plate.
Equilibrate overlay mixture at room temperature prior to addition of molten TSA.
- Remove TSA plate with stabbed strain from incubator. Melt solid TSA in microwave and keep at 55 °C water bath until ready to use. Add 3 ml of molten TSA with 1.5% agar cooled to 55 °C to overlay mixture.
- Prepare the overlay mixture immediately before pipetting, the mixture will begin to solidify very quickly. NOTE: If the overlay begins to solidify before plating, the results will be uninterpretable.
- Mix the overlay mixture by pipetting up and down several times with a 5 ml pipet being careful not to introduce bubbles into the mixture. Slowly pipette overlay mixture directly on to the stabbed TSA plate. Keep the plate on the benchtop for few minutes allowing the agar to harden.
- Do not rotate the plate to distribute the overlay mixture, this will pull growth from the stabbed culture into the overlay.
Carefully place TSA plate with overlay into 37 °C incubator containing 5% CO2 O/N (approximately 16-18 hr).
Observe the plates after overnight growth. The plates should appear opaque from growth of the overlay strain except in areas where inhibition or pheromone signaling has occurred. These will appear as either clear or blue zones around the stabbed strain, respectively. NOTE: Although measurements of the diameter of blue or clearing can be determined for quantitative assessment, differences in the diameter can also vary depending on the amount of bacteria that was stabbed into the plate. The overlay assay is a qualitative assay, any quantitative assessment should be interpreted with caution. Any visible clearing as compared to the negative control is regarded as activity.
Representative Results
There are two possible outcomes in the overlay assay. It should be possible to see growth of the stabbed strain after overnight incubation. In Figure 1A, the overlay strain is sensitive to the bacteriocins being produced. A zone of clearance can be visualized surrounding the stabbed strain. Additionally, swirling of the stabbed strain into the overlay mixture is possible and can be seen in the Figure 1A. In Figure 1B, the overlay strain is immune to the bacteriocins being secreted as noted by an absence of a halo surrounding the stab. It is also possible that the stabbed strain is not secreting bacteriocins. For the signaling assay, two different outcomes are possible. The stabbed strain is able to secrete pheromone that interacts with the histidine kinase of the overlay strain, as seen through the breakdown of X-gal as shown in Figure 2A. If the pheromone does not activate transcription of the blp locus in the reporter strain, or if the stabbed strain does not secrete pheromone, breakdown of X-gal will not occur as shown in Figure 2B.
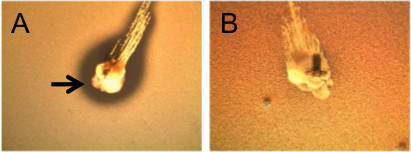 Figure 1. Appearance of blp mediated inhibition and immunity in an overlay assay. (A) Strain P133, a bacteriocin producer was spiked into a TSA plate and allowed to grow for 6 hr. Strain P250, a bacteriocin negative strain, was inoculated into the overlay. Plates were photographed after O/N incubation. A zone of clearance is indicated by the arrow. (B) Strain P133 was spiked into a TSA plate and incubated for 6 hr. Strain 130, containing a frameshift mutation in blpA, was inoculated into the overlay. After O/N incubation, plates were photographed.
Figure 1. Appearance of blp mediated inhibition and immunity in an overlay assay. (A) Strain P133, a bacteriocin producer was spiked into a TSA plate and allowed to grow for 6 hr. Strain P250, a bacteriocin negative strain, was inoculated into the overlay. Plates were photographed after O/N incubation. A zone of clearance is indicated by the arrow. (B) Strain P133 was spiked into a TSA plate and incubated for 6 hr. Strain 130, containing a frameshift mutation in blpA, was inoculated into the overlay. After O/N incubation, plates were photographed.
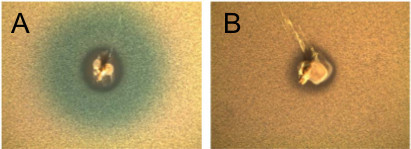 Figure 2. Pheromone mediated activation of blp locus in signaling assay. (A) Strain P133, a BlpC type R6 secretor, was spiked into a TSA plate and incubated for 6 hr. Strain P981, a BlpC type R6 reporter with a deletion in BlpC, was inoculated into the overlay. Photographs were taken after O/N incubation. (B) P133 was spiked into a TSA plate. After incubation for 6 hr, P845, a BlpC type P164 reporter with a deletion in BlpC, was inoculated into the overlay. Photographs were taken after O/N growth.
Figure 2. Pheromone mediated activation of blp locus in signaling assay. (A) Strain P133, a BlpC type R6 secretor, was spiked into a TSA plate and incubated for 6 hr. Strain P981, a BlpC type R6 reporter with a deletion in BlpC, was inoculated into the overlay. Photographs were taken after O/N incubation. (B) P133 was spiked into a TSA plate. After incubation for 6 hr, P845, a BlpC type P164 reporter with a deletion in BlpC, was inoculated into the overlay. Photographs were taken after O/N growth.
Discussion
This overlay assay is a rapid way to determine the range of activity for bacteriocin producers and to demonstrate competitive interactions that may occur among pneumococcal strains. The overlay assay can be adapted to use for screening multiple strains for bacteriocin production on a single plate. We have successfully screened large strain collections with this assay by using a 48-pin replicator to inoculate SBA plates from one half of a 96-well plate. After overnight incubation, growth is transferred to the TSA plate using the replicator and piercing the agar surface of the plate with the pins of the replicator.
Other bacterially secreted antimicrobial peptides that are regulated in a density dependent manner like lantibiotics can also be examined using the overlay technique7,12,13. This is especially useful when purification of a bacteriocin or lantibiotic is difficult. Though supernatants can be tested for antimicrobial activity, expression of the blp derived bacteriocins in broth is very limited or nonexistent (data not shown). The overlay assay allows for growth to a higher density which is not achieved when organisms are grown in liquid. Growth on a solid medium might also more closely reflect the in vivo growth conditions of the nasopharynx.
The overlay assay can also be used to examine the ability of the stabbed strain to secrete the bacteriocin specific signaling pheromone. This requires construction of a reporter strain which expresses the pheromone receptor protein and in which a pheromone responsive promoter is placed upstream of the lacZ gene. Appreciation of pheromone secretion by a stabbed strain is particularly useful as a readout of blp locus activity if the stabbed strain does not inhibit the overlay strain. The inability to inhibit the overlay strain could be the result of a non-functional blp locus (such as in a strain with a transporter mutation) or because the bacteriocins secreted are not active against the particular overlay strain tested. The reporter strain overlay provides additional information that the locus in the producer has a fully functional two component system and BlpA transporter allowing for upregulation and secretion of the pheromone. This implies that any encoded bacteriocins are also secreted.
There are some limitations to this assay. The overlay assay is a qualitative assay, so comparisons between the inhibitory activity or pheromone secretion of different strains have to be made carefully. Growth rates and conditions should be considered when comparing different strains because the outcome of the assay relies heavily on the growth rate of the overlay and stabbed strains. The addition of catalase to both the plate and the overlay is important in order to remove the inhibitory effects of pneumococcal H2O2 production on the growth of the overlay strain, in particular when using catalase negative organism in the overlay. Given the known diversity of genomic content in the pneumococcal population, any inhibitory activity seen with this assay cannot be automatically attributed to the blp locus without deletional analysis.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work has been supported by Elizabeth E. Kennedy Research Award.
References
- Dawid S, Roche AM, Weiser JN. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 2007;75(1):443–451. doi: 10.1128/IAI.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa N, et al. Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1. Appl Environ Microbiol. 2012;78(3):900–903. doi: 10.1128/AEM.06497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan TJ, Dawid S. The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol. 2013;195(7):1561–1572. doi: 10.1128/JB.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux T, Nuhn M, Hakenbeck R, Reichmann P. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J Bacteriol. 2007;189(21):7741–7751. doi: 10.1128/JB.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann P, Hakenbeck R. Allelic variation in a peptide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol Lett. 2000;190(2):231–236. doi: 10.1111/j.1574-6968.2000.tb09291.x. [DOI] [PubMed] [Google Scholar]
- Saizieu A, et al. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182(17):4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour A, Philip K, Muniandy S. Enhanced production, purification, characterization and mechanism of action of salivaricin 9 lantibiotic produced by Streptococcus salivarius NU10. PLoS One. 2013;8(10):77751. doi: 10.1371/journal.pone.0077751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wladyka B, et al. Isolation, biochemical characterization, and cloning of a bacteriocin from the poultry-associated Staphylococcus aureus strain CH-91. Appl Microbiol Biotechnol. 2013;97(16):7229–7239. doi: 10.1007/s00253-012-4578-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea EF, et al. Bactofencin a, a new type of cationic bacteriocin with unusual immunity. 2013;4(6):00498–00413. doi: 10.1128/mBio.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MR, et al. Conserved mutations in the pneumococcal bacteriocin transporter gene, blpA, result in a complex population consisting of producers and cheaters. MBio. 2011;2(5) doi: 10.1128/mBio.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochan TJ, Dawid S. The HtrA protease of Streptococcus pneumoniae controls density-dependent stimulation of the bacteriocin blp locus via disruption of pheromone secretion. J Bacteriol. 2013;195:1561–1572. doi: 10.1128/JB.01964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdick DA, et al. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc Natl Acad Sci U S A. 2003;100(7):4316–4321. doi: 10.1073/pnas.0230516100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol. 2009;75(17):5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


