Abstract
Brain morphometry in recent decades has increased our understanding of the neural bases of psychiatric disorders by localizing anatomical disturbances to specific nuclei and subnuclei of the brain. At least some of these disturbances precede the overt expression of clinical symptoms and possibly are endophenotypes that could be used to diagnose an individual accurately as having a specific psychiatric disorder. More accurate diagnoses could significantly reduce the emotional and financial burden of disease by aiding clinicians in implementing appropriate treatments earlier and in tailoring treatment to the individual needs. Several methods, especially those based on machine learning, have been proposed that use anatomical brain measures and gold-standard diagnoses of participants to learn decision rules that classify a person automatically as having one disorder rather than another. We review the general principles and procedures for machine learning, particularly as applied to diagnostic classification, and then review the procedures that have thus far attempted to diagnose psychiatric illnesses automatically using anatomical measures of the brain. We discuss the strengths and limitations of extant procedures and note that the sensitivity and specificity of these procedures in their most successful implementations have approximated 90%. Although these methods have not yet been applied within clinical settings, they provide strong evidence that individual patients can be diagnosed accurately using the spatial pattern of disturbances across the brain.
Keywords: Machine learning, brain morphometry, automated diagnosis, cortical thickness, psychiatric disorders, cross validation, support vector machines
Morphometry is the study of forms or shapes. It quantifies anatomical features of interest and then correlates those features with other characteristics of the individual, including clinical, behavioral, and genetic measures.(Bookstein, 1997) Many Magnetic Resonance Imaging (MRI) studies have applied the principles of morphometry to understand the association of morphological features with various psychiatric disorders (Fig. 1).
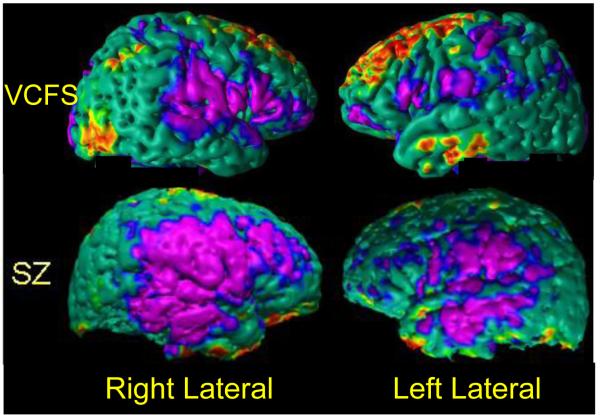
Fig. 1.
Spatial Patterns of Abnormalities in Local Volumes across cerebral cortex in participants with various neuropsychiatric disorders. Top Row: 44 individuals with velo-cardio-facial syndrome (VCFS); and Second Row: 81 Schizophrenia (SZ) patients. These abnormalities were detected by comparing each group of patients with a group of healthy individuals who were age- and sex-matched with the patient groups. Affected populations have distinct spatial patterns of abnormalities that can be used to train a classifier to diagnose individual people within that population. The P-values of abnormalities are color-encoded and displayed on the template brain. Purple: regions of significant (P-value < 0.0001) local volume reductions; Red: regions of significant (P-value < 0.0001) local volume increases in the affected populations. VCFS=Velo-Cardio-Facial Syndrome; SZ=Schizophrenia.
MRI generates images of the brain with high signal to noise ratios (SNRs) and exquisite contrast between soft tissues to yield definable boundaries for brain regions that have long been hoped would be useful in discriminating patients who have one clinical diagnosis from patients who have a differing diagnosis. Each brain region, however, consists of several subnuclei that have specific information processing functions and may contribute differentially to the disease process of interest. Soft tissue contrast and SNR, moreover, are generally insufficient to distinguish visual boundaries of the various subnuclei of most brain regions, and therefore initial morphometric studies contented themselves with measuring overall volumes of brain regions. Although these rather simplistic approaches of studying psychiatric illness using overall conventional volumes of brain regions improved our understanding of the anatomical disturbances present in persons who have specific psychiatric disorders, opposing volumetric changes in differing subnuclei of a single brain region likely substantially reduced the ability of the overall volume of that brain region to discriminate diagnostic groups. Recent advances in methods for analyzing MR images, however, have provided substantial information about constituent subnuclei and their roles in the disease process of interest.(Bansal et al., 2005, Bansal et al., 2007, Csernansky et al., 2004, Davatzikos et al., 2005, Thompson and Toga, 1996b, Thompson and Toga, 1996a, Peterson et al., 2009, Plessen et al., 2006) These advanced imaging techniques permit the detection, comparison, and correlation of anatomical disturbances within subnuclei, thereby facilitating more accurate diagnoses of psychiatric disorders using MR images.
The accurate diagnosis of patients using clinical interviews and family history can be difficult, especially early in the course of illness when the full complex of symptoms have yet to become manifest. Overlapping diagnostic criteria, the presence of comorbid disorders, and the variable expression of psychiatric disorders further complicates accurate diagnosis. Accumulated evidence strongly suggests that psychiatric disorders have unique spatial patterns of anatomical disturbances, and that these disturbances, together with connectivity measures, metabolite concentrations, and functional disturbances in the brain are detectable even before the overt expression of symptoms. By quantifying brain disturbances in affected patients or in persons at increased risk for developing a psychiatric illness and comparing those measures with persons without an illness, those patients might be accurately diagnosed as having a disorder or not, or as having one disorder rather than another.
Although various computer algorithms have been developed and applied to brain measures to attempt to classify the brains of persons who have psychiatric illnesses, the most common type of algorithm developed for this purpose has been supervised machine learning. A supervised method for machine-based learning and pattern classification learns decision rules and classification boundaries using a labeled set of data. Those data are so-called “feature vectors”, features of the image captured in the form of multi-dimensional variables that encode the location and magnitude of the brain measure of interest. Supervised machine learning then associates or links each of these vector variables with a previously known set of numerically coded diagnostic labels or classes. The learning algorithm develops decision rules and boundaries that classify individual brains as having a given diagnostic label based on the imaging feature vectors that are present in each brain. The algorithm that has been “trained” using the previously known labels then should, when tested rigorously, apply the learned rules and boundaries to the classification of new brains that did not participate in the training of the algorithm. The feature vectors can be virtually any brain measure, but in the examples we will provide, they are most commonly measures of cortical thickness or local volumes of gray matter or white matter at each point (or “voxel”, a volumetric pixel in three dimensions) in the brain. The diagnostic labels are usually previously established clinical diagnoses. A decision rule or classification boundary can be represented as a function that maps the feature vector to the given diagnostic label. The performance of a classifier is typically assessed by dividing labeled data into two sets, a training set in which the classifier is trained, and a test set in which the trained algorithm is applied to classify a set of brains that were not involved in training the classifier. The correct diagnoses (labels) are known in the test dataset, thereby permitting the accuracy of classification to be discerned and quantified easily when applying the trained algorithm to the test dataset.
If previously established diagnostic labels are unavailable when developing the classifier, other methods can be employed to discover natural groupings in the feature vectors. These methods are termed unsupervised learning algorithms (or, alternatively, data mining or clustering procedures). The validity of the natural groupings that are identified by these unsupervised learning procedures must then be established by applying statistical methods, such as linear discriminant analysis, to assess the statistical significance of differences in clinical and behavioral measures across the naturalistic groupings.
Extant methods for machine-based classification of an individual brain using imaging data(Barnes et al., 2004, Davatzikos et al., 2008, Duchesnay et al., 2007, eFigueiredo et al., 1995, Fan et al., 2005, Herholz et al., 2002, Jack et al., 2004, Kawasaki et al., 2007, Klöppel et al., 2008a, Lao et al., 2004, Lerch et al., 2006, Liu et al., 2004, Lochhead et al., 2004, Mourao-Miranda et al., 2005, Teipel et al., 2007, Wahlund et al., 2005) can generally be characterized as supervised. Imaging data from all participants are first spatially warped, or “normalized”, to a template brain to bring all corresponding points into a common spatial register (termed “template space”) and to compute the feature vectors of interest for each of the brains. The quantitative features are typically computed from images by applying various technical procedures to extract feature vectors from the imaging dataset, procedures that include volume preserved warping (VPW)(Xu et al., 2007), mass preserved warping(Haker et al., 2001), voxel based morphometry (VBM)(Ashburner and Friston, 2000), deformation based morphometry(Paus et al., 2001), surface morphometry(Bansal et al., 2007, Thompson and Toga, 1996b), and spherical harmonics(Chung et al., 2008). Of these various procedures, the most commonly employed has probably been VBM, which generates point-wise (or “voxel-wise”) maps of measures at each voxel in the brain. It is an automated method that analyzes the nonlinear deformation field used for the spatial normalization of one particular brain into a common template space and that then generates a voxel-wise map of the local changes in brain volume that are required to normalize the brain to the template brain (i.e., to bring all points in one brain into register with the corresponding points in the template brain). These measures of local changes needed for spatial localization are fine-grained in their spatial resolution (typically on the order of a cubic millimeter), and those measures can then be compared across groups of participants.
We will first review the principles and procedures for machine learning and how they can be applied to the diagnosis of psychiatric illnesses. Each of the various components of these procedures is relatively independent of the others and can be modified to influence significantly the accuracy of classification. We will also review several previous attempts at localizing and associating disturbances in the brain with specific psychiatric disorders and the specific components of machine learning procedures that best exploit these disturbances for more accurately diagnosing illness. Finally, we will discuss the strengths and limitations of extant machine learning algorithms. These methods provide sufficient preliminary evidence that morphometric data adequately localize disturbances in a specific disorder and that machine learning procedures in the foreseeable future will likely yield useful tools for aiding the clinical diagnosis of patients.
A: Machine Learning
Machine learning can be partitioned into components and procedures, including participants, imaging modality, brain regions, brain measures, classifiers, and statistical validations. Each of these components is described briefly in the subsections below.
I: Selection of Participants to which the Machine Learning Procedures are applied
The participants recruited into an imaging study are undoubtedly the most crucial determinant of how generalizable the performance of a machine learning-based classification will be in the larger population of individuals affected with the disorder that is being characterized and classified. Machine learning techniques are usually applied to imaging data that have been acquired in case-control studies(Stolley and Schlesselman, 1982, MacMaster et al., 2008, Plant et al., 2010, Klöppel et al., 2008b, Patel et al., 2008, Piven et al., 1995), which usually group-match participants on demographic characteristics such as age, sex, handedness, ethnicity, and socioeconomic status in the affected and healthy participants. In addition to matching groups on these variables, these studies also usually control for the effects of these demographic variables on imaging measures by controlling statistically for them and other potential confounders. Controlling for these effects, either by the selection of subjects who enter the study or statistically, is expected to allow the findings to generalize to the entire population of affected individuals. Imaging studies are expensive to conduct, however, and therefore the number of participants who can be included is usually limited to a few dozen participants – indeed, the largest imaging studies have had at most only a few hundred affected and healthy participants combined. The limited number of participants limits the ability to stratify across the key demographic features of the population, and if those demographic influences on brain structure and function overlap those features that are the basis for classification within the machine learning algorithms, or if they influence performance of those algorithms, then the trained machine classifications will likely be unable to diagnose accurately a new person from the larger population of affected individuals. To take an extreme example, if the classification boundary and decision rules were learned using data only from females(Savio et al., 2009) and if the features that enter the classifier include female-specific features that differ from males, then the classifier might not reasonably be expected to generalize to classify males. Thus, the selected participants on whom the classifier is trained should be as representative as possible of affected individuals in the larger population if the algorithm is to generalize to provide accurate classification of new individuals.
II: Imaging Modality
In vivo brain imaging data are acquired using one or more imaging modalities, including MRI, computed tomography (CT), positron emission tomography (PET), and single photon emission computed tomography (SPECT). Of these modalities, MRI is usually preferred for studies of machine learning-based classification because it is noninvasive and provides exquisite detail of brain structure and function, including high resolution, high contrast, and excellent SNR, at relatively low cost. In addition, MRI is perhaps the most versatile among the various imaging modalities because it can probe differing aspects of the tissue within the same scanner: anatomical MRI provides detailed information about the structure and shape of various brain regions(Naidich et al., 2009); functional MRI (fMRI) quantifies the change in concentration of oxygenated hemoglobin in the blood as a proxy for changes in neural activity(Buxton, 2009); diffusion tensor imaging (DTI) measures the diffusion of water molecules in the brain, which can be used to infer connectivity along the axons that connect one brain region to another(Mori, 2007); magnetic resonance spectroscopy (MRS) quantifies the concentration of various chemicals and metabolites in the brain(Atlas, 2008); and arterial spin labeling (ASL) measures perfusion of the blood in the brain by tagging the water molecules as they flow into the brain(Salvolini and Scarabino, 2006). These MR modalities provide complementary information about the brain, which may be differentially affected by disturbances in the brain associated with specific neuropsychiatric disorder.(Hao et al., 2011) Therefore, simultaneous use of brain measures from these modalities in principle should diagnose an individual more accurately than using measures only from a single MR modality. However, investigators thus far have used data from either anatomical MRI or fMRI alone, but not both together, for diagnostic classification. The accuracy of diagnostic classification using data from a single modality compared to other modalities, or to data from all modalities simultaneously, is unknown.
III: Methods for Delineating Brain Regions
Because morphometry is the study of shapes and forms, and because machine learning algorithms are applied to measures of regional brain morphology, the accurate delineation of brain regions is an important prerequisite for the accurate measure of feature vectors that the algorithm classifies. Inaccurate delineation of brain regions will ultimately undermine the accuracy of diagnostic classification using machine learning algorithms, and in many instances will prove fatal to those efforts. Delineation of a brain region by an expert in brain anatomy remains the gold-standard for definition of brain regions, but manual delineation is tedious and can be prone to drifts in measures over time and to variability across experts. Studies(Peterson, 2000, Peterson et al., 2007, Peterson et al., 2001a, Peterson et al., 2000, Peterson et al., 1993a, Peterson et al., 1993b, Peterson et al., 2001b, Peterson et al., 2003, Peterson et al., 2009, Plessen et al., 2006) that employ manual delineation should ensure accurate definition by extensively documenting all protocols for region definition, maintaining high reproducibility of measures across experts and across time by reassessing the reliability of the expert at predefined time intervals, and employing quality control checks on the definitions of each expert by an independent rater.
Because of the time, training, and expense required to implement gold-standard manual delineations of brain regions, most studies employ automated tools for region delineations. For example, the automated labeling pathway (ALP) platform combines a series of publicly available software packages, including Analysis of Functional Neuro-Images (AFNI), Brain Extraction Tool (BET)(Smith, 2002), FMRIB’s Linear Image Registration Tool (FLIRT), and Insight Segmentation and Registration Toolkit (ITK)(Yoo, 2004) to isolate brain from non-brain tissue(Wu et al., 2006), to spatially co-register each brain to a template brain using nonlinear deformations (in the ITK(Chen et al., 1999) algorithm), to segment brain tissue into gray and white matter, and to label brain regions. Labeling of brain regions can be aided using either the automated anatomical labeling (AAL) atlas containing 90 manually traced regions(Tzourio-Mazoyer et al., 2002), the Brodmann atlas that has 82 regions(Rorden and Brett., 2000) or locally generated regions defined from functional MRI studies(Carter et al., 2000). Automatically delineated regions, however, typically have large errors when compared with gold-standard manual definitions, especially near the outer boundary of the brain region being delineated, and they therefore tend to increase variance in the computed morphometric feature on which the classifier usually operates. These errors in region definition undermine the statistical power to detect brain features that discriminate groups when participant numbers are relatively small, as they usually are in imaging studies.
IV: Brain Measures
Feature vectors are morphological measures that can be compared statistically across groups of participants. The features that typically have been used to study persons with psychiatric disorders include volumes of entire brain regions, voxel-wise maps of thickness of the cortical mantel, voxel-wise maps of gray matter density (i.e., amount of gray matter per unit volume), local volumes of gray and white matter, and measures of local variation in the deformation fields that spatially normalize a brain to the template brain. Although the dimensionality of a feature vector (i.e. the number of features in the vector) in voxel-wise maps is typically on the order of hundreds of thousands, an imaging study comprises imaging data from at most several hundred participants. Because imaging data are available for only a limited number of participants, the probability distribution of features in a high-dimensional Euclidean space, called the feature space, cannot be discerned reliably. If the dimensionality of the feature space is not reduced using some sort of data reduction procedure, then data from an exorbitantly large number of participants would be required to learn accurate classification boundaries. For example, to estimate a multivariate Gaussian distribution of 10 features, data from more than 800,000 participants are required to ensure that at the mode the estimated distribution is close the true distribution.(Silverman, 1986) The dimensionality of the feature space therefore must be reduced significantly to learn the decision rules and classification boundaries that are valid in the affected population. Some of the popular methods for dimensionality reduction include (a) principal component analysis (PCA), which generates components that are uncorrelated with one another(Jolliffe, 2002), and (b) independent component analyses (ICA), which generates components that are statistically independent(Hyvarinen et al., 2001). Although statistically independent components are uncorrelated, components that are uncorrelated may not necessarily be statistically independent. In the reduced feature space, a machine learning algorithm compares two feature vectors using either their Euclidean distance, which computes the dissimilarity between feature vectors as the length of the line segment connecting them in the feature space, or the Mahalanobis distance, which accounts for the correlation among features and their variances to compute the distance between the feature vectors(Mahalanobis, 1936).
V: Choice of a Classifier
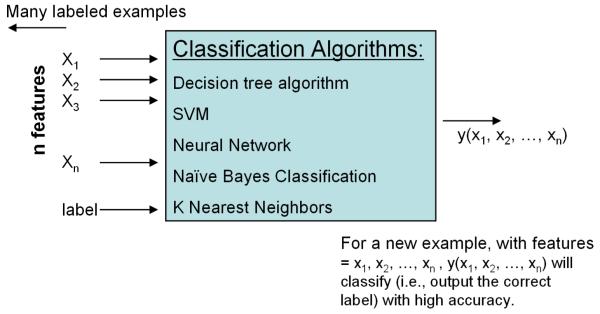
We now briefly review the various classifiers that have been used to diagnose patients as having one of several psychiatric disorders.(Mitchell, 1999) The last decade has witnessed an explosion in the use of classifiers in a wide range of disciplines that includes economics, astronomy, molecular biology, and diverse medical specialties such as cardiology and radiology. The use of classifiers in neuroimaging, however, is just beginning. A classifier is a computer algorithm that learns the most concise and accurate decision rules that best discriminate data among classes and applies the learned rules to data that it has not yet seen (Fig. 2).
Fig. 2. Classification Algorithm.
The input data for a classification algorithm is a set of labeled feature vectors (i.e. xi, a feature vector along with its associated class or diagnostic label).The classification algorithm generates a classification rule or decision boundary that best separates the feature vectors that belong to different classes or diagnostic labels. The learned decision rule is then applied to classify new feature vectors from a new participant as belonging to one of the various diagnostic classes.
Decision Tree Classifiers
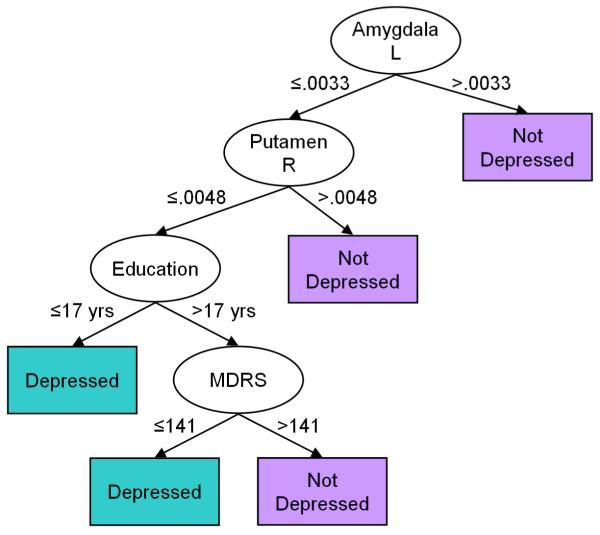
Decision trees are a popular tool for classifying data.(Hunt et al., 1966) They create a model tree that best predicts the diagnosis of a patient based on a feature vector in the patient’s brain. In a decision tree, a node represents a feature, an edge between two nodes represents a combination of the features at the two nodes, and a leaf represents a diagnosis. A new patient is diagnosed by first computing its features and then using the computed features to traverse the tree from the root node to a leaf that determines the diagnosis for that individual (Fig. 3).
Fig. 3.
A Hypothetical Decision Tree for Classifying an Individual as Depressed or Healthy using four features: the volume of the left amygdala and right putamen, educational level, and Mattis Dementia Rating Scale (MDRS). These four features are computed and used to classify a new individual by traversing down the tree. Starting at the root node, the node labeled Amygdala L, if the volume of the left amygdala is greater than the specified threshold then the individual is classified as healthy and the algorithm stops. Otherwise, it moves down along the edge labeled ≤ .0033 to the next node and compares the volume of the right putamen to the threshold: If the volume is greater than the threshold then the algorithm moves to the leaf node labeled Not Depressed and the individual is classified as healthy. Otherwise the process is repeated until a leaf node is reached and a diagnosis is assigned to the individual. The decision tree was obtained by personal communications with the authors(Dutta et al., 2007). L, Left; R, Right; MDRS, Mattis Dementia Rating Scale.
A decision tree is learned by assigning a feature to a node that best partitions the set of brains into two or more subsets, such that the majority of the brains in each subset belong to one or the other diagnostic labels. The feature assigned to the node is removed from the feature vector, and then each subset is recursively partitioned into further subsets using the remaining features in the vector. The recursive partitioning stops when all brains in a subset have the same diagnosis.
Decision tree algorithms have several limitations, including: (1) the learning algorithm may not generalize well to the larger population of affected people, (2) the optimal tree can be learned only by searching over all possible feature assignments to the nodes, and (3) small changes in the feature values of the training set may affect partitioning of the set into subsets, thereby producing large changes in the structure of the learned tree. Therefore, noise in the feature vectors can significantly affect the performance of the classifier. Decision tree classifiers, however, become increasingly stable and more accurate with an increasing number of participants in the training set, thereby producing a more accurate diagnostic classification of new participants.
Naive Bayes’ Classifiers
A naive Bayes’ classifier assumes that the features in feature vectors are statistically independent, and it uses this assumption to compute probabilities of a diagnosis from given sets of feature vectors. The estimated probabilities, in turn, are applied to diagnose an individual.(Domingos and Pazzani, 1997, Rish, 2001, Hand and Yu, 2001) Bayes’ theorem describes the relationships of the marginal, conditional, and joint probabilities of features and diagnostic labels that are used to learn the classification rule. A naive Bayes’ classifier may outperform other sophisticated classifiers because the assumption that the features are independent reduces the number of parameters of the probability distributions that must be estimated. The parameters in a naive Baye’s classifier therefore can be estimated accurately using fewer feature vectors, thereby increasing its performance as compared to that of other classifiers.
AdaBoost Classifiers
AdaBoost is a meta-algorithm that constructs a diagnostic classifier with high accuracy (i.e.,AdaBoost is a strong classifier) by linearly combining several classifiers that have low accuracy (i.e., separately they are weak classifiers).(Freund and Schapire, 1997) AdaBoost classifiers reduce both bias and variance in classification accuracy, and they maximize the distance between the decision boundary and the training feature vectors.(Freund and Schapire, 1997) Furthermore, AdaBoost classifiers are easy to implement for a wide variety of learning tasks, and they usually generalize well to the affected population. However, AdaBoost classifiers are sensitive to noise and outliers in the training data set and may not significantly improve accuracy if one of the classifiers in the linear combination is a strong classifier.
Support Vector Machine (SVM) Classifiers
A support vector machine (SVM) is a linear classifier(Cortes and Vapnik, 1995, Vapnik, 1999) -- i.e., it constructs a linear classification boundary in the feature space that best separates feature vectors in the training set. The classification boundary is an optimal hyperplane (a plane in feature spaces of dimension greater than two) that maximizes the distance between the hyperplane and the feature vectors that are closest to the hyperplane. The feature vectors that define the hyperplane are called the “support vectors” (Fig. 4).
Fig.4.
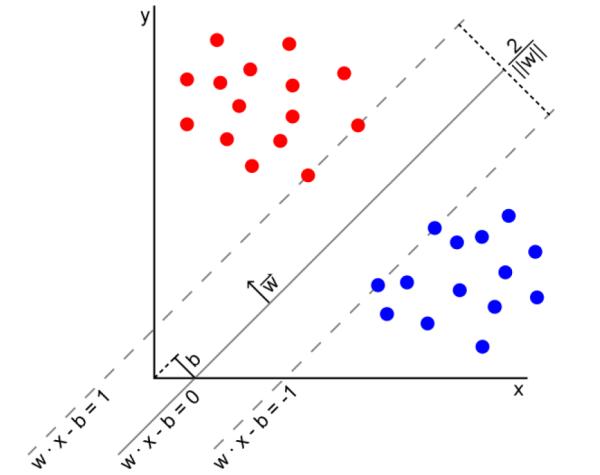
An Example Support Vector Machine (SVM) that separates the training samples that belong to one of two classes (red dots and blue dots) in the feature space using a maximum-margin hyperplane. Because the dimension of the feature space is 2 in this example, the hyperplane is a straight line separating the samples. The hyperplane is only estimated by few features called the support vectors, and therefore, small variations in support vectors may cause large variations in the estimated decision boundary. Furthermore, feature vectors other than the support vector do not influence the hyperplane. Thus, small changes in the location of the support vectors can significantly change the orientation of the hyperplane in the feature space (the n-dimensional Euclidean space of the features), thereby affecting the performance of the SVM in diagnosing a new patient not in the training set.
Support vector machines have been extended to classifying a feature vector among more than two diagnostic classes and for computing nonlinear separating planes by first mapping the features to a higher dimensional space using nonlinear functions and then estimating the optimal hyperplane that best separates the features in the higher dimensional space.(Aizerman et al., 1964) Although an SVM may perform well in classifying a new feature vector that was not used to train the SVM, the correct choice of the optimal mapping to the higher dimensional space and the formidable computational complexity remain the greatest limitations of SVM for diagnostic classification.(Burges, 1998) In addition, the decision boundary learned by an SVM is defined by the support vectors only, and therefore if the data are sparse, the classification boundary is highly sensitive to the noise in these vectors, such that even small variations in support vectors can lead to a large variation in the decision boundary. Furthermore, feature vectors other than support vectors do not define the decision boundary, and therefore, an SVM classifier ignores a large amount of valuable data, thereby undermining the generalizability of the learned boundary to the larger population of affected individuals.
VI: Validation Procedures
The performance of a classifier in diagnosing a new patient is evaluated by computing (1) misclassification rates using cross-validation procedures, and (2) prediction error using bootstrap procedures and Akaike’s Information Criteria (AIC). Prediction error measures the accuracy of a classifier in predicting the diagnostic class for an individual from its feature vector. Prediction error is computed as the expected value of the squared difference in the feature vector of the person and its predicted value by the classifier.(Efron and Tibshirani, 1993) In classification problems, the prediction error computes the probability of an incorrect classification, which is the probability that the predicted diagnosis differs from the actual diagnosis of a patient. A model with a lower prediction error will generalize better to the population than the model with a higher prediction error. The AIC computes a prediction error by adjusting the error using the complexity c (i.e., the number of diagnostic classes) of the learned model and an estimate of the residual variance of feature vectors in each class.
Bootstrapping
A bootstrap procedure estimates prediction error by (a) creating B sets of features with replacement from the original set of feature vectors, (b) generating B models from each of these B sets, and then (c) using the original set to compute the prediction error for each of the B models. The overall prediction error is computed as the average of the B prediction errors, one for each of the B models.(Efron and Tibshirani, 1993) However, this simple bootstrap procedure is biased, meaning that it underestimates the prediction error. An improved bootstrap procedure can be used to compute an unbiased estimate of the prediction error by first estimating the bias in the simple bootstrap procedure and then adding the estimated bias to generate the improved estimate of the prediction error.(Efron and Tibshirani, 1993)
Cross-Validation
A cross-validation procedure computes the sensitivity and specificity of a classifier by first dividing the set of feature vectors into two sets: a training set and a test set. Then it learns the classification rules using the feature vectors in the training set and evaluates the learned rules using the feature vectors in the test set. A method for cross-validation therefore divides the data into two parts: one part to generate the model, and the other to assess the model’s accuracy. An algorithm accurately diagnoses or classifies a participant if the person’s true diagnosis as identified by clinical experts matches the diagnosis that the algorithm assigns; otherwise the participant is misclassified. The misclassification rates for a classifier are computed using a three-step algorithm: (1) The set of N feature vectors is divided into n = N / k sets containing roughly equal numbers of feature vectors, where k is a user-specified number between 1 and N/2; (2) one of the n sets is selected as the test set, and the classification rules are learned using the feature vectors in the remaining n −1 sets. The feature vectors in the test set are used to compute the prediction error using the learned classification rules; (3) the second step is repeated using each of the n sets as the test set, yielding n estimates of the prediction errors, which are averaged to estimate the overall prediction error. For example, in a procedure termed “Leave-One-Out” (LOO) cross-validation, the validation procedure is repeated N times by treating every one of the N feature vectors as the test feature, training the classifier using the remaining features, and computing the misclassification rates by counting the number of feature vectors that were misclassified. The average misclassification rate is computed for each diagnosis by dividing the number of misclassified individuals by the total number of individuals who have that clinical diagnosis. A procedure termed “split-half” cross-validation, on the other hand, partitions the set into two halves, with an equal number of feature vectors. By dividing the set into several random halves, the split-half procedure can be repeated several times to compute the average and standard deviation of misclassification rates across these several applications of split-half cross-validation procedures.
B: Examples of classifiers for diagnosing psychiatric illnesses using MRI
MRI has enabled us to localize brain disturbances to specific brain regions, and these disturbances have been analyzed using machine learning algorithms to diagnose an individual as having a specific disorder or not. We will briefly describe neuroimaging findings in Alzheimer’s Disease (AD), Major Depressive Disorder (MDD), and Autism Spectrum Disorder (ASD) and the classifiers that have been developed to diagnose an individual as having one of these disorders.
I: Alzheimer’s Disease
Alzheimer’s Disease (AD) is a neurodegenerative disease characterized by progressive impairment of memory and other cognitive functions. Brain MRI measures have been demonstrated to be more sensitive indicators of AD than cognitive assessment alone.(Scheltens and Korf, 2000) Several MRI studies have identified severe localized volume loss, presumably representing synaptic and neuronal degeneration, in hippocampus(Hyman et al., 1984, Bobinski et al., 1996, Braak and Braak, 1991a, Braak and Braak, 1991b, Brion et al., 1994, Mesulam, 2000, Ball et al., 1985), entorhinal cortex(Hyman et al., 1984, Bobinski et al., 1996, Janke et al., 2001, Ball et al., 1985), amygdala(Braak and Braak, 1991a, Braak and Braak, 1991b, Brion et al., 1994, Mesulam, 2000), parahippocampal formation(Braak and Braak, 1991a, Braak and Braak, 1991b, Brion et al., 1994, Mesulam, 2000), and temporoparietal association cortices(DeCarli, 2000). Volume loss in the anterior hippocampus and amygdala occur even before the onset of clinical symptoms and in individuals who are at genetic risk of developing AD.(Lehtovirta et al., 2000, Fox et al., 1996) Furthermore, in the earlier stages of AD, hippocampal volume has been shown to be 1.75 standard deviations smaller than those in age-matched healthy participants.(Jack Jr. et al., 2000) MRI findings are possibly a sensitive measure of dementia(Scheltens and Korf, 2000), and early atrophy of hippocampus and the ventromedial temporal lobe is consistent with the presence of early memory impairment(Scheltens and Fox, 2002, Erkinjuntti et al., 1993, Braak et al., 1993). Hippocampal atrophy, however, is not unique to AD, as it has been associated with Parkinson’s dementia(Laakso et al., 1996) and otherwise healthy individuals who have impairments in recent memory(De Leon et al., 1999).
A number of procedures for machine learning using anatomical MRI data have been developed to diagnose individuals as having AD or not. Two studies(Zhang et al., 2011, Hinrichs et al., 2009) used data from both anatomical MRI and PET for classification. All of these procedures applied an SVM to learn the decision boundary in a feature space, and most validated performance of the classifier using an LOO cross-validation. Two studies applied 10-fold cross-validation(Zhang et al., 2011, Hinrichs et al., 2009), and another(Cuingnet et al., 2010) applied split-half validation by dividing the participants into a a training set or a test set. These procedures classified individuals with high sensitivity (SN ± SD = 83% ± 14%, range = 57%-100%) and specificity (SP ± SD = 88% ± 9%, range = 66%-97%) (Table 1).
Table 1.
Recent Procedures for Diagnosing an Individual as having Alzheimer’s Disorder or not using anatomical MRI data. Two studies used PET imaging data in addition to MRI data. Features extracted from these imaging data typically consisted of voxel-wise maps intensities across the entire brain, therefore yielding a high dimensional feature space. In all studies, the decision boundary within the feature space was learned using support vector machine (SVM), and the classification accuracy was typically assessed using leave-one-out (LOO) cross-validation. SN=sensitivity; SP=specificity; MCI=mild cognitive impairment; MCIc=MCI participants who converted to AD; MCInc=MCI participants who did not convert to AD; GM=gray matter; WM=white matter; CSF=cerebrospinal fluid; ROI=region of interest; PET=positron emission tomography; FDG-PET=18fluorodeoxyglucose-PET; HC=healthy controls; SPHARM=spherical harmonics; SPHARM-PDM=spherical harmonics-point distribution model; LOO=leave-one-out.
| Study | Participants | Features | Performance |
|---|---|---|---|
| Fan, 2008(Fan et al., 2008) |
56 with AD, 88 with MCI, and 66 healthy controls |
Voxel-wise maps of localized volumes in (1) GM, (2) WM, and (3) CSF. |
LOO cross-validation. (1) AD vs HC: Accuracy=94.3%; (2) MCI vs HC: Accuracy=81.8%; (3) AD vs MCI: Accuracy=74.3%. |
| Klöppel, 2008(Klöppel et al., 2008b) |
Group 1: 20 with AD and 20 healthy; Group 2: 14 with AD and 14 healthy. with histopathological confirmation; and Group 3: 33 with mild AD and 57 healthy. |
Voxel-wise measure of gray matter intensity |
LOO cross-validation. Group 1: SN=95%; SP=95%; Group 2: SN=100%; SP=85.7%; Group 3: SN=60.6%; SP=93%. |
| Gerardin, 2009(Gerardin et al., 2009) |
23 with AD; 23 with MCI; 25 healthy controls |
SPHARM coefficients for the shape of hippocampus |
LOO cross-validation. (1) AD vs HC: SN=96%; SP=92%; (2) MCI vs HC: SN=83%, SP=84%. |
| Hinrichs, 2009(Hinrichs et al., 2009) |
77 with AD and 82 healthy controls | Voxel-wise maps for (1) gray matter probability, and (2) FDG-PET intensity |
10-fold cross-validation. SN=78.5%; SP=81.8%. |
| Magnin, 2009(Magnin et al., 2009) |
16 with AD, and 22 healthy controls | Percent gray matter within 90 ROIs across the brain. |
Bootstrap validation. SN=91.5%, SP=96.6%. |
| Misra, 2009(Misra et al., 2009) |
103 with MCI who converted to AD (MCIc); and 76 MCI who did not convert to AD (MCInc). |
Voxel-wise maps of variations in localized volumes within (1) GM, (2) WM, and (3) CSF. |
LOO cross-validation. Accuracy (number of correctly classified) >75%. |
| Cuingnet, 2011(Cuingnet et al., 2011) |
162 elderly controls (HC); 137 with AD; 76 with MCI who converted to AD (MCIc); 134 with MCI who did not convert to AD (MCInc) |
(1) voxel-wise probability maps of GM, WM, and CSF; (2) Voxel-wise measure of cortical thickness; (3) Hippocampal volume and shape using spherical armonics |
LOO cross-validation. SN and SP varied across 28 different methods tested: (1) HC vs AD: SN>63%; SP>77%. The best SN=81% and SP=95% (Table 4(Cuingnet et al., 2011)); (2) HC vs MCIc: SN>22%; SP>73%. The best SN=65% and SP=94% (Table 5(Cuingnet et al., 2011)); (3) MCInv vs MCIc: SN>0%, SP>61%. The best SN=57% and SP=78% (Table 6(Cuingnet et al., 2011)). |
| Oliveira, 2010(Oliveira et al., 2010) |
14 with AD, and 20 healthy controls | Volume of 45 brain regions delineated automatically using FreeSurfer |
LOO cross-validation. SN=92.8%; SP=85%. |
| Zhang, 2011(Zhang et al., 2011) |
51 with AD, 99 with MCI, and 52 healthy controls |
For 93 ROIs, average measures of GM volume, and intensity in PET; and three proteins in CSF. |
10-fold cross-validation. (1) AD vs HC: SN=93%, SP=93.3%;(2) MCI vs HC: SN=81.8%, SP=66%. |
II: Major Depressive Disorder
Major Depressive Disorder (MDD) is defined by the presence of depressed mood and associated disturbances in sleep, appetite, energy, pleasure, self-esteem, and concentration that persist for more than two weeks. Using MRI, atrophy in cortex(Morris and Rapoport, 1990) (Ballmaier et al., 2004a, Pantel et al., 1997, Rabins et al., 1991), frontal lobe(Kumar et al., 2000), orbitofrontal cortex (Ballmaier et al., 2004b, Lai et al., 2000, Lee et al., 2003), gyrus rectus and anterior cingulate (Ballmaier et al., 2004b), hippocampus(Shah et al., 1998, Sheline et al., 1999, Steffens et al., 2000, Steffens et al., 2002, Krishnan et al., 1992, Steffens and Krishnan, 1998), amygdala(Sheline et al., 1999, Steffens et al., 2000), and lesions in the basal ganglia(Rabins et al., 1991, Steffens and Krishnan, 1998, Tupler et al., 2002), especially in the caudate (Krishnan et al., 1992) and putamen (Steffens and Krishnan, 1998, Tupler et al., 2002), have been associated with MDD. MRI studies have also identified differences between early versus late-onset MDD (Ballmaier et al., 2004b, Ballmaier et al., 2004a), with late-onset illness showing less frontal and more temporal and parietal atrophy. In addition, several studies using semi-quantitative ratings(Butters et al., 2004, Greenwald et al., 1998) and semi-automated measures(Taylor et al., 2003) have reported an increased prevalence of white matter hyperintensities in periventricular and subcortical regions, especially in the elderly.(Salloway et al., 1996) Diffusion tensor imaging (DTI) has shown decreased anisotropy in water diffusion, and therefore possible loss of integrity in neural pathways across white matter, of the prefrontal cortex.(Alexopoulos et al., 2002, Taylor et al., 2004, Hickie et al., 1995) Patients with subsyndromal depression had similar patterns of atrophy in the prefrontal cortex as those with MDD.(Kumar et al., 1998) Although several studies have associated disturbances with MDD using anatomical MRI, only one fMRI study in 19 patients with MDD and 19 healthy controls has attempted to diagnose an individual as having MDD using an affect task of sad faces.(Fu et al., 2007) This latter study applied SVM to the voxel-wise maps of activation across the brain and reported high sensitivity (=84%) and specificity (=89%) in LOO cross-validation analyses. However, because the study employed fMRI data from only 38 participants to learn the classification rules, and then computed sensitivity and specificity using LOO cross-validation, the method may not generalize well to classify an individual in clinical populations.
III: Autism Spectrum Disorders (ASDs)
ASDs are characterized by abnormalities in reciprocal social interaction and communication, as well as the presence of restricted interests and behaviors. These disorders usually manifest before the age of three years. Imaging studies have demonstrated disturbances in the cortex, corpus callosum(Piven et al., 1997), basal ganglia(Sears et al., 1999), and cerebellum(Piven et al., 1992), and they have reported correlations among gray matter volume, Intelligence Quotient (IQ), and measures of symptom severity(Rojas et al., 2006). Previous attempts to diagnose an individual as having ASD have applied either SVM(Ecker et al., 2010b, Ecker et al., 2010a) or LPBoost(Singh et al., 2008) to voxel-wise maps of (1) localized volume change in GM, WM, and CSF(Ecker et al., 2010b), (2) cortical thickness and average convexity, mean curvature, local area, and distortion of the cortical surface(Ecker et al., 2010b), or (3) cortical thickness(Singh et al., 2008). These methods evaluated classification accuracy, which was greater than 85%, using methods for either LOO(Ecker et al., 2010b), leave-two-out(Ecker et al., 2010a), or 9-fold(Singh et al., 2008) cross-validation. The classification rules, however, were learned using imaging data from fewer than 40 participants, whereas the dimensionality of the feature space was very large, on the order of thousands of voxels across the entire brain. In addition, the several brain regions that contributed to classifying an individual were small, typically fewer than 100 voxels, and they were randomly scattered across the entire brain. Although the small, randomly scattered regions may have classified the small number of participants with high accuracy, disturbances in the brains of individuals with ASDs are unlikely to be randomly distributed in this way in small regions across the brain. Thus, because these methods used brain regions that may not be representative of the disturbances in ASDs, and because they used a small number of participants that may not be representative of the larger population of people with ASDs, the learned classification rules cannot yet be used within clinical settings to diagnose a patient as having an ASD (Table 2).
Table 2.
Recent Procedures for Diagnosing an Individual as having Autism Spectrum Disorders (ASDs) or not using anatomical MRI data. The classification accuracy was computed using methods for leave-k-out (LOO) cross-validation. SN=sensitivity; SP=specificity; GM=gray matter; WM=white matter; CSF=cerebrospinal fluid; LOO=leave-one-out; SVM=support vector machine.
| Study | Participants | Features | Performance |
|---|---|---|---|
| Ecker 2010a(Ecker et al., 2010a) |
20 with ASD and 20 healthy controls | Five morphological measures brain surfaces: (1) convexity and concavity, (2) mean curvature, (3) metric distortion, (4) cortical thickness, and (5) pial area |
SN>85%, SP>85% of a SVM classifier computed using leave- two-out cross validation |
| Ecker 2010b(Ecker et al., 2010b) |
22 with ASD and 22 healthy controls | Locally averaged voxels from GM, WM, and CSF |
SN=88%, SP=86% of a SVM classifier computed using LOO cross validation |
| Singh 2008(Singh et al., 2008) |
16 with ASD and 11 healthy controls | Cortical thickness | Accuracy (number of correctly classified) ~90% of a LPBoost classifier computed using 9-fold cross validation |
C: Discussion
Many sophisticated imaging and statistical methods have been developed to diagnose an individual automatically as having a specific neuropsychiatric disorder. These methods learned classification rules and decision boundaries by applying support vector machines (SVMs) to selected image features, such as a voxel-wise map of gray matter intensities, gray matter volumes, white matter volumes, or volumes and shape features of various subcortical regions of the brain. Classification accuracy was evaluated using either leave-one-out (LOO) or 10-fold cross-validation, with reported sensitivities and specificities frequently being greater than 90%. Most of these machine learning algorithms were applied to diagnose an individual as having Alzheimer’s Disease (AD). However, these methods may not generalize well in the affected populations because the classification rules were learned from small groups of participants and from image features that may not be clinically relevant, and the classification accuracies were not assessed in clinical settings. Furthermore, although LOO cross-validation is a widely accepted method for evaluating the performance of classification algorithms, it may have overestimated the sensitivity and specificity of the classifiers, especially because data were available for only a small number of participants. Therefore, more rigorous validation and statistical procedures, in particular split-half validation, should be used to evaluate accurately the performance of these classifiers. Thus, although the classification methods used thus far in imaging studies have shown that brain morphometry has the potential to diagnosis individuals accurately, evaluation of the performance of the classifiers using more representative features and more rigorous validation procedures are essential to ensure accurate and valid performance estimates and to improve the generalizability of the learned classification rules within the affected population.
All attempts thus far to use imaging-based automated classifiers to diagnose psychiatric disorders have assessed performance of the classifiers against the “gold standard” clinical diagnoses, though exactly how golden those standards are, is questionable. For example, even when those clinical diagnoses are made rigorously using structured or semi-structured diagnostic interviews, those diagnoses may not be perfect. Moreover, and possibly more problematically, the unquestionable etiological and neurobiological heterogeneity of psychiatric diagnoses has for some time brought the categorical boundaries for clinical diagnoses under increasing suspicion and criticism as being invalid. The extent to which those diagnoses are invalid is the extent to which the assessment of performance of imaging-based classifiers will be undermined. Certainly the performance of a classifier can be no better than the quality of the gold standard against which it is evaluated, because even if the brain-based classifier were to diagnose patients more accurately than do symptom-based clinical diagnoses, we could never know it, given that we have nothing better or more golden than the clinical diagnosis by which to judge the brain-based classifier. Some kind of independent gold standard would be needed to evaluate and validate both the brain-based classification and clinical diagnosis, such as their ability to predict future clinical course, treatment response, behavioral or cognitive profiles, or comorbid illnesses. Nevertheless, the difficulties that the etiological and neurobiological heterogeneity pose for the validity of symptom-based clinical diagnoses does not entirely doom or preclude their utility as a benchmark for evaluating imaging-based classifiers, given that whatever degree of validity they have provides the opportunity to demonstrate the diagnostic potential of a putative classifier and can provide proof-of-concept support for the entire enterprise of imaging-based classification. Also certainly, it is impossible for classifiers consistently perform at a level that is statistically significantly better than chance if symptom-based clinical diagnoses are entirely invalid. Furthermore, the presence of neurobiological heterogeneity for any single clinical diagnoses does not mean that those neurobiological subtypes have nothing in common with one another. Indeed, the morphological signatures in the brain of those neurobiological subtypes may as a whole have more in common with one another than they have with the morphological signatures of other symptom-based clinical diagnoses. Those subtypes, in fact, may very well share certain features that constitute in essence a “final common pathway” that is responsible for those features producing overlapping symptom complexes and that have caused those symptom complexes to be classified as a single clinical disorder in the first place. The fact that the best performing imaging-based classifiers consistently report diagnostic accuracies in the 80-90% range suggests that the “gold standard” clinical diagnoses must be at least that good themselves in terms of having brain features that are more alike than they are like the brain features of the comparator group.
Methods for diagnostic classification typically have used SVMs to generate classification boundaries and decision rules using imaging data from participants who have gold-standard clinical diagnoses already available, and they have used those classifiers to diagnose individuals who were not included in the datasets used to generate the diagnostic algorithms. Reported classification accuracies generally have exceeded 85% and typically have been estimated using either LOO cross-validation or 10-fold cross-validation. LOO cross-validation is sensitive to noise in the imaging data, especially if the feature space consists of voxelwise maps of brain measures and if the imaging data are available for only a small number of participants. Although 10-fold cross-validation in principle should be more robust and more accurate in assessing the accuracy of diagnostic classification than LOO cross-validation, the limited number of participants in these studies (typically fewer than 100 combined across diagnostic groups) likely rendered performance of these two methods comparable. The limitations of cross-validation techniques undermine the accuracy estimates of these diagnostic methods and argue for use of more rigorous validation procedures, such as split-half cross-validation, to assess diagnostic accuracy of the classifiers.
The accuracy of diagnostic classifications using imaging data alone is limited by the features of the image that are used to generate the classification boundaries and decision rules. Although the computational and statistical complexities of classifiers have increased rapidly in the last two decades, their classification accuracy has typically not surpassed 85-90%. Further improvements in diagnostic accuracy will require more accurate localization and measurement of anatomical disturbances that are both robust and specific to particular psychiatric illnesses. Classifiers have used increasingly fine-grained features from imaging data across the entire brain in an effort to capture anatomical disturbances that are more circuit-based and distributed across multiple brain regions.(Peterson, 2010) Using fine-grained, voxel-wise maps of local volume disturbances prohibitively increases noise in the imaging features and increases the dimensionality of the feature space, thereby requiring data from many more participants to generate models for diagnostic classification that are accurate and generalizable. Therefore, using features that are computed at an appropriate level of spatial resolution might be essential to help reduce the dimensionality of the feature space and make more tractable the study of a sufficient sample of participants to generate classifiers that generalize to the larger population of affected individuals.
The sampling frames used to identify patients and healthy control participants in the attempts of research studies thus far to use classifiers to diagnose patients on the basis of features in brain images also likely will prove to limit the generalizability and practical utility of these approaches to diagnosis. Patients in these studies have been identified from samples of convenience, usually from psychiatric clinics, and therefore they are not truly representative of all affected people in the general population.(Berkson, 1946) Similarly, the healthy participants, although usually recruited to match the patient group demographically, have been screened so that they do not have any psychiatric disorder or neurological disease, and therefore also are not representative of the general community. The generalizability of a classifier therefore is limited by the demographic, clinical, and behavioral characteristics of the participants used to generate the classification boundaries. For example, excluding participants who fear the tight spaces of the MRI scanner may yield a non-representative sample of research participants in a study of generalized anxiety disorders. Similarly, excluding patients with DSM-IV Axis 1 comorbodities would create a sample that is not representative of the population affected with Bipolar Disorder (BPD). Thus, for a classifier to diagnose accurately a patient within clinical settings, and for performance estimates of the classifier generated in research settings to estimate likely performance within clinical settings, the classifier must be generated using imaging data from participants that are as representative as possible of affected people in the general population.
The greatest challenge in generating classifiers in research studies that can be applied to the affected population in clinical settings is the availability of imaging data from only a small number of participants. The limitation of sample size can perhaps be overcome by the growing trend to establish national repositories of imaging data from federally funded imaging studies. Pooling data into one repository should significantly increase the imaging data available for the training and testing of diagnostic classifiers. Large samples of pooled data also afford the possibility of being more representative of both the affected and healthy populations across the country. An example of such a public database is Alzheimer’s Disease Neuroimaging Initiative (ADNI), which has acquired imaging data in 895 elderly persons who are healthy, with Alzheimer’s disease, or with mild cognitive impairment. Merging imaging data across multiple laboratories has its own challenges, however, including differing ascertainment procedures for recruiting participants, differing manufacturers and performance of the MRI scanners, differing SNR and contrast in the images, differing patterns of inhomogeneity in image intensity across the imaging volume, and differing patterns of geometric distortion in the images. In addition, differing methods for processing the images and for defining brain regions have large effects on the imaging measures that enter classifiers. Each of these confounding factors reduces the accuracy and statistical power of the diagnostic classifiers and therefore must be carefully addressed to generate valid decision rules and classification boundaries in datasets combined across sites in multi-site imaging studies.
The presence of differing anatomical subtypes of a single, phenotypically defined illness presumably will reduce the accuracy of classifiers that are based upon identifying common anatomical features that discriminate between two or more phenotypically identified disorders. A classification procedure that is trained using imaging data from one anatomical subtype perhaps cannot accurately classify a participant from a differing anatomical subtype unless those two subtypes share more imaging features with one another than they do with the conditions against which they are being compared and classified. Because anatomical subtypes of neuropsychiatric disorders have not yet been identified, increasing the number of participants seems to be the only available recourse at present to reduce the adverse effects of biological heterogeneity on the performance of a diagnostic classifier.
The performance of a classifier can be improved significantly by reducing errors when measuring the feature vectors that are used to train and test the classifier. Errors in delineating brain regions as well as errors in localizing and quantifying disturbances across multiple regions across the brain will increase measurement error in the feature vectors and thereby will undermine the accuracy of any given classifier. Automated methods for delineating brain regions, for example, or failing to account adequately for artifactual variations in image intensity across the brain, typically introduce large measurement errors near the surfaces of brain regions, particularly for small regions located deep in the brain. Yet current methods for classification typically employ automated methods for delineating brain regions because manual delineation is labor-intensive and expensive. However, manual delineation of brain regions is at present the only way to minimize errors sufficiently in the feature vectors to improve substantially the accuracy of the diagnostic algorithms.
Although current methods for making computer-based diagnoses using brain images alone cannot yet be applied within clinical settings, a growing number of studies thus far provides strong proof-of-concept evidence that these techniques can and will be used to diagnose individuals with high accuracy in the near future. Although the accuracy of diagnosis using these algorithms thus far has generally not exceeded 90%, imaging datasets from much larger and more representative populations, when subjected to advanced image processing and statistical techniques that provide precise and fine-grained features across the entire brain, will be able to generalize better to the larger population of affected individuals and therefore will make the automated diagnosis of psychiatric illness using brain images alone clinically realistic and logistically feasible.
Key Points.
Numerous recent studies using advanced imaging technologies provide strong evidence that individuals can be correctly diagnosed using appropriate brain imaging measures and automated, machine-based methods for classification.
Nevertheless, the automated diagnosis of an individual as having a specific psychiatric disorder or not using only MRI data in routine clinical settings still faces considerable challenges.
The generalizability of learned classification rules to affected populations is limited by the availability of imaging data from a relatively small numbers of participants and the high dimensionality of imaging measures.
Imaging measures that encode the nature and locations of disturbances in the brain for specific disorders will be necessary for the accurate clinical diagnosis of individual patients in clinical settings.
Rigorous, split-half cross-validations procedures should be used alongside other less stringent, but more common, validation procedures to assess performance of classifiers within real-world, clinical settings.
Acknowledgements
This work was supported in part by NIMH grants MH036197, MH068318, MH16434, MH089582, 1P50MH090966, and K02-74677, NIDA grants DA017820 and DA027100, and the Brain & Behavior Research Foundation (formerly NARSAD).
Glossary
- Classification Boundaries
are hypersurfaces that partition the feature spaces into regions of features such that brains within each region have the same diagnosis.
- Cross-Validation Procedures
compute the sensitivity and specificity of a classifier first by generating the classifier using a set of features called the training set, and then by evaluating the performance of the classifier using an independent set of features called the test set.
- Euclidean Distance
is a measure of dissimilarity between two feature vectors. It is computed as the length of the line segment connecting the two features in the feature space.
- Feature Vector
is a vector of features that encode the location and magnitude of the brain measures used to generate a classifier.
- Independent Component Analysis
generates components (i.e. linear combinations of features) that are pairwise statistically independent.
- Machine Learning
aims at generating decision rules and classification boundaries within a feature space that permit assigning a diagnostic label to each brain based on its specific set of features.
- Mahalanobis Distance
is a measure of dissimilarity between two feature vectors that accounts for correlations among features and their variances in the feature space.
- Morphometry
studies forms and shapes by quantifying their features of interests and correlating these features with clinical, behavioral, and genetic measures of individuals.
- Principal Component Analysis
generates components (i.e. linear combinations of the features) that are uncorrelated with one another.
- Supervised Machine Learning
generates decision rules and classification boundaries using a set of features from brains with known clinical diagnosis.
- Template Brain
is a brain of an individual that is morphologically most representative of the brains of individuals in a study.
- Unsupervised Machine Learning
generates naturalistic groupings of brains using their features without a priori knowing their clinical diagnosis.
References
- AIZERMAN M, BRAVERMAN E, ROZONOER L. Theoretical foundations of the potential function method in pattern recognition learning. Automation and Remote Control. 1964;25:821–837. [Google Scholar]
- ALEXOPOULOS GS, KIOSSES DN, CHOI SJ, MURPHY CF, LIM KO. Frontal white matter microstructure and treatment response of late-life depression: a preliminary study. American Journal of Psychiatry. 2002;159:1929–1932. doi: 10.1176/appi.ajp.159.11.1929. [DOI] [PubMed] [Google Scholar]
- ASHBURNER J, FRISTON KJ. Voxel-Based Morphometry—The Methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- ATLAS SW. Magnetic resonance imaging of the brain and spine. Lippincott Williams & Wilkins; 2008. [Google Scholar]
- BALL M, FISMAN M, HACHINSKI V, BLUME W, FOX A, KRAL VA, KIRSHEN AJ, FOX H, MERSKEY H. A new definition of Alzheimer’s disease: a hippocampal dementia. The Lancet. 1985;1:14–16. doi: 10.1016/s0140-6736(85)90965-1. [DOI] [PubMed] [Google Scholar]
- BALLMAIER M,A, KUMAR A, THOMPSON PM, NARR KL, LAVRETSKY H, ESTANOL L, DELUCA H, TOGA AW. Localizing gray matter deficits in late-onset depression using computational cortical pattern matching methods. American Journal of Psychiatry. 2004a;161:2091–2099. doi: 10.1176/appi.ajp.161.11.2091. [DOI] [PubMed] [Google Scholar]
- BALLMAIER M, TOGA AW, BLANTON RE, SOWELL ER, LAVRETSKY H, PETERSON J, PHAM D, KUMAR A. Anterior cingulate, gyrus rectus, and orbitofrontal abnormalities in elderly depressed patients: an MRI-based parcellation of the prefrontal cortex. American Journal of Psychiatry. 2004b;161:99–108. doi: 10.1176/appi.ajp.161.1.99. [DOI] [PubMed] [Google Scholar]
- BANSAL R, STAIB LH, WANG Y, PETERSON BS. ROC-based assessments of 3D cortical surface-matching algorithms. NeuroImage. 2005;24:150–162. doi: 10.1016/j.neuroimage.2004.08.054. [DOI] [PubMed] [Google Scholar]
- BANSAL R, STAIB LH, XU D, ZHU H, PETERSON BS. Statistical Analysis of Brain Surfaces Using Gaussian Random Fields on 2D Manifold. IEEE Transactions on Medical Imaging. 2007;26:46–57. doi: 10.1109/TMI.2006.884187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARNES J, SCAHILL RI, BOYES RG, FROST C, LEWIS EB, ROSSOR CL. Differentiating AD from aging using semiautomated measurement of hippocampal atrophy rates. NeuroImage. 2004;23:574–581. doi: 10.1016/j.neuroimage.2004.06.028. [DOI] [PubMed] [Google Scholar]
- BERKSON J. Limitations of the application of fourfold table analysis to hospital data. Biometrics. 1946;2:47–53. [PubMed] [Google Scholar]
- BOBINSKI M, WEGIEL J, WISNIEWSKI HM, TARNAWSKI M, BOBINSKI M, REISBERG B, DE LEON MJ, MILLER DC. Neurofibrillary pathology--correlation with hippocampal formation atrophy in Alzheimer disease. Neurobiology of Aging. 1996;17:909–919. doi: 10.1016/s0197-4580(97)85095-6. [DOI] [PubMed] [Google Scholar]
- BOOKSTEIN FL. Morphometric tools for landmark data: Geometry and biology. Cambridge University Press; 1997. [Google Scholar]
- BRAAK H, BRAAK E. Demonstration of amyloid deposits and neurofibrillary changes in whole brain sections. Brain Pathology. 1991a;1:213–216. doi: 10.1111/j.1750-3639.1991.tb00661.x. [DOI] [PubMed] [Google Scholar]
- BRAAK H, BRAAK E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica. 1991b;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- BRAAK H, BRAAK E, BOHL J. Staging of Alzheimer-related cortical destruction. European Neurology. 1993;33:403–408. doi: 10.1159/000116984. [DOI] [PubMed] [Google Scholar]
- BRION J, OCTAVE JN, COUCK AM. Distribution of the phosphorylated microtubule-associated protein tau in developing cortical neurons. Neuroscience. 1994;63:895–909. doi: 10.1016/0306-4522(94)90533-9. [DOI] [PubMed] [Google Scholar]
- BURGES CJC. A tutorial on support vector machines for pattern recognition. Data Mining and Knowledge Discovery. 1998;2:121–167. [Google Scholar]
- BUTTERS MA, WHYTE EM, NEBES RD, BEGLEY AE, DEW MA, MULSANT BH, ZMUDA MD, BHALLA R, MELTZER CC, POLLOCK BG, REYNOLDS CF, III, BECKER JT. The nature and determinants of neuropsychological functioning in late-life depression. Archives of General Psychiatry. 2004;61:587–595. doi: 10.1001/archpsyc.61.6.587. [DOI] [PubMed] [Google Scholar]
- BUXTON RB. Introduction to Functional Magnetic Resonance Imaging: Principles and Techniques. Cambridge University Press; 2009. [Google Scholar]
- CARTER CS, MACDONALD AM, BOTVINICK M, ROSS LL, STENGER VA, NOLL D, COHEN JD. Parsing executive processes: strategic vs. evaluative functions of the anterior cingulate cortex. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1944–1948. doi: 10.1073/pnas.97.4.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN W, ZHU X-H, THULBORN KR, UGURBIL K. Retinotopic mapping of lateral geniculate nucleus in humans using functional magnetic resonance imaging. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2430–2434. doi: 10.1073/pnas.96.5.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHUNG MK, DALTON KM, DAVIDSON RJ. Tensor-Based Cortical Surface Morphometry via Weighted Spherical Harmonic Representation. IEEE Transactions on Medical Imaging. 2008;27:1143–1151. doi: 10.1109/TMI.2008.918338. [DOI] [PubMed] [Google Scholar]
- CORTES C, VAPNIK V. Support-Vector Networks. Machine Learning. 1995;20 [Google Scholar]
- CSERNANSKY JG, SCHINDLER MK, SPLINTER NR, WANG L, GADO M, SELEMON LD, RASTOGI-CRUZ D, POSENER JA, THOMPSON PA, MILLER MI. Abnormalities of thalamic volume and shape in schizophrenia. Am. J. Psychiatry. 2004;161:896–902. doi: 10.1176/appi.ajp.161.5.896. [DOI] [PubMed] [Google Scholar]
- CUINGNET R, GERARDIN E, TESSIERAS J, AUZIAS G, LEHÉRICY S, HABERT M-O, CHUPIN M, BENALI H, COLLIOT O. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. NeuroImage. 2010 doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- CUINGNET R, GERARDIN E, TESSIERAS J, AUZIAS G, LEHÉRICY S, HABERT M-O, CHUPIN M, BENALI H, COLLIOT O. Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. NeuroImage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- DAVATZIKOS C, FAN Y, WU X, SHEN D, RESNICK SM. Detection of prodromal Alzheimer’s disease via pattern classification of MRI. Neurobiol. Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVATZIKOS C, SHEN D, GUR RC, WU X, LIU D, FAN Y, HUGHETT P, TURETSKY BI, GUR RE. Whole-brain morphometric study of schizophrenia reveals a spatially complex set of focal abnormalities. Arch. General Psychiatry. 2005;62:1218–1227. doi: 10.1001/archpsyc.62.11.1218. [DOI] [PubMed] [Google Scholar]
- DE LEON MJ, CONVIT A, DE SANTI S, BOBINSKI M. Structural neuroimaging: Early diagnosis and staging of Alzheimer’s disease. Alzheimer’s Disease and Related Disorders. 1999;14:105–126. [Google Scholar]
- DECARLI C. Part IV. Neuroimaging in dementing disorders. Disease-a-Month. 2000;46:706–724. doi: 10.1016/s0011-5029(00)90031-2. [DOI] [PubMed] [Google Scholar]
- DOMINGOS P, PAZZANI M. On the optimality of the simple Bayesian classifier under zero-one loss. Machine Learning. 1997;29:103–137. [Google Scholar]
- DUCHESNAY E, CACHIA A, ROCHE A, RIVIÈRE D, COINTEPAS Y, PAPADOPOULOS-ORFANOS D, ZILBOVICIUS M, MARTINOT J-L, RÉGIS J, MANGIN JF. Classification Based on Cortical Folding Patterns. IEEE Trans on Medical Imaging. 2007;26:553–565. doi: 10.1109/TMI.2007.892501. [DOI] [PubMed] [Google Scholar]
- DUTTA P, WU M, TAMBURO R, BUTTERS M, REYNOLDS C, AIZENSTEIN H. Tensor-based morphometry and classifier algorithms for the identification of structural brain changes in geriatric depression. International Society for Magnetic Resonance in Medicine Conference.2007. [Google Scholar]
- ECKER C, MARQUAND A, MOURÃO-MIRANDA J, JOHNSTON P, DALY EM, BRAMMER MJ, MALTEZOS S, MURPHY CM, ROBERTSON D, WILLIAMS SCR, MURPHY DG. Describing the Brain in Autism in Five Dimensions—Magnetic Resonance Imaging-Assisted Diagnosis of Autism Spectrum Disorder Using a Multiparameter Classification Approach. The Journal of Neuroscience. 2010a;11:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECKER C, ROCHA-REGO V, JOHNSTON P, MOURAO-MIRANDA J, MARQUAND A, DALY EM, BRAMMER MJ, MURPHY C, MURPHY DG, CONSORTIUM’ TMA. Investigating the predictive value of whole-brain structural MR scans in autism: A pattern classification approach. NeuroImage. 2010b;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- EFIGUEIREDO RJ, SHANKLE WR, MACCATO A, DICK MB, MUNDKUR P, MENA I. Neural-network-based classification of cognitively normal, demented, Alzheimer disease and vascular dementia from single photon emission with computed tomography image data from brain. Proc Natl Acad Sci. 1995;92:5530–5534. doi: 10.1073/pnas.92.12.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFRON B, TIBSHIRANI RJ. An Introduction to the Bootstrap. Chapman & Hall; 1993. [Google Scholar]
- ERKINJUNTTI T, LEE DH, GAO F, STEENHUIS R, ELIASZIW M, FRY R, MERSKEY H, HACHINSKI VC. Temporal lobe atrophy on magnetic resonance imaging in the diagnosis of early Alzheimer’s disease. Archives of Neurology. 1993;50:305–310. doi: 10.1001/archneur.1993.00540030069017. [DOI] [PubMed] [Google Scholar]
- FAN Y, BATMANGHELICH N, CLARK CM, DAVATZIKOS C. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008;39:1731–1743. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN Y, SHEN D, DAVATZIKOS C. Classification of structural images via high-dimensional image warping, robust feature extraction, and {SVM}. Med Image Comput Comput Assist Interv Int Conf; 2005. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- FOX NC, WARRINGTON EK, FREEBOROUGH PA, HARTIKAINEN P, KENNEDY AM, STEVENS JM, ROSSOR MN. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study. Brain. 1996;119:2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- FREUND Y, SCHAPIRE RE. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. Journal of Computer and System Sciences. 1997;55:119–139. [Google Scholar]
- FU CHY, MOURAO-MIRANDA J, COSTAFREDA SG, KHANNA A, MARQUAND AF, WILLIAMS SCR, BRAMMER MJ. Pattern Classification of Sad Facial Processing: Toward the Development of Neurobiological Markers in Depression. Biol Psychiatry. 2007;63:656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- GERARDIN E, CHETELAT G, CHUPIN M, CUINGNET R, DESGRANGES B, KIM HS, NIETHAMMER M, DUBOIS B, LEHERICY S, GARNERO L, EUSTACHE F, COLLIOT O. Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. NeuroImage. 2009;47:1476–1486. doi: 10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREENWALD BS, KRAMER-GINSBERG E, KRISHNAN KR, ASHTARI M, AUERBACH C, PATEL M. Neuroanatomic localization of magnetic resonance imaging signal hyperintensities in geriatric depression. Stroke. 1998;29:613–617. doi: 10.1161/01.str.29.3.613. [DOI] [PubMed] [Google Scholar]
- HAKER S, TANNENBAUM A, KIKINIS R. Mass Preserving Mappings and Image Registration. In: NIESSEN WJ, VIERGEVER MA, editors. Proceedings of the 4th International Conference on Medical Image Computing and Computer-Assisted Intervention; London: Springer-Verlag; 2001. pp. 120–127. [Google Scholar]
- HAND DJ, YU K. Idiot’s Bayes - not so stupid after all? International Statistical Review. 2001;69:385–399. [Google Scholar]
- HAO X, XU D, BANSAL R, DONG Z, LIU J, WANG Z, KANGARLU A, LIU F, DUAN Y, SHOVA S, GERBER AJ, PETERSON BS. Multimodal Magnetic Resonance Imaging: The Coordinated Use of Multiple, Mutually Informative Probes to Understand Brain Structure and Function. Human Brain Mapping. 2011 doi: 10.1002/hbm.21440. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERHOLZ K, SALMON E, PERANI D, BARON JC, HOLTHOFF V, FROLICH L. Discrimination between Alzheimer dementia and controls by automated analysis of multicenter FDG PET. NeuroImage. 2002;17:302–316. doi: 10.1006/nimg.2002.1208. [DOI] [PubMed] [Google Scholar]
- HICKIE I, SCOTT E,P,M, WILHELM K, AUSTIN MP, BENNETT B. Subcortical hyperintensities on magnetic resonance imaging: clinical correlates and prognostic significance in patients with severe depression. Biological Psychiatry. 1995;37:151–160. doi: 10.1016/0006-3223(94)00174-2. [DOI] [PubMed] [Google Scholar]
- HINRICHS C, SINGH V, XU G, JOHNSON SC. MKL for Robust Multi-modality AD Classification. International Conference on Medical Image Computing and Computer Assisted Intervention; London, UK: 2009. pp. 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNT EB, MARIN J, STONE PJ. Experiments in in induction. Academic Press; New York: 1966. [Google Scholar]
- HYMAN B, VAN HORSEN GW, DAMASIO AR, BARNES CL. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- HYVARINEN A, KARHUNEN J, OJA E. Independent Component Analysis. John Wiley & Sons, Inc.; 2001. [Google Scholar]
- JACK CR, SHIUNG MM, GUNTER JL, O’BRIEN PC, WEIGAND SD, KNOPMAN DS. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACK CR, JR., PETERSEN RC, XU Y, O’BRIEN PC, SMITH GE, IVNIK RJ, BOEVE BF, TANGALOS EG, KOKMEN E. Rates of hippocampal atrophy in normal aging, mild cognitive impairment, and Alzheimer’s disease. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANKE A, DE ZUBICARAY G, ROSE SE, GRIFFIN M, CHALK JB, GALLOWAY GJ. 4D deformation modeling of cortical disease progression in Alzheimer’s dementia. Magnetic Resonance in Medicine. 2001;46:661–666. doi: 10.1002/mrm.1243. [DOI] [PubMed] [Google Scholar]
- JOLLIFFE IT. Principal Component Analysis. Springer; 2002. [Google Scholar]
- KAWASAKI Y, SUZUKI M, KHERIF F, TAKAHASHI T, ZHOU SY, NAKAMURA K. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. NeuroImage. 2007;34:235–242. doi: 10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- KLÖPPEL S, STONNINGTON CM, CHU C, DRAGANSKI B, SCAHILL RI, ROHRER JD, FOX NC, JACK CR, ASHBURNER J, FRACKOWIAK RSJ. Automatic Classification of MR Scans in Alzheimer’s Disease. Brain. 2008a;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLÖPPEL S, STONNINGTON CM, CHU C, DRAGANSKI B, SCAHILL RI, ROHRER JD, FOX NC, JACK CR, JR., ASHBURNER J, FRACKOWIAK RSJ. Automatic classification of MR scans in Alzheimer’s disease. Brain: A Journal of Neurology. 2008b;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRISHNAN KR, MCDONALD WM, ESCALONA PR, DORAISWAMY PM, NA C, HUSAIN MM, FIGIEL GS, BOYKO OB, ELLINWOOD EH, NEMEROFF CB. Magnetic resonance imaging of the caudate nuclei in depression. Preliminary observations. Archives of General Psychiatry. 1992;49:553–557. doi: 10.1001/archpsyc.1992.01820070047007. [DOI] [PubMed] [Google Scholar]
- KUMAR A, BILKER W, JIN Z, UDUPA J. Atrophy and high intensity lesions: complementary neurobiological mechanisms in late-life major depression. Neuropsychopharmacology. 2000;22:264–274. doi: 10.1016/S0893-133X(99)00124-4. [DOI] [PubMed] [Google Scholar]
- KUMAR A, JIN Z, BILKER W, UDUPA J, GOTTLIEB G. Late-onset minor and major depression: early evidence for common neuroanatomical substrates detected by using MRI. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7654–7658. doi: 10.1073/pnas.95.13.7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAAKSO MP, PARTANEN K, RIEKKINEN P, LEHTOVIRTA M, HELKALA EL, HALLIKAINEN M, HANNINEN T, VAINIO P, SOININEN H. Hippocampal volumes in Alzheimer’s disease, Parkinson’s disease with and without dementia, and in vascular dementia: An MRI study. Neurology. 1996;46:678–681. doi: 10.1212/wnl.46.3.678. [DOI] [PubMed] [Google Scholar]
- LAI T-J, PAYNE ME, BYRUM CE, STEFFENS DC, K.R.R. K. Reduction of orbital frontal cortex volume in geriatric depression. Biological Psychiatry. 2000;48:971–975. doi: 10.1016/s0006-3223(00)01042-8. [DOI] [PubMed] [Google Scholar]
- LAO Z, SHEN D, XUE Z, KARACALI B, RESNICK S, DAVATZIKOS C. Morphological classification of brains via high-dimensional shape transformations and machine learning methods. NeuroImage. 2004;21:46–57. doi: 10.1016/j.neuroimage.2003.09.027. [DOI] [PubMed] [Google Scholar]
- LEE SH, PAYNE ME, STEFFENS DC, MCQUOID DR, LAI T-J, PROVENZALE JM, KRISHNAN KRR. Subcortical lesion severity and orbitofrontal cortex volume in geriatric depression. Biological Psychiatry. 2003;54:529–533. doi: 10.1016/s0006-3223(03)00063-5. [DOI] [PubMed] [Google Scholar]
- LEHTOVIRTA M, LAAKSO MP, FRISONI GB, SOININEN H. How does the apolipoprotein E genotype modulate the brain in aging and in Alzheimer’s disease? A review of neuroimaging studies. Neurobiology of Aging. 2000;21:293–300. doi: 10.1016/s0197-4580(00)00120-2. [DOI] [PubMed] [Google Scholar]
- LERCH JP, PRUESSNER J, ZIJDENBOS AP, COLLINS DL, TEIPEL SJ, HAMPEL H, EVANS A. Automated cortical thickness measurements from MRI can accurately separate Alzheimer’s patients from normal elderly controls. Neurobiol Aging. 2006;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- LIU Y, TEVEROVSKIY L, CARMICHAEL O, KIKINIS R, SHENTON M, CARTER CS, STENGER VA, DAVIS S, AIZENSTEIN H, BECKER J, LOPEZ O, MELTZER C. In: Discriminative MR Image Feature Analysis for Automatic Schizophrenia and Alzheimer’s Disease Classification. BARILLOT C, HAYNOR DR, HELLIER P, editors. Springer-Verlag GmbH; Saint-Malo, France: 2004. pp. 393–401. [Google Scholar]
- LOCHHEAD RA, PARSEY RV, OQUENDO MA, MANN JJ. Regional brain gray matter volume differences in patients with bipolar disorder as assessed by optimized voxel-based morphometry. Biol Psychiatry. 2004;55:1154–1162. doi: 10.1016/j.biopsych.2004.02.026. [DOI] [PubMed] [Google Scholar]
- MACMASTER FP, MIRZA Y, SZESZKO PR, KMIECIK LE, EASTER PC, TAORMINA SP, LYNCH M, ROSE M, MOORE GJ, ROSENBERG DR. Amygdala and Hippocampal Volumes in Familial Early Onset Major Depressive Disorder. Biological Psychiatry. 2008;63:385–390. doi: 10.1016/j.biopsych.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAGNIN B, MESROB L, KINKINGNEHUN S, PELEGRINI-ISSAC M, COLLIOT O, SARAZIN M, DUBOIS B, LEHERICY S, BENALI H. Support vector machine-based classification of Alzheimer’s disease from whole-brain anatomical MRI. Neuroradiology. 2009;51:73–83. doi: 10.1007/s00234-008-0463-x. [DOI] [PubMed] [Google Scholar]
- MAHALANOBIS PC. On the generalized distance in statistics. Proc. of the National Institute of Sciences of India. 1936;2:49–55. [Google Scholar]
- MESULAM M. A plasticity-based theory of the pathogenesis of Alzheimer’s disease. Annals of the New York Academy of Sciences. 2000;924:42–52. doi: 10.1111/j.1749-6632.2000.tb05559.x. [DOI] [PubMed] [Google Scholar]
- MISRA C, FAN Y, DAVATZIKOS C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. NeuroImage. 2009;44:1415–1422. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITCHELL TM. Machine Learning and Data Mining. Communications of the ACM. 1999;42:31–36. [Google Scholar]
- MORI S. Introduction to Diffusion Tensor Imaging. Elsevier; 2007. [Google Scholar]
- MORRIS P, RAPOPORT SI. Neuroimaging and affective disorder in late life: a review. Canadian Journal of Psychiatry - Revue Canadienne de Psychiatrie. 1990;35:347–354. doi: 10.1177/070674379003500415. [DOI] [PubMed] [Google Scholar]
- MOURAO-MIRANDA J, BOKDE AL, BORN C, HAMPEL H, STETTER M. Classifying brain states and determining the discriminating activation patterns: Support Vector Machine on functional MRI data. NeuroImage. 2005;28:980–995. doi: 10.1016/j.neuroimage.2005.06.070. [DOI] [PubMed] [Google Scholar]
- NAIDICH TP, DUVERNOY HM, DELMAN BN, SORENSEN AG, KOLLIAS SS, HAACKE EM. Duvernoy’s Atlas of the Human Brain Stem and Cerebellum: High-Field MRI, Surface Anatomy, Internal Structure, Vascularization and 3 D Sectional Anatomy. Springer; 2009. [Google Scholar]
- OLIVEIRA PJ, NITRINI R, BUSATTO G, BUCHPIGUEL C, SATO J, AMARO EJ. Use of SVM methods with surface-based cortical and volumetric subcortical measurements to detect Alzheimer’s disease. The Journal of Alzheimers Disease. 2010;19:1263–1272. doi: 10.3233/JAD-2010-1322. [DOI] [PubMed] [Google Scholar]
- PANTEL J, SCHRÖDER J, ESSIG M, POPP D, DECH H, KNOPP MV, SCHAD LR, EYSENBACH K, BACKENSTRASS M, FRIEDLINGER M. Quantitative magnetic resonance imaging in geriatric depression and primary degenerative dementia. Journal of Affective Disorders. 1997;42:69–83. doi: 10.1016/s0165-0327(96)00105-x. [DOI] [PubMed] [Google Scholar]
- PATEL T, POLIKAR R, DAVATZIKOS C, CLARK CM. EEG and MRI Data Fusion for Early Diagnosis of Alzheimer’s Disease. 30th Annual International IEEE EMBS Conference; Vancouver, BC, Canada. 2008. [DOI] [PubMed] [Google Scholar]
- PAUS CW, CHUNG MK, WORSLEY KJ, PAUS T, CHERIF C, COLLINS DL, GIEDD JN, RAPOPORT JL, EVANS AC. A Unified Statistical Approach to Deformation-Based Morphometry. NeuroImage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- PETERSON BS. Neuroimaging studies of Tourette Syndrome. A decade of progress. In: COHEN DJ, GOETZ CG, JANKOVIC J, editors. Advances in Neurology. Tourette Syndrome and Associated Disorders. Lippincott Williams & Wilkins; Philadelphia: 2000. pp. 179–196. [PubMed] [Google Scholar]
- PETERSON BS. Form determines function: new methods for identifying the neuroanatomical loci of circuit-based disturbances in childhood disorders. J Am Acad Child Adolesc Psychiatry. 2010;49:533–538. doi: 10.1016/j.jaac.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERSON BS, CHOI HA, HAO X, AMAT J, ZHU H, WHITEMAN R, LIU J, XU D, BANSAL R. Morphology of the Amygdala and Hippocampus in Children and Adults with Tourette Syndrome. Archives General Psychiatry. 2007 doi: 10.1001/archpsyc.64.11.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERSON BS, FEINEIGLE PA, STAIB LH, GORE JC. Automated measurement of latent morphological features in the human corpus callosum. Human Brain Mapping. 2001a;12:232–245. doi: 10.1002/1097-0193(200104)12:4<232::AID-HBM1018>3.0.CO;2-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERSON BS, LECKMAN JF, TUCKER D, SCAHILL L, STAIB L, ZHANG H, KING RA, COHEN DJ, GORE JC, LOMBROSO P. Preliminary Findings of Antistreptococcal Antibody Titers and Basal Ganglia Volumes in Tic, Obsessive-compulsive, and Attention-Deficit/Hyperactivity Disorders. Arch Gen Psychiatry. 2000;57:364–372. doi: 10.1001/archpsyc.57.4.364. [DOI] [PubMed] [Google Scholar]
- PETERSON BS, RIDDLE MA, COHEN DJ, KATZ L, SMITH JC, LECKMAN JF. Human basal ganglia volume asymmetries on magnetic resonance images. Mag. Reson. Imaging. 1993a;11:493–498. doi: 10.1016/0730-725x(93)90468-s. [DOI] [PubMed] [Google Scholar]
- PETERSON BS, RIDDLE MA, COHEN DJ, KATZ LD, SMITH JC, HARDIN MT, LECKMAN JF. Reduced basal ganglia volumes in tourette’s syndrome using three-dimensional reconstruction techniques from magnetic resonance images. Neurology. 1993b;43:941–949. doi: 10.1212/wnl.43.5.941. [DOI] [PubMed] [Google Scholar]
- PETERSON BS, STAIB L, SCAHILL L, ZHANG H, ANDERSON C, LECKMAN JF, GORE JC, ALBERT J, WEBSTER R. Regional brain and ventricular volumes in Tourette syndrome. Arch. Gen. Psychiatry. 2001b;58:427–440. doi: 10.1001/archpsyc.58.5.427. [DOI] [PubMed] [Google Scholar]
- PETERSON BS, THOMAS P, KANE MJ, OTHERS A. Basal ganglia volumes in patients with Gilles de la Tourette syndrome. Arch Gen Psychiatry. 2003;60:415–424. doi: 10.1001/archpsyc.60.4.415. [DOI] [PubMed] [Google Scholar]
- PETERSON BS, WARNER V, BANSAL R, ZHU H, HAO X, LIU J, DURKIN K, ADAMS PB, WICKRAMARATNE P, WEISSMAN MM. Cortical thinning in persons at increased familial risk for major depression. Proc Natl Acad Sci USA. 2009;106:6273–6278. doi: 10.1073/pnas.0805311106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PIVEN J, ARNDT S, BAILEY J, HAVERCAMP S, ANDREASEN NC, PALMER P. An MRI Study of Brain Size in Autism. American Journal of Psychiatry. 1995;152:1145–1149. doi: 10.1176/ajp.152.8.1145. [DOI] [PubMed] [Google Scholar]
- PIVEN J, BAILEY J, RANSON BJ, ARNDT S. An MRI study of the corpus callosum in autism. The American Journal of Psychiatry. 1997;154:1051–1056. doi: 10.1176/ajp.154.8.1051. [DOI] [PubMed] [Google Scholar]
- PIVEN J, NEHME E, SIMON J, BARTA P, PEARLSON G, FOLSTEIN SE. Magnetic resonance imaging in autism: measurement of the cerebellum, pons, and fourth ventricle. Biological Psychiatry. 1992;31:491–504. doi: 10.1016/0006-3223(92)90260-7. [DOI] [PubMed] [Google Scholar]
- PLANT C, TEIPEL SJ, OSWALD A, BOEHM C, MEINDL T, MOURAE-MIRANDA J, BOKDE AW, HAMPEL H, EWERS M. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer’s disease. NeuroImage. 2010;50:162–174. doi: 10.1016/j.neuroimage.2009.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PLESSEN KJ, BANSAL R, ZHU H, WHITEMAN R, QUACKENBUSH GA, HUGDAHL K, PETERSON BS. Hippocampus and amygdala morphology in Attention-Deficit/Hyperactivity Disorder. Arch Gen Psychiatry. 2006;63:795–807. doi: 10.1001/archpsyc.63.7.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RABINS PV, PEARLSON GD, AYLWARD E, KUMAR AJ, DOWELL K. Cortical magnetic resonance imaging changes in elderly inpatients with major depression. American Journal of Psychiatry. 1991;148:617–620. doi: 10.1176/ajp.148.5.617. [DOI] [PubMed] [Google Scholar]
- RISH I. An empirical study of the naive Bayes classifier. IJCAI Workshop on Empirical Methods in Artificial Intelligence. 2001 [Google Scholar]
- ROJAS DC, PETERSON E, WINTERROWD E, REITE ML, ROGERS SJ, TREGELLAS JR. Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry. 2006;6 doi: 10.1186/1471-244X-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RORDEN C, BRETT M. Stereotaxic display of brain lesions. Behavioural Neurology. 2000;12:191–200. doi: 10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- SALLOWAY S, MALLOY P, KOHN R, GILLARD E, DUFFY J, ROGG J, TUNG G, RICHARDSON E, THOMAS C, WESTLAKE R. MRI and neuropsychological differences in early- and late-life-onset geriatric depression. Neurology. 1996;46:1567–1574. doi: 10.1212/wnl.46.6.1567. [DOI] [PubMed] [Google Scholar]
- SALVOLINI U, SCARABINO T. High field brain MRI: use in clinical practice. Springer; 2006. [Google Scholar]
- SAVIO A, GARCÍA-SEBASTIÁN M, GRAÑA M, VILLANÚA J. Results of an Adaboost approach on Alzheimer’s Disease detection on MRI. 3rd International Work-Conference on The Interplay Between Natural and Artificial Computation: Part II: Bioinspired Applications in Artificial and Natural Computation.2009. pp. 114–123. [Google Scholar]
- SCHELTENS P, FOX NC. Structural magnetic resonance imaging in the practical assessment of dementia: beyond exclusion. The Lancet Neurology. 2002;1:13–21. doi: 10.1016/s1474-4422(02)00002-9. [DOI] [PubMed] [Google Scholar]
- SCHELTENS P, KORF ES. Contribution of neuroimaging in the diagnosis of Alzheimer’s disease and other dementias. Current Opinion in Neurology. 2000;13:391–396. doi: 10.1097/00019052-200008000-00005. [DOI] [PubMed] [Google Scholar]
- SEARS LL, VEST C, MOHAMED S, BAILEY J, RANSON BJ, PIVEN J. An MRI study of the basal ganglia in autism. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 1999;23:613–624. doi: 10.1016/s0278-5846(99)00020-2. [DOI] [PubMed] [Google Scholar]
- SHAH PJ, EBMEIER KP, GLABUS MF, GOODWIN GM. Cortical grey matter reductions associated with treatment-resistant chronic unipolar depression. Controlled magnetic resonance imaging study. British Journal of Psychiatry. 1998;172:527–532. doi: 10.1192/bjp.172.6.527. [DOI] [PubMed] [Google Scholar]
- SHELINE YI, SANGHAVI M, MINTUN MA, GADO MH. Depression duration but not age predicts hippocampal volume loss in medically healthy women with recurrent major depression. Journal of Neuroscience. 1999;19:5034–5043. doi: 10.1523/JNEUROSCI.19-12-05034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SILVERMAN BW. Density Estimation: for Statistics and Data Analysis. Chapman & Hall/CRC; 1986. [Google Scholar]
- SINGH V, MUKHERJEE L, CHUNG MK. Cortical Surface Thickness as a Classifier: Boosting for Autism Classification. In: METAXAS D, editor. MICCAI 2008. 2008. pp. 999–1007. [DOI] [PubMed] [Google Scholar]
- SMITH SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFFENS DC, BYRUM CE, MCQUOID DR, GREENBERG DL, PAYNE ME, T.F. B, J.R. M, KRISHNAN KR. Hippocampal volume in geriatric depression. Biological Psychiatry. 2000;48:301–309. doi: 10.1016/s0006-3223(00)00829-5. [DOI] [PubMed] [Google Scholar]
- STEFFENS DC, KRISHNAN KR. Structural neuroimaging and mood disorders: recent findings, implications for classification, and future directions. Biological Psychiatry. 1998;43:705–712. doi: 10.1016/s0006-3223(98)00084-5. [DOI] [PubMed] [Google Scholar]
- STEFFENS DC, PAYNE ME, GREENBERG DL, BYRUM CE, WELSH-BOHMER KA, WAGNER HR, MACFALL JR. Hippocampal volume and incident dementia in geriatric depression. American Journal of Geriatric Psychiatry. 2002;10:62–71. [PubMed] [Google Scholar]
- STOLLEY PD, SCHLESSELMAN JJ. Case-control studies: design, conduct, analysis. Oxford University Press; Oxford: 1982. [Google Scholar]
- TAYLOR WD, MACFALL JR, PAYNE ME, MCQUOID DR, PROVENZALE JM, STEFFENS DC, KRISHNAN KRR. Late-life depression and microstructural abnormalities in dorsolateral prefrontal cortex white matter. American Journal of Psychiatry. 2004;161:1293–1296. doi: 10.1176/appi.ajp.161.7.1293. [DOI] [PubMed] [Google Scholar]
- TAYLOR WD, MACFALL JR, STEFFENS DC, PAYNE ME, PROVENZALE JM, KRISHNAN KR. Localization of age-associated white matter hyperintensities in latelife depression. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2003;27:539–544. doi: 10.1016/S0278-5846(02)00358-5. [DOI] [PubMed] [Google Scholar]
- TEIPEL SJ, BORN C, EWERS M, BOKDE AL, REISER MF, MOLLER HJ. Multivariate deformation-based analysis of brain atrophy to predict Alzheimer’s disease in mild cognitive impairment. NeuroImage. 2007;38:13–24. doi: 10.1016/j.neuroimage.2007.07.008. [DOI] [PubMed] [Google Scholar]
- THOMPSON PM, TOGA AW. Detection, visualization and animation of abnormal anatomic structure with a deformable probabilistic brain atlas based on random vector field transformations. Medical Image Analysis. 1996a;1:271–294. doi: 10.1016/s1361-8415(97)85002-5. [DOI] [PubMed] [Google Scholar]
- THOMPSON PM, TOGA AW. A surface-based technique for warping 3-dimensional images of the brain. IEEE Transactions on Medical Imaging. 1996b;15:1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- TUPLER LA, KRISHNAN KR, MCDONALD WM, DOMBECK CB, D’SOUZA S, STEFFENS DC. Anatomic location and laterality of MRI signal hyperintensities in late-life depression. Journal of Psychosomatic Research. 2002;53:665–676. doi: 10.1016/s0022-3999(02)00425-7. [DOI] [PubMed] [Google Scholar]
- TZOURIO-MAZOYER N, LANDEAU B, PAPATHANASSIOU D, CRIVELLO F, ETARD O, DELCROIX N, MAZOYER B, JOLIOT M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- VAPNIK V,N. The Nature of Statistical Learning Theory. Springer; 1999. [DOI] [PubMed] [Google Scholar]
- WAHLUND LO, ALMKVIST O, BLENNOW K, ENGEDAHL K, JOHANSSON A, G. W. Evidence-based evaluation of magnetic resonance imaging as a diagnostic tool in dementia workup. Top Magn Reson Imagin. 2005;16:427–437. doi: 10.1097/01.rmr.0000245463.36148.12. [DOI] [PubMed] [Google Scholar]
- WU M, CARMICHAEL O, LOPEZ-GARCIA P, CARTER CS, AIZENSTEIN HJ. Quantitative Comparison of Neuroimage Registration for fMRI Analyses by AIR, SPM, and a Fully Deformable Model. Human Brain Mapping. 2006;27:747–754. doi: 10.1002/hbm.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU D, HAO X, BANSAL R, PLESSEN KJ, GENG W, PETERSON BS. Unifying the analyses of anatomical and diffusion tensor images using volume-preserved warping. Journal of Magnetic Resonance Imaging. 2007;25:612–624. doi: 10.1002/jmri.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOO TS. Insight into Images: Principles and Practice for Segmentation, Registration, and Image Analysis. A K Peters/CRC Press; 2004. [Google Scholar]
- ZHANG D, Y. W, ZHOU L, YUAN H, SHEN D. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]