Abstract
The epithelium lining the epididymis in the male reproductive tract maintains a luminal environment that promotes sperm cell maturation. This process is dependent on the coordinated expression of many genes that encode proteins with a role in epithelial transport. We previously generated genome-wide maps of open chromatin in primary human epididymis epithelial (HEE) cells to identify potential regulatory elements controlling coordinated gene expression in the epididymis epithelium. Subsequent in silico analysis identified transcription factor-binding sites (TFBS) that were over-represented in the HEE open chromatin, including the motif for paired box 2 (PAX2). PAX2 is a critical transcriptional regulator of urogenital tract development, which has been well studied in the kidney but is unexplored in the epididymis. Due to the limited lifespan of primary HEE cells in culture, we investigated the role of PAX2 in an immortalized HEE cell line (REP). First, REP cells were evaluated by DNase I digestion followed by high-throughput sequencing and the PAX2-binding motif was again identified as an over-represented TFBS within intergenic open chromatin, though on fewer chromosomes than in the primary HEE cells. To identify PAX2-target genes in REP cells, RNA-seq analysis was performed after siRNA-mediated depletion of PAX2 and compared with that with a non-targeting siRNA. In response to PAX2-represssion, 3135 transcripts were differentially expressed (1333 up-regulated and 1802 down-regulated). Novel PAX2 targets included multiple genes encoding proteins with predicted functions in the epididymis epithelium.
Keywords: epididymis epithelium, open chromatin, PAX2, transcriptional network
Introduction
Immature spermatozoa leaving the testis are not competent to fertilize an oocyte; passage through the epididymis confers this competency and is essential for their mature functions. Properties of the epididymis epithelium that are central to the normal sperm maturation process include those that maintain the luminal environment. The epididymal fluid is regulated by membrane proteins, which control water content, pH, ion secretion and absorption and protein secretion. Mutations in ion channels, such as the cystic fibrosis transmembrane conductance regulator (CFTR), (Traystman et al., 1994; van der Ven et al., 1996) and chloride:bicarbonate exchangers, including solute carrier family 26 A3 (SCL26A3, Hoglund et al., 2006), can perturb epididymis epithelial function, as can the loss of transcription factors (TFs) such as forkhead box i1 (Blomqvist et al., 2006). However, the molecular mechanisms that coordinate the expression and response to biological stimuli of the many functionally important genes in the epididymis epithelium are less well studied. This paucity of information is largely due to the lack of robust cellular models to investigate the regulation of human epididymal function in vitro and the difficulty in obtaining human epididymal tissue. To address this problem, we previously established cultures of immature primary human epididymis epithelial (HEE) cells (Harris and Coleman, 1989) and immortalized these cells with an origin-defective SV-40 (Coleman and Harris, 1991) to produce REP cells. Although these cells do not reflect the full, differentiated properties of adult human epididymis epithelium in vivo, in the absence of other reagents, they are of value. Our goal is to determine transcriptional networks that control gene expression and hence the function of the human epididymis epithelium. Transcription factor-binding sites (TFBS) may be revealed by inspection of the DNA sequence in regions of open chromatin. In a previous analysis of immature primary HEE cells, we used DNase I digestion followed by high-throughput sequencing (DNase-seq) to map chromatin accessibility genome wide. Bioinformatic analysis of the sequence within open chromatin peaks was then used to predict over-represented TFBS in HEE-selective regions of open chromatin. Among a number of cell-type-selective TFs identified in this analysis, the paired box gene family member 2 (PAX2) was chosen for further investigation since it is known to be critical for the development of the urogenital tract (Torres et al., 1995) but has not been studied in the human epididymis. PAX2 is expressed in mouse epididymis epithelium (Oefelein et al., 1996) from birth to adulthood and is an abundant TF in HEE and REP cells. Since primary HEE cells are not suitable for long-term analyses due to their limited replicative potential, we chose to use the REP cell line to investigate PAX2 targets in epididymis epithelial cells. First, DNase-seq was used to identify open chromatin genome-wide in REP cells and to confirm that PAX2 TFBS were also over-represented in open chromatin in this cell type. Next, we identified target genes for PAX2 in REP cells by depletion of this factor with a specific siRNA compared with a non-targeting siRNA. Target genes that were positively or negatively regulated by PAX2 depletion were then subjected to gene ontology process enrichment analysis, revealing multiple pathways relevant to epididymis epithelial function.
Materials and Methods
Cell culture
The REP cell line, immortalized by origin-defective simian virus 40 (SV40 ori-) transformation of immature primary HEE cells, was cultured as previously described (Coleman and Harris, 1991). For the experiments to test androgen receptor (AR) function, cells were cultured in phenol-free CMRL-1066 medium (androgen-free) containing 10% dextran coated-charcoal (C6241, Sigma), stripped FBS for 72 h prior to stimulation with vehicle, testosterone (200 nM, T1500, Sigma) or the synthetic androgen R1881 (1 nM, methyltrienolone, NLP005005MG, PerkinElmer, Waltham, MA) for a further 16 h.
Western blotting and immunofluorescence
Western blots were done by standard protocol as described previously (Leir and Harris, 2011). The following primary antibodies were used: PAX2 (PRB-276P, Covance Emeryville, CA), AR (sc-816, Santa Cruz, Dallas, TX) and beta-tubulin (T4026, Sigma). Immunofluorescence was performed by standard protocols. Cells grown on coverslips, were fixed in 3.7% paraformaldehyde, permeabilized in 0.5% Triton X-100 and then blocked in 1% BSA, prior to immunostaining with AR antibody. Alexa Fluor®488-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) was used to detect AR localization. Nuclei were counterstained with DAPI. Cells on coverslips were mounted on glass slides with FluorSavereagent (EMD Millipore, Billerica, MA) and examined with a fluorescent microscope (Leica DMR).
Microarray analysis
RNA was isolated by Trizol extraction from REP cells treated with vehicle or R1881 for 16 h in two independent experiments. Total RNA was purified by Millipore Microcon YM-100 filter centrifugation and shipped to MoGene for gene expression analysis on Agilent SurePrint G3 8 × 60 K (version 2) arrays. Data from the two microarrays were analyzed using Nexus 3.0 software (BioDiscovery, Inc., Hawthorne, CA, USA). In brief, microarray text files were processed using the default settings for Agilent FE Single Channel files (quantile normalization), incorporating the batch correction option for the two samples. Probes expressed at a very low level in both samples (log2 value <4.5 in 90% of samples) were removed (34036 probes were retained). Differentially expressed probes were determined by comparing the averages of each group (i.e. R1881 treatment versus vehicle control).
DNase-seq
DNase-seq libraries were prepared from two independent cultures of DNase I-digested REP cells and sequenced separately on an Illumina Hi-Seq machine. DNase peak calls from these two libraries were combined and used for further analysis. Comparative analysis of DNase-seq data used primary HEE cells (Bischof et al., 2013) and five cell types (FibroP, GM12878, K652, HepG2 and HUVEC) (Song et al., 2011) from the Encyclopedia of DNA Elements Consortium (http://genome.ucsc.edu/ENCODE/). All genome data coordinates refer to the hg19 genome and their GEO accession numbers are listed in Supplementary data, Table SIII. REP data are available at GEO (GSE59783).
The peak calls for the DNase-seq data were made with the F-seq application to give a discrete number of DNase I hypersensitive sites (DHS) within the REP cells (Boyle et al., 2008). These sites were determined by F-seq by fitting the data to a gamma distribution to calculate P-values. P-values below a 0.05/0.01 threshold in contiguous regions were considered significant. All DHS identified on chromosome Y were removed from the analysis to allow for comparison across both male and female cell types. After producing the number and location of DHS for the REP cells, the DHS of five non-related cell types (FibroP, GM12878, K562, HepG2 and HUVEC) were intersected with the REP data for comparison. Those sites from the REP cells that were not overlapped by any other data sets were designated REP-selective sites. The REP sites that were overlapped by a DHS in all of the cell types comprised the list of ‘REP-ubiquitous sites’. The REP-selective sites were also compared with those sites from HEE cells.
DHS overlapping gene annotation
The genomic indices of REP-selective DHS, ubiquitous DHS (i.e. DHS occurring in REP cells that were overlapped by DHS from all background cell types), and all DHS were intersected with the genomic indices of all human genes, their promoters (2 kb upstream of transcriptional start sites), exons, introns and intergenic sequence. The human gene annotation was derived from a list of all human Entrez genes downloaded from NCBI. These Entrez genes were each linked to their representative RefSeq sequences and the RefSeq indices were downloaded from UCSC's genome browser (http://www.ncbi.nlm.nih.gov, http://genome.ucsc.edu).
Clover analysis
The Clover application was used to search for motifs common to DHS within promoter and intergenic sequence near genes. Clover is an open-source application that identifies motifs (from an input set) that are statistically over- or under-represented in a set of sequences with respect to a set of background sequences (Frith et al., 2004). The promoter regions were defined as 2 kb upstream of the transcription start site of all human genes. The intergenic regions that were searched included any genomic region >2 kb but <10 kb from the start or stop of gene transcription.
SiRNA-mediated depletion of PAX2
Cells at 40–50% confluence were transfected with non-targeting control- or PAX2-targeting siRNA (sc-37007 and sc-38745, Santa Cruz) using RNAiMax Lipofectamine 2000 reagent (Life Technologies, Grand Island, NY) at a final concentration of 20 nM. After 24 h, media was replaced with fresh media. At 72 h post transfection, the cells were washed in PBS and immediately harvested for RNA extraction using TRIzol® (Life Technologies) or for whole cell lysate (using NET Buffer (Leir and Harris, 2011)).
Quantitative RT–PCR (RT-qPCR)
The TaqMan® reverse transcription kit (Life Technologies) was used to make cDNA from total RNA and RT-qPCR was then used to measure gene expression levels. The primers used are listed in Supplementary data, Table SII.
Library preparation for RNA-seq
Total RNA yield and purity was assessed by NanoDrop (Thermo Scientific, Waltham, MA, USA). RNA libraries were prepared from 2 μg of total RNA from four biological replicates from control- and PAX2-siRNA-transfected cells, using the TruSeq™ RNA sample preparation kit as per the manufacturer's protocol (Illumina, San Diego, CA, USA). Sequencing was performed on an Illumina Hi-Seq 2000.
RNA-Seq data analysis
The eight sets of RNA-seq data (4× control and 4× PAX2-knockdown) generated 3.2 × 108 aligned sequences, or ∼4 × 107 reads each. First, all sequences were mapped back to human genome (hg19) using the default setting of TopHat (v2.0.9) (Trapnell et al., 2012). The aligned reads were then processed by Cufflinks (v2.1.1) to assemble expressed genes and transcripts against the UCSC hg19 gene structure annotation and to estimate expression levels for each transcript. The assembly files for each of the eight samples were then merged to create a single transcriptome annotation and differentially expressed genes identified by Cuffdiff. Samples were then normalized and expressed as fragments per kilobase of transcript per million fragments mapped (FPKM) and both a test statistic (P-value) and an adjusted P-value (q-value) were calculated. Genes with a fold change ≥1.5 and an FPKM ≥1.5 were considered differentially expressed. Data are available at GEO (http://www.ncbi.nlm.nih.gov/geo/ GSE59783). Biological processes associated with differentially expressed genes were identified using DAVID.
Statistical analysis
Data are expressed as mean ± SD. The GraphPad Prism 6 software was used to analyze statistical differences between experimental groups using Student's t-test and the values of P < 0.05 were considered significant.
Results
Characterization of REP cells
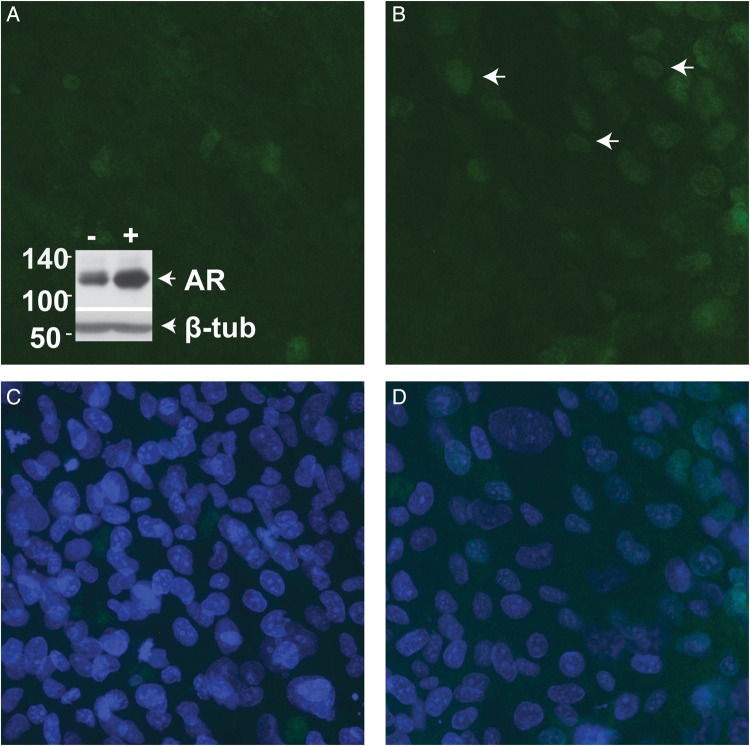
Since REP cells were derived from primary cultures of immature epididymis by SV40 ori immortalization, we first demonstrated their utility for studies of epididymis epithelial function. A key TF in this epithelium is the AR, which was shown by western blot to be expressed in both vehicle- and androgen R1881 (1 nM)-treated REP cells (Fig. 1 inset). Moreover, functional activity of the AR was confirmed by immunofluorescence (Fig. 1A–D), showing its nuclear accumulation in response to testosterone (200 nM, 16 h) (white arrows in Fig. 1B) in comparison with faint cytoplasmic staining in vehicle-treated cells (Fig. 1A). To investigate potential AR target genes, microarray analysis of RNA extracted from vehicle and R1881-stimulated REP cells (in duplicate) was performed. This revealed 92 genes that were differentially expressed (by at least 1.5-fold, P < 0.01) in response to R1881 treatment in both replicas (Supplementary data, Table SI).
Figure 1.
REP cells express the AR and it relocates to the nucleus upon ligand stimulation. (A–D) Confocal microscopy confirmed functional activity of the AR by its nuclear accumulation (B, white arrows) in response to testosterone (200 nM, 16 h) compared with the faint and diffuse AR staining in vehicle-treated cells (A). (C and D show DAPI staining of the same sections). A, Inset, western blot shows AR protein (110 kDa) expression in REP cells with vehicle (−) or R1881 (+) treatment at 1 nM for 16 h. Magnification× 400.
Genome-wide mapping of open chromatin in REP cells
Cis-acting regulatory elements controlling gene expression are often associated with regions of open chromatin, which coincide with DHS. DNase-seq was used to map open chromatin genome-wide in REP cells. This analysis identified a similar number of DHS (126 084) in REP cells (Table I) to that reported previously in primary HEE cells (132 814 sites) (Bischof et al., 2013). To identify cis-regulatory elements that were selective for REP cells, REP DHS were intersected with DHS from five different cell types, generated by the ENCODE consortium (Song et al., 2011) and non-overlapping sites recorded. The five ENCODE data sets were from skin fibroblasts (FibroP), a lymphoblastoid cell line (GM12878), an erythroleukemia cell line (K562), a liver carcinoma cell line (HepG2) and human umbilical vein endothelial cells (HUVEC). Of the sites, 18% (22 644) were REP selective and 26% (32 434) were ubiquitous in that they intersected with DHS identified in the ENCODE cell lines. The genomic overlap between the REP DHS and the other five cell types was equivalent to that observed in primary HEE cells (45–58%) (Bischof et al., 2013) (Supplementary data, Fig. S1), and the distribution of cell-type selective (25.3%) and ubiquitous (23.8%) sites was similar. Of note was the high number of overlapping selective-sites (62 115 sites; 65%) between the REP and the primary HEE cells demonstrating extensive similarity between the two cell types despite the immortalization process. The numbers of DHS in each cell type are shown in Table I.
Table I.
Number of open chromatin sites in REP cells, HEE cells and the five other cell types used for comparison.
| DNase-seq cell lines | Number of open chromatin sites |
|---|---|
| REP | 126 084 |
| HEE | 132 814 |
| FibroP (normal fibroblasts from patients with Parkinson's) | 139 663 |
| GM12878 (lymphoblastoid) | 124 321 |
| HepG2 (liver carcinoma) | 115 765 |
| HUVEC (human umbilical vein endothelial cell) | 126 284 |
| K562 (erythroleukemia cell) | 112 025 |
Next, we mapped the location of the three categories of REP DHS (REP-selective, ubiquitous and all DHS) across different genomic elements according to the following criteria: promoter (2 kb 5′ to the transcriptional start site), exon 1, intron 1, other genic, 2 kb 3′ to the transcriptional stop site and intergenic (Supplementary data, Fig. S2). The REP-selective DHS showed a similar distribution to that observed in HEE-selective DHS (Bischof et al., 2013). The majority were located in other genic or intergenic sites, that is within gene bodies or between genes rather than in gene promoters.
REP-selective DHS are enriched for binding sites for transcription factors relevant to epididymis epithelial function
To search for over-represented TFBS predicted in open chromatin peaks in REP cells, we performed Clover analysis (Frith et al., 2004) on promoter (Supplementary data, Table SIV) and intergenic DHS (Supplementary data, Table SV) regions. Within these categories, data were analyzed in three groups: all DHS, REP-selective DHS and ubiquitous DHS. The comparisons between the intergenic sites are the most informative since the representation of motifs is markedly different in the REP-selective and ubiquitous sites (Supplementary data, Table SV). In silico prediction of TFBS that are over-represented in REP-selective DHS on 10–23 chromosomes identified multiple TFs relevant to the differentiated function of the epididymis epithelium. These included hepatocyte nuclear factor 4 alpha (HNF4α), SMAD family member 4 (SMAD4), sterol regulatory element-binding transcription factor 2 (SREBP2) and nuclear factor, erythroid 2-like 2 (NFE2L2). TFBS that are highly over-represented on a similar number of chromosomes in both REP-selective sites and ubiquitous-sites include PAX2, AR, signal transducer and activator of transcription 6, interleukin-4 induced (STAT6), E74-like factor 5 (ets domain TF) (ELF5) and ets variant 4 (PEA3).
Transcriptional profile of the REP epididymis cell line and its regulation by the PAX2 transcription factor
One of the TFs identified in the Clover analysis of over-represented TFBS in both REP cells (Supplementary data, Table SV) and HEE cells (Bischof et al., 2013) is PAX2. PAX2 is known to have an important role in regulating gene expression and epithelial function in the urogenital tract. However, it has not been well studied in the epididymis epithelium, so we chose to investigate its role in controlling gene expression in REP cells.
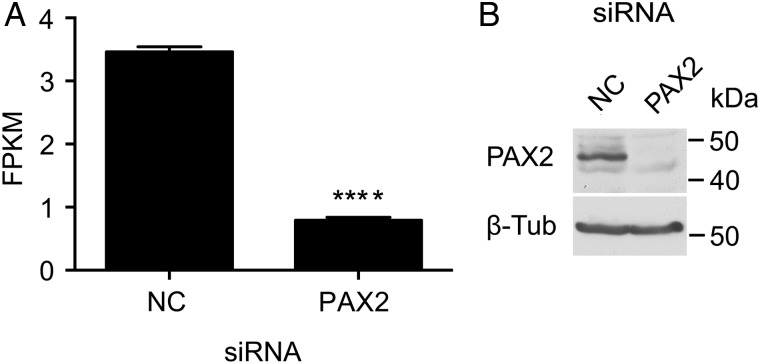
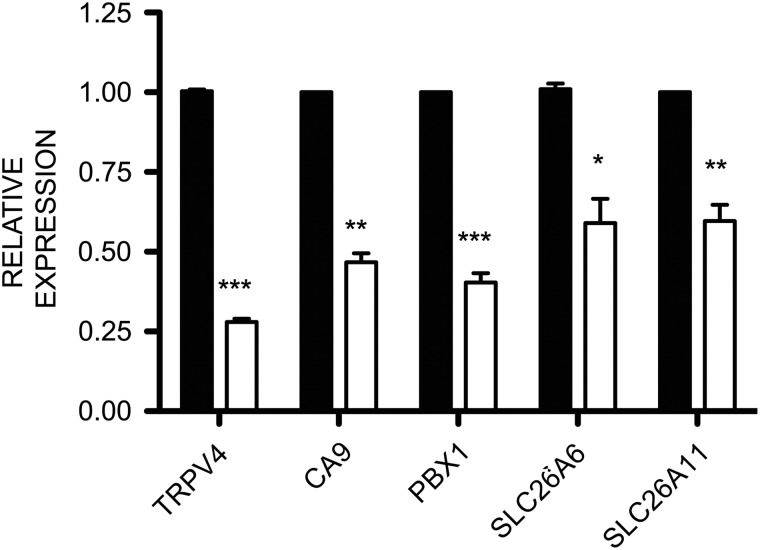
RNA-seq analysis was performed on four biological replicas of REP cells transfected with non-targeting control siRNA or PAX2-specific siRNA. Efficacy of siRNA-mediated depletion of PAX2 in REP cells is shown in Fig. 2. RNA-seq libraries were subjected to 100 base pair, paired end sequencing (Hi-Seq), yielding ∼4 × 107 reads per sample. RNA-seq data were analyzed by TopHat and Cufflinks (Trapnell et al., 2012) to obtain estimates of the expression level of genes and transcripts. The MultiDimensional Scaling plot in Supplementary data, Fig. S3 demonstrates that the PAX2- and negative control siRNA-treated samples clustered together as two independent groups. PAX2 depletion in REP cells differentially regulated the expression of 3135 genes by at least 1.5-fold (FPKM ≥ 1.5). Of these, 1333 were up-regulated and 1802 were down-regulated (Supplementary data, Table SVI). To confirm the RNA-seq data, qRT–PCR was used to measure gene expression in independent samples of PAX2- or negative control siRNA-treated REP cells. Down-regulation of five genes was confirmed by qRT–PCR including transient receptor potential cation channel, subfamily V, member 4 (TRPV4, P < 0.001), carbonic anhydrase IX (CA9, P < 0.01), pre-B-cell leukemia homeobox 1 (PBX1, P < 0.001) and solute carrier family 26 (anion exchanger), members -6 and -11 (SLC26A6, P < 0.05 and SLC26A11, P < 0.01) in PAX2-depleted cells (all n = 3, Fig. 3 and Table III).
Figure 2.
Efficacy of siRNA-mediated depletion of PAX2 in REP cells. (A) RNA-seq demonstrates PAX2 mRNA is depleted 4.5-fold in REP cells after specific siRNA transfection, measured in mean FPKM (± SD, n = 4), in comparison with negative control siRNA (NC). (B) Western blot probed with anti-PAX2 antibody shows depletion of PAX2 protein (43 kDa) after specific siRNA transfection, in comparison with negative control siRNA (NC). ****P < 0.0001 versus NC siRNA-transfected cells.
Figure 3.
Quantitative RT–PCR validation of differentially expressed genes in PAX2-knockdown versus control REP cells. cDNA was synthesized from total RNA and qRT–PCR analysis confirmed the down-regulation of transient receptor potential cation channel, subfamily V, member 4 (TRPV4, P < 0.001), carbonic anhydrase IX (CA9, P < 0.01), pre-B-cell leukemia homeobox 1 (PBX1, P < 0.001), solute carrier family 26 (anion exchanger), members 6 and −11 (SLC26A6, P < 0.05 and SLC26A11, P < 0.01) in PAX2-depleted cells (white bars). Data are expressed relative to beta-2 microglobulin (mean ± SD, n = 3) in comparison with negative control siRNA (NC, black bars). *P < 0.05, **P < 0.01, **P < 0.001 and ***P < 0.0001 versus NC siRNA-transfected cells.
Table III.
Functionally relevant proteins encoded by the identified PAX2-regulated genes.
| Gene symbol | Gene name | RNA-Seq (FPKM) | Relevant function |
|---|---|---|---|
| PAX2 | Paired box 2 | −4.5 | |
| PBX1 | Pre-B-cell leukemia homeobox 1 | −3.2 | Development |
| GREM1 | Gremlin 1, DAN family BMP antagonist | +2.1 | |
| MAPK8IP1 | Mitogen-activated protein kinase 8 interacting 1 protein | −2.9 | Apoptosis |
| CLDN2 | Claudin 2 | −6.5 | Cell junctional complexes |
| CLDN4 | Claudin 4 | −2.2 | |
| CLDN11 | Claudin 11 | −2.2 | |
| JAM2 | Junctional adhesion molecule 2 | −2.0 | |
| JAM3 | Junctional adhesion molecule 3 | −2.8 | |
| CGN | Cingulin | −2.6 | |
| PVRL1 | Poliovirus receptor-related 1 (herpesvirus entry mediator C) | −2.0 | |
| CDH6 | Cadherin 6, type 2, K-cadherin (fetal kidney) | −7.5 | |
| PDGFA | Platelet-derived growth factor alpha polypeptide | −2.1 | Growth factor signaling |
| PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | −2.0 | |
| TGFB3 | Transforming growth factor, beta 3 | −1.9 | |
| TGFBR2 | Transforming growth factor, beta receptor 2 | +1.7 | |
| TGFBR1 | Transforming growth factor, beta receptor 1 | +2.2 | |
| TGFB2 | Transforming growth factor, beta 2 | +2.3 | |
| RBP1 | Retinol-binding protein 1, cellular | −2.2 | Vitamin A signaling |
| STRA6 | Stimulated by retinoic acid 6 | −13.1 | |
| CRABP1 | Cellular retinoic acid-binding protein 1 | −4.6 | |
| CRABP2 | Cellular retinoic acid-binding protein 2 | −3.4 | |
| VDR | Vitamin D (1,25-dihydroxyvitamin D3) receptor | −2.9 | Vitamin D signaling |
| CYP27A1 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | −5.0 | |
| CYP24A1 | Cytochrome P450, family 24, subfamily A, polypeptide 1 | +2.8 | |
| ATP1A1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | −2.0 | Transport of ions and other molecules |
| ATP1A3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | −5.2 | |
| AQP3 | Aquaporin 3 (Gill blood group) | +2.2 | |
| ATP6V0A2 | ATPase, H+ transporting, lysosomal V0 subunit a2 | −1.8 | |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21 kDa, V0 subunit b | −1.6 | |
| ATP6V0E2 | ATPase, H+ transporting V0 subunit e2 | −2.9 | |
| CELSR3 /SLC26A6 | Cadherin, EGF LAG seven-pass G-type receptor 3/solute carrier family 26 (anion exchanger), member 6 | −1.8 | |
| SLC26A11 | Solute carrier family 26 (anion exchanger), member 11 | −2.0 | |
| CA9 | Carbonic anhydrase IX | −3.0 | |
| CA11 | Carbonic anhydrase XI | −2.0 | |
| CA12 | Carbonic anhydrase XII | −3.8 | |
| TRPV4 | Transient receptor potential cation channel, subfamily V, member 4 | −3.7 | |
| ARNT2 | Aryl hydrocarbon receptor nuclear translocator 2 | −2.1 | Hypoxia |
| VEGFA | Vascular endothelial growth factor A | −1.6 | |
| SOD3 | Superoxide dismutase 3, extracellular | +1.5 |
Identification of PAX2-targeted pathways in REP epididymis cells
Entrez Gene IDs for the 3207 genes differentially expressed by 1.5-fold in PAX2-depleted cells were evaluated by the DAVID program (Huang da et al., 2009a, b). Relevant statistically significant DAVID ontologies/pathways (P < 0.05) are listed in Table II. These include a number of critical pathways in epididymis epithelial function.
Table II.
Statistically over-represented processes, relevant to epididymis epithelial function from DAVID analysis of the 3135 genes differentially expressed by at least 1.5-fold (FPKM ≥ 1.5) in PAX2-knockdown versus control REP cells.
| No. | Category | Term | P-value |
|---|---|---|---|
| 1 | GOTERM_BP_FAT | GO:0051270∼regulation of cell motion | 1.5 × 106 |
| 2 | GOTERM_BP_FAT | GO:0051272∼positive regulation of cell motion | 1.6 × 106 |
| 3 | GOTERM_CC_FAT | GO:0070161∼anchoring junction | 9.6 × 106 |
| 4 | GOTERM_BP_FAT | GO:0030335∼positive regulation of cell migration | 1.0 × 105 |
| 5 | GOTERM_BP_FAT | GO:0040017∼positive regulation of locomotion | 1.3 × 105 |
| 6 | GOTERM_BP_FAT | GO:0042127∼regulation of cell proliferation | 1.5 × 105 |
| 7 | GOTERM_BP_FAT | GO:0051252∼regulation of RNA metabolic process | 1.9 × 105 |
| 8 | GOTERM_BP_FAT | GO:0030334∼regulation of cell migration | 2.5 × 105 |
| 9 | GOTERM_BP_FAT | GO:0006355∼regulation of transcription, DNA-dependent | 4.0 × 105 |
| 10 | GOTERM_BP_FAT | GO:0070482∼response to oxygen levels | 4.1 × 105 |
| 11 | GOTERM_CC_FAT | GO:0005912∼∼adherens junction | 5.6 × 105 |
| 12 | GOTERM_BP_FAT | GO:0048545∼response to steroid hormone stimulus | 5.7 × 105 |
| 13 | GOTERM_MF_FAT | GO:0046872∼metal ion binding | 6.5 × 105 |
| 14 | GOTERM_BP_FAT | GO:0008284∼positive regulation of cell proliferation | 7.1 × 105 |
| 15 | GOTERM_CC_FAT | GO:0016323∼basolateral plasma membrane | 7.4 × 105 |
| 16 | GOTERM_BP_FAT | GO:0009725∼response to hormone stimulus | 9.0 × 105 |
| 17 | GOTERM_BP_FAT | GO:0040012∼regulation of locomotion | 1.1 × 104 |
| 18 | GOTERM_BP_FAT | GO:0006350∼transcription | 1.2 × 104 |
| 19 | GOTERM_CC_FAT | GO:0005924∼cell-substrate adherens junction | 1.2 × 104 |
| 20 | GOTERM_CC_FAT | GO:0031252∼cell leading edge | 1.5 × 104 |
| 21 | GOTERM_MF_FAT | GO:0043169∼cation binding | 1.6 × 104 |
| 22 | GOTERM_CC_FAT | GO:0005625∼soluble fraction | 1.9 × 104 |
| 23 | GOTERM_MF_FAT | GO:0046914∼transition metal ion binding | 2.0 × 104 |
| 24 | GOTERM_BP_FAT | GO:0010033∼response to organic substance | 2.4 × 104 |
| 25 | GOTERM_MF_FAT | GO:0043167∼ion binding | 2.8 × 104 |
First, transcriptional pathways that reflect both the nuclear location and the ability of PAX2 to bind DNA in vitro (Torban and Goodyer, 1998; Soofi et al., 2012) were evident, for example GO:0006350, P = 1.2 × 10−4 and GO:0045449, P = 3.0 × 10−4, among others with lower but still highly significant P values. Next, there were pathways associated with developmental processes, consistent with the critical role of PAX2 in urogenital tract development (Torres et al., 1995); these include GO:0051094, P = 1.8 × 10−3 and GO:0048754, P = 9.0 × 10−3. The role of PAX2 in epithelial cell proliferation and cell survival was illustrated by highly significant association with GO:0042127, P = 1.5 × 10−5 and GO:0008284, P = 7.1 × 10−5 and is consistent with the function of PAX2 in promoting proliferation and suppressing apoptosis in response to renal injury (Torban and Goodyer, 1998; Zhang et al., 2004; Cohen et al., 2007). PAX2 also appears to have an important role in cell adhesion (GO:0070161, P = 9.6 × 10−6 and GO:0005912, P = 5.6 × 10−5) and cell migration (GO:0051272, P = 1.6 × 10−6, GO:0030335, P = 1.0 × 10−5, and GO:0040017, P = 1.3 × 10−5), among other related pathways with a P-value of <10−4. Pathways associated with cell and substrate adherens junctions (GO:0005912, P = 5.6 × 10−5 and GO:0005924, P = 1.2 × 10−4) and cell–cell junctions (GO:0005911, P = 3.5 × 10−3 and GO:0030054, P = 5.7 × 10−3) were also identified.
Another major group of pathways that are influenced by PAX2 depletion involves signaling by growth factors and hormones. This is important since luminal fluid growth factors are known to interact with their receptors in the epididymis epithelium to regulate gene transcription via the activation of MAP kinase- or PI3 kinase pathways (Kirby et al., 2003). Processes relevant to MAPKKK cascades identified in PAX2-depeleted cells include GO:0000165, P = 1.7 × 10−3 and GO:0043408, P = 6.5 × 10−3. Steroid hormones (androgens and glucocorticoids) and their nuclear receptors (AR and GR) also play important roles in the epididymis (Silva et al., 2011). The observation of high P-value pathways associated with steroid hormone signaling after PAX2 depletion including GO:0048545, P = 5.7 × 10−5, and GO:0009725, P = 9.0 × 10−5 is consistent with this role. Another signaling pathway of specific interest in the epididymis, which is altered on PAX2 depletion, is retinoic acid signaling. Pathways of response to vitamins including vitamin A (retinol) were identified in the DAVID analysis, including GO:0033273, P = 3.0 × 10−4, GO:0033189, P = 1.7 × 10−2 and GO:0032526, P = 1.6 × 10−2.
Finally, another process potentially relevant to epididymis epithelial function that was altered after PAX2 depletion was hypoxia (GO:0070482, P = 4.1 × 10−5 and GO:0001666, P = 4.0 × 10−5).
Identification of novel regulatory elements in PAX2-target genes
The definition of PAX2-target genes by RNA-seq after specific siRNA-depletion led us to inspect the open chromatin maps for individual genes to determine whether they contained predicted PAX2-binding peaks of open chromatin. This approach facilitates dissection of the molecular basis of PAX2 interactions in gene expression. To illustrate this we examined two of the genes that were down-regulated by loss of PAX2 and were subsequently confirmed by qRT–PCR, TRPV4 and PBX1. For clarity in this analysis we concentrated on REP-selective DHS, though many important regulatory elements, particularly those at gene promoters, would be found in multiple cell types. TRPV4 contained a peak of open chromatin in its first intron that was predicted by in silico analysis to encompass a PAX2 motif (Fig. 4A arrow). The first intron is a common site for critical cis-regulatory elements. In the much larger PBX1 locus, multiple REP-selective intronic DHS were identified, several of which coincided with HEE-selective sites. Of particular interest are two sites seen in REP and HEE that contain PAX2 motifs (Fig. 4B arrows marking gray bars).
Figure 4.
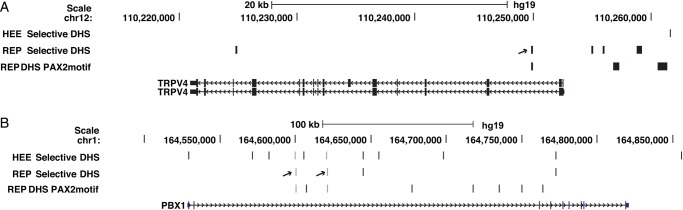
Identification of novel candidate cis-regulatory elements in PAX2-regulated genes in REP cells. REP-selective open chromatin maps are shown above each gene, together with HEE-selective maps, and REP DHS predicted in silico to contain PAX2-binding motifs. (A) TRPV4 locus, arrow denotes REP-selective DHS in the first intron that contains a PAX2 motif. (B) PBX1 locus, grey bars marked by arrows show two REP-selective sites that also seen in HEE cells and that contain PAX2 motifs.
Discussion
Genome-wide analysis of transcriptional networks in different cell types has the power to identify regulatory elements to further our understanding of tissue-specific biological processes. Previous DNase-seq analysis in immature primary HEE cells identified the binding site for PAX2, a critical TF for urogenital tract development (Torres et al., 1995), to be over-represented in HEE-selective regions of open chromatin. In light of the limited replicative potential of HEE cells, we investigated PAX2-regulated genes in the HEE-derived cell line, REP. We confirmed PAX2 expression in these cells and also detected a functional AR, which is pivotal for epididymis formation and function (O'Hara et al., 2011). The effect of the synthetic androgen, R1881, on gene expression was confirmed by microarray analysis. Although few known AR targets were identified, AR-regulated genes have not been well characterized in HEE cells. We further characterized the REP cells, by performing DNase-seq analysis of these cells and comparing the data with results from primary HEE cells (Bischof et al., 2013). Specifically, DNase-seq mapped chromatin accessibility genome wide. Bioinformatic analysis of the sequences within open chromatin peaks was then used to predict over-represented TFBS in REP-selective regions of open chromatin and identified HNF4α, SMAD4, SREBP2 and NFE2L2 among other factors. These TFs have not previously been studied in human epididymis epithelium; however, in the mouse kidney, Hnf4α was shown to be a collaborating factor for the AR (Pihlajamaa et al., 2014) and Nfe2l2 has a role in mouse spermatogenesis (Nakamura et al., 2010; Yu et al., 2012). SMAD4 is an important downstream effector of TGF-β signaling, which is involved in many cellular functions, including proliferation, apoptosis, development and the cell cycle (Massague, 2012). Specific to epididymis function, TGF-β was shown to modulate blood–epididymis-barrier permeability via effects on tight junctions (Stammler et al., 2013). SREBP2 regulates the de novo synthesis of cholesterol, a precursor of important hormones in the epididymis. NFE2L2 binds antioxidant response elements in target gene promoters to regulate gene expression, which may be relevant to the secreted antioxidant enzymes/molecules that protect spermatozoa from oxidative stress (Vernet et al., 2004). Equally, over-represented in both REP-selective sites and ubiquitous sites were TFBS for PAX2, AR, STAT6, ELF5 and PEA3, all of which are relevant to the biology of the epididymis epithelium. STAT6 may play a role in the epididymis epithelial response to inflammation (Turner et al., 2011); ELF5 may regulate a number of genes in glandular epithelial cells (Lapinskas et al., 2004) and PEA3 (also known as ETV4) may play a role in urogenital tract development (Kuure et al., 2010). The critical importance of PAX2 in the urogenital tract (Torres et al., 1995) has already been mentioned and our aim was to determine its targets in the epididymis epithelium.
Identification of PAX2-regulated genes
Among the genes that are differentially expressed by at least 1.5-fold (FPKM ≥ 1.5) in response to PAX2 depletion in REP cells are many with predicted importance in epididymis epithelial function (Table III, Supplementary data, Table SVI).
Development
PAX2-regulated genes relevant to development include Gremlin 1, DAN Family BMP Antagonist 1 (GREM1), a bone morphogenic protein antagonist which was up-regulated (2.1-fold) in PAX2-depleted cells. Like PAX2, GREM1 participates in kidney branching morphogenesis (Michos et al., 2007). In response to renal injury, several studies support pro-proliferative and anti-apoptotic roles for PAX2 (Torban and Goodyer, 1998; Zhang et al., 2004; Cohen et al., 2007). Although the anti-apoptotic mechanism is not yet defined, the anti-apoptotic role of MAPK8IP1 (Bonny et al., 2000) and the ability of its encoded protein (JNK-interacting protein-1) to interact with PAX2 (Cai et al., 2002) may suggest a novel role for MAPK8IP1 in the anti-apoptotic mechanism of PAX2.
Cell junctional complexes
PAX2-regulated genes associated with cell–cell junctions, that are important for sperm maturation, include several members of the Claudin family (CLDN2, -4 and -11), the junctional adhesion molecule (JAM) family (JAM2 and -3) and the Cingulin (CGN) gene. Claudins are known to be important in the epididymis, where they contribute to the blood–epididymis barrier (Cyr et al., 2007). Knockdown of CLDN1, −3, −4 or −7 in cultured human epididymal cell lines dramatically decreased their trans-epithelial electrical resistance, a measure of epithelial barrier function (Dube et al., 2010). Adherens junctions are important for cell adhesion and consist of two adhesive multi-protein complexes: the nectin–afadin complex and the cadherin–catenin complex. Of these, the expression of both the PVRL1 gene (which encodes Nectin-1) and the CDH6 gene (which encodes Cadherin-6) were down-regulated by PAX2 depletion.
Signaling pathways
PAX2 depletion also modulated several ligands and receptors of the PDGF and TGF-β signaling pathways. Perhaps of relevance to this is the phenotype of Pdfg-α-null mice, which exhibit disruption of the epididymis epithelium at post-natal Day 25 (Basciani et al., 2004). Several genes encoding components of the retinoic acid signaling pathway, including stimulated by retinoic acid 6 (STRA6), retinol-binding protein 1, cellular (RBP1) and cellular retinoic acid-binding proteins 1 and 2 (CRABP1 and CRABP2), were also down-regulated by PAX2 depletion. The importance of retinoic acid in maintaining integrity and function of the epididymis epithelium was described previously (Robaire and Hinton, 2002).
Transport of ions and other molecules
PAX2-regulated genes associated with water and ion transport include ATPase, Na+/K+ transporting, alpha 1 and alpha 3 polypeptide (ATP1A1 and ATP1A3) and aquaporin 3 (AQP3). The ATP1A members encode subunits of the Na+/K+-ATPase ion pump that facilitates water movement across the epididymis (Ilio and Hess, 1992), AQP3 was detected in rat epididymis epithelial cells (Hermo et al., 2004) where it probably contributes to water reabsorption from the epididymis lumen. In the airway, AQP3 protein is thought to functionally interact with CFTR, another protein that is critical to ion transport across the epididymis epithelium (Schreiber et al., 1999). PAX2 depletion also down-regulated several genes encoding different subunits of the V-ATPase proton pump, which maintains an acidic epididymal lumen required for sperm maturation (Jensen et al., 1999; Isnard-Bagnis et al., 2003; Pastor-Soler et al., 2003). These include ATPase, H+ transporting, lysosomal V0 subunit a2, b and e2 (ATP6V0A2, ATP6V0B and ATP6V0E2). Maintenance of an acidic luminal milieu in the epididymis is also dependent on the carbonic anhydrase family, of which CA9 and CA11 were both repressed following PAX2 depletion in REP cells. Luminal acidification is in turn fine tuned by bicarbonate transport across the epididymis epithelium (Shum et al., 2008), which can be regulated by solute carrier family 26 anion exchanger (SLC26A) family members. PAX2 depletion was seen to down-regulate SLC26A6 and SLC26A11, which encode two bicarbonate transporters. SL26A6 and CFTR are known to function together in the pancreatic duct, where mutations in CFTR can be measured as defects in bicarbonate transport (Ko et al., 2002; Chernova et al., 2003; Ko et al., 2004) and SLC26A6 can also mediate bicarbonate transport under the control of CFTR in other tissues (Shcheynikov et al., 2008). In human efferent ducts, SLC26A6 is coexpressed with CFTR and the sodium–hydrogen exchanger 3 (NHE3 or SLC9A3) (Kujala et al., 2007).
Hypoxia
There is increasing interest in the role of hypoxia on male fertility, in part related to transient existence at high altitude and this correlates with aberrant function of the epididymis (Vargas et al., 2011). Hence, the identification of expression level changes in hypoxia-associated genes [e.g. the aryl hydrocarbon receptor nuclear translocator 2 (ARNT2) and vascular endothelial growth factor A (VEGFA)] after PAX2 depletion is of interest and is consistent with a report of PAX2 up-regulation in renal epithelial cell lines exposed to hypoxia (Luu et al., 2009). In response to hypoxia, ARNT2 interacts with HIF1-alpha in the nucleus to bind to hypoxia-responsive elements in enhancers and promoters of oxygen-responsive genes such as VEGFA. A role for VEGF in non-endothelial cells is supported by the detection of ligands and receptors of VEGF in basal cells of the human epididymis (Ergun et al., 1998) and in renal epithelial cells (Tufro et al., 1999; Kanellis et al., 2000; Villegas and Tufro, 2002).
The data reported here reveal multiple genes and processes regulated by the PAX2 TF in a human epididymal cell line, which are relevant to the biological functions of the epididymis epithelium. These results will open new pathways of investigation that may facilitate novel therapeutic approaches to male infertility.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
J.A.B., R.Y., S.-H.L. and A.H. conceived and designed experiments. J.A.B., R.Y., S.-H.L., L.S., G.E.C. and A.H. acquired, analyzed and interpreted the data. J.A.B., R.Y. and A.H. wrote the article. J.A.B., R.Y., S.-H.L., G.E.C. and A.H. revised article for intellectual content. J.A.B., R.Y., S.-H.L., L.S., G.E.C. and A.H. approved the version to be published.
Funding
This work was supported by National Institutes of Health (R01HD068901 to A.H) and the Cystic Fibrosis Foundation (Harris11G0 to A.H.).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We thank Lisa Jones and Dr Wanda O'Neal for RNA-seq library generation, Jared Bischof, Drs Austin Gillen and Hong Dang for bioinformatics assistance and Dr Samantha Gadd for microarray analysis.
References
- Basciani S, Mariani S, Arizzi M, Brama M, Ricci A, Betsholtz C, Bondjers C, Ricci G, Catizone A, Galdieri M, et al. Expression of platelet-derived growth factor (PDGF) in the epididymis and analysis of the epididymal development in PDGF-A, PDGF-B, and PDGF receptor beta deficient mice. Biol Reprod. 2004;70:168–177. doi: 10.1095/biolreprod.103.019232. [DOI] [PubMed] [Google Scholar]
- Bischof JM, Gillen AE, Song L, Gosalia N, London D, Furey TS, Crawford GE, Harris A. A genome-wide analysis of open chromatin in human epididymis epithelial cells reveals candidate regulatory elements for genes coordinating epididymal function. Biol Reprod. 2013;89:104. doi: 10.1095/biolreprod.113.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist SR, Vidarsson H, Soder O, Enerback S. Epididymal expression of the forkhead transcription factor Foxi1 is required for male fertility. EMBO J. 2006;25:4131–4141. doi: 10.1038/sj.emboj.7601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonny C, Oberson A, Steinmann M, Schorderet DF, Nicod P, Waeber G. IB1 reduces cytokine-induced apoptosis of insulin-secreting cells. J Biol Chem. 2000;275:16466–16472. doi: 10.1074/jbc.M908297199. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Lechner MS, Nihalani D, Prindle MJ, Holzman LB, Dressler GR. Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J Biol Chem. 2002;277:1217–1222. doi: 10.1074/jbc.M109663200. [DOI] [PubMed] [Google Scholar]
- Chernova MN, Jiang L, Shmukler BE, Schweinfest CW, Blanco P, Freedman SD, Stewart AK, Alper SL. Acute regulation of the SLC26A3 congenital chloride diarrhoea anion exchanger (DRA) expressed in Xenopus oocytes. J Physiol. 2003;549:3–19. doi: 10.1113/jphysiol.2003.039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T, Loutochin O, Amin M, Capolicchio JP, Goodyer P, Jednak R. PAX2 is reactivated in urinary tract obstruction and partially protects collecting duct cells from programmed cell death. Am J Physiol Renal Physiol. 2007;292:F1267–F1273. doi: 10.1152/ajprenal.00281.2006. [DOI] [PubMed] [Google Scholar]
- Coleman L, Harris A. Immortalization of male genital duct epithelium: an assay system for the cystic fibrosis gene. J Cell Sci. 1991;98(Pt 1):85–89. doi: 10.1242/jcs.98.1.85. [DOI] [PubMed] [Google Scholar]
- Cyr DG, Gregory M, Dube E, Dufresne J, Chan PT, Hermo L. Orchestration of occludins, claudins, catenins and cadherins as players involved in maintenance of the blood-epididymal barrier in animals and humans. Asian J Androl. 2007;9:463–475. doi: 10.1111/j.1745-7262.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- Dube E, Dufresne J, Chan PT, Hermo L, Cyr DG. Assessing the role of claudins in maintaining the integrity of epididymal tight junctions using novel human epididymal cell lines. Biol Reprod. 2010;82:1119–1128. doi: 10.1095/biolreprod.109.083196. [DOI] [PubMed] [Google Scholar]
- Ergun S, Luttmer W, Fiedler W, Holstein AF. Functional expression and localization of vascular endothelial growth factor and its receptors in the human epididymis. Biol Reprod. 1998;58:160–168. doi: 10.1095/biolreprod58.1.160. [DOI] [PubMed] [Google Scholar]
- Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Coleman L. Ductal epithelial cells cultured from human foetal epididymis and vas deferens: relevance to sterility in cystic fibrosis. J Cell Sci. 1989;92(Pt 4):687–690. doi: 10.1242/jcs.92.4.687. [DOI] [PubMed] [Google Scholar]
- Hermo L, Krzeczunowicz D, Ruz R. Cell specificity of aquaporins 0, 3, and 10 expressed in the testis, efferent ducts, and epididymis of adult rats. J Androl. 2004;25:494–505. doi: 10.1002/j.1939-4640.2004.tb02820.x. [DOI] [PubMed] [Google Scholar]
- Hoglund P, Hihnala S, Kujala M, Tiitinen A, Dunkel L, Holmberg C. Disruption of the SLC26A3-mediated anion transport is associated with male subfertility. Fertil Steril. 2006;85:232–235. doi: 10.1016/j.fertnstert.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ilio KY, Hess RA. Localization and activity of Na+,K(+)-ATPase in the ductuli efferentes of the rat. Anat Rec. 1992;234:190–200. doi: 10.1002/ar.1092340206. [DOI] [PubMed] [Google Scholar]
- Isnard-Bagnis C, Da Silva N, Beaulieu V, Yu AS, Brown D, Breton S. Detection of ClC-3 and ClC-5 in epididymal epithelium: immunofluorescence and RT-PCR after LCM. Am J Physiol Cell Physiol. 2003;284:C220–C232. doi: 10.1152/ajpcell.00374.2001. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Stuart-Tilley AK, Peters LL, Lux SE, Alper SL, Breton S. Immunolocalization of AE2 anion exchanger in rat and mouse epididymis. Biol Reprod. 1999;61:973–980. doi: 10.1095/biolreprod61.4.973. [DOI] [PubMed] [Google Scholar]
- Kanellis J, Fraser S, Katerelos M, Power DA. Vascular endothelial growth factor is a survival factor for renal tubular epithelial cells. Am J Physiol Renal Physiol. 2000;278:F905–F915. doi: 10.1152/ajprenal.2000.278.6.F905. [DOI] [PubMed] [Google Scholar]
- Kirby JL, Yang L, Labus JC, Hinton BT. Characterization of fibroblast growth factor receptors expressed in principal cells in the initial segment of the rat epididymis. Biol Reprod. 2003;68:2314–2321. doi: 10.1095/biolreprod.102.011270. [DOI] [PubMed] [Google Scholar]
- Ko SB, Shcheynikov N, Choi JY, Luo X, Ishibashi K, Thomas PJ, Kim JY, Kim KH, Lee MG, Naruse S, et al. A molecular mechanism for aberrant CFTR-dependent HCO(3)(-) transport in cystic fibrosis. EMBO J. 2002;21:5662–5672. doi: 10.1093/emboj/cdf580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SB, Zeng W, Dorwart MR, Luo X, Kim KH, Millen L, Goto H, Naruse S, Soyombo A, Thomas PJ, et al. Gating of CFTR by the STAS domain of SLC26 transporters. Nat Cell Biol. 2004;6:343–350. doi: 10.1038/ncb1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala M, Hihnala S, Tienari J, Kaunisto K, Hastbacka J, Holmberg C, Kere J, Hoglund P. Expression of ion transport-associated proteins in human efferent and epididymal ducts. Reproduction. 2007;133:775–784. doi: 10.1530/rep.1.00964. [DOI] [PubMed] [Google Scholar]
- Kuure S, Chi X, Lu B, Costantini F. The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development. 2010;137:1975–1979. doi: 10.1242/dev.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas EJ, Palmer J, Ricardo S, Hertzog PJ, Hammacher A, Pritchard MA. A major site of expression of the ets transcription factor Elf5 is epithelia of exocrine glands. Histochem Cell Biol. 2004;122:521–526. doi: 10.1007/s00418-004-0713-x. [DOI] [PubMed] [Google Scholar]
- Leir SH, Harris A. MUC6 mucin expression inhibits tumor cell invasion. Exp Cell Res. 2011;317:2408–2419. doi: 10.1016/j.yexcr.2011.07.021. [DOI] [PubMed] [Google Scholar]
- Luu VD, Boysen G, Struckmann K, Casagrande S, von Teichman A, Wild PJ, Sulser T, Schraml P, Moch H. Loss of VHL and hypoxia provokes PAX2 up-regulation in clear cell renal cell carcinoma. Clin Cancer Res. 2009;15:3297–3304. doi: 10.1158/1078-0432.CCR-08-2779. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- Nakamura BN, Lawson G, Chan JY, Banuelos J, Cortes MM, Hoang YD, Ortiz L, Rau BA, Luderer U. Knockout of the transcription factor NRF2 disrupts spermatogenesis in an age-dependent manner. Free Radic Biol Med. 2010;49:1368–1379. doi: 10.1016/j.freeradbiomed.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara L, Welsh M, Saunders PT, Smith LB. Androgen receptor expression in the caput epididymal epithelium is essential for development of the initial segment and epididymal spermatozoa transit. Endocrinology. 2011;152:718–729. doi: 10.1210/en.2010-0928. [DOI] [PubMed] [Google Scholar]
- Oefelein M, Grapey D, Schaeffer T, Chin-Chance C, Bushman W. Pax-2: a developmental gene constitutively expressed in the mouse epididymis and ductus deferens. J Urol. 1996;156:1204–1207. doi: 10.1016/s0022-5347(01)65751-3. [DOI] [PubMed] [Google Scholar]
- Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003;278:49523–9. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajamaa P, Sahu B, Lyly L, Aittomaki V, Hautaniemi S, Janne OA. Tissue-specific pioneer factors associate with androgen receptor cistromes and transcription programs. EMBO J. 2014;33:312–326. doi: 10.1002/embj.201385895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaire B, Hinton B. The Epididymis: From Molecules to Clinical Practice. XIII edn. New York: Kluwer Academic/Plenum Publishers; 2002. [Google Scholar]
- Schreiber R, Nitschke R, Greger R, Kunzelmann K. The cystic fibrosis transmembrane conductance regulator activates aquaporin 3 in airway epithelial cells. J Biol Chem. 1999;274:11811–11816. doi: 10.1074/jbc.274.17.11811. [DOI] [PubMed] [Google Scholar]
- Shcheynikov N, Yang D, Wang Y, Zeng W, Karniski LP, So I, Wall SM, Muallem S. The Slc26a4 transporter functions as an electroneutral Cl−/I−/HCO3− exchanger: role of Slc26a4 and Slc26a6 in I− and HCO3− secretion and in regulation of CFTR in the parotid duct. J Physiol. 2008;586:3813–3824. doi: 10.1113/jphysiol.2008.154468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum WW, Da Silva N, McKee M, Smith PJ, Brown D, Breton S. Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell. 2008;135:1108–1117. doi: 10.1016/j.cell.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva EJ, Queiroz DB, Rodrigues A, Honda L, Avellar MC. Innate immunity and glucocorticoids: potential regulatory mechanisms in epididymal biology. J Androl. 2011;32:614–624. doi: 10.2164/jandrol.111.013565. [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soofi A, Levitan I, Dressler GR. Two novel EGFP insertion alleles reveal unique aspects of Pax2 function in embryonic and adult kidneys. Dev Biol. 2012;365:241–250. doi: 10.1016/j.ydbio.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stammler A, Muller D, Tabuchi Y, Konrad L, Middendorff R. TGFbetas modulate permeability of the blood-epididymis barrier in an in vitro model. PloS one. 2013;8:e80611. doi: 10.1371/journal.pone.0080611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torban E, Goodyer P. What PAX genes do in the kidney. Exp Nephrol. 1998;6:7–11. doi: 10.1159/000020498. [DOI] [PubMed] [Google Scholar]
- Torres M, Gomez-Pardo E, Dressler GR, Gruss P. Pax-2 controls multiple steps of urogenital development. Development. 1995;121:4057–4065. doi: 10.1242/dev.121.12.4057. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traystman MD, Schulte NA, MacDonald M, Anderson JR, Sanger WG. Mutation analysis for cystic fibrosis to determine carrier status in 167 sperm donors from the Nebraska Genetic Semen Bank. Hum Mutat. 1994;4:271–275. doi: 10.1002/humu.1380040407. [DOI] [PubMed] [Google Scholar]
- Tufro A, Norwood VF, Carey RM, Gomez RA. Vascular endothelial growth factor induces nephrogenesis and vasculogenesis. J Am Soc Nephrol. 1999;10:2125–2134. doi: 10.1681/ASN.V10102125. [DOI] [PubMed] [Google Scholar]
- Turner TT, Mammen T, Kavoussi P, Lysiak JJ, Costabile RA. Cytokine responses to E. coli-induced epididymitis in the rat: blockade by vasectomy. Urology. 2011;77:1507. doi: 10.1016/j.urology.2011.02.037. e9–14. [DOI] [PubMed] [Google Scholar]
- van der Ven K, Messer L, van der Ven H, Jeyendran RS, Ober C. Cystic fibrosis mutation screening in healthy men with reduced sperm quality. Hum Reprod. 1996;11:513–517. doi: 10.1093/humrep/11.3.513. [DOI] [PubMed] [Google Scholar]
- Vargas A, Bustos-Obregon E, Hartley R. Effects of hypoxia on epididymal sperm parameters and protective role of ibuprofen and melatonin. Biol Res. 2011;44:161–167. [PubMed] [Google Scholar]
- Vernet P, Aitken RJ, Drevet JR. Antioxidant strategies in the epididymis. Mol Cell Endocrinol. 2004;216:31–39. doi: 10.1016/j.mce.2003.10.069. [DOI] [PubMed] [Google Scholar]
- Villegas G, Tufro A. Ontogeny of semaphorins 3A and 3F and their receptors neuropilins 1 and 2 in the kidney. Gene Expr Patterns. 2002;2:151–155. doi: 10.1016/s0925-4773(02)00305-2. [DOI] [PubMed] [Google Scholar]
- Yu B, Lin H, Yang L, Chen K, Luo H, Liu J, Gao X, Xia X, Huang Z. Genetic variation in the Nrf2 promoter associates with defective spermatogenesis in humans. J Mol Med. 2012;90:1333–1342. doi: 10.1007/s00109-012-0914-z. [DOI] [PubMed] [Google Scholar]
- Zhang SL, Guo J, Moini B, Ingelfinger JR. Angiotensin II stimulates Pax-2 in rat kidney proximal tubular cells: impact on proliferation and apoptosis. Kidney Int. 2004;66:2181–2192. doi: 10.1111/j.1523-1755.2004.66008.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.