Abstract
Unwanted sedimentation and attachment of a number of cells onto the bottom channel often occur on relatively large-scale inlets of conventional microfluidic channels as a result of gravity and fluid shear. Phenomena such as sedimentation have become recognized problems that can be overcome by performing microfluidic experiments properly, such as by calculating a meaningful output efficiency with respect to real input. Here, we present a dual-inlet design method for reducing cell loss at the inlet of channels by adding a new “ upstream inlet ” to a single main inlet design. The simple addition of an upstream inlet can create a vertically layered sheath flow prior to the main inlet for cell loading. The bottom layer flow plays a critical role in preventing the cells from attaching to the bottom of the channel entrance, resulting in a low possibility of cell sedimentation at the main channel entrance. To provide proof-of-concept validation, we applied our design to a microfabricated flow cytometer system (μFCS) and compared the cell counting efficiency of the proposed μFCS with that of the previous single-inlet μFCS and conventional FCS. We used human white blood cells and fluorescent microspheres to quantitatively evaluate the rate of cell sedimentation in the main inlet and to measure fluorescence sensitivity at the detection zone of the flow cytometer microchip. Generating a sheath flow as the bottom layer was meaningfully used to reduce the depth of field as well as the relative deviation of targets in the z-direction (compared to the x-y flow plane), leading to an increased counting sensitivity of fluorescent detection signals. Counting results using fluorescent microspheres showed both a 40% reduction in the rate of sedimentation and a 2-fold higher sensitivity in comparison with the single-inlet μFCS. The results of CD4+ T-cell counting also showed that the proposed design results in a 25% decrease in the rate of cell sedimentation and a 28% increase in sensitivity when compared to the single-inlet μFCS. This method is simple and easy to use in design, yet requires no additional time or cost in fabrication. Furthermore, we expect that this approach could potentially be helpful for calculating exact cell loading and counting efficiency for a small input number of cells, such as primary cells and rare cells, in microfluidic channel applications.
INTRODUCTION
Microfluidic technologies for cell manipulation have played an essential role in various fields of cell biology and biomedical research such as cancer treatment, intra-cellular drug delivery, and flow cytometry because of the ability to precisely control the cellular environment, analyze cellular information at the single-cell level, and provide additional capabilities to the point-of-care testing (POCT) systems.1–5 Most microfluidic chips for analysis of both relatively stationary cells (e.g., cell imaging) and relatively dynamic cells (e.g., flow cytometry) lie horizontally, and the sample inlet and outlet ports are perpendicular to the microchannel inside the microchip. This is because most microfluidic chips are fabricated by photolithography, which is primarily a 2-dimensional process. As a result, bonding between upper and lower substrates is required to close the microchannels within the microchip. The main reason why this type of microchips is preferred in microfluidic research is that it facilitates an inter-connection between the microchip and its sample delivery tubes on the stage of the inverted fluorescence microscopes that are commonly used for cell analysis in laboratories. However, horizontally placed microchips have some disadvantages related to gravity, such as a geometric change from a macroscale inlet to a microscale channel entrance, resulting in a remarkable cell loss due to unwanted cell sedimentation at the inlet area. Recently, low-cost three-dimensional (3D) fabrication methods of a microfluidic device were developed using a layered paper and a paper–PDMS composite.6,7 The devices using those fabrication methods can bring new function and capabilities to current microfluidic systems, such as fluids flowing vertically and flexible microfluidic devices. However, those techniques cannot directly apply to this study because the current fabrication methods can produce microchannels at a minimal width of 100 μm.
Traditional flow cytometers use 3D hydrodynamic focusing to align target cells into a single line within a glass capillary or a quartz cuvette, the so-called “flow cell.”8,9 In addition, sample injection and the flow cell have the same direction, perpendicular to the ground. These features increase sensitivity of the detection of signals from the target cells, and also prevent clogging by exerting gravity on the cells. The microfluidic flow cytometry approach is known to overcome some of the drawbacks of conventional flow cytometry, such as (1) complications related to instrument size (e.g., difficulty of downsizing), (2) the difficulty associated with dealing with a small sample volume, and (3) inflexibility in combining with other cellular assays. However, several challenges remain to be overcome, such as out-of-plane 3D hydrodynamic focusing, insensitive detection of scattering signals, and unwanted cell clogging and sedimentation at the sample inlet channel.
Among those challenges, the issue of increasing the sensitivity of signal detection has been solved by using microfluidic technologies that can enable several 3D hydrodynamic focusing techniques and microstructures, such as multi-layer microstructures,10,11 single-layer microstructures with multiple fluidic ports,12,13 and external forces (e.g., dielectrophoretic force).14,15 Several designs to prevent the cell clogging that often occurs when cells enter microchannels have been suggested16–18 but are unsatisfactory for total cell counting per real input of cells in microfluidics. In general, the standard microchips can easily cause erroneous results in cell counting and complete fouling of the microchannels during cell loading. A specially designed microstructure, like the “asymmetric” inlet design, or a particular cell-to-channel ratio (lower than 0.3) can be used to decrease the probability of microchannel clogging. Few studies on the effect of cell sedimentation at the inlet channel on total cell loss in microchannel networks (i.e., the cell loss from the start time of cell loading at inlet channels to the end time of collection at outlet channels) have been reported.
In this paper, we present a design strategy to minimize the sedimentation of cells at the entrance to main channels and increase cell-loading efficiency throughout the entire microchannel network. We also discuss why the dual inlet design is needed by highlighting current fluid force theories and address how to decrease total cell loss due to cell sedimentation. To provide the proof-of-concept validation of our design (i.e., minimization of cell sedimentation and enhancement of detection sensitivity), we applied this idea to a microfabricated flow cytometer that has the capability of affordable 3D hydrodynamic focusing by a single-layer microstructure.
MATERIALS AND METHODS
Setup of the flow cytometry system and its peripherals
To quantify the sedimentation rate, we measured the fluorescent signals from human blood samples and polystyrene microspheres using a microfabricated flow cytometer and optics system similar to that previously reported.3,19 Briefly, the system consists of a 488-nm laser of 20 mW, an excitation band-pass filter (482 ± 16 nm), a 506-nm dichroic mirror for separating excitation and emission light paths, an objective lens (20X LCPlanFL, Olympus, Japan), an emission filter (536 ± 20 nm), and a slit of width 250 μm positioned in front of photo-multipliers (PMTs; H6780–20, Hamamatsu, Japan). The system was operated by an imbedded controller, and a 16-bit data acquisition board (USB-6218, National Instruments (NI), USA) was used to record the PMT signals. Data acquisition was accomplished using a custom-made program written in C++ language with NI-DAQ library (ver. 8.8, NI). Data were saved in the Flow Cytometry Standard (FCS) file format compatible with commercial FACS data files, and were subsequently expressed as histograms. Every sample was delivered by using a peristaltic pump (400/M, Watson Marlow, USA). The flow rate was selected from the standard protocol of flow cytometry in microchannels (data not shown here).
Preparation of a microfabricated flow cytometer microchip
Microchannel patterns shown in Figure 1 were etched on a 4″ quartz wafer of 50 μm depth by conventional photolithography. On the other quartz wafer, through-holes were punched by sand blasting for connection with sample delivery tubes. These two wafers were bonded by thermo-compression to close each other and, the bonded wafer was diced into 60 microchips. Design parameters were optimized to detect white blood cells (WBCs; ∼10 μm diameter on average): the height of the microchannel was 50 μm; the channel width ranged from 50 μm to 250 μm; and the detection area located below the sample inlet, the so-called expansion channel, had a 250–μm circular shape as previously reported.19 The microchannel with above parameters allows WBCs to focus into the single file, since the channel can make the width of sample flow about 10 μm. Briefly, the detection area was carefully designed so that stagnation zones are not generated, and the expansion ratio was optimized for detection of CD4+ T lymphocytes and 10 μm polystyrene microspheres. Here, the best ratio of expansion was the 5 times of the width of the narrowest channel. Therefore, the width of the optimized detection area was 50 μm at its entrance and 250 μm at its detection spot.
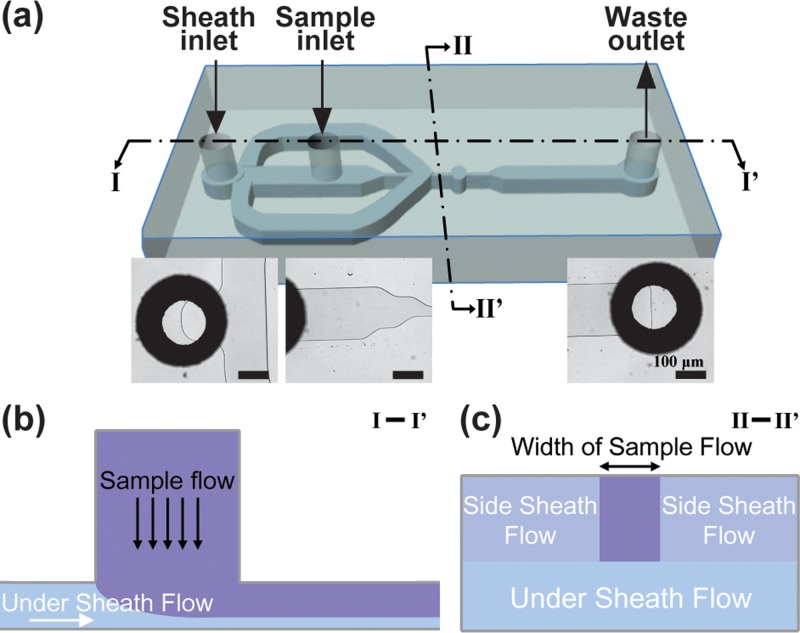
FIG. 1.
Schematic illustration of the dual inlet design. (a) Schematic view of the microfluidic flow cytometry channel using the proposed design strategy. Insets show bottom-up microscopic images at sheath inlet (left), sample inlet (middle), and waste outlet (right), respectively. The scale bar is 100 μm. The design has one sheath inlet (located outside) and one sample flow inlet (located at the middle) for cell loading and focusing. (b) Cross-sectional views of the microchannel. The cross sectional area of I-I′ (b) shows the role of the sheath flow in preventing sedimentation, and the cross sectional area of II-II′ (c) shows the 3D focusing effect using the single sheath inlet. In this study, the width of sample flow is about 10 μm so that WBCs can align into a single-file focusing flow.
Preparation of human white blood cells for flow cytometric test
To enumerate CD4+ T-cells in human blood by microchip flow cytometry, whole blood samples were obtained from healthy donors at the Korea University Ansan Hospital in Ansan, South Korea. The samples were collected into EDTA Vacutainer tubes (BD, USA), maintained at room temperature (RT) and processed within 24 h of collection. The samples were treated with a conventional lyse-wash protocol using an erythrocyte lysis solution (FACS lysing solution, BD, USA). To separate leukocytes from whole blood, 5 ml of erythrocytes lysis solution (1×) was added to 500 μl of whole blood, and then the sample was mixed by gentle shaking. After a 10-min incubation period at RT, the tube was centrifuged at 1500 rpm (i.e., 250g) for 5 min, the supernatant was removed, and 5 ml of 1× phosphate buffered saline (PBS) was added. After gentle tapping and vortexing, the sample was centrifuged at 1500 rpm for 5 min and the pellet was re-suspended in 300 μl PBS. Twenty microliters of anti-CD4 monoclonal antibody labeled with FITC was added to 100 μl of the leukocyte sample and incubated at 4 °C for 30 min in the dark. The sample was centrifuged at 1500 rpm for 5 min and washed with 1 ml PBS twice and the final pellet was resuspended in 300 μl PBS. We also used 10–μm fluorescent polystyrene microspheres at a concentration of 8000 beads per μl to quantitatively measure the sedimentation rate.
RESULTS
We first describe the forces determining cell sedimentation in theory, and what factors should be considered to solve the problem in design and experimentation. We then report the main results of our study, showing how our system addresses these issues. We also include examples of CD4 counting as a representative example.
The concept of dual inlet is firmly supported by fluid force theories
Blood cells are heavier than the plasma and saline solution, therefore they naturally settle to the bottom of the solution. In general, cell sedimentation occurs when the density of cells is greater than that of stationary fluids, or in a curved channel due to the centrifugal or Coriolis forces exerted on the cells. Despite the low Reynolds number of general microfluidic conditions (data not shown), cells moving through a straight microchannel does not generate a situation of sedimentation because of the short time scale. The main forces acting on buoyant particles flowing in microchannels are lift, Stokes' drag, and gravity. Basically, the drag force (FD) based on Stokes' law acting on particles in microfluidics can be expressed as . The lift (FL) and the gravitational force (FG) on the particle in fluid can be represented as , and , respectively, where “a” denotes the particle diameter, the fluid viscosity, U the mean flow velocity, L the characteristic length (which is in general a hydraulic diameter of the microchannel), the particle density, and the density of the fluid. According to the above equations, the sedimentation occurs if the density of particles in solution is much higher than the density of the solution, or the lift and drag forces are not sufficient to generate particle movement against the viscous resistance to the direction of the outlet. In general, mammalian cells and microbeads used in microfluidic experiments have a slightly higher density than that of the surrounding fluids, so the sedimentation rate is most strongly related to the velocity of the particles. Inertia of a cell is generally assumed to be negligible. Therefore, we also do not consider the inertia effect of the cell in this study in order to simplify the above equations. However, in some cases with highly deformable cells like red blood cells (RBCs), inertia of the cells needs to be considered to better understand blood flow at low Reynolds number.20
The dual inlet generates a sheath flow as the bottom layer, leading to a low possibility of cell sedimentation
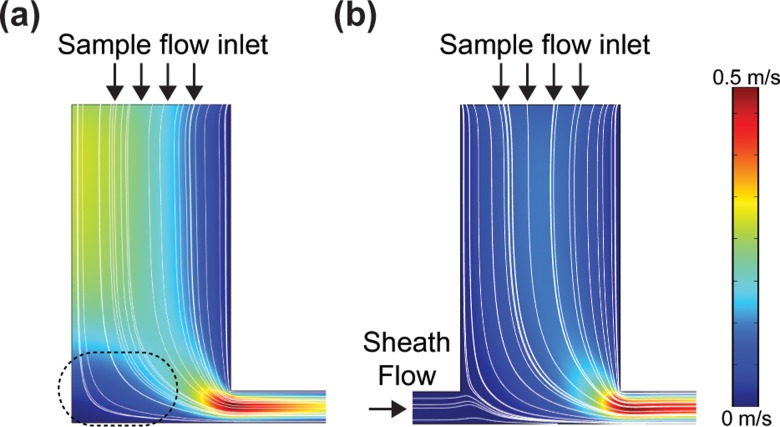
To investigate how stagnation areas were generated in the inlets, numerical simulation was conducted using COMSOL Multiphysics1 (v4.0, Comsol Group, USA). The simulation condition was as follows: density of fluid, 1000 kg m−3; dynamic viscosity of fluid, 0.001005 Pa s; and pressure difference between the inlet and the outlet, 1 kPa. Simulation results indicate that the single inlet structure has a stagnation area that generates a low-flow velocity field whereas the dual inlet has hardly any stagnation area, as shown in Figure 2.
FIG. 2.
COMSOL simulation results for streamlines in the single and dual inlets. Calculation of (a) single inlet (2D focusing) and (b) dual inlet (3D focusing) using COMSOL Multiphysics1 (v4.0, COMSOL group, USA) to investigate whether stationary areas would be generated at the inlet bottom. The simulation was conducted under the following conditions: density of fluid, 1000 kg m−3, dynamic viscosity of fluid, 0.001005 Pa s, and pressure difference between the inlet and outlet, 1 kPa. The simulation results show that the single inlet structure (with almost 2D focusing) generates a non-uniform stationary area, leading to a relatively lower velocity field(indicated by dotted ellipsoid), whereas the dual inlet structure (similar to 3D focusing) has a uniform velocity profile due to intake of additional sheath flow.
The perpendicular interconnection between the microchannel and the sample inlet that is often used in conventional 2D hydrodynamic focusing can generate stagnation areas at the bottom corners where it has very low flow velocity (Figure 2(a)). Therefore, the cells and particles in these stagnation areas cannot attain enough drag force to move into the flow channel. However, the “dual inlet” (i.e., an upstream inlet and a main inlet) of the proposed design strategy generates hardly any stagnation areas at the bottom of the microchannel (Figure 2(b)).
As shown in Figure 1, to achieve a low sedimentation rate, we also designed a microchannel to generate a sheath flow as the bottom layer towards the sample inlet, reducing the possibility of cells contacting to the bottom and ensuring that any sedimented cells were easily flushed to the main channel. Unlike previous 2D hydrodynamic focusing techniques that can produce only side sheath flows, the proposed technique can be used to generate an additional sheath flow enabling 3D hydrodynamic focusing (called “under sheath flow” in Figures 2(b) and 2(c)) without additional layers of “out-of-plane” microstructures or external powers such as dielectrophoretic force.
The dual inlet increases both cell loading and counting efficiency
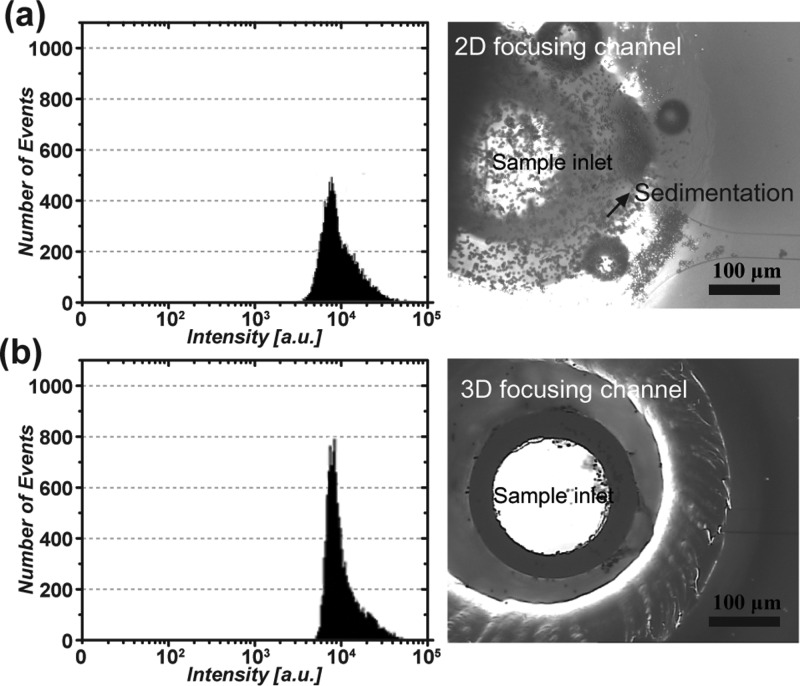
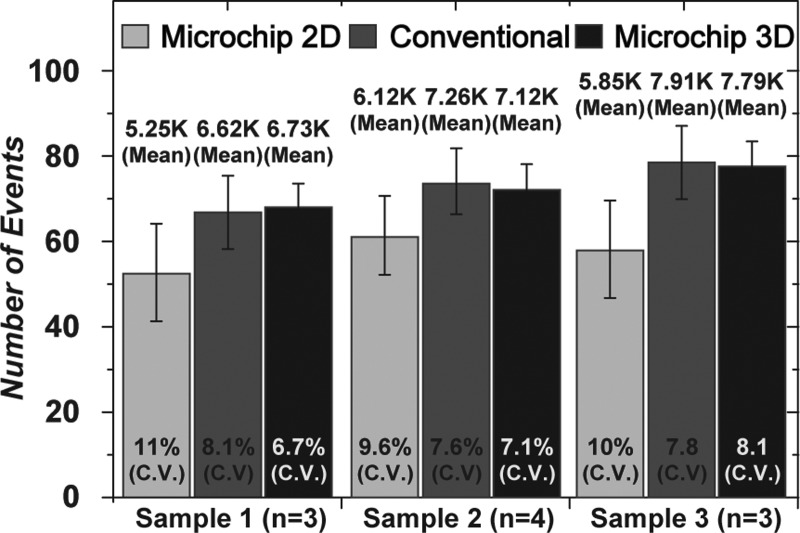
To quantitatively compare the sedimentation rate between the previous and proposed hydrodynamic focusing techniques, microfluidic flow cytometry tests were performed using 10-μm fluorescent microbeads. Microscopic images of each inlet show that a sample inlet in the 2D channel has a high sedimentation rate of microspheres compared to that of the proposed inlet, as shown in Figure 3. The counting results were approximately 10 000 and 14 000 for 2D and 3D focusing, respectively. Therefore, the number of cells was increased by approximately 40% using the proposed dual inlet. This result demonstrates that the proposed inlet can reduce the rate of cell sedimentation and minimize cell loss during flow cytometric experiments. In addition, the coefficient of variation (CV; defined as the ratio of the standard deviation to the average) of the counting result from dual inlet (3.2%) was lower than that of the single inlet system (7.1%). This is because the dual inlet design reduced the depth of field and the deviation of targets in the z-direction, resulting in a large increase in detection sensitivity of fluorescent signals. Figure 4 shows that the counting result for CD4+ T-cells was also improved compared to both the single inlet microchannel-type flow cytometer and conventional flow cytometry system. As expected, the average total number of counted cells measured by the dual inlet microchip (7213 counts) and conventional FACS (7263 counts) was higher than that of the single inlet system (5740 counts). The CV value of the counted cells from the dual inlet (7.3%) and conventional FACS (7.8%) was lower than that of the single inlet system (10.2%).
FIG. 3.
Counting results for microspheres in microfabricated flow cytometers. Comparison of counting results using 10-μm microbeads for (a) the single inlet (2D focusing) and (b) the dual inlet (3D focusing) channel. The counting number was approximately 10 000 counts at the single inlet (top left, histogram) compared with 14 000 counts at the dual inlet (bottom left, histogram). Note that we used the same concentration of microspheres for both. This result clearly indicates that the total number of loaded samples can be efficiently preserved without loss at the dual inlet. Note that the x-axis denotes the fluorescence intensity measured in a log-scale level, and the y-axis denotes the counting number. The microscopic images show the single sample inlet, in which a lot of the microspheres sedimented (top right), and the dual sample inlet with an additional sheath inlet that prevented sedimentation of microspheres (bottom right).
FIG. 4.
CD4+ T-cell counting results. Graph comparing CD4+ T-cell counting results for the single inlet, called “microchip 2D focusing” (light gray), conventional FACS (dark gray), and the dual inlet, called “microchip 3D focusing” (black). Note that we used FACS Calibur (BD, USA) for conventional FACS. The counting results of CD4+ T-cells indicate that the dual inlet (average 7213 counts) and the conventional system (average 7263 counts) were similar, whereas the original 2D focusing system (average 5740 counts) exhibited lower counts.
DISCUSSION
The simple dual inlet can overcome a known dilemma in microchannels
Some critical factors for cell manipulation in microfluidic channels should be discussed in more depth. Basically, when the height of the microchannel is low, the sedimentation rate of cells increases and the detection sensitivity of fluorescent signals also increase. Conversely, when the height of the microchannel is high, the sedimentation rate of cells decreases, and detection sensitivity of fluorescent signals decreases. To overcome this dilemma, we proposed a simple new microchannel design strategy and simultaneously achieved a low rate of cell sedimentation and high detection sensitivity in flow cytometry tests. Moreover, this technique requires only one reservoir to drive 3D hydrodynamic focusing and minimize cell sedimentation with a single microstructure-layer.
The cell loading efficiency can be calibrated with actual cell number with the aid of a dual inlet
Cell loading efficiencies have often been interpreted as the ratio of the number of cells inside a designated area to the number of cells in the whole area. In many applications of microfluidics, the number of cells is counted after the microchannel entrance therefore there is no consideration of cell loss at the inlet, where it often occurs. It is possible that the loading efficiency at the meaningful area might be very low if the input number was a few million cells. Similarly, many researchers assume that all of the cells enter the microchannels successfully without loss. This would not be a big problem if the number of cells is excessive, but is another issue that should be addressed if the number of cells is small. For example, if a small number of rare cells or primary cells is involved, such cell loss might become a critical issue for successful cell screening and analyses in microfluidics. It should be noted that cell counting would be not a big issue even in the case of rare cells if researchers used a conventional cell counting glass or chip. In this study, however, we aimed to address the issue of cell loss in microfluidics and proposed the design of dual inlet that can enable us to perform cell loading at the actual input cell concentration. Actually, the cell loading efficiency can be calibrated and re-defined as the ratio of the number of final output cells to the number of initial input cells. In other words, the definition of cell loading efficiency can be improved by considering not the area but the actual cell number. This is one of the main reasons why the dual inlet is needed for cell loading, especially for microfluidics experiments involving a small number of cells.
Implication of using the dual inlet for cell analysis in microfluidics
There have been many studies on 3D hydrodynamic focusing of moving cell targets in microfluidics, especially in flow cytometry. Most researchers have focused more on improving detection sensitivity rather than cell loading efficiency because they have enough cells for counting. However, it is a quite different story if the cell number is not sufficient. In general, conventional FACS machines work well for cell counting and sorting, even if they have some limitations in handling a small number of cells. During the sorting process in particular, the sorted cells are often not collected correctly, but instead scattered inside the FACS tube. This could be a serious issue even in the case of single-cell screening and analysis. Since microfluidic technologies have a miniaturized workspace for handling a small number of cells, they may play a key role in solving these issues related to small cell number. The dual inlet potentially has an important implication in microfluidic technologies for analysis of single cells or a few cells. Some modified designs of the dual inlet may be more appropriate than the proposed one. For example, the “dual” function may apply well to an inlet, but not always to an outlet. Although the outlet has the same fluidic environment as the inlet, there is the additional issue of collecting cells from the outlet. Since collection is also very important, further studies on design and process in microfluidics should be performed to prevent cell loss at the outlet. In microfluidics, the collection chamber itself can become a work space for cell analysis, unlike the FACS tube. This might be one solution to collection issues in microfluidics. For cell counting alone without sorting, collection is no longer of interest because the counting has been already performed with whole cell population before reaching the outlet. Here, we aimed to focus more on the role of the dual inlet in preventing cell loss in microfluidics than the 3D focusing techniques mentioned previously. Our findings suggest that use of the dual inlet has potentially beneficial implications for cell analysis in microfluidics.
Other applications of the use of dual inlet in microfluidics
Can the concept of the dual inlet be applied only to cell counting? The answer to this question is NO. For example, microchannel-type electroporation has certain benefits such as uniform electric field strength and capability of direct visualization.21,22 However, there is a large fundamental problem related to the fact that the external voltage is applied only inside the microchannel because of its relatively higher channel resistance. In other words, the cells at the inlet and outlet are hardly affected by the electric field strength, resulting in no electroporation. If the dual inlet is used in microfluidic electroporation,23,24 most of the cells would enter the main channel and experience successful electroporation, and then move to the outlet. This concept would allow all the cells to be electroporated in contrast to a mixture of non-electroporated and electroporated cells at the outlet. Similarly, the dual inlet can be used for massive parallel cell loading over a microfluidic network in a valve-controlled manner, and could provide on-chip diagnostic applications at an affordable cost level. Some questions remain regarding whether the dual inlet is meaningful when using RBCs in microfluidics. In general, the RBCs are flexible in shape and abundant. Thus, the use of dual inlet may have little effect on the loading efficiency, but might be potentially beneficial for parallel buffer dilution during loading.
Limitations of the proposed method and system
The proposed technique has successfully demonstrated the affordable 3D focusing requiring only one reservoir to drive 3D hydrodynamic focusing with a single microstructure-layer. However, the geometry of microchannel should be altered according to the applicable sizes of cells or particles. When samples with higher size heterogeneity are injected into the proposed device, the CV value of fluorescent signals from the heterogeneous cell populations would be higher. The best design parameters for the heterogeneous sample under the present circumstances are to use parameters of the smallest size of samples in order to minimize the CV value. The cell loading efficiency of a microfluidic chip depends on many parameters, such as the flow velocity, channel geometry, and cell density. Above mentioned, the density of mammalian cells and microbeads, generally used in microfluidic experiments, is slightly higher than the density of the surrounding solution. While the sedimentation can occur in the conventional cell loading channel if the velocity of flow is too low, the proposed microfluidic channel can prevent the sedimentation at the same conditions. However, if samples contain cells with much higher density (e.g., cells conjugated with large magnetic beads), the sedimentation can be occurred at the very low flow velocity.
CONCLUSIONS
In this work, we propose a method for improving cell loading in microfluidics by simply reconstructing a dual inlet geometry. To provide the proof-of-concept validation for the dual inlet, we performed cell loading and counting experiments using human white blood cells and polystyrene microspheres in a microfabricated flow cytometer. Our experimental results showed that the dual inlet is simple in design and can be easily used to generate a vertical sheath flow, resulting in a low rate of cell loss. Furthermore, we discussed the potential role of the dual inlet in microfluidic channel applications by specifying key examples such as microchannel electroporation and diagnostics. Overall, we believe that this method has potential benefit for improving several microchannel applications ranging from single cell analysis to massive parallel handling in microfluidics.
ACKNOWLEDGMENTS
This work was supported by a grant from the Kyung Hee University in 2013 (KHU-20130431). This research was also supported by the National Research Foundation of Korea (NRF 2014023680) and Basic Science Research Program through the NRF, funded by the Ministry of Science, ICT and Future Planning (2014001331).
References
- 1.Yun H., Kim K., and Lee W. G., “ Cell manipulation in microfluidics,” Biofabrication 5, 022001 (2013). 10.1088/1758-5082/5/2/022001 [DOI] [PubMed] [Google Scholar]
- 2.Whitesides G. M., “ The origins and the future of microfluidics,” Nature 442, 368–373 (2006). 10.1038/nature05058 [DOI] [PubMed] [Google Scholar]
- 3.Yun H., Bang H., Min J., Chung C., Chang J. K., and Han D.-C., “ Simultaneous counting of two subsets of leukocytes using fluorescent silica nanoparticles in a sheathless microchip flow cytometer,” Lab Chip 10, 3243–3254 (2010). 10.1039/c0lc00041h [DOI] [PubMed] [Google Scholar]
- 4.Patra B., Chen Y.-H., Peng C.-C., Lin S.-C., Lee C.-H., and Tung Y.-C., “ A microfluidic device for uniform-sized cell spheroids formation, culture, harvesting and flow cytometry analysis,” Biomicrofluidics 7, 054114 (2013). 10.1063/1.4824480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Y. L., Hu J., Genin G. M., Lu T. J., and Xu F., “ BioPen: Direct writing of functional materials at the point of care,” Sci. Rep. 4, 4872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez A. W., Phillips S. T., and Whitesides G. M., “ Three-dimensional microfluidic devices fabricated in layered paper and tape,” Proc. Natl. Acad. Sci. U.S.A. 105, 19606–19611 (2008). 10.1073/pnas.0810903105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Y. L., Wang W., Hu J., Huang G., Wang S., Lee W. G.et al. , “ Benchtop fabrication of three-dimensional reconfigurable microfluidic devices from paper–polymer composite,” Lab Chip 13, 4745–4749 (2013). 10.1039/c3lc50919b [DOI] [PubMed] [Google Scholar]
- 8.Chapman G. V., “ Instrumentation for flow cytometry,” J. Immunol. Methods 243, 3–12 (2000). 10.1016/S0022-1759(00)00224-6 [DOI] [PubMed] [Google Scholar]
- 9.Brown M. and Wittwer C., “ Flow cytometry: principles and clinical applications in hematology,” Clin. Chem. 46, 1221–1229 (2000). [PubMed] [Google Scholar]
- 10.Arakawa T., Shirasaki Y., Aoki T., Funatsu T., and Shoji S., “ Three-dimensional sheath flow sorting microsystem using thermosensitive hydrogel,” Sens. Actuators, A 135, 99–105 (2007). 10.1016/j.sna.2006.06.074 [DOI] [Google Scholar]
- 11.Yang R., Feeback D. L., and Wang W., “ Microfabrication and test of a three-dimensional polymer hydro-focusing unit for flow cytometry applications,” Sens. Actuators, A 118, 259–267 (2005). 10.1016/j.sna.2004.09.001 [DOI] [Google Scholar]
- 12.Rosenauer M., Buchegger W., Finoulst I., Verhaert P., and Vellekoop M., “ Miniaturized flow cytometer with 3D hydrodynamic particle focusing and integrated optical elements applying silicon photodiodes,” Microfluid. Nanofluid. 10, 761–771 (2011). 10.1007/s10404-010-0707-z [DOI] [Google Scholar]
- 13.Mao X., Lin S.-C. S., Dong C., and Huang T. J., “ Single-layer planar on-chip flow cytometer using microfluidic drifting based three-dimensional (3D) hydrodynamic focusing,” Lab Chip 9, 1583–1589 (2009). 10.1039/b820138b [DOI] [PubMed] [Google Scholar]
- 14.Leu T.-S., Chen H.-Y., and Hsiao F.-B., “ Micro sorters with 3D focusing and continuous sorting,” in 2nd IEEE International Conference on Nano/Micro Engineered and Molecular Systems, 2007. NEMS'07 (2007), pp. 608–613. [Google Scholar]
- 15.Nikolic-Jaric M., Cabel T., Salimi E., Bhide A., Braasch K., Butler M.et al. , “ Differential electronic detector to monitor apoptosis using dielectrophoresis-induced translation of flowing cells (dielectrophoresis cytometry),” Biomicrofluidics 7, 024101 (2013). 10.1063/1.4793223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J., Chung S., Yun H., Cho K., Chung C., Han D.et al. , “ Asymmetric nozzle structure for particles converging into a highly confined region,” Curr. Appl. Phys. 6, 992–995 (2006). 10.1016/j.cap.2005.07.004 [DOI] [Google Scholar]
- 17.Sharp K. and Adrian R., “ On flow-blocking particle structures in microtubes,” Microfluid. Nanofluid. 1, 376–380 (2005). 10.1007/s10404-005-0043-x [DOI] [Google Scholar]
- 18.To K., Lai P., and Pak H., “ Jamming of granular flow in a two-dimensional hopper,” Phys. Rev. Lett. 86, 71–74 (2001). 10.1103/PhysRevLett.86.71 [DOI] [PubMed] [Google Scholar]
- 19.Bang H., Yun H., Lee W. G., Park J., Lee J., Chung S.et al. , “ Expansion channel for microchip flow cytometers,” Lab Chip 6, 1381–1383 (2006). 10.1039/b604578b [DOI] [PubMed] [Google Scholar]
- 20.Luo Z. Y., Wang S. Q., He L., Xu F., and Bai B. F., “ Inertia-dependent dynamics of three-dimensional vesicles and red blood cells in shear flow,” Soft Matter 9, 9651–9660 (2013). 10.1039/c3sm51823j [DOI] [PubMed] [Google Scholar]
- 21.Lee W. G., Bang H., Yun H., Min J., Chung C., Chang J. K.et al. , “ An impulsive, electropulsation-driven backflow in microchannels during electroporation,” Lab Chip 8, 224–226 (2008). 10.1039/b714371k [DOI] [PubMed] [Google Scholar]
- 22.Lee W. G., Demirci U., and Khademhosseini A., “ Microscale electroporation: Challenges and perspectives for clinical applications,” Integr. Biol. 1, 242–251 (2009). 10.1039/b819201d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun H. and Hur S. C., “ Sequential multi-molecule delivery using vortex-assisted electroporation,” Lab Chip 13, 2764–2772 (2013). 10.1039/c3lc50196e [DOI] [PubMed] [Google Scholar]
- 24.Kim K., Kim J. A., Lee S.-G., and Lee W. G., “ Seeing the electroporative uptake of cell-membrane impermeable fluorescent molecules and nanoparticles,” Nanoscale 4, 5051–5058 (2012). 10.1039/c2nr30578j [DOI] [PubMed] [Google Scholar]