Kitamura et al. identify NLRC4 as causing familial cold autoinflammatory syndrome using whole exome sequencing on a family with multiple affected family members. They identify a mutation in the NOD domain and show that the mutant protein increases Nlrc4 oligomerization and is associated with increased IL-1β. Transgenic mice with the same NLRC4 mutation are shown to develop a similar FCAS-like syndrome.
Abstract
Autoinflammatory syndromes cause sterile inflammation in the absence of any signs of autoimmune responses. Familial cold autoinflammatory syndrome (FCAS) is characterized by intermittent episodes of rash, arthralgia, and fever after exposure to cold stimuli. We have identified a missense mutation in the NLRC4 gene in patients with FCAS. NLRC4 has been known as a crucial sensor for several Gram-negative intracellular bacteria. The mutation in NLRC4 in FCAS patients promoted the formation of NLRC4-containing inflammasomes that cleave procaspase-1 and increase production of IL-1β. Transgenic mice that expressed mutant Nlrc4 under the invariant chain promoter developed dermatitis and arthritis. Inflammation within tissues depended on IL-1β–mediated production of IL-17A from neutrophils but not from T cells. Our findings reveal a previously unrecognized link between NLRC4 and a hereditary autoinflammatory disease and highlight the importance of NLRC4 not only in the innate immune response to bacterial infections but also in the genesis of inflammatory diseases.
Autoinflammatory syndrome is characterized by inflammatory responses in the absence of autoimmunity or infections, and it is generally caused by hyperactivation of innate immune cells (Chen and Nuñez, 2010; Park et al., 2012). Several studies, including those from our group, have identified the causative genes for familial autoinflammatory syndromes (McDermott et al., 1999; Jéru et al., 2008; Masters et al., 2009; Agarwal et al., 2010; Kitamura et al., 2011; Liu et al., 2012; Park et al., 2012). Among these genes, mutations in NLRP3 cause autoinflammatory syndromes, including familial cold autoinflammatory syndrome (FCAS), Muckle-Wells syndrome (MWS), and neonatal onset multisystem inflammatory disease (NOMID; Hoffman et al., 2001; Jéru et al., 2008; Masters et al., 2009; Aksentijevich and Kastner, 2011; Park et al., 2012). These diseases are named cryopyrin-associated periodic syndromes (CAPS). FCAS, the mildest of the CAPS, is characterized by rash, fever, and arthralgia by exposure to cold stimuli. Patients with MWS have more frequent inflammatory episodes and they frequently develop progressive sensorineural hearing loss and systemic amyloidosis. NOMID is the most severe of the three syndromes and is characterized by severe chronic inflammation involving the joints and nervous system. However, there are still significant numbers of CAPS without any mutations in NLRP3 (Aksentijevich et al., 2007).
Heterozygous mutations in NLRP3 result in overactivation of caspase 1. This enzyme cleaves the precursors of IL-1β and IL-18 (members of the IL-1 family of cytokines) into their active forms (Masters et al., 2009; Aksentijevich and Kastner, 2011). The recombinant IL-1 receptor antagonist anakinra, canakinumab, and the IL-1 receptor type I fusion protein rilonacept have induced clinical response in CAPS, demonstrating that signaling via the IL-1 receptor is crucial for the pathogenesis of CAPS (Aksentijevich and Kastner, 2011; Dinarello and van der Meer, 2013). Recent studies have provided evidence that heterozygous mutations in NLRP12 cause FCAS-like symptoms (Jéru et al., 2008). The mutations are reported to inhibit NF-κB or activate caspase 1, depending on the genetic variation (Jéru et al., 2008; Jéru et al., 2011).
In the current study, we used exome resequencing to analyze candidate genes of patients in one Japanese family with cold-induced urticaria and arthritis but without mutations in NLRP3 or NLRP12. We identified a heterozygous missense mutation in NLRC4. The mutant NLRC4 activates caspase-1 and this activation results in increased production of IL-1β. Expression of the mutant Nlrc4 in mice causes severe dermatitis, arthritis, and splenomegaly with augmented infiltration of neutrophils as well as cold-induced exanthema. The inflammation depended on IL-1β and IL-17A produced by neutrophils but not T cells. These data indicate that NLRC4 is a causative gene for this disease and highlight the crucial roles of NLRC4 not only in the innate immune response to bacterial infections but also in the pathogenesis of human inflammatory diseases.
RESULTS
Linkage and exome analyses of a Japanese family with a history of FCAS revealed a missense mutation in NLRC4
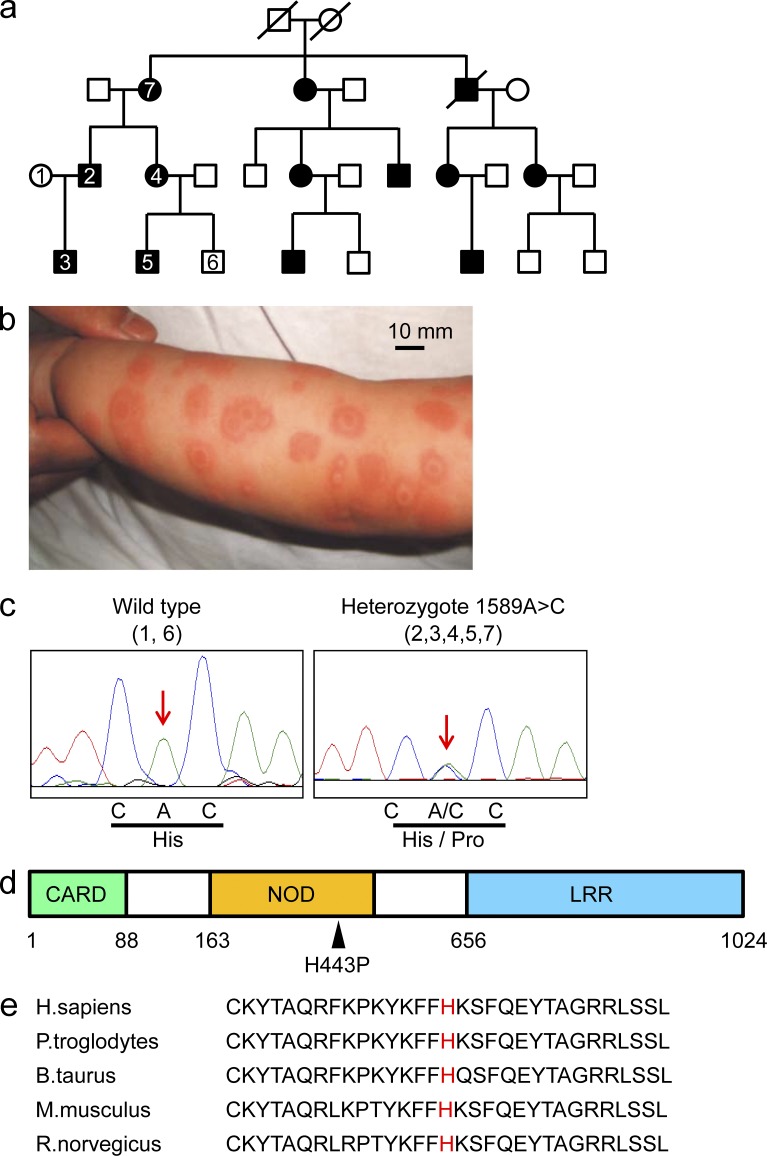
Members of a non-consanguineous Japanese family (Fig. 1 a) experienced episodes of high fever, urticarial-like rash (Fig. 1 b), and arthralgia that began at ∼2–3 mo of age. These symptoms have frequently been induced by exposure to cold stimuli, and the urticarial-like rash after cold exposure has not been accompanied by itching. Such patients have not developed splenomegaly or bone erosion. In most cases, the symptoms have been resolved without treatment in this family, although patients have taken nonsteroidal anti-inflammatory drugs to reduce joint pain. The laboratory findings of patient 3, from whom a blood sample was obtained during an episode of urticarial-like rash when he was 7 mo old, are shown in Table 1. We undertook SNP array and linkage analyses to identify the causal gene. Parametric linkage analysis of five affected and two unaffected family members was performed. A maximum LOD score of 1.143 was obtained throughout the whole genome, with a total critical region of 243 Mbp (Table 2), but an LOD score >2.3 was not detected.
Figure 1.
NLRC4 is a causative gene for FCAS. (a) The pedigree of a Japanese family with FCAS. The genomes of the patients or healthy members with a number inside of the square or circle were evaluated. (b) An image of the urticarial-like rash that patient number 3 developed at the age of 7 mo. Bar, 10 mm. (c) The genotypes of family members #1 and 6 (wild-type; left) or #2, 3, 4, 5, and 7 (heterozygote; right) are shown. The red arrows indicate position 1589. (d) The structure of NLRC4 consists of a caspase recruitment domain (CARD), a nucleotide-binding oligomerization domain (NOD), and a leucine-rich repeat (LRR). The black arrowhead indicates the mutation from histidine to proline at position 443 of NLRC4. (e) The amino acid sequence at position 443 (red) of NLRC4 in five species.
Table 1.
Laboratory findings of patient #3
| WBC | 11,800/µl |
| Hb | 13.3 g/dl |
| Ht | 32.1% |
| PLT | 52.9 × 104/µl |
| TP | 6.4 g/dl |
| Alb | 4.2 g/dl |
| GOT | 28 IU/liter |
| GPT | 21 IU/liter |
| LDH | 220 IU/liter |
| γ-GTP | 15 IU/liter |
| Ferritin | 61 ng/ml |
| CRP | 0.29 mg/dl |
| IgG | 354 mg/dl |
| IgA | 40 mg/dl |
| IgM | 66 mg/dl |
| PT-INR | 0.84 |
| APTT | 55.5 s |
| Fibrinogen | 449 mg/dl |
| D-dimer | 1.2 mg/dl |
| ANA | Negative |
| CH50 | 53.0 U/ml |
| IL-1β | <10 pg/ml |
| TNF | 4.6 pg/ml |
| IL-6 | 3.0 pg/ml |
| Soluble IL-2 receptor | 1,818 U/ml |
Table 2.
Linkage analysis
| Chromosome | HLOD | Distance | Position (start) | Position (end) |
| Mb | ||||
| 2 | 1.143 | 37,899,000 | 8,247,000 | 46,146,000 |
| 3 | 1.143 | 25,141,000 | 0 | 25,141,000 |
| 4 | 1.143 | 12,916,000 | 138,293,000 | 151,209,000 |
| 5 | 1.143 | 62,984,000 | 54,059,000 | 117,043,000 |
| 6 | 1.143 | 5,070,000 | 15,539,000 | 20,609,000 |
| 8 | 1.143 | 3,152,000 | 14,762,000 | 17,914,000 |
| 8 | 1.143 | 19,073,000 | 107,941,000 | 127,014,000 |
| 12 | 1.143 | 13,013,000 | 89,504,000 | 102,517,000 |
| 12 | 1.143 | 8,741,000 | 124,637,000 | 0 |
| 13 | 1.143 | 9,413,000 | 98,949,000 | 108,362,000 |
| 14 | 1.143 | 14,817,000 | 58,142,000 | 72,959,000 |
| 16 | 1.143 | 30,822,000 | 56,372,000 | 87,194,000 |
| Total 12 loci | 243,041,000 |
To identify the critical gene, genomic DNA of the family members (Fig. 1 a; five patients and two healthy family members) was evaluated by whole exome sequencing. A total of 3.7 Gbp of sequences was aligned to the exome target. DNA variants were filtered against the dbSNP build 134, the 1000 genomes project, the NHLBI Exome Sequencing project, and then classified by the predicted function to include all missense, nonsense, frameshift, or splice-site alleles. Exome analysis on linkage analysis–based 243 Mbp candidate regions identified three rare changes in NLRC4, ALK, and DTNB (Table 3). We did not detect any mutations or mosaicism in the exon or exon-intron boundary region of NLRP3. The variation in NLRC4, but not those in ALK and DTNB, was located in a region that is conserved among species and that encodes a functional domain (Table 3). Furthermore, after additional testing of 200 genomes obtained from healthy Japanese donors, we detected the same variations in ALK and DTNB (allele frequency 1/100 in both genes; unpublished data). Those data strongly suggested that NLRC4 was the causative gene for FCAS in this family.
Table 3.
Variants by exome analysis
| Gene | Variants | Functional domain | Amino acid | Homology in six species |
| ALK | SNV | + | R510W | 3/6 |
| NLRC4 | SNV | + | H443P | 5/5 |
| DTNB | Insertion | − | In-frame at c-terminal | - |
SNV: single nucleotide variation.
The mutation in NLRC4 was heterozygous in all of the patients we studied. It caused an A>C transversion at nucleotide 1589 in exon 4 (Fig. 1 c) and changed the predicted amino acid from a histidine to a proline at position 443 (Fig. 1, c and d). The missense mutation was detected in all affected patients but not in healthy members of the family. Amino acid 443 is located in the nucleotide binding domain (Fig. 1 d) and is conserved among five species (Fig. 1 e).
The mutation in NLRC4 increased its oligomerization and resulted in hyperactivation of caspase-1
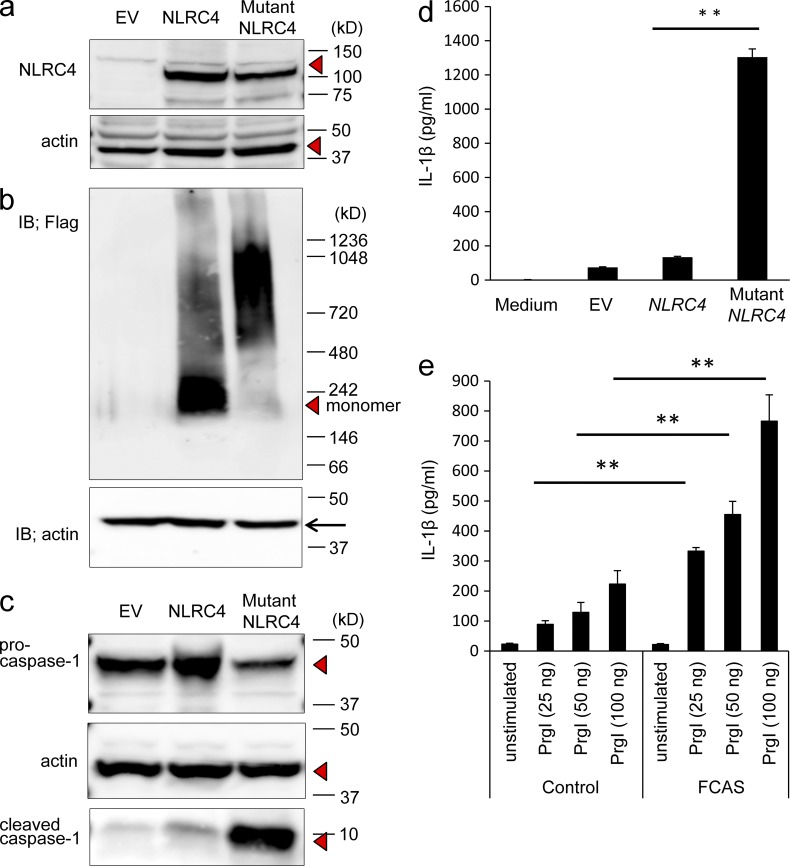
We first asked if the mutation in NLRC4 affected the expression of NLRC4 protein. The 3× Flag-tagged cDNA of wild-type or mutant NLRC4 was transfected into 293T cells and the lysates from those transfected cells were subjected to SDS-PAGE. The expression levels of wild-type and mutant NLRC4 proteins were similar; this finding suggested that the missense mutation did not affect the expression of NLRC4 (Fig. 2 a). As the activation of NLRC4 induces its oligomerization (Kofoed and Vance, 2011), we wished to determine if the missense mutation increased this process. The 3× Flag-tagged cDNA of wild-type or mutant NLRC4 was transfected into 293T cells and the lysates from those transfected cells were subjected to Blue Native-PAGE. A marked shift of wild-type NLRC4 to an oligomeric complex (∼1,000 kD) was detected in cells transfected with mutant NLRC4, whereas the monomeric form predominated in cells transfected with wild-type NLRC4 (Fig. 2 b). These data indicated that the missense mutation in NLRC4 facilitated the formation of oligomers.
Figure 2.
A mutation in NLRC4 induces overproduction of IL-1β through activation of caspase-1. (a) Flag-tagged wild-type, mutant NLRC4 expression vectors or empty vector (EV) were transfected into 293T cells. 2 d later, after the cell lysates were subjected to SDS-PAGE, Western blotting was performed with an anti-Flag or anti-actin antibody. Red triangle indicates NLRC4. The data in the figure are representative of three independent experiments. (b) Cell lysates from 293T cells transfected with empty vector (EV), flag-tagged wild-type, or mutant NLRC4 were subjected to Blue Native PAGE and the expression of the tagged proteins or actin was evaluated by Western blotting with an anti-Flag or actin antibody, respectively. The data in the figure are representative of three independent experiments. (c) 293T cells were transfected with a CASP1 expression vector together with empty vector (EV), an expression vector for wild-type or mutant NLRC4. 24 h later, the expression of procaspase-1, cleaved active caspase-1 (10 kD), and actin was evaluated by Western blotting. Red triangles indicate procaspase-1, cleaved active caspase-1, and actin. The data in the figure are representative of five independent experiments. (d) 293T cells were transfected with expression vectors for CASP1 and IL1B together with empty vector (EV), an expression vector for wild-type or mutant NLRC4. 24 h later, the concentration of IL-1β in the supernatant was evaluated by ELISA. The data are shown as means ± SD (**, P < 0.01; n = 5). The data in the figure are representative of three independent experiments. (e) Peripheral blood mononuclear cells were transfected with three concentrations of Prgl and cultured for 48 h. The concentrations of IL-1β in the supernatants were measured by ELISA. The data shown are means ± SD (**, P < 0.01). The data in the figure are representative of three independent experiments.
To determine if the missense mutation in NLRC4 resulted in increased cleavage of procaspase-1, a vector expressing either the wild-type or mutant NLRC4 and a CASP1 expression vector were transfected into 293T cells. Cells transfected with wild-type NLRC4 contained less of the 10 kD species (active form of caspase-1) than those transfected with mutant NLRC4 contained (Fig. 2 c). Because the cleavage of procaspase-1 is required for the production of IL-1β, we measured IL-1β production in 293T cells transfected with vectors expressing wild-type or mutant NLRC4 and CASP1 and IL1B. Cells overexpressing mutant NLRC4 secreted significantly more IL-1β than did cells overexpressing wild-type NLRC4 (Fig. 2 d). This finding indicated that the missense mutation in NLRC4 augmented cleavage of procaspase-1 and as a result, the secretion of IL-1β was increased.
NLRC4 is activated in response to bacterial type III secretory systems, including PrgI (Rayamajhi et al., 2013). We transfected PrgI in peripheral blood mononuclear cells from FCAS patient and a healthy donor and evaluated the production of IL-1β. The production of IL-1β was higher in FCAS patients compared with the healthy donor-derived cells at three different concentrations of PrgI (Fig. 2 e).
The mutation in Nlrc4 caused an autoinflammatory syndrome in mice
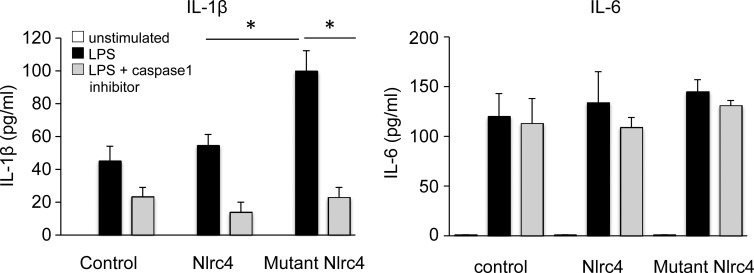
We next sought to assess if the equivalent of the NLRC4 1589A>C mutation caused FCAS-like inflammatory responses in mice. First, we tested if the equivalent mutation in mouse Nlrc4 also augmented IL-1β secretion. BM-derived macrophages (BMMCs) transfected with a control vector or with a vector encoding wild-type or mutant Nlrc4 were stimulated with LPS and the secretion of IL-1β was measured. BMMCs that expressed the mutant Nlrc4 produced a greater amount of IL-1β than did those transfected with the wild-type Nlrc4 vector or the control vector. In addition, the production of IL-1β was inhibited by the addition of a caspase-1 inhibitor (Fig. 3). In contrast, the amount of IL-6 secreted by LPS-stimulated BMMCs overexpressing mutant Nlrc4 was similar to that of cells overexpressing wild-type Nlrc4, and the secretion of IL-6 was not inhibited by treatment with a caspase-1 inhibitor (Fig. 3). These data indicated that the mutant NLRC4 affected the secretion of IL-1β and that this activity depended on the cleavage of caspase-1.
Figure 3.
BM-derived macrophages that overexpress mutant Nlrc4 secrete IL-1β. BM-derived macrophages (BMMC) infected with control retrovirus, retroviruses encoding wild-type, or mutant Nlrc4 were stimulated with 10 µg/ml LPS for 1 d in the absence (black) or presence (gray) of a caspase-1 inhibitor. As the control, unstimulated BMMCs were used (white). The concentrations of IL-1β and IL-6 in the supernatant were measured by ELISA. The data shown are means ± SD (*, P < 0.05; n = 5). The data shown in this figure are representative of three independent experiments.
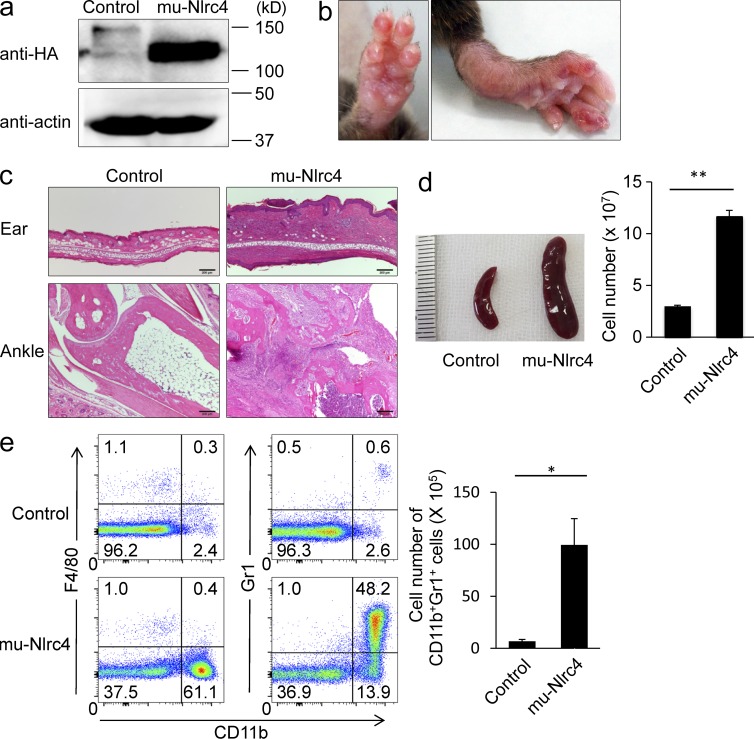
We then established transgenic mice (mu-Nlrc4 mice) in which expression of a 3× HA-tagged mutant Nlrc4 cDNA was driven by an invariant chain promoter (van Santen et al., 2000). The transgene was highly expressed in the spleen of mu-Nlrc4 mice (Fig. 4 a). The mu-Nlrc4 mice exhibited dermatitis and swollen joints starting at 3 wk of age (Fig. 4 b). Histological analysis of mu-Nlrc4 mice revealed severe cell infiltration in the skin and joints and erosion of the bone (Fig. 4 c). The mu-Nlrc4 mice exhibited splenomegaly and the total cell number in the spleen was approximately fivefold higher in mu-Nlrc4 mice than in control mice (Fig. 4 d). Phenotypical analysis of the spleen revealed a 15-fold increase in the number of CD11b+Gr1+F4/80− cells in mu-Nlrc4 mice as compared with control mice (Fig. 4 e). Similar phenotypes were detected in six independent transgenic lines. We evaluated the expression levels of total Nlrc4 protein in B cells of wild-type and three mu-Nlrc4 lines (line #1 was used in experiments of all figures; Table 4). The expression levels of mutant Nlrc4 was positively correlated with the number of total spleen cells, T cells (CD3+), and neutrophils (CD11b+Gr1+; Table 4).
Figure 4.
A mutation in Nlrc4 causes autoinflammation in mice. (a) The expression of HA or actin in the spleen of C57BL/6 or mu-Nlrc4 mice was evaluated by Western blotting. The data in the figure are representative of two independent experiments. (b) The foot of a mu-Nlrc4 mouse at the age of 5 mo is shown. (c) Sections of ankle joints and ear skin from C57BL/6 mice (left) or mu-Nlrc4 mice (right) at the age of 12 wk were stained with hematoxylin and eosin. Bars, 200 µm. (d) Spleen size and the total number of spleen cells was counted. The data are shown as means ± SD (**, P < 0.01; n = 5). The data in the figure are representative of three independent experiments. (e) The spleen cells from C57BL/6 or mu-Nlrc4 mice at the age of 12 wk were stained with anti-Gr1, anti-F4/80, and anti-CD11b and analyzed with a flow cytometer. The total number of Gr1+CD11b+ cells is shown. The data are shown as means ± SD (*, P < 0.05; n = 5). The data in the figure are representative of three independent experiments.
Table 4.
Characteristics of three lines of mutant Nlrc4 transgenic mice
| Mouse line | Expression of Nlrc4 (fold increases compared with wild type) | Spleen cells (×107) | T cells (×107) | CD11b+Gr1+ cells (×107) |
| Wild type | 1 | 5.2 ± 0.3 | 0.8 ± 0.1 | 0.05 ± 0.01 |
| Line #1 | 3.1 ± 0.2 | 21.1 ± 1.4 | 3.4 ± 0.4 | 0.59 ± 0.2 |
| Line #2 | 5.2 ± 0.3 | 32.1 ± 3.3 | 5.4 ± 2.2 | 1.2 ± 0.2 |
| Line #3 | 4.2 ± 0.3 | 27.8 ± 4.5 | 4.5 ± 0.4 | 0.89 ± 0.1 |
The data were obtained at the age of 16 wk. The data in line #1 are used in all figures.
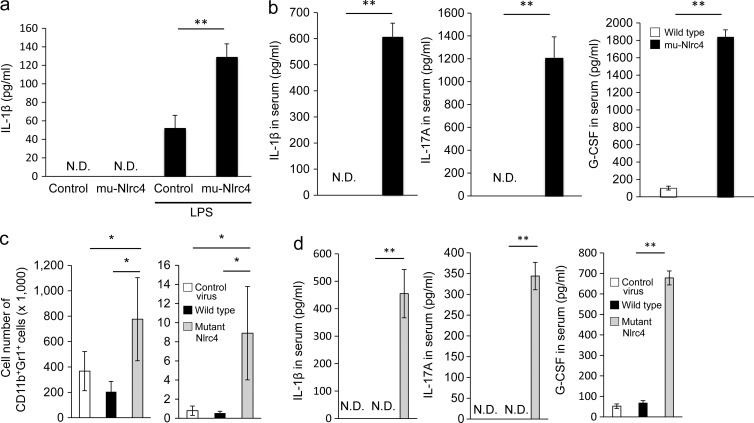
We next compared the production of cytokines in the spleens of mu-Nlrc4 and control mice. Splenocytes of mu-Nlrc4 mice produced IL-1β when stimulated with LPS, whereas the production of IL-1β by splenocytes of control mice was limited (Fig. 5 a). The serum levels of IL-1β, IL-17A, and G-CSF were much higher in mu-Nlrc4 mice than in control mice (Fig. 5 b). To determine whether the overexpression of wild-type Nlrc4 might also up-regulate proinflammatory cytokines, we transplanted BM cells transduced with the wild-type or the mutant Nlrc4 gene into irradiated recipient mice. The number of CD11b+Gr1+ cells in the spleen and peritoneal cavity was higher in mutant Nlrc4 chimeras than in control chimeric mice (Fig. 5 c). The serum levels of IL-1β, IL-17A, and G-CSF were higher in C57BL/6 mice reconstituted with mutant Nlrc4-transduced BM cells than those with wild-type Nlrc4 or control virus transduced BM cells (Fig. 5 d).
Figure 5.
Cytokine production in mu-Nlrc4 mice or mice reconstituted with mutant Nlrc4 transduced BM cells. (a) Splenocytes from wild-type or mu-Nlrc4 mice were stimulated with LPS for 24 h, and the concentration of IL-1β in each supernatant was evaluated by ELISA. The data shown are means ± SD (**, P < 0.01). N.D.: not detected. The data in the figure are representative of three independent experiments. (b) The concentrations of IL-1β, IL-17A, and G-CSF in the serum of wild-type (white) and mu-Nlrc4 (black) mice at the age of 8 wk were evaluated by ELISA. For all panels, the data shown are means ± SD (**, P < 0.01; n = 5). N.D.: not detected. The data in the figure are representative of three independent experiments. (c) Intraperitoneal cells or splenocytes from C57BL/6 mice transplanted with BM cells infected with control virus (white), wild-type (black), or mutant Nlrc4 (gray)-encoding virus were evaluated for the number of Gr1+CD11b+ cells 2 mo after the transplantation. The data shown are means ± SD (*, P < 0.05; n = 5 in each group). The data in the figure are representative of three independent experiments. (d) Irradiated C57BL/6 mice were reconstituted with BM cells infected with control virus (white), wild-type (black), or mutant Nlrc4 (gray)-encoding virus. Serum IL-1β, IL-17A, and G-CSF were evaluated by ELISA 6 wk after BM transplantation. For all panels, the data shown are means ± SD (**, P < 0.01; n = 5). N.D.: not detected. The data in the figure are representative of two independent experiments.
Cold exposure of mu-Nlrc4 mice increased autoinflammation
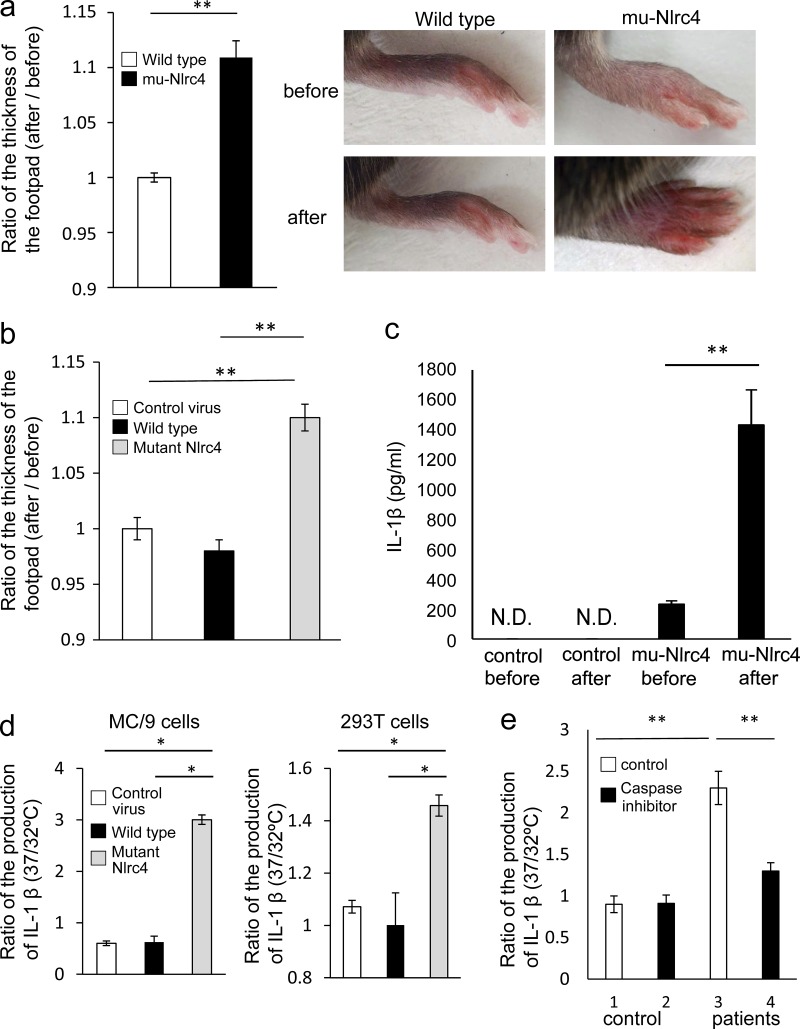
To determine if mu-Nlrc4 mice exposed to cold stimuli would exhibit a heightened inflammatory response, the right feet of mu-Nlrc4 mice were cooled for 5 min and then the thickness of the footpads was evaluated. Cooling the footpads of control mice did not change their color or thickness, but the footpads of mu-Nlrc4 mice increased in thickness and displayed an exanthema-like color change after being cooled (Fig. 6 a). The foot pad swelled more in C57BL/6 mice reconstituted with mutant Nlrc4-transduced BM cells than in those with wild-type Nlrc4- or control virus-transduced BM cells (Fig. 6 b). In addition, cooling of the whole body of the mice at 4°C for 1 min increased IL-1β in mu-Nlrc4 but not control mice (Fig. 6 c). As for cell level assay, LPS-stimulated MC/9 cells overexpressing mutant Nlrc4 or 293T cells overexpressing mutant NLRC4 produced more IL-1β when cultured at 32°C than at 37°C, whereas MC/9 cells overexpressing wild-type Nlrc4 or 293T cells overexpressing wild-type NLRC4 did not increase IL-1β in response to 32°C stimulation (Fig. 6 d). Peripheral blood mononuclear cells from an FCAS patient produced IL-1β in response to 32°C stimulation, whereas control cells did not increase IL-1β production in response to 32°C stimulation (Fig. 6 e). The induction of IL-1β by cold stimuli was inhibited by the addition of caspase inhibitor (Fig. 6 e). These data demonstrated that cells harboring mutant Nlrc4 activated Nlrc4-dependant inflammasomes upon exposure to cold stimuli, and this activation resulted in tissue inflammation.
Figure 6.
Cold exposure of mu-Nlrc4 mice increases autoinflammation. (a) The footpads of C57BL/6 (wild type; white) and mu-Nlrc4 (black) mice were exposed to 4°C for 5 min. The thickness of each footpad was measured before and after exposure to the cold stimulus. The data are shown as the ratio of the thickness after exposure to the thickness before exposure. The data are shown as means ± SD (**, P < 0.01; n = 5). The data in the figure are representative of three independent experiments. (b) Irradiated C57BL/6 mice were reconstituted with BM cells infected with control virus (white), wild-type (black), or mutant Nlrc4 (gray)-encoding virus. The footpads were exposed to 4°C for 5 min. The thickness of each footpad was measured before and after exposure to the cold stimulus. The data are shown as the ratio of the thickness after exposure to the thickness before exposure. The data are shown as means ± SD (**, P < 0.01; n = 5). The data in the figure are representative of three independent experiments. (c) The entire bodies of control C57BL/6 and mu-Nlrc4 mice were exposed to 4°C for 1 min and kept for 3 min at room temperature, and then the serum concentrations of IL-1β were measured. The data are shown as means ± SD. N.D., not detected. (**, P < 0.01; n = 5). The data in the figure are representative of three independent experiments. (d) MC/9 cells infected with control retrovirus (white), retroviruses encoding wild-type (black), or mutant Nlrc4 (gray) were cultured at 32 or 37°C for 48 h. 293T cells were transfected with control vector (white), vectors encoding wild-type (black), or mutant NLRC4 (gray) and 48 h later, cells were washed and cultured at 32 or 37°C for 6 h. The levels of IL-1β in the supernatants were evaluated by ELISA. The data are shown as the mean ratio of the IL-1β level in cells cultured at 32°C to the level in cells cultured at 37°C. The data are shown as means ± SD (*, P < 0.05). The data in the figure are representative of three independent experiments. (e) Peripheral mononuclear cells from an FCAS patient or a healthy control were cultured at 32°C for 48 h in the presence (black) or absence (white) of a caspase inhibitor or cultured at 37°C for 48 h. The level of IL-1β in the supernatant was evaluated by ELISA. The data are shown as the mean ratio of the IL-1β level in cells cultured at 32°C to the level in cells cultured at 37°C. The data are shown as means ± SD (**, P < 0.01). The data in the figure are representative of two independent experiments.
The increase of neutrophils in mu-Nlrc4 mice depended on IL-1β or IL-17A
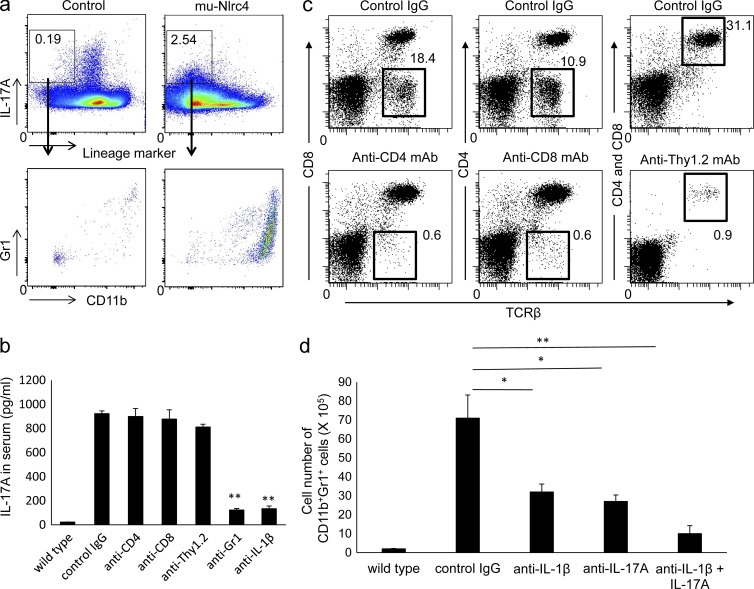
Th17 cells that produce IL-17A contribute to various inflammatory disorders (Korn et al., 2009). We assessed which circulating cells were responsible for the production of IL-17A. The major IL-17A producers in mu-Nlrc4 mice were in the TCR-β−TCR-γδ−B220−CD11c−NK1.1− population, whereas those in control mice were in a lineage-positive fraction (Fig. 7 a). Most of the lineage marker–negative cells that produced IL-17A in mu-Nlrc4 mice were Gr1+CD11b+ or Gr1intCD11b+ cells that included neutrophils, monocytes, and macrophages (Fig. 7 a).
Figure 7.
The accumulation of neutrophils in mu-Nlrc4 mice depends on IL-1β and IL-17A. (a) Spleen cells from C57BL/6 or mu-Nlrc4 mice at the age of 12 wk were stimulated with 25 ng/ml PMA and 1 µg/ml ionomycin in the presence of 2 µM monensin for 5 h and then stained with anti–TCR-β, anti–TCR-γδ, anti-B220, anti-CD11c, anti-NK1.1, anti-Gr1, and anti-CD11b antibodies. The cells were fixed and stained with anti–IL-17A antibody. The expression of IL-17A was analyzed by flow cytometry. The number in each rectangle indicates the percentage of IL-17A–positive cells in the lineage-negative fraction. The data in the figure are representative of five independent experiments. (b) mu-Nlrc4 mice at the age of 12 wk were injected with control IgG, anti-CD4, anti-CD8, anti-Thy1.2, anti-Gr1, or anti–IL-1β antibody five times at 3-d intervals. The serum IL-17A levels 2 d after final antibody treatment was measured by ELISA. Control sera from C57BL/6 (wild type) mice was used. The data shown are means ± SD (**, P < 0.01; n = 5 for all). The data in the figure are representative of three independent experiments. (c) mu-Nlrc4 mice at the age of 12 wk were injected twice with control rat IgG, anti-CD4, anti-CD8, or anti-Thy1.2 at 3 d intervals. The spleen cells from mice 1 d after the final injection were stained with anti-CD4 and anti–TCR-β (for anti-CD8–treated mice), anti-CD8 and anti–TCR-β (for anti-CD4–treated mice), or anti-CD4, anti-CD8, and anti–TCR-β antibodies (for anti-Thy1.2–treated mice). The data in the figure are representative of two independent experiments. (d) mu-Nlrc4 mice at the age of 12 wk were injected with control IgG, anti–IL-1β, anti–IL-17A, or anti–IL-1β, and anti–IL-17A antibody five times at 3-d intervals. The number of Gr1+CD11b+ cells in the spleen 2 d after the final antibody treatment was evaluated by flow cytometry. The data shown are means ± SD (*, P < 0.05; **, P < 0.01; n = 5 for all). The data in the figure are representative of three independent experiments.
To determine which cells were responsible for IL-17A production in mu-Nlrc4 mice, we depleted CD4+, CD8+, Thy1.2+, or Gr1+ cells. The depletion of CD4+, CD8+, or Thy1.2+ cells did not affect the IL-17A level (Fig. 7 b), although the depletion of CD8−TCR-β+, CD4−TCR-β+, or CD4+CD8+TCR-β+ cells was efficient (Fig. 7 c). However depletion of Gr1+ cells and suppression of anti–IL-1β reduced the IL-17A level (Fig. 7 b), suggesting that IL-1β was required for IL-17A production and neutrophils were major producers of IL-17A in mu-Nlrc4 mice. We then treated mu-Nlrc4 mice with anti–IL-1β, anti–IL-17A, or anti–IL-1β plus anti–IL-17A antibody to evaluate which cytokine was involved in neutrophil accumulation. The number of CD11b+Gr1+ cells in mu-Nlrc4 mice was decreased by treating the mice with anti–IL-1β or anti–IL-17A (Fig. 7 d). The combination of anti–IL-1β and anti–IL-17A further decreased the number of CD11b+Gr1+ cells (Fig. 7 d).
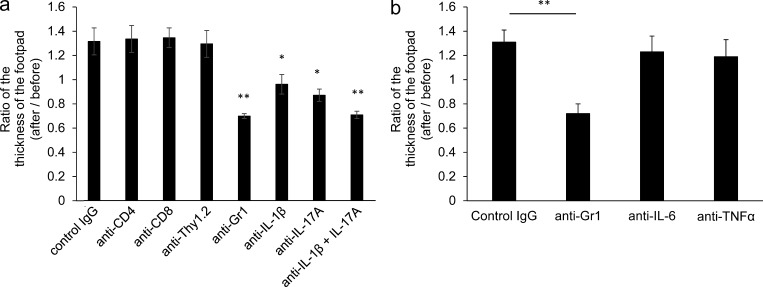
To evaluate which cells or cytokines contributed to inflammation in mu-Nlrc4 mice, the animals were treated with intraperitoneal injections of anti-CD4+, anti-CD8+, anti-Thy1.2+, or anti-Gr1+ antibodies. Alternatively, intraperitoneal injections contained anti–IL-1β, anti–IL-17A, or anti–IL-1β plus anti–IL-17A antibodies. Footpad swelling after antibody treatment was subsequently measured (Fig. 8 a). The depletion of CD4+, CD8+, or Thy1.2+ cells did not affect swelling. In contrast, depletion of Gr1+ cells suppressed the footpad swelling (Fig. 8 a). Footpad swelling in mu-Nlrc4 mice was inhibited by treating the mice with anti–IL-1β or anti–IL-17A, and the combination of anti–IL-1β plus anti–IL-17A further decreased the number of CD11b+Gr1+ cells (Fig. 8 a). The treatment of mu-Nlrc4 mice with anti–IL-6 or TNF did not inhibit footpad swelling (Fig. 8 b). These data demonstrated that the accumulation of neutrophil was mediated by IL-1β, IL-17A, and neutrophils.
Figure 8.
The inflammation in mu-Nlrc4 mice depends on IL-1β, IL-17A, and neutrophils. (a) The mu-Nlrc4 mice at the age of 8 wk were injected with control IgG, anti-CD4, anti-CD8, anti-Thy1.2, anti-Gr1, anti–IL-1β, anti–IL-17A, or anti–IL-1β, and anti–IL-17A antibody five times at 3-d intervals. The ratio of left footpad thickness before and after antibody treatment was calculated. The data shown are means ± SD (*, P < 0.05; **, P < 0.01; n = 5 for all). The data in the figure are representative of three independent experiments. (b) The mu-Nlrc4 mice at the age of 8 wk were injected with control IgG, anti-Gr1, anti–IL-6, or anti-TNF antibody five times at 3-d intervals. The ratio of left footpad thickness before and after antibodies treatment was calculated. The data shown are means ± SD (**, P < 0.01; n = 5 for all). The data in the figure are representative of three independent experiments.
DISCUSSION
Previous studies have identified mutations in NLRP3 and NLRP12 in patients with FCAS (Hoffman et al., 2001; Jéru et al., 2008). However, not all patients with FCAS carry mutations in these two genes (Aksentijevich et al., 2007). In the current study, we used linkage and exome resequencing to analyze genetic variations in patients with FCAS in one Japanese family and discovered a missense mutation in NLRC4, a gene which has been known as a crucial sensor for several Gram-negative intracellular bacteria (Davis et al., 2011; Kofoed and Vance, 2011; Lamkanfi, 2011; Zhao et al., 2011). The mutant NLRC4 activated caspase-1 and this activation resulted in increased secretion of IL-1β. Transgenic mice that express the mutant Nlrc4 develop dermatitis and arthritis accompanied by bone erosion. The inflammatory responses induced by the mutant Nlrc4 protein depend on IL-1β and IL-17A acting in concert with neutrophil infiltration, and the production of IL-17A depends on neutrophils but not on T cells. These findings highlight the roles of NLRC4 not only in innate responses to bacterial infections but also in autoinflammatory responses.
The NLRC4 inflammasome is crucial for the responses to several Gram-negative intracellular bacteria including Pseudomonas aeruginosa, Shigella flexneri, and Salmonella enterica (Miao et al., 2010; Kofoed and Vance, 2011; Zhao et al., 2011). NLRC4 can be activated by the flagellin protein expressed by these bacteria and by the basal body rod component of the Salmonella type III secretion system (Franchi et al., 2012). The recognition of flagellin or basal body rod component requires proteins of the NAIP family (Kofoed and Vance, 2011; Zhao et al., 2011; Rayamajhi et al., 2013; Yang et al., 2013). Our present data show that a mutation in NLRC4 increased the formation of NLRC4 inflammasomes in the absence of flagellin or basal body rod component-mediated stimulation. As the mutation is located in the NOD domain, it might increase oligomerization of NLRC4 by increasing the binding affinity for other NLRC4 monomers. Indeed, we detected increased oligomerization of the mutant NLRC4 without any exogenous stimuli in addition to increased production of IL-1β from peripheral blood mononuclear cells from FCAS patients than those from control in response to bacterial product. An interesting question is whether intrinsic ligands are required for the oligomerization of mutant NLRC4 or whether there are intrinsic ligands that are able to activate wild-type NLRC4. These questions could be at least partly addressed by comparing the abilities of wild-type and mutant NLRC4 proteins to self-assemble in a cell-free system. In addition, a recent study has revealed the structure of NLRC4 (Hu et al., 2013). Thus, a comparison of the structures of the wild-type and mutant NLRC4 proteins might help reveal the activation mechanisms for mutant NLRC4 as well as wild-type NLRC4.
The mu-Nlrc4 mice developed severe arthritis with bone erosion, dermatitis, and splenomegaly together with an increased number of CD11b+Gr1+ neutrophil and activated T cells. Although mu-Nlrc4 mice developed an urticarial-like dermatitis upon exposure to a cold stimulus, it is important to note that these mice spontaneously developed an inflammatory syndrome even without exposure to cold stimuli. Previous studies have reported that mice harboring a mutation in NLRP3 also spontaneously develop an inflammatory disorder (Meng et al., 2009). Therefore, these data suggest that a cold environment is one of the factors that trigger the appearance of the syndrome but also that it is not required to cause the disease. As for the mechanisms of autoinflammation in mu-Nlrc4 mice, neutrophil infiltration and footpad inflammation were suppressed by blocking the function of IL-1β and IL-17A or depleting neutrophils but not CD4+, CD8+, or Thy1.2+ cells. Furthermore, IL-17A production was inhibited by suppressing IL-1β, or depleting neutrophils but not CD4+ and CD8+ or Thy1.2+ cells. Collectively, those data suggest that increased production of IL-1β by the mutation in NLRC4 would up-regulate the production of IL-17A from neutrophils, which would further recruit neutrophils leading to progressive inflammation. The mice harboring a mutation in NLRP3 spontaneously develop an inflammatory disorder (Meng et al., 2009) that depends on Th17. Therefore, mutations in NLRP3 and NLRC4 likely lead to production of IL-17A through different molecular networks. Previous studies have not discovered mutations in NLRC4 in American patients with CAPS who do not have mutations in NLRP3 (Aksentijevich et al., 2007). Our discovery of a new mutation in NLRC4 in one Japanese family will facilitate the screening of NLRC4 in patients with CAPS. Furthermore, the finding that expression of the mutant Nlrc4 in mice gives rise to spontaneous and severe autoinflammatory symptoms, including splenomegaly and infiltration of neutrophils into the joints, suggests that NLRC4 may be involved in non-FCAS types of autoinflammatory syndromes that are currently not associated with known genetic mutations. In addition, two recent papers reported that the heterozygous mutation in NLRC4 causes macrophage activation syndrome or neonatal-onset enterocolitis, periodic fever, and fatal or near-fatal episodes of autoinflammation (Canna et al., 2014; Romberg et al., 2014). Furthermore, it has been reported that polymorphisms of NLRC4 are associated with atopic dermatitis (Macaluso et al., 2007). Therefore, it will be important to characterize the NLRC4 genotypes/phenotypes association in patients with an autoinflammatory syndrome as well as in patients with inflammatory disorders. In conclusion, our findings reveal a previously unrecognized link between NLRC4 and FCAS, and they highlight the importance of NLRC4 not only in the innate immune response to bacterial infections but also in the genesis of inflammatory diseases.
MATERIALS AND METHODS
Patients.
Genomic DNA was extracted from peripheral blood after the Ethical Committee of the University of Tokushima approved the human genome research protocol and informed consent was obtained from each patient. The study was conducted in accordance with the principles of the Declaration of Helsinki. The cell number or serum levels of enzymes or cytokines in peripheral blood was counted by automated cell counter and by standard ELISA (SRL).
SNP microarray and linkage analysis.
SNP genotyping was conducted with an Illumina Human 660W Quad system (Illumina). DNA was prepared in accordance with the manufacturer’s instructions and hybridized onto the array. Parametric linkage analysis, which used a fully penetrant dominant model, was conducted with the Merlin software package (Abecasis et al., 2002).
Exome resequencing and DNA sequencing.
A paired-end library was prepared from genomic DNA and hybridized to biotinylated cRNA oligonucleotide baits from the SureSelect Human All Exon kit (Agilent Technologies). The library was sequenced with paired-end, 75 bp reads on one lane of an Illumina Hiseq2000 sequencing systems. The sequence reads were aligned to the human reference sequence of the UCSC Genome Browser (hg19). The entire targeted exome covered by >10 reads was 90%. DNA variants located in the candidate region were filtered against the databases of dbSNP build 134, the 1000 Genomes Project, and the NHLBI Exome Sequencing Project, and missense, nonsense, frameshift, and splice-site alleles were evaluated. The mutation in NLRC4 was sequenced by using a Prism 3100 genetic analyzer (Applied Biosystems).
Flow cytometry and antibodies.
Cells were resuspended in staining buffer at a density of 2 × 106 cells/ml. The cells were incubated with mAb anti-FcγRII/III and then with antibodies against CD4 (GK1.5), CD8 (53–6.7), CD11b (M1/70), CD11c (N418), CD44 (IM7), CD62L (MEL-14), TCR-β (H57-597), B220 (RA3-6B2), F4/80 (BM8), NK1.1 (PK136), TCR-γδ (GL3), or Gr1 (RB6-8C5; BioLegend). After gating out cells that were positive for 7-AAD, the fluorescence intensity of 105 cells was measured with a FACS Canto II flow cytometer (BD) and analyzed with the FACS Diva (BD) or FlowJo (Tree Star) software programs. For intracellular staining, cells were fixed with an intracellular staining kit (eBioscience), permeabilized, and stained with PE-conjugated anti–IL-17A antibody (BioLegend).
Western blotting and Blue Native PAGE.
Cell pellets were lysed in cold RIPA buffer (Wako Pure Chemical Industries) and protease inhibitor (Roche). The lysates were boiled in SDS loading dye. The samples were resolved by SDS-PAGE and the blots were incubated with anti-Flag (F1804; Sigma-Aldrich), anti-HA (3724; Cell Signaling Technology), or anti–caspase-1 (Santa Cruz Biotechnology, Inc.) antibodies, followed by incubation with peroxidase-conjugated goat anti–mouse IgG (Thermo Fisher Scientific) or goat anti–rabbit IgG (Bio-Rad Laboratories) antibodies. As a control, membranes were probed with polyclonal anti-actin (Sigma-Aldrich) and HRP-conjugated goat anti–rabbit IgG antibodies (Bio-Rad Laboratories). In some experiments, membranes were blotted with anti-Flag and anti-actin, followed by HRP-conjugated anti–mouse and rabbit IgG. The bands were detected with ECL Prime Chemiluminescent Substrate (GE Healthcare) and with the ImageQuant LAS-4000 mini system (GE Healthcare). Blue Native PAGE was performed by using a Novex Native PAGE Bis-Tris Gel System (Invitrogen) in accordance with the manufacturer’s protocols. In brief, the cells were resuspended in 1× Native PAGE sample buffer (Invitrogen) containing 1% digitonin and protease inhibitors (Roche) and then incubated on ice for 10 min. After centrifugation, 25 µg of the supernatant was resuspended with 1.0 µl of 5% G250 sample additive and 10 µl of 1× Native PAGE Sample Buffer (Invitrogen). The samples were loaded onto 3–12% Bis-Tris Native PAGE gels and then electrophoresed in a 1× Native PAGE Running buffer system (Invitrogen). NativeMark protein standard (Invitrogen) was used as a molecular weight marker. ECL prime Chemiluminescent Substrate (GE Healthcare) was added to the membranes, after which they were analyzed with the Lumino Image Analyzer (GE Healthcare).
Animal husbandry and generation of transgenic mice.
Female C57BL/6 mice were purchased from Japan SLC (Hamamatsu). The mutant mouse Nlrc4 cDNA tagged with 3xHA was cloned into pDOI6 (van Santen et al., 2000; provided by D. Mathis, C. Benoist [Harvard Medical School, Cambridge, MA], and S. Ishido [Showa Pharmaceutical University, Tokyo, Japan]). The linearized transgene construct, generated from the plasmid by BamHI digestion, was microinjected into fertilized eggs from C57BL/6N mice at the RIKEN CDB facility in Kobe, Japan (LARGE, 2014, accession no. CDB0496T). The genotypes of the transgenic mice were determined by PCR (forward primer: 5′-GCATGTCTATCTTTTACCCATACGATGTTCCTG-3′, reverse primer: 5′-TTAAGCAGTCACTAGTTTAAAGGTGCCTTTAATAGCG-3′). All mice were maintained under specific pathogen-free conditions in the animal research center of the University of Tokushima and all experiments were performed in accordance with our institution’s guidelines for animal care and use.
Exposure of mice or cells to cold stimuli.
Either the foot or the whole body of mice was exposed to 4°C for 5 min or 4°C for 1 min, followed by room temperature (24–25°C) for 3 min, respectively. The size of the footpads was measured before and after exposure. As for cold stimuli for cells, the cultured cells were kept at 32°C for 48 h and cytokines in the supernatant was measured.
Cell culture.
293T cells were maintained in DMEM supplemented with 10% FBS. MC/9 cells were provided by N. Kambe (Chiba University, Chiba, Japan) and maintained in DMEM supplemented with 10% FBS and 10 ng/ml. The 293T cells were transfected with human NLRC4 or mutant NLCR4 using GeneJuice (Millipore) according to the manufacturer’s protocol. GeneJuice was used to generate retroviruses by transfecting each vector encoding GFP into Plat-E cells (provided by T. Kitamura, University of Tokyo, Tokyo, Japan; Morita et al., 2000). Retroviruses carrying wild-type or mutant Nlrc4 and control viruses were used to infect MC/9 cells by incubating MC/9 cells with virus-containing medium for 24 h. After expansion of cells, GFP-positive MC/9 cells were enriched with a cell sorter (FACSAriaIII; BD). Peripheral blood mononuclear cells were transfected with different concentration of Prgl by Nucleofector (Lonza) according to the manufacturer’s protocol.
Antibody treatment of mice.
The mu-Nlrc4 mice were intraperitoneally injected with rat IgG, anti-CD4 (GK1.5; 200 µg/dose), anti-CD8 (53–6.72; 150 µg/dose), anti-Thy1.2 (30-H12; 150 µg/dose), anti-Gr1 (RB6-8C5; 150 µg/dose), anti–IL-1β (B122; 150 µg/dose), or anti–IL-17A (17F3; 150 µg/dose), anti–IL-6 (MP5-20F3; 200 µg/dose), or anti-TNF (XT3.11; 200 µg/dose) antibody five times at 3-d intervals.
Vectors and cDNA.
The cDNAs of human NLRC4 (provided by F. Takeshita, Daiichi-Sankyo, Tokyo, Japan) and mouse Nlrc4 (provided by R. Vance, University of California, Berkeley, Berkeley, CA) were cloned into the pKE004 or pCIneo vectors, respectively. The A>C mutation at position 1589 in exon 4 of NLRC4 was inserted by PCR-based mutagenesis and the mutant NLRC4 cDNA was cloned into the pCIneo vector. The human CASP1 and IL1B cDNAs (Komune et al., 2011) were provided by Y. Yanagi and T. Ichinohe (Kyushu University, Fukuoka, Japan). The Salmonella PrgI was cloned into the pcDNA3.1.
Isolation, culture, and transfection of BM-derived macrophages.
Mouse BM cells were recovered by flushing the dissected femurs and tibias with RPMI 1640 medium. After lysis of the RBCs with NH4Cl, the cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum, 2 mM l-glutamine, 40 U/ml penicillin, 40 µg/ml streptomycin, 55 µM 2-mercaptoethanol, and 20% culture supernatant from 10 ng/ml M-CSF (R&D Systems) for 7 d. GeneJuice (EMD Millipore) was used to generate retroviruses by transfecting each vector encoding GFP into Plat-E cells (provided by T. Kitamura; Morita et al., 2000). Retroviruses carrying wild-type or mutant Nlrc4 and control viruses were used to infect BM cells 2 times on days 0 and 1. The cells were stimulated with 10 µg/ml LPS for 20 h, and in some experiments, they were treated with a caspase-1 inhibitor (R&D Systems).
ELISA.
The concentration of IL-1β and IL-6 was measured with the mouse IL-1β or IL-6 ELISA Ready-Set-Go kit, respectively (eBioscience). The concentration of IL-17 was measured with a biotin-conjugated anti–mouse IL-17A antibody and a purified anti–mouse IL-17A antibody (BioLegend). The concentration of G-CSF was measured with the Mouse G-CSF Quantikine ELISA kit (R&D Systems).
BM transplantation.
Recipient female C57BL/6 mice were irradiated with 9.5 Gy when they were 8 wk old. Lineage-negative BM cells from C57BL/6 mice were purified with a mouse Lineage Cell Depletion kit (Miltenyi Biotec). After purification, the cells were infected 3× with control viruses or with viruses carrying either the wild-type or mutant Nlrc4 cDNA, as described previously (Morita et al., 2000). Approximately 5–7 × 105 BM cells were injected into irradiated mice 1 d after they were irradiated.
Histological analysis.
The tissues were fixed in 10% formalin, embedded in paraffin, and sectioned. The tissue sections were stained with hematoxylin and eosin.
Statistical analysis.
The statistical significance of between-group differences was evaluated by an unpaired, two-tailed, Student’s t test. A p-value of <0.05 was considered significant.
Acknowledgments
We thank Drs. C. Benoist, S. Ishido, N. Kambe, R. Vance, Y. Yanagi, T. Ichinohe, F. Takeshita, and T. Kitamura for providing reagents. We would also like to thank Ms. K. Takahashi, C. Kinouchi, Y. Shinomiya, and C. Tomari for their technical and editorial assistance.
This study was supported in part by a grant from Core Research and Evolutional Science and Technology, Japan Science and Technology Corporation.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CAPS
- cryopyrin-associated periodic syndromes
- FCAS
- familial cold autoinflammatory syndrome
- NOMID
- neonatal onset multisystem inflammatory disease
References
- Abecasis G.R., Cherny S.S., Cookson W.O., and Cardon L.R.. 2002. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Agarwal A.K., Xing C., DeMartino G.N., Mizrachi D., Hernandez M.D., Sousa A.B., Martínez de Villarreal L., dos Santos H.G., and Garg A.. 2010. PSMB8 encoding the β5i proteasome subunit is mutated in joint contractures, muscle atrophy, microcytic anemia, and panniculitis-induced lipodystrophy syndrome. Am. J. Hum. Genet. 87:866–872 10.1016/j.ajhg.2010.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksentijevich I., and Kastner D.L.. 2011. Genetics of monogenic autoinflammatory diseases: past successes, future challenges. Nat Rev Rheumatol. 7:469–478 10.1038/nrrheum.2011.94 [DOI] [PubMed] [Google Scholar]
- Aksentijevich I., D Putnam C., Remmers E.F., Mueller J.L., Le J., Kolodner R.D., Moak Z., Chuang M., Austin F., Goldbach-Mansky R., et al. . 2007. The clinical continuum of cryopyrinopathies: novel CIAS1 mutations in North American patients and a new cryopyrin model. Arthritis Rheum. 56:1273–1285 10.1002/art.22491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S.W., de Jesus A.A., Gouni S., Brooks S.R., Marrero B., Liu Y., DiMattia M.A., Zaal K.J., Sanchez G.A., Kim H., et al. . 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140–1146 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.Y., and Nuñez G.. 2010. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10:826–837 10.1038/nri2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.K., Wen H., and Ting J.P.. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu. Rev. Immunol. 29:707–735 10.1146/annurev-immunol-031210-101405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A., and van der Meer J.W.. 2013. Treating inflammation by blocking interleukin-1 in humans. Semin. Immunol. 25:469–484 10.1016/j.smim.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Muñoz-Planillo R., and Núñez G.. 2012. Sensing and reacting to microbes through the inflammasomes. Nat. Immunol. 13:325–332 10.1038/ni.2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H.M., Mueller J.L., Broide D.H., Wanderer A.A., and Kolodner R.D.. 2001. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29:301–305 10.1038/ng756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Yan C., Liu P., Huang Z., Ma R., Zhang C., Wang R., Zhang Y., Martinon F., Miao D., et al. . 2013. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 341:172–175 10.1126/science.1236381 [DOI] [PubMed] [Google Scholar]
- Jéru I., Duquesnoy P., Fernandes-Alnemri T., Cochet E., Yu J.W., Lackmy-Port-Lis M., Grimprel E., Landman-Parker J., Hentgen V., Marlin S., et al. . 2008. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc. Natl. Acad. Sci. USA. 105:1614–1619 10.1073/pnas.0708616105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jéru I., Le Borgne G., Cochet E., Hayrapetyan H., Duquesnoy P., Grateau G., Morali A., Sarkisian T., and Amselem S.. 2011. Identification and functional consequences of a recurrent NLRP12 missense mutation in periodic fever syndromes. Arthritis Rheum. 63:1459–1464 10.1002/art.30241 [DOI] [PubMed] [Google Scholar]
- Kitamura A., Maekawa Y., Uehara H., Izumi K., Kawachi I., Nishizawa M., Toyoshima Y., Takahashi H., Standley D.M., Tanaka K., et al. . 2011. A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Invest. 121:4150–4160 10.1172/JCI58414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komune N., Ichinohe T., Ito M., and Yanagi Y.. 2011. Measles virus V protein inhibits NLRP3 inflammasome-mediated interleukin-1β secretion. J. Virol. 85:13019–13026 10.1128/JVI.05942-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Lamkanfi M.2011. Emerging inflammasome effector mechanisms. Nat. Rev. Immunol. 11:213–220 10.1038/nri2936 [DOI] [PubMed] [Google Scholar]
- LARGE (Laboratory for Animal Resources and Genetic Engineering), RIKEN Center for Developmental Biology. Mutant Mice Generated in Center for Developmental Biology (CDB). Available at: http://www.cdb.riken.jp/arg/TG%20mutant%20mice%20list.html (accessed October 24, 2014).
- Liu Y., Ramot Y., Torrelo A., Paller A.S., Si N., Babay S., Kim P.W., Sheikh A., Lee C.C., Chen Y., et al. . 2012. Mutations in proteasome subunit β type 8 cause chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature with evidence of genetic and phenotypic heterogeneity. Arthritis Rheum. 64:895–907 10.1002/art.33368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso F., Nothnagel M., Parwez Q., Petrasch-Parwez E., Bechara F.G., Epplen J.T., and Hoffjan S.. 2007. Polymorphisms in NACHT-LRR (NLR) genes in atopic dermatitis. Exp. Dermatol. 16:692–698 10.1111/j.1600-0625.2007.00589.x [DOI] [PubMed] [Google Scholar]
- Masters S.L., Simon A., Aksentijevich I., and Kastner D.L.. 2009. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu. Rev. Immunol. 27:621–668 10.1146/annurev.immunol.25.022106.141627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott M.F., Aksentijevich I., Galon J., McDermott E.M., Ogunkolade B.W., Centola M., Mansfield E., Gadina M., Karenko L., Pettersson T., et al. . 1999. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 97:133–144. [DOI] [PubMed] [Google Scholar]
- Meng G., Zhang F., Fuss I., Kitani A., and Strober W.. 2009. A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 30:860–874 10.1016/j.immuni.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., and Aderem A.. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11:1136–1142 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Kojima T., and Kitamura T.. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Park H., Bourla A.B., Kastner D.L., Colbert R.A., and Siegel R.M.. 2012. Lighting the fires within: the cell biology of autoinflammatory diseases. Nat. Rev. Immunol. 12:570–580 10.1038/nri3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg N., Al Moussawi K., Nelson-Williams C., Stiegler A.L., Loring E., Choi M., Overton J., Meffre E., Khokha M.K., Huttner A.J., et al. . 2014. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46:1135–1139 10.1038/ng.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen H., Benoist C., and Mathis D.. 2000. A cassette vector for high-level reporter expression driven by a hybrid invariant chain promoter in transgenic mice. J. Immunol. Methods. 245:133–137 10.1016/S0022-1759(00)00276-3 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]