Klein et al. find that frequently arising antibodies that normally fail to control HIV-1 infection can synergize with passively transferred bNAbs to prevent the emergence of bNAb viral escape variants.
Abstract
Antibody-mediated immunotherapy is effective in humanized mice when combinations of broadly neutralizing antibodies (bNAbs) are used that target nonoverlapping sites on the human immunodeficiency virus type 1 (HIV-1) envelope. In contrast, single bNAbs can control simian–human immunodeficiency virus (SHIV) infection in immune-competent macaques, suggesting that the host immune response might also contribute to the control of viremia. Here, we investigate how the autologous antibody response in intact hosts can contribute to the success of immunotherapy. We find that frequently arising antibodies that normally fail to control HIV-1 infection can synergize with passively administered bNAbs by preventing the emergence of bNAb viral escape variants.
Broadly neutralizing antibodies (bNAbs) targeting HIV-1 are of special interest because of their ability to prevent and treat HIV-1 and SHIV infection in humanized mice and nonhuman primates (NHPs), respectively (Klein et al., 2012; Moldt et al., 2012; Barouch et al., 2013; Klein et al., 2013; Shingai et al., 2013). Although there was excellent agreement between humanized mice and macaques in passive protection experiments (Balazs et al., 2012; Pietzsch et al., 2012; Shingai et al., 2013), immunotherapy of established infection was far more effective in macaques infected with SHIVAD8 or SHIVSF162P3 than in humanized mice infected with HIV-1YU2. Humanized mice treated with single mAbs showed only a transient drop in viremia with rapid escape caused by selection of antibody-resistant mutants (Klein et al., 2012; Horwitz et al., 2013). In contrast, passive transfer experiments of single bNAbs in macaques produced a profound decrease in viremia (Barouch et al., 2013; Shingai et al., 2013), and prolonged control (Barouch et al., 2013), with only occasional viral escape (Shingai et al., 2013). This disparity could be due in part to the host immune system, which is present in the macaques but defective in humanized mice. However, how the host immune system might enhance passive antibody therapy is not known. Here, we investigate the role of the autologous antibody response in suppressing the emergence of viral bNAb escape variants in vivo.
RESULTS AND DISCUSSION
Simultaneous administration of three bNAbs (tri-mix) targeting the CD4-binding site (CD4bs; 3BNC117; Scheid et al., 2011), the V1/V2-loop (PG16; Walker et al., 2009), and the V3-stem (10–1074; Mouquet, 2012) effectively suppresses viremia in HIV-1YU2–infected humanized mice without the emergence of viral escape variants (Horwitz et al., 2013). The humanized mice used in the experiments are NOD Rag1−/− IL2RγNULL mice that are reconstituted with human hematopoietic stem cells. These mice support the development of human T lymphocytes that can be infected with HIV-1 but they do not produce significant antibody responses to the pathogen (Baenziger et al., 2006; Klein et al., 2012).
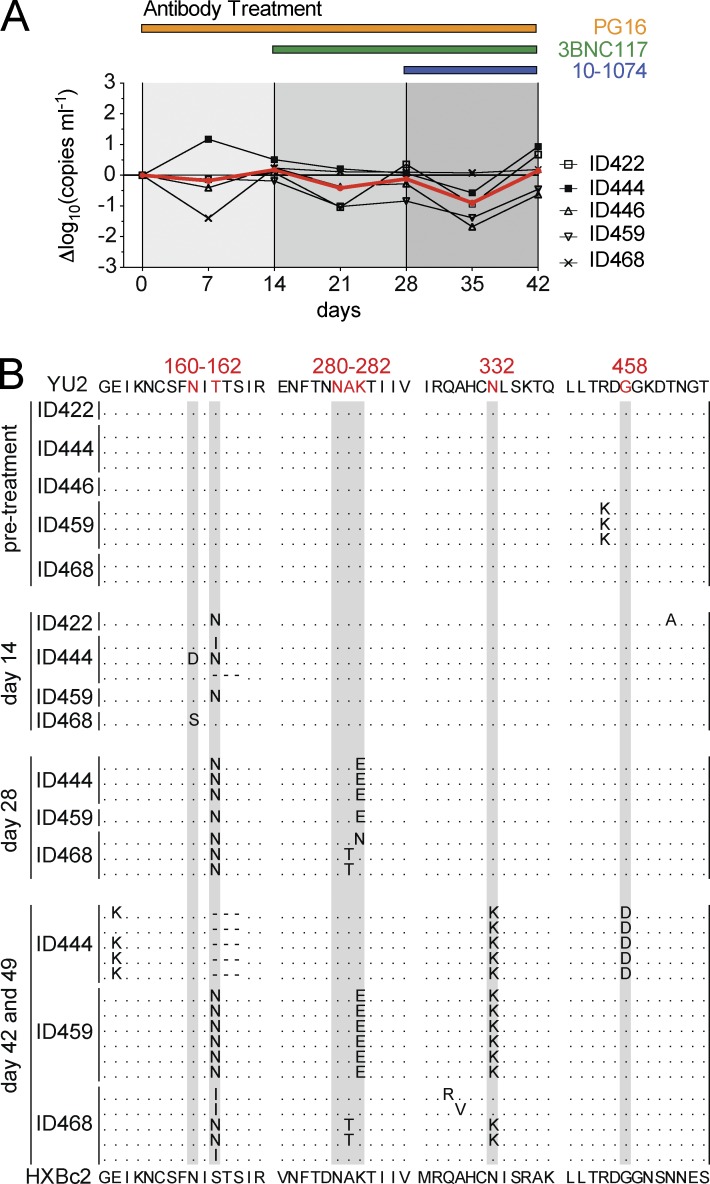
To determine whether HIV-1 can escape from all three antibodies when they are administered sequentially, we treated HIV-1YU2–infected mice with bNAbs starting with PG16 alone, and added 3BNC117 after 14 d, and 10–1074 after 28 d (Fig. 1 A). We found a transient reduction (0.18 log10 to 0.78 log10) of the viral load shortly after each antibody was administered, followed by rapid rebound to baseline viremia (day 42; +0.14 log10 compared with day 0; Fig. 1 A). Thus, sequential antibody administration differs from co-administration of the same tri-mix in that sequential therapy fails to control viral replication.
Figure 1.
Sequential treatment of HIV-1YU2–infected humanized mice with bNAbs selects for triple-escape mutants. (A) HIV-1YU2–infected mice were sequentially treated with PG16 (orange), PG16, and 3BNC117 (green) and finally with the tri-mix consisting of PG16, 3BNC117, and 10–1074 (blue) as indicated. Graph shows the log10 change in viral RNA copies in plasma plotted on the y-axis and time in days after starting treatment on the x-axis. The red line shows the mean of changes in viral load. Individual mice IDs are listed at the right. (B) gp120 envelope sequence analysis before and after 14, 28, and 42–49 d of treatment revealed the emergence of HIV-1YU2 escape variants at the respective target sites of the bNAbs (i.e., PNGS at position N160 for PG16; 280–282 and 458 for 3BNC117; PNGS at position N332 for 10–1074). Each dotted line represents an independent sequence and changes to gp120YU2 are shown in bold. Red letters and gray highlights indicate regions corresponding to known escape sites as identified in previous monotherapy experiments (Klein et al., 2012; Horwitz et al., 2013). Residues in HIV-1YU2 (top) were numbered according to HXBc2 (bottom). Presented data were obtained from five treated mice in a single experiment, and sequence information was retrieved and analyzed from at least three mice at each indicated time point.
Failure to suppress viremia with sequential tri-mix administration suggested that this form of therapy selects for viral variants that are resistant to all three antibodies (Fig. 1 A). Consistent with this idea, viral envelope sequence analysis at day 0, 14, 28, and 42–49 revealed sequential development of specific antibody-resistant HIV-1YU2 escape variants (Fig. 1 B). For example, 14 d after starting PG16 therapy, all gp120 sequences analyzed carried mutations at position N160 or T162 that remove the epitope targeted by PG16 (Fig. 1 B). Sequential addition of 3BNC117 and 10–1074 selected for viral escape variants that carry mutations in all three antibody target sites (Fig. 1 B; Klein et al., 2012; Horwitz et al., 2013). Thus, sequential triple bNAb therapy selects for HIV-1YU2 variants that are resistant to all three bNAbs.
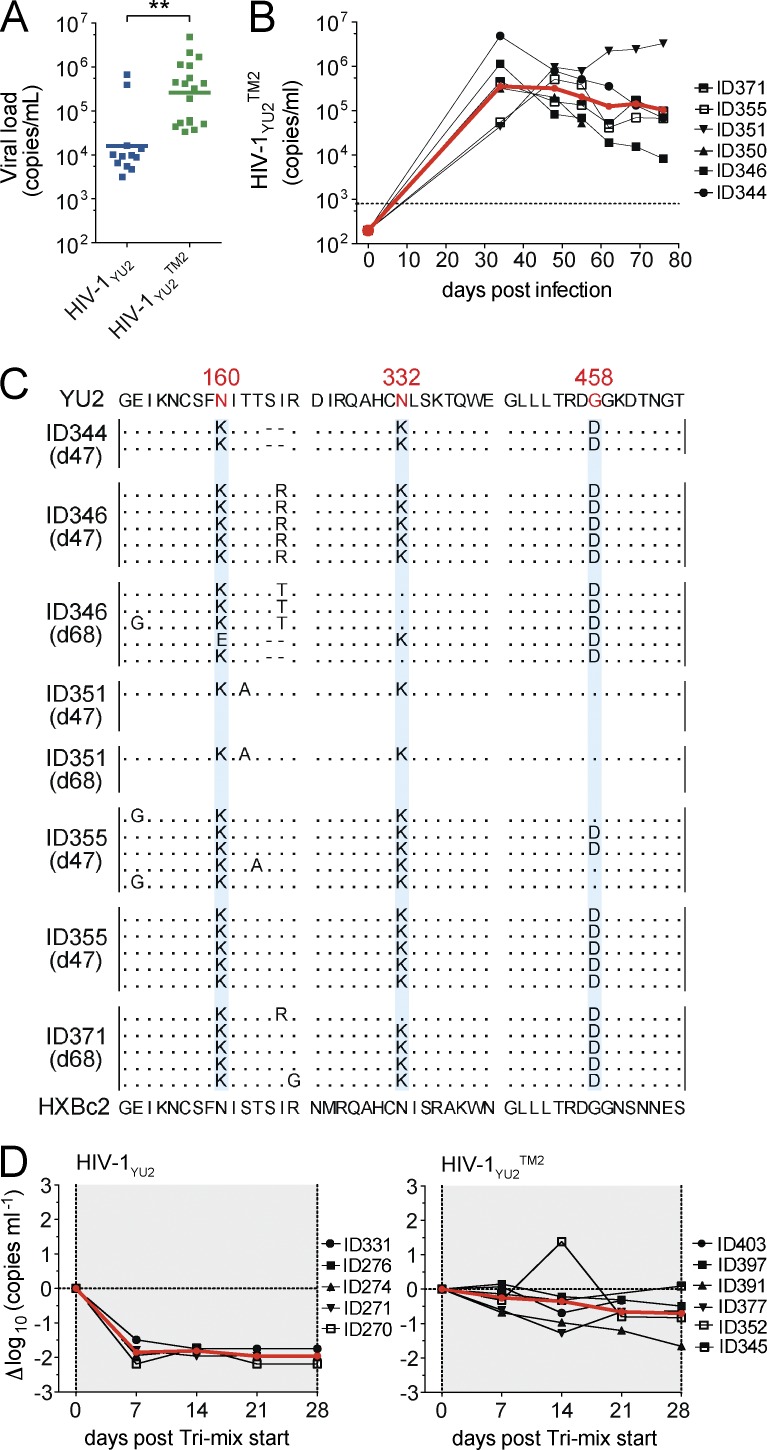
To determine whether tri-mix resistant HIV-1YU2 retains infectivity we compared infection with WT HIV-1YU2 and a variant harboring the N160K, N332K, and G458D mutation (HIV-1YU2TM2). 34 d after infection, HIV-1YU2– and HIV-1YU2TM2–infected mice showed geometric mean viral loads of 4.2 log10 and 5.42 log10, respectively (P = 0.0003; Fig. 2 A). Viremia was long lasting in HIV-1YU2TM2–infected mice (Fig. 2 B) and in most cases the N160K, N332K, and G458D mutations were maintained even in the absence of antibody selection pressure (Fig. 2 C). Finally, HIV-1YU2TM2–infected mice were resistant to tri-mix therapy (Fig. 2 D). We concluded that HIV-1YU2 can escape from sequential tri-mix therapy in vivo without measurable loss of infectivity or impaired viral fitness in humanized mice.
Figure 2.
Infection of humanized mice with HIV-1YU2 triple mutant (HIV-1YU2TM2). (A) Viral loads in humanized mice infected with WT HIV-1YU2 (blue) and HIV-1YU2TM2 (green) at day 34 after infection. P-value was determined using a two-sided Mann-Whitney U test. **, P < 0.001. Plot shows data of 30 infected mice of 1 representative experiment of 2 performed. (B) Graph shows viral RNA copies/ml (y-axis) versus days after infection (x-axis) for HIV-1YU2TM2–infected humanized mice. Each black line represents a single mouse, and the red line represents the geometric mean. (C) env sequence analysis of individual mice that are shown in (B). Time of sequence analysis is indicated in days after infection. (D) Tri-mix (PG16, 3BNC117, 10–1074) therapy in mice infected with HIV-1YU2 (left) and HIV-1YU2TM2 (right). Changes in viral load in Δlog10 (y-axis) plotted against days after infection (x-axis) compared with baseline (day 0). Each black line represents a single mouse and the red line illustrates the mean. Treatment response in mice infected with HIV-1YU2 or HIV-1YU2TM2 was analyzed in parallel in a single experiment and each group consisted of at least five individual mice (D).
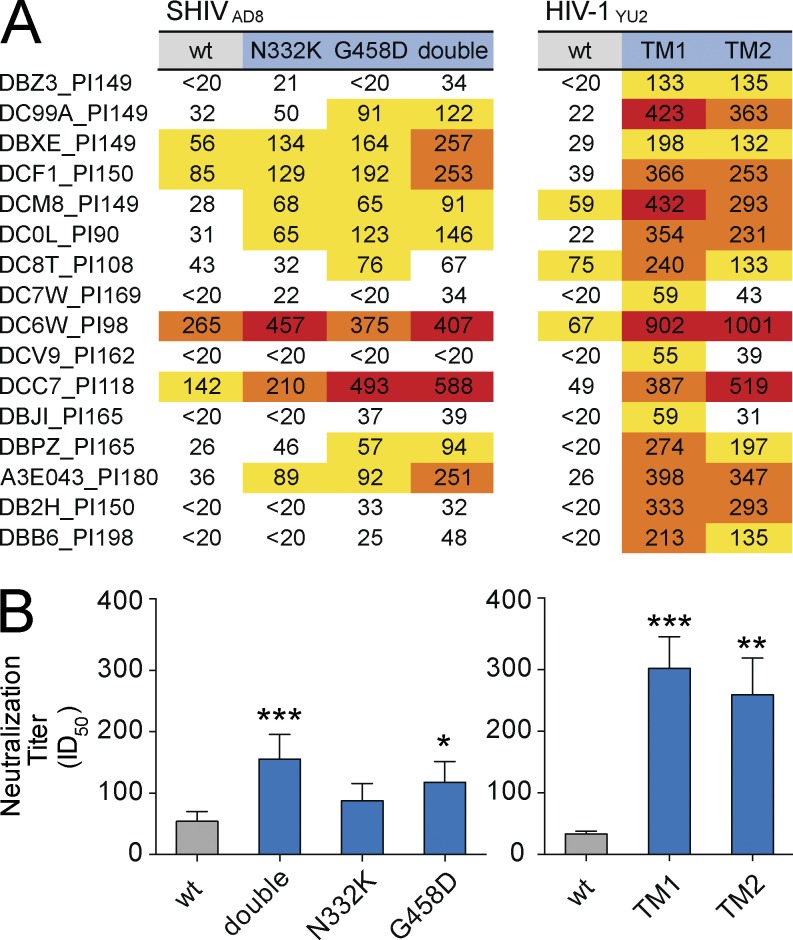
To determine whether chronically SHIVAD8-infected NHPs or HIV-1–infected humans harbor antibodies that might neutralize bNAb-resistant variants, we assayed plasma/serum samples for neutralizing activity against escape variants in vitro. Plasma samples from chronically SHIVAD8-infected macaques (Shingai et al., 2012, 2013) were tested against WT SHIVAD8 and SHIVAD8 variants. The SHIVAD8 variants carried either an N332K mutation (SHIVAD8N332K) that rendered the virus resistant to the bNAb 10–1074 or the G458D mutation (SHIVAD8G458D) that strongly reduced (200-fold) sensitivity to the bNAb 3BNC117. In addition, we included an SHIVAD8 variant that harbored both mutations (N332K-G458D; SHIVAD8DM). Although plasma from 16 SHIVAD8-infected macaques showed varying levels of neutralizing activity against the WT virus SHIVAD8 (Fig. 3 A; ID50), a significant increase in neutralizing activity was detected for bNAb-resistant variants that carried the G458D and the double N332K-G458D mutation (Fig. 3 B). Moreover, this effect was not specific for SHIVAD8 since even more striking differences were observed for WT HIV-1YU2 and HIV-1YU2 triple bNAb escape mutants TM1 (N160K, N332K, and N280Y) and TM2 (N160K, N332K, and G458D; Fig. 3, A and B). Thus, macaques chronically infected with SHIVAD8 frequently carry antibodies that neutralize the bNAb escape mutants studied.
Figure 3.
Increased neutralizing NHP serum activity against bNAb escape variants. (A) Table shows serum neutralization (ID50) of NHPs after established infection with SHIVAD8 (days post infection [PI] as indicated in NHP ID). Neutralizing activity was measured against SHIVAD8 WT (gray), N332K, and G458D single mutants (blue), as well as an SHIVAD8 double mutant (double; N332K and G458D, blue). NHP plasma was also tested against WT HIV-1YU2 (gray) and HIV-1YU2TM1 (N160K, N332K, N280Y, blue) and HIV-1YU2TM2 (TM2; N160K, N332K, G458D, blue). (B) Bars represent the mean and SEM of the respective data shown in A. Statistical analysis was performed using a Friedman test followed by Dunn multiple comparison test. Significant differences compared with WT are indicated by asterisks (*, P < 0.05; **, P < 0.001; ***, P < 0.0001). All neutralizing activities were measured in duplicate.
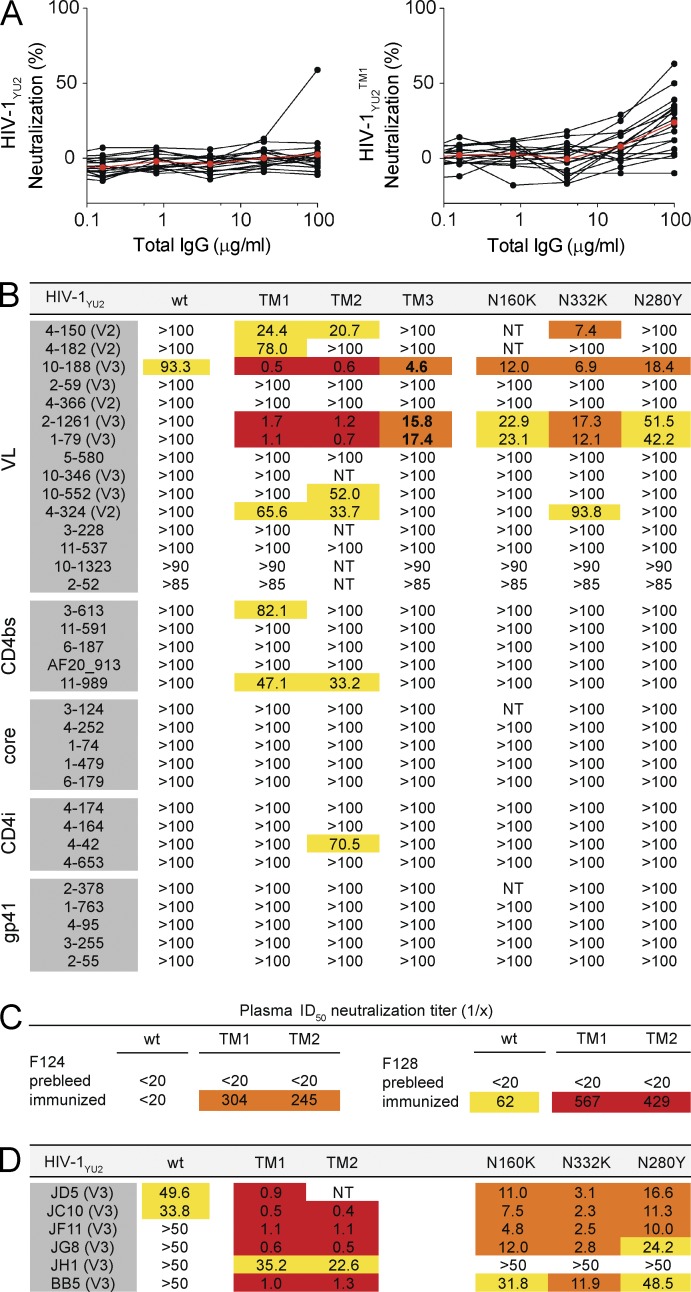
To determine whether chronically infected humans also have similar antibodies, we tested a collection of 17 serum samples from HIV-1–infected individuals. Similar to macaques, purified IgGs from infected humans showed little or no neutralization against WT HIV-1YU2, but increased activity against HIV-1YU2TM1 (Fig. 4 A; mean percentage of neutralization at 100 µg/ml total IgG: 2.5 vs. 23.9; P = 0.0004, two-sided Mann-Whitney U test). Thus, most humans infected with HIV-1 carry antibodies that have demonstrable in vitro neutralizing activity against the studied bNAb-resistant variants of HIV-1YU2.
Figure 4.
Increased neutralizing activity against bNAb escape variants. (A) Graph shows percentage of neutralizing activity (y-axis) of purified IgG (µg/ml; x-axis) from 17 serum samples of HIV-1–infected individuals measured against WT HIV-1YU2 (left) or HIV-1YU2TM1 (right; N160K, N332K, N280Y). Individual samples and the mean are shown in black and red, respectively. (B) Table shows the IC50 antibody concentrations (µg/ml) for the indicated antibodies against HIV-1YU2 (WT), HIV-1YU2TM1 (TM1; N160K, N332K, N280Y), HIV-1YU2TM2 (TM2; N160K, N332K, G458D), HIV-1YU2TM3 (TM3; N162I, N332K, N279K), HIV-1YU2N160K (N160K), HIV-1YU2N332K (N332K), and HIV-1YU2N280Y (N280Y). The epitopes targeted by the antibodies are indicated on the left and in parentheses. (C) Values reflect ID50 neutralization titers against HIV-1YU2 (WT), HIV-1YU2TM1 (TM1), and HIV-1YU2TM2 (TM2) for plasma samples from two macaques (F124, F128) before and after immunization with gp140-F (YU2). (D) IC50 concentrations (µg/ml) against HIV-1YU2 and HIV-1YU2 mutants for six randomly selected V3 loop-directed mAbs obtained from gp140-F immunized macaques as described in B. Neutralization activity is color-coded and IC50-values of mAbs (B are D) are highlighted in red (<2 µg/ml), orange (2–20 µg/ml), yellow (>20 µg/ml), and white (IC50 is not reached at concentration tested). ID50-titers of plasma are highlighted in red (>400), orange (200–400), yellow (50–200), and white (<50). Neutralizing activities of plasma samples, antibodies, or purified IgGs were measured in duplicate.
Individuals infected with HIV-1 develop up to 60 different clones of B cells, producing antibodies that have neutralizing activity against easy to neutralize (tier-1) HIV-1 strains but have weak or no activity against more resistant tier-2 primary isolates (Zolla-Pazner, 2005; Scheid et al., 2009). To determine whether these antibodies with limited activity (tier-1 strains only; here termed tier-1 neutralizing antibodies) are active against viruses that carry bNAb escape mutations, we assayed a panel of 34 tier-1 neutralizing mAbs directed against the V2 or the V3 loop, the CD4bs, the CD4-induced site (CD4i), and gp41 (Scheid et al., 2009; Pietzsch et al., 2010; Mouquet et al., 2011). Each of these antibodies was tested against WT HIV-1YU2 and bNAb-resistant variants of HIV-1YU2 with mutations that naturally arose in in vivo experiments (single mutations: N160K, N332K, and N280Y; HIV-1YU2 triple mutations: TM1-3; Fig. 4 B; Klein et al., 2012; Horwitz et al., 2013). Only one of the tier-1 neutralizing antibodies showed activity against the WT HIV-1YU2 at a very high concentration (IC50 = 93 µg/ml; Fig. 4 B). In contrast, three V2 loop-, four V3 loop-, two CD4bs-, and one CD4i-directed antibodies showed activity against the HIV-1YU2 mutant viruses with IC50s as low as 0.5 µg/ml (Fig. 4 B). The three V2 loop-directed antibodies were active against HIV-1YU2N332K, HIV-1YU2TM1, and HIV-1YU2TM2. The four V3 loop-directed antibodies showed activity against HIV-1YU2N280Y, HIV-1YU2N160K, HIV-1YU2N332K, as well as all three HIV-1YU2TMs. Finally, at least one of each of the antibodies targeting CD4bs and CD4i was active against the HIV-1YU2TMs in vitro (Fig. 4 B).
To determine whether antibodies that neutralize bNAb escape variants can also be elicited by vaccination, we analyzed plasma from NHPs immunized with YU2 gp140-F trimers (Sundling et al., 2010). Although plasma samples from the vaccinated animals showed little or no neutralizing activity against HIV-1YU2 (WT; Fig. 4 C), there was a significant increase in neutralizing activity against the HIV-1YU2 triple mutants (HIV-1YU2TM1, HIV-1YU2TM2; Fig. 4 C). To determine whether macaque antibodies recognizing the V3 loop might account for this activity, we tested 6 different anti-V3 mAbs isolated from these immunized macaques against WT HIV-1YU2, HIV-1YU2N280Y, HIV-1YU2N160K, HIV-1YU2N332K, and two HIV-1YU2TMs (Fig. 4 D). 2 of the 6 antibodies had low levels of activity against HIV-1YU2 (WT; IC50s of 33.8 and 49.6 µg/ml), whereas the other four did not reach an IC50 when measured up to a concentration of 50 µg/ml. In contrast, 5 out of these 6 V3 loop-directed antibodies showed activity against the HIV-1YU2 single mutants. Moreover, neutralization activity was strongly increased against both HIV-1YU2TMs as reflected by IC50s in the range of 1 µg/ml (Fig. 4 D). Thus, although tier-1 neutralizing human and macaque anti–HIV-1 antibodies do not neutralize WT HIV-1YU2, they are active against bNAb escape variants in TZM-bl assays in vitro.
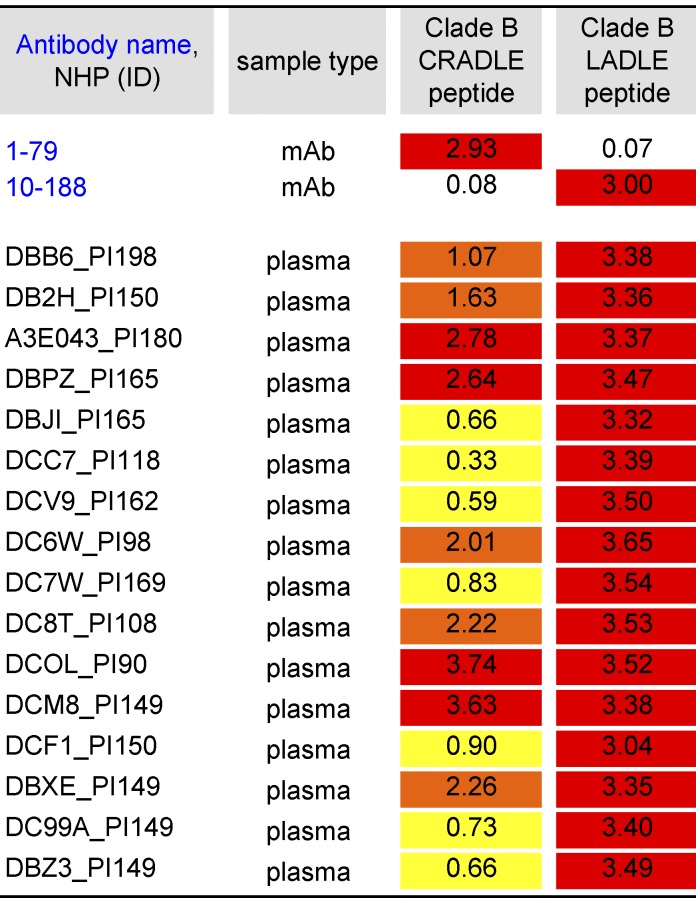
Of the tested human tier-1 neutralizing antibodies, 10–188 and 1–79 demonstrated the best neutralizing activity against the triple mutated HIV-1YU2 (Fig. 4 B). Both antibodies recognize the crown of the V3 loop. Notably, 10–188 and 1–79 bind to different regions of the V3 loop: 1–79 targets the hydrophobic V3 core at aa 307, 309, and 317 (CRADLE-type; Burke et al., 2009; Almond et al., 2010; Jiang et al., 2010; and unpublished data) and 10–188 targets the V3 β-turn at aa 312–314 (LADLE-type; Burke et al., 2009). To determine, whether similar antibodies are present in NHPs chronically infected with SHIVAD8, we measured reactivity of plasma samples against selected peptides by ELISA (Fig. 5; Totrov et al., 2010). All tested NHP sera showed activity against both peptides, demonstrating the presence of CRADLE- and LADLE-type antibodies in SHIVAD8-infected NHPs (Fig. 5).
Figure 5.
Presence of V3-specific antibodies in plasma of SHIVAD8-infected NHPs. Values reflect OD405 measurements (values >0.3 were considered positive). PI, post infection (days). mAbs were measured at 10 µg/ml. Plasma samples were measured at a titer of 1:100. ID50 titers of plasma are highlighted in red (>2.5), orange (1–2.5), and yellow (0.3–1).
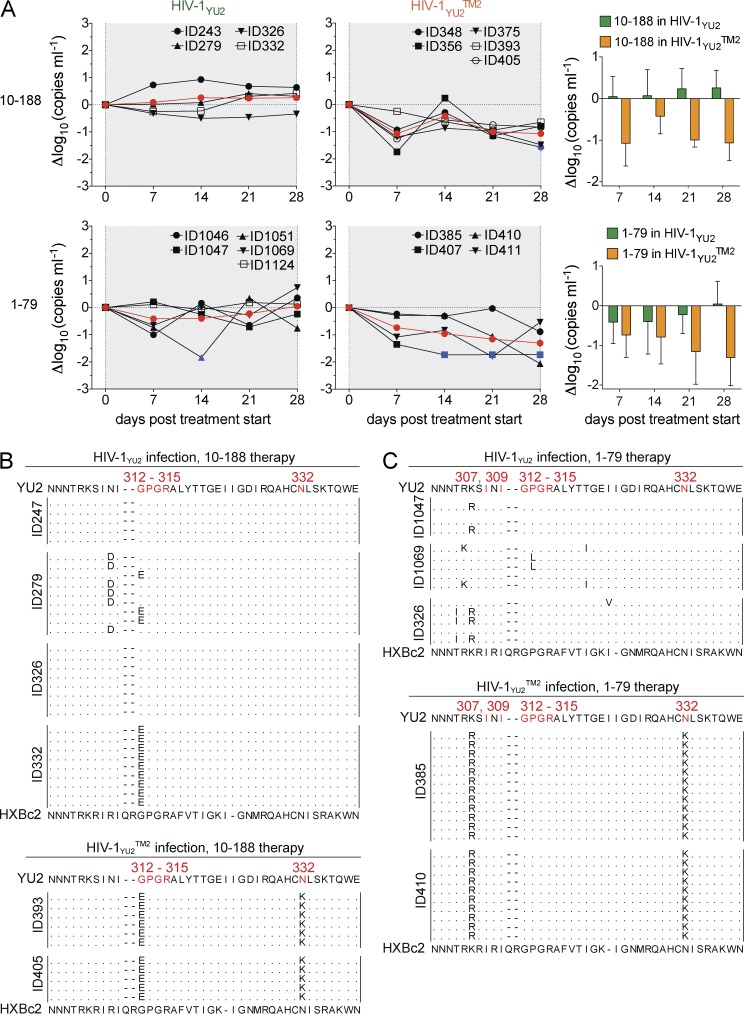
To examine the possibility that HIV-1–directed tier-1 neutralizing antibodies can actively suppress viremia in vivo, we infected humanized mice with HIV-1YU2 (WT) or with HIV-1YU2TM2. Infected mice were treated with either of the tier-1 neutralizing human V3 loop-directed antibodies 10–188 or 1–79. Although only a small effect on the viral load was detected in HIV-1YU2–infected mice (Fig. 6 A, left), a reduction in viremia was observed in mice infected with the mutant virus HIV-1YU2TM2 (Fig. 6 A, middle and right). We conclude that the tier-1 neutralizing anti-V3 loop antibodies 10–188 and 1–79 do not alter the viral load in HIV-1YU2–infected mice, but can suppress HIV-1YU2TM2 infection in vivo.
Figure 6.
Suppression of HIV-1YU2TM2 but not HIV-1YU2 viremia by V3-loop antibodies 10–188 and 1–79. (A, left and middle) Graphs show changes in viral load in response to 10–188 or 1–79 treatment in Δlog10 (y-axis) plotted against days after infection (x-axis) compared with baseline (day 0) for mice infected with HIV-1YU2 or HIV-1YU2TM2. Each black line represents a single mouse, and the red line illustrates the mean. Blue colored symbols indicate a viral load measurement below 200 copies/ml. (A, right) Statistical analysis to determine group differences between HIV-1 YU2 (green) and HIV-1YU2TM2 (orange) using repeated measures ANOVA; 10–188, P = 0.003; 1–79, P = 0.032. (B and, C) gp120 sequences obtained from HIV-1YU2– or HIV-1YU2TM2–infected mice treated with 10–188 (B) or 1–79 (C) between days 14 and 28. Residues in HIV-1YU2 (top) were numbered according to HXBc2 (bottom). Treatment response of 10–188 and 1–79 in mice infected with HIV-1YU2 or HIV-1YU2TM2 was analyzed in a single experiment in which each group consisted of at least four individual mice and sequence data were obtained from at least two mice per group.
To determine whether 10–188 and 1–79 exerted selective pressure on HIV-1YU2 and/or HIV-1YU2TM2, we cloned and sequenced cDNA encoding gp120 from HIV-1YU2– and HIV-1YU2TM2–infected mice treated with these antibodies. Although 10–188 showed little activity against HIV-1YU2 in vitro (IC50 93 µg/ml) and had no measurable effect on the viral load in HIV-1YU2–infected mice, 2 out of the 4 mice showed mutations in the β-turn in the crown of the V3 loop, which is the epitope targeted by 10–188 (Zolla-Pazner and Cardozo, 2010; Mouquet et al., 2011; Fig. 6 B, top). In addition, all gp120 sequences analyzed from HIV-1YU2TM2–infected mice (2 animals) carried mutations in the same region (Fig. 6 B, bottom). In the case of HIV-1YU2–infected mice treated with 1–79, we also detected mutations in the V3 crown in some but not all of the sequences in the 3 animals we analyzed, indicating incomplete selection (Fig. 6 C). In contrast, all sequences obtained from HIV-1YU2TM2–infected mice treated with 1–79 harbored the same K305R mutation (Fig. 6 C).
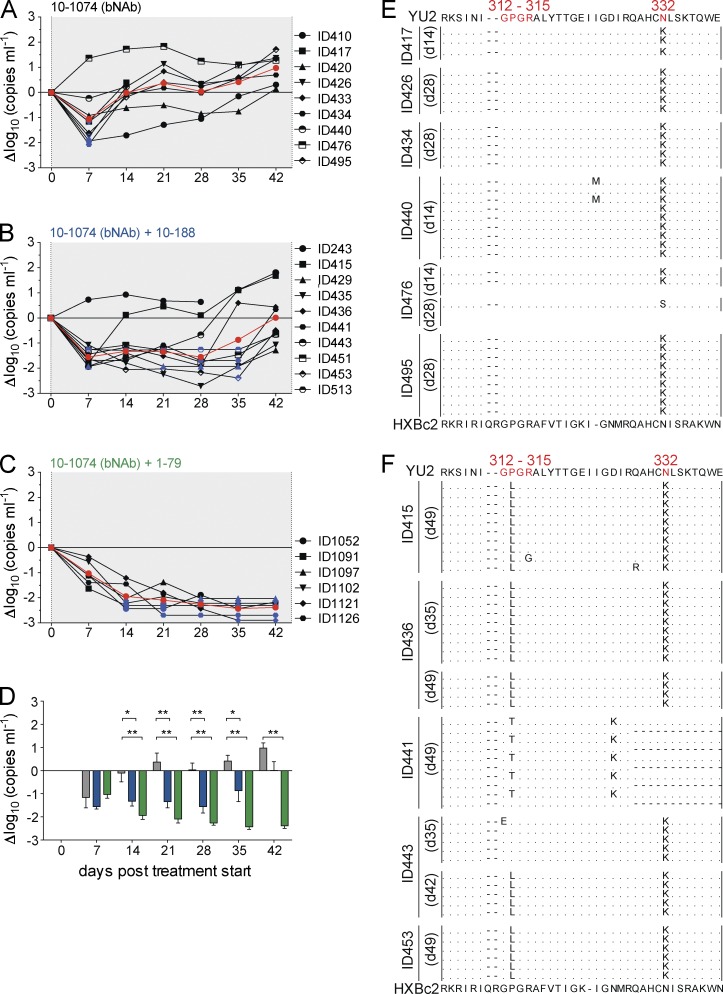
HIV-1YU2TM2 is an engineered virus that carries bNAb escape mutations that naturally occurred in mice treated with single antibodies (Klein et al., 2012; Horwitz et al., 2013). However, it is not a naturally arising strain that might also contain compensatory mutations as part of a heterogeneous swarm. To determine whether combination immunotherapy with a bNAb and a tier-1 neutralizing antibody can delay or suppress emergence of resistant HIV-1 variants during an established infection with a naturally arising strain, we infected humanized mice with HIV-1YU2 (WT) and treated them with the bNAb 10–1074 alone (Fig. 7 A) or in combination with one of the tier-1 neutralizing antibodies 10–188 (Fig. 7 B) or 1–79 (Fig. 7 C). Notably, the humanized mice used in our experiments are unable to make a significant humoral immune response to the HIV-1 envelope, and therefore lack any autologous neutralizing antibodies (Baenziger et al., 2006; Klein et al., 2012). In contrast to monotherapy with any of the three antibodies alone (Figs. 6 A and 7 A), both combinations produced a prolonged and sustained drop in viremia (Fig. 7, B–D). Moreover, the combination of 10–1074 and 1–79 continued to suppress viremia below the limit of detection, with no viral escape during 6 wk of therapy (Fig. 7 C). Escape from 10–1074 monotherapy was associated with N332K/S in 6 out of 6 mice analyzed (Fig. 7 E). When rebound occurred with the combination of 10–1074 and 10–188, it was associated with selection of mutations in both the 10–1074 and 10–188 target sites at position 332 and the β-turn of the V3 crown (position 312–315), respectively (Fig. 7 F).
Figure 7.
Synergy between tier-1 neutralizing V3 antibodies (10–188 or 1–79) and a bNAb (10–1074) in controlling viremia in vivo. (A–D) Graphs show changes in viral load (in Δlog10; y-axis) in HIV-1YU2–infected mice plotted against days after starting antibody treatment (x-axis) with 10–1074 alone (A), the combination of 10–1074 and 10–188 (B), and the combination of 10–1074 and 1–79 (C). Individual mice are plotted as black lines, and the mean is shown in red. Blue symbols indicate a viral load measurement below 200 copies/ml. (D) Changes of log10 viral loads (mean and SEM) of mice treated with 10–1074 (gray), 10–1074 + 10–188 (blue), and 10–1074 + 1–79 (green). Significant differences between treatment groups were determined by using repeated measures ANOVA with a Bonferroni post-hoc test. *, P < 0.05; **, P < 0.001; ***, P < 0.0001). Animals that died before day 42 after treatment start were excluded from statistical analysis. (E) gp120 sequences 14–28 d after the start of 10–1074 treatment. (F) gp120 sequences 35–49 d after the start of treatment with 10–1074 and 10–188. Residues in HIV-1YU2 (top) were numbered according to HXBc2 (bottom). Treatment combinations were analyzed on 6–10 animals per group in a single round experiment. Sequence data were obtained from at least five individual mice of each analyzed group.
We conclude that tier-1 neutralizing antibodies with little demonstrable activity against primary isolates in vitro can make significant contributions to control HIV-1 infection when combined with potent bNAbs in vivo.
Most individuals infected with HIV-1 develop antibodies that neutralize autologous but not primary heterologous viral strains (Doria-Rose et al., 2009; Simek et al., 2009). As a result of the rapid rate of viral evolution in the host, the effect of these antibodies on effectively controlling viremia is limited (Schmitz et al., 2003; Miller et al., 2007; Gaufin et al., 2009a,b; Huang et al., 2010). Nevertheless, these antibodies put selective pressure on HIV-1 as indicated by the emergence of antibody-resistant escape variants (Wei et al., 2003). Escape from tier-1 neutralizing antibodies frequently involves changes that indirectly conceal the epitope (Ly and Stamatatos, 2000; Wei et al., 2003; Pinter et al., 2004; Blish et al., 2008; Bunnik et al., 2008; Bosch et al., 2010; O’Rourke et al., 2010). For example, the glycan at position N332 can shield the V3 loop from tier-1 neutralizing anti-V3 loop antibodies, but does not alter the antibody target sequence directly (Wei et al., 2003; McCaffrey et al., 2004). Similarly, glycans at position N276 or N301 can shield the CD4bs from tier-1 neutralizing anti-CD4bs antibodies (Koch et al., 2003; McGuire et al., 2013). These changes are often favored over direct changes in the target site that might interfere with viral fitness. For example, a glycan can be added to shield the V3 loop without reducing infectivity. This shielding mechanism appears to be preferred over escape mutations in the highly conserved crown of the V3-loop that could interfere with co-receptor binding and alter viral fitness (Zolla-Pazner and Cardozo, 2010). However, the same glycans that shield V3 also make important contributions to the epitopes of some of the most potent bNAbs, such as 10–1074 or PGT121 (Walker et al., 2011; Mouquet et al., 2012). These antibodies have profound effects on viremia in NHPs but less so in immune compromised humanized mice when used as a single reagent (Klein et al., 2012; Barouch et al., 2013; Shingai et al., 2013; Fig. 7 A). Our data suggest that one of the reasons that escape from 10–1074 or PGT121 immunotherapy in macaques is difficult might be because the autologous antibodies, which are present in macaques but not in humanized mice, prevent escape by the N332 mutation. Interestingly, several of the 10–1074-escape variants in SHIVAD8-infected NHPs carried a mutation that removes the potential N-linked glycosylation site (PNGS) at position 332 but also generated a new PNGS at position 334 (Shingai et al., 2013). Therefore, the glycan was shifted from N332 to N334, allowing escape from 10–1074 as well as retaining a glycan at the V3-stem that is likely to protect the V3-loop from autologous antibodies.
Our experiments indicate that HIV-1 infection is in part more readily controlled during immunotherapy in immunocompetent hosts because escape from bNAbs can create holes in the glycan shield that render the virus susceptible to otherwise ineffective autologous antibodies that are present in nearly all infected individuals. Thus, although antibodies with tier-1 neutralizing activity, such as V3 loop-directed monoclonals (Hioe et al., 2010), generally display weak and sporadic neutralizing activity against most tier-2 viruses, they can effectively synergize with bNAbs in anti–HIV-1 immunotherapy.
MATERIALS AND METHODS
Human samples.
Serum samples from HIV-1–infected individuals were collected under informed consent and in accordance with the Institutional Review Board (IRB; protocol number 09–281, University of Cologne, Cologne, Germany). All samples were heat-inactivated for 1 h at 56°C and the IgG fraction was purified with Protein G–Sepharose 4 Fast Flow (GE Healthcare). Sterile filtration and buffer exchange to PBS was performed for all IgGs before testing for neutralizing activity.
Nonhuman primate samples.
Plasma samples were obtained from nonhuman primates (NHPs; Macaca mulatta) 90–198 wk after infection with SHIVAD8 (Shingai et al., 2012, 2013) or from uninfected NHPs (Macaca mulatta) before and after immunization with soluble YU2 gp140-F trimers (Yang et al., 2002), as previously described (Sundling et al., 2010). SHIVAD8–infected macaques were housed in a biosafety level 2 National Institute of Allergy and Infectious Disease (NIAID) facility and cared for in accordance with standards of the American Association for Accreditation of Laboratory Animal Care (AAALAC) in AAALAC-accredited facilities. All animal procedures were performed according to NIAID animal protocol LMM32, approved by the Institutional Animal Care and Use Committees of NIAID/NIH. The animals used for immunizations were housed at the AAALAC-accredited Astrid Fagraeus Laboratory animal facility in Stockholm in compliance with the guidelines of the Swedish Board of Agriculture. Isolation of mAbs from NHPs was performed essentially as previously described (Sundling et al., 2012a) but by sorting for total Env-specific memory B cells and using optimized primers for amplification of NHP VDJ sequences (Sundling et al., 2012b). For antibody expression, equal amounts of heavy- and light-chain plasmid DNAs were transfected into FreeStyle 293F cells and the secreted IgGs were purified by protein A–Sepharose columns (GE Healthcare). The specificities of these V3 loop-directed antibodies were determined by ELISA binding analyses using a set of different YU2 Env probes.
Humanized mice.
NOD Rag1−/− IL2RγNULL mice were humanized with hematopoietic stem cells (HSCs), as previously described (Klein et al., 2012). In brief, human fetal livers were procured from Advanced Bioscience Resources, Inc. and HSCs were isolated using a CD34+ isolation kit (Stem Cell Technologies, Inc.). Mice (NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ) were obtained from The Jackson Laboratory and bred and maintained at the Comparative Bioscience Center of The Rockefeller University. Between 1–5-d-old mice were irradiated with 100 cGy and injected with 2 × 105 human HSCs. HSCs used to reconstitute the mice were obtained from human fetal liver tissues of 12 different donors. In the experiments shown, up to 12 humanized mice shared the same donor. Engraftment was evaluated by FACS analysis of the peripheral blood as previously described (Klein et al., 2012) and mice with successful reconstitution of human lymphocytes were challenged with HIV-1YU2 or HIV-1YU2TM2. All experiments were performed under approval of the Institutional Review Board and the Institutional Animal Care and Use Committee of The Rockefeller University.
HIV-1 infection.
HIV-1YU2 and HIV-1YU2TM2 were produced by transiently transfecting HEK 293T/17 cells using a construct consisting of HIV-1NL4/3 backbone carrying the HIV-1YU2 envelope (Zhang et al., 2002). HIV-1YU2TM2 harbors the mutations N160K, N332K, and G458D that were introduced by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies). The concentration of the virus was determined by measuring p24 using the Alliance HIV-1 p24 Antigen ELISA kit (PerkinElmer). Humanized mice were infected by intraperitoneal injection of HIV-1YU2 or HIV-1YU2TM2 viral supernatant containing 110 ng of p24.
Production and administration of mAbs.
For mouse treatment experiments we used the following mAbs: PG16 (V1/V2; Walker et al., 2009), 10–1074 (V3-stem; Mouquet et al., 2012), 3BNC117 (CD4bs; Scheid et al., 2011), 10–188 (V3-crown; Mouquet et al., 2011), and 1–79 (V3-crown; Scheid et al., 2009). With the exception of 3BNC117, all antibodies were produced by transiently transfected HEK 293-6E cells with equal amounts of immunoglobulin heavy and light chain expression vectors. After 7 d, the supernatant was harvested and antibodies were concentrated by ammonium sulfate precipitation. IgG was purified with Protein G–Sepharose 4 Fast Flow. 3BNC117 was produced in CHO cells by Celldex Therapeutics, Inc. All antibodies were filtered (Ultrafree-CL Centrifugal Filters, 0.22 µm; Millipore) and administered s.c. to humanized mice. For treatment of HIV-1–infected mice, 1 mg (each) of 3BNC117, PG16, and 10–1074 was injected as loading dose followed by 0.5 mg of each antibody/mouse twice a week. 10–188 and 1–79 were injected at 4 mg/mouse for the loading dose followed by 2 mg/mouse twice a week.
HIV-1 plasma viral load.
Plasma viral load in HIV-1–infected humanized mice was determined as previously described (Klein et al., 2012).
HIV-1 envelope glycoprotein sequence analysis.
Total RNA from 100 µl EDTA-plasma was extracted using the MinElute Virus Spin kit (QIAGEN) and reverse transcribed using SuperScript III Reverse transcription (Invitrogen). For cDNA synthesis, the primer was either 5′-TAGCAATAGTTGTGTGGACC-3′ or 5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′. Amplification of HIV-1 gp120 sequences was performed by a double-nested PCR using the Expand High Fidelity PCR System (Roche). Primers for first round PCR were FW_5′-TAGCAATAGTTGTGTGGAC-3′ and Rev_5′-ATTGTTCTGCTGTTGCACTA-3′ and for second round PCR FW_5′-AGAAAGAGCAGAAGACAGTGGC-3′ and Rev_5′-TACCGTCAGCGTTATTGACG-3′. Cycling parameters in first and second PCR were 92°C, 2 min; (92°C, 20 s; 57°C, 30 s; and 68°C, 1:40 min) over 35 cycles with additional 20 s/cycle elongation time (starting after cycle 15) followed by 10 min at 68°C. After the second round of PCR, 0.5 µl of Taq Polymerase was added to each reaction and incubated for 15 min at 72°C. PCR products were gel purified and cloned into the pCR 4-TOPO vector (Invitrogen) and transfected into MAX Efficiency Stbl2 Competent Cells (Life Technologies). Individual colonies were sequenced with 5 different primers and assembled consensus sequences aligned to gp120-YU2 (available from GenBank under accession no. M93258). All analyses were performed using Geneious Pro software (Biomatters Ltd.) and residues were numbered according to HXBc2 (http://www.hiv.lanl.gov/content/sequence/LOCATE/locate.html).
Peptide ELISA.
ELISA plates (StreptaWell; Roche) were coated at 37°C for 1.5 h with 1 µg/ml biotinylated peptides (Clade B CRADLE peptide, Cyclic Biotin-RCRIHIGPGRAFYACG-OH; Clade B LADLE peptide, Cyclic Biotin-ACQAFYASSPRKSIHIGACA-OH). Plates were washed 6 times with PBS containing 0.05% Tween-20, pH 7.4, and incubated for 1.5 h (37°C) with plasma that was diluted 1:100 in RPMI-1640 (Sigma-Aldrich) containing 15% fetal bovine serum. After washing plates (six times) alkaline phosphatase-conjugated goat anti–human IgG (1:2,000) was added for 1.5 h (37°C). Plates were washed and 10% diethanolamine substrate was added for 30 min followed to analyze at A405 nm. All reagents were added in a volume of 100 µl/well and each sample was run in duplicate.
Neutralization.
Neutralizing activities of NHP plasma samples, purified IgGs, and mAbs were determined in a TZM.bl neutralization assay (Li et al., 2005; Seaman et al., 2010). Mutations in HIV-1 and SHIV envelope glycoprotein sequences were produced using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions and used for pseudovirus production. SHIV and HIV-1 pseudovirus mutants included the single mutations SHIVAD8 (N332K), SHIVAD8 (G458D), HIV-1YU2 (N160K), HIV-1YU2 (N332K), and HIV-1YU2 (N280K), as well as the double and triple mutations SHIVAD8 (N160K, N332K), HIV-1YU2TM1 (N160K, N332K, N280Y), HIV-1YU2TM2 (N160K, N332K, G458D), and HIV-1YU2TM3 (N162I, N332K, and N279K).
Statistics.
Statistical significance between IgG neutralizing activity against HIV-1YU2wt and HIV-1YU2TM2 (Fig. 4 A), and differences in viral load between HIV-1YU2wt and HIV-1YU2TM2–infected mice (Fig. 2 A) was determined by performing a two-sided Mann-Whitney U test in which P < 0.05 was considered significant. Statistical analysis for comparing neutralizing activity of NHP sera against SHIV and HIV-1 variants was performed by Friedman test followed by Dunn multiple comparison. Significant differences in log10 changes of viral load between mice treated with 10–1074, 10–1074 + 10–188, and 10–1074 + 1–79, as well as HIV-1YU2- and HIV-1YU2TM2–infected mice treated with 10–188 or 1–79 were determined by using repeated measures ANOVA with a Bonferroni post-hoc test considering P < 0.05 significant. Sample size of humanized mice in antibody treatment experiments was estimated based on previous results testing single antibodies in HIV-1–infected humanized mice (Horwitz et al., 2013; Klein et al., 2012). All analyses were performed using GraphPad Prism version 5.0b for Mac OS X, GraphPad Software.
Acknowledgments
We thank all HIV-1–infected individuals who participated in this study; Gisela Kremer for patient coordination; Reha-Baris Incesu for IgG purification; Israel Tojal Silva for sequence alignments; and Johannes Scheid and Hugo Mouquet for providing antibody plasmids.
F. Klein was supported by the Stavros Niarchos Foundation and the Robert Mapplethorpe Foundation. G.B. Karlsson Hedestam and G. Phad were supported by grants from the Swedish Research Council, the International AIDS Vaccine Initiative and Karolinska Institutet. C. Lehmann was supported by the Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) grant 01KI1017. C. Lehmann and G. Fätkenheuer were supported by the the German Centre for Infection Research (DZIF), partner site Bonn-Cologne, Cologne, Germany. This work was supported in part by the Bill and Melinda Gates Foundation with Comprehensive Antibody Vaccine Immune Monitoring Consortium Grant 1032144 (to M.S. Seaman) and Collaboration for AIDS Vaccine Discovery Grants 1040753 (to P.J. Bjorkman) and 38619 (to M.C. Nussenzweig). This work was also supported in part by grant #UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), AI 100663-01 and CHAVI-ID Award UM1AI100663 (to M.C. Nussenzweig), the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (to M.A. Martin), Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery, AI 100148-01 (to P.J. Bjorkman and M.C. Nussenzweig), and National Institutes of Health grant P01 AI100151 and R01 HL59725 (to S. Zolla-Pazner), and research funds from the Department of Veterans Affairs (to S. Zolla-Pazner). P.J. Bjorkman and M.C. Nussenzweig are Howard Hughes Medical Institute Investigators.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- bNAb
- broadly neutralizing antibody
- CD4bs
- CD4 binding site
- NHP
- nonhuman primate
- PNGS
- potential N-linked glycosylation site
- SHIV
- simian–human immunodeficiency virus
References
- Almond D., Kimura T., Kong X., Swetnam J., Zolla-Pazner S., and Cardozo T.. 2010. Structural conservation predominates over sequence variability in the crown of HIV type 1’s V3 loop. AIDS Res. Hum. Retroviruses. 26:717–723 10.1089/aid.2009.0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baenziger S., Tussiwand R., Schlaepfer E., Mazzucchelli L., Heikenwalder M., Kurrer M.O., Behnke S., Frey J., Oxenius A., Joller H., et al. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice. Proc. Natl. Acad. Sci. USA. 103:15951–15956 10.1073/pnas.0604493103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs A.B., Chen J., Hong C.M., Rao D.S., Yang L., and Baltimore D.. 2012. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 481:81–84 10.1038/nature10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S., et al. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 503:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blish C.A., Nguyen M.A., and Overbaugh J.. 2008. Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med. 5:e9 10.1371/journal.pmed.0050009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch K.A., Rainwater S., Jaoko W., and Overbaugh J.. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology. 398:115–124 10.1016/j.virol.2009.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik E.M., Pisas L., van Nuenen A.C., and Schuitemaker H.. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 10.1128/JVI.00757-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke V., Williams C., Sukumaran M., Kim S.S., Li H., Wang X.H., Gorny M.K., Zolla-Pazner S., and Kong X.P.. 2009. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: implications for design of V3-based immunogens. Structure. 17:1538–1546 10.1016/j.str.2009.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Klein R.M., Manion M.M., O’Dell S., Phogat A., Chakrabarti B., Hallahan C.W., Migueles S.A., Wrammert J., Ahmed R., et al. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J. Virol. 83:188–199 10.1128/JVI.01583-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufin T., Gautam R., Kasheta M., Ribeiro R., Ribka E., Barnes M., Pattison M., Tatum C., MacFarland J., Montefiori D., et al. 2009a. Limited ability of humoral immune responses in control of viremia during infection with SIVsmmD215 strain. Blood. 113:4250–4261 10.1182/blood-2008-09-177741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaufin T., Pattison M., Gautam R., Stoulig C., Dufour J., MacFarland J., Mandell D., Tatum C., Marx M.H., Ribeiro R.M., et al. 2009b. Effect of B-cell depletion on viral replication and clinical outcome of simian immunodeficiency virus infection in a natural host. J. Virol. 83:10347–10357 10.1128/JVI.00880-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe C.E., Wrin T., Seaman M.S., Yu X., Wood B., Self S., Williams C., Gorny M.K., and Zolla-Pazner S.. 2010. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE. 5:e10254 10.1371/journal.pone.0010254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz J.A., Halper-Stromberg A., Mouquet H., Gitlin A.D., Tretiakova A., Eisenreich T.R., Malbec M., Gravemann S., Billerbeck E., Dorner M., et al. 2013. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. USA. 110:16538–16543 10.1073/pnas.1315295110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.H., Bonsall D., Katzourakis A., Thomson E.C., Fidler S.J., Main J., Muir D., Weber J.N., Frater A.J., Phillips R.E., et al. 2010. B-cell depletion reveals a role for antibodies in the control of chronic HIV-1 infection. Nat. Commun. 1:102 10.1038/ncomms1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Burke V., Totrov M., Williams C., Cardozo T., Gorny M.K., Zolla-Pazner S., and Kong X.P.. 2010. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 17:955–961 10.1038/nsmb.1861 [DOI] [PubMed] [Google Scholar]
- Klein F., Halper-Stromberg A., Horwitz J.A., Gruell H., Scheid J.F., Bournazos S., Mouquet H., Spatz L.A., Diskin R., Abadir A., et al. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 492:118–122 10.1038/nature11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F., Mouquet H., Dosenovic P., Scheid J.F., Scharf L., and Nussenzweig M.C.. 2013. Antibodies in HIV-1 vaccine development and therapy. Science. 341:1199–1204 10.1126/science.1241144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M., Pancera M., Kwong P.D., Kolchinsky P., Grundner C., Wang L., Hendrickson W.A., Sodroski J., and Wyatt R.. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology. 313:387–400 10.1016/S0042-6822(03)00294-0 [DOI] [PubMed] [Google Scholar]
- Li M., Gao F., Mascola J.R., Stamatatos L., Polonis V.R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K.M., et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 10.1128/JVI.79.16.10108-10125.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly A., and Stamatatos L.. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769–6776 10.1128/JVI.74.15.6769-6776.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey R.A., Saunders C., Hensel M., and Stamatatos L.. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279–3295 10.1128/JVI.78.7.3279-3295.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A.T., Hoot S., Dreyer A.M., Lippy A., Stuart A., Cohen K.W., Jardine J., Menis S., Scheid J.F., West A.P., et al. 2013. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J. Exp. Med. 210:655–663 10.1084/jem.20122824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.J., Genescà M., Abel K., Montefiori D., Forthal D., Bost K., Li J., Favre D., and McCune J.M.. 2007. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J. Virol. 81:5024–5035 10.1128/JVI.02444-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B., Rakasz E.G., Schultz N., Chan-Hui P.Y., Swiderek K., Weisgrau K.L., Piaskowski S.M., Bergman Z., Watkins D.I., Poignard P., and Burton D.R.. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA. 109:18921–18925 10.1073/pnas.1214785109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Klein F., Scheid J.F., Warncke M., Pietzsch J., Oliveira T.Y., Velinzon K., Seaman M.S., and Nussenzweig M.C.. 2011. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS ONE. 6:e24078 10.1371/journal.pone.0024078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H., Scharf L., Euler Z., Liu Y., Eden C., Scheid J.F., Halper-Stromberg A., Gnanapragasam P.N., Spencer D.I., Seaman M.S., et al. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc. Natl. Acad. Sci. USA. 109:E3268–E3277 10.1073/pnas.1217207109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke S.M., Schweighardt B., Phung P., Fonseca D.P., Terry K., Wrin T., Sinangil F., and Berman P.W.. 2010. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J. Virol. 84:11200–11209 10.1128/JVI.00790-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J., Scheid J.F., Mouquet H., Klein F., Seaman M.S., Jankovic M., Corti D., Lanzavecchia A., and Nussenzweig M.C.. 2010. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 207:1995–2002 10.1084/jem.20101176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J., Gruell H., Bournazos S., Donovan B.M., Klein F., Diskin R., Seaman M.S., Bjorkman P.J., Ravetch J.V., Ploss A., and Nussenzweig M.C.. 2012. A mouse model for HIV-1 entry. Proc. Natl. Acad. Sci. USA. 109:15859–15864 10.1073/pnas.1213409109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A., Honnen W.J., He Y., Gorny M.K., Zolla-Pazner S., and Kayman S.C.. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 10.1128/JVI.78.10.5205-5215.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Feldhahn N., Seaman M.S., Velinzon K., Pietzsch J., Ott R.G., Anthony R.M., Zebroski H., Hurley A., et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 458:636–640 10.1038/nature07930 [DOI] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 333:1633–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J.E., Kuroda M.J., Santra S., Simon M.A., Lifton M.A., Lin W., Khunkhun R., Piatak M., Lifson J.D., Grosschupff G., et al. 2003. Effect of humoral immune responses on controlling viremia during primary infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 77:2165–2173 10.1128/JVI.77.3.2165-2173.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G., et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 10.1128/JVI.02108-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M., Donau O.K., Schmidt S.D., Gautam R., Plishka R.J., Buckler-White A., Sadjadpour R., Lee W.R., LaBranche C.C., Montefiori D.C., et al. 2012. Most rhesus macaques infected with the CCR5-tropic SHIV(AD8) generate cross-reactive antibodies that neutralize multiple HIV-1 strains. Proc. Natl. Acad. Sci. USA. 109:19769–19774 10.1073/pnas.1217443109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M., Nishimura Y., Klein F., Mouquet H., Donau O.K., Plishka R., Buckler-White A., Seaman M., Piatak M. Jr, Lifson J.D., et al. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 503:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek M.D., Rida W., Priddy F.H., Pung P., Carrow E., Laufer D.S., Lehrman J.K., Boaz M., Tarragona-Fiol T., Miiro G., et al. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 10.1128/JVI.00110-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C., Forsell M.N., O’Dell S., Feng Y., Chakrabarti B., Rao S.S., Loré K., Mascola J.R., Wyatt R.T., Douagi I., and Karlsson Hedestam G.B.. 2010. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J. Exp. Med. 207:2003–2017 10.1084/jem.20100025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C., Li Y., Huynh N., Poulsen C., Wilson R., O’Dell S., Feng Y., Mascola J.R., Wyatt R.T., and Karlsson Hedestam G.B.. 2012a. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci. Transl. Med. 4:42ra96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C., Phad G., Douagi I., Navis M., and Karlsson Hedestam G.B.. 2012b. Isolation of antibody V(D)J sequences from single cell sorted rhesus macaque B cells. J. Immunol. Methods. 386:85–93 10.1016/j.jim.2012.09.003 [DOI] [PubMed] [Google Scholar]
- Totrov M., Jiang X., Kong X.P., Cohen S., Krachmarov C., Salomon A., Williams C., Seaman M.S., Abagyan R., Cardozo T., et al. 2010. Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp120 V3 loop on a CTB scaffold. Virology. 405:513–523 10.1016/j.virol.2010.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Phogat S.K., Chan-Hui P.Y., Wagner D., Phung P., Goss J.L., Wrin T., Simek M.D., Fling S., Mitcham J.L., et al. Protocol G Principal Investigators. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 326:285–289 10.1126/science.1178746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.M., Huber M., Doores K.J., Falkowska E., Pejchal R., Julien J.P., Wang S.K., Ramos A., Chan-Hui P.Y., Moyle M., et al. Protocol G Principal Investigators. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 477:466–470 10.1038/nature10373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., et al. 2003. Antibody neutralization and escape by HIV-1. Nature. 422:307–312 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- Yang X., Lee J., Mahony E.M., Kwong P.D., Wyatt R., and Sodroski J.. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634–4642 10.1128/JVI.76.9.4634-4642.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.J., Hatziioannou T., Zang T., Braaten D., Luban J., Goff S.P., and Bieniasz P.D.. 2002. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 76:6332–6343 10.1128/JVI.76.12.6332-6343.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S.2005. Improving on nature: focusing the immune response on the V3 loop. Hum. Antibodies. 14:69–72. [PubMed] [Google Scholar]
- Zolla-Pazner S., and Cardozo T.. 2010. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10:527–535 10.1038/nri2801 [DOI] [PMC free article] [PubMed] [Google Scholar]