Ye et al. identify cytosolic carboxypeptidase CCP6 as a protein required for the regulation of bone marrow megakaryopoiesis in mice. The authors find that Mad2 (a core component of spindle checkpoint in mitosis) is a substrate of CCP6 in megakaryocytes and is polyglutamylated by proteins TTLL6 and TTLL4, subsequently affecting the activity of Aurora B kinase. Mad2 is thus additionally implicated in megakaryopoiesis regulation.
Abstract
Bone marrow progenitor cells develop into mature megakaryocytes (MKs) to produce platelets for hemostasis and other physiological functions. However, the molecular mechanisms underlying megakaryopoiesis are not completely defined. We show that cytosolic carboxypeptidase (CCP) 6 deficiency in mice causes enlarged spleens and increased platelet counts with underdeveloped MKs and dysfunctional platelets. The prominent phenotypes of CCP6 deficiency are different from those of CCP1-deficient mice. We found that CCP6 and tubulin tyrosine ligase-like family (TTLL) members TTLL4 and TTLL6 are highly expressed in MKs. We identify Mad2 (mitotic arrest deficient 2) as a novel substrate for CCP6 and not CCP1. Mad2 can be polyglutamylated by TTLL4 and TTLL6 to modulate the maturation of MKs. CCP6 deficiency causes hyperglutamylation of Mad2 to promote activation of Aurora B, leading to suppression of MK maturation. We reveal that Mad2 polyglutamylation plays a critical role in the regulation of megakaryopoiesis.
Megakaryocytes (MKs) are the BM precursors of platelets that are derived from hematopoietic stem cells (HSCs). MKs are a fairly rare subset of nucleated cells in normal BM (Wen et al., 2011; Machlus and Italiano, 2013). During the process of MK development, MK progenitors, including the MK/erythroid progenitor (MEP), burst-forming unit MK (BFU-MK), and CFU-MK undergo proliferation and endomitosis, differentiating into mature polyploid MKs to generate platelets into blood stream (Nakorn et al., 2003; Wen et al., 2011; Boitano et al., 2012; Thon and Italiano, 2012). Platelets are indispensable for the physiological processes such as hemostasis, angiogenesis, wound healing, inflammation, and innate immunity (Wong et al., 2013). Dysplastic megakaryopoiesis is related to the pathogenesis of the myeloproliferative neoplasms (MPNs; Wen et al., 2011). MK hyperproliferation and platelet alteration are the specific features of essential thrombocythemia (ET) and primary myelofibrosis (PMF; Papadantonakis et al., 2012; Thon and Italiano, 2012; Tefferi, 2013). However, the molecular mechanisms underlying megakaryopoiesis are incompletely defined.
Endomitosis, a modified form of mitosis, is one of the most characteristic features in MK maturation (Gao et al., 2012). During the endomitotic process, the DNA is repeatedly replicated without a cytoplasmic division, resulting in 8N to 128N polyploidy of MKs. The spindle assembly checkpoint (SAC) plays a critical role in the regulation of DNA polyploidization. Mad2 is one of the core conserved components of the spindle checkpoint in mitosis. Mad2 binds directly to Cdc20, together with other Mad and Bub family partners, monitoring the spindle organization and inhibiting the onset of anaphase in a cell cycle (Shah and Cleveland, 2000; Chao et al., 2012). Mad2 deletion causes a premature onset of anaphase leading to increased polyploidy and aneuploidy (Michel et al., 2001). Besides its checkpoint function, Mad2 was reported to be involved in the regulation of kinetochore-microtubule (k-MT) attachment stabilization and Aurora B activity. Mad2 depletion significantly increases the quantity of Aurora B to the centromere and promotes the kinase activity of Aurora B (Kabeche and Compton, 2012). Aurora B kinase activity is required for cytokinesis and mitosis. A recent report showed that Aurora B exerts a crucial role in the endomitosis of MKs (Lordier et al., 2010). MK-specific Aurora B transgenic mice display an elevated MK number and mildly decreased DNA ploidy (Zhang et al., 2004).
Polyglutamylation is a unique posttranslational modification that gives rise to variable lengths of glutamate side chains onto the γ-carboxyl groups of glutamic acid residues in the primary sequence of target proteins (Janke and Bulinski, 2011). The well-known targets of polyglutamylation are tubulins and nucleosome assembly proteins (NAPs; Eddé et al., 1990; Regnard et al., 2000). Polyglutamylation modification on tubulins has been defined to regulate the interaction between MTs and their partners (Sirajuddin et al., 2014), modulating MT-related processes such as stability of centrosomes (Bobinnec et al., 1998), motility of cilia and flagella (Gagnon et al., 1996; Kubo et al., 2010; Suryavanshi et al., 2010; Bosch Grau et al., 2013), neurite outgrowth (Ikegami et al., 2006), and neurodegeneration (Rogowski et al., 2010). Polyglutamylation is catalyzed by polyglutamylases, members of the tubulin tyrosine ligase-like (TTLL) enzyme family (Janke et al., 2005; van Dijk et al., 2007), which are restrictedly expressed in specific tissues or cell types.
Deglutamylation is hydrolyzed by a family of cytosolic carboxypeptidases (CCPs; Kalinina et al., 2007; Rodriguez de la Vega et al., 2007; Rogowski et al., 2010). Purkinje cell degeneration (pcd) mice have several mutations of CCP1 gene (Fernandez-Gonzalez et al., 2002). WT CCP1 can rescue the Purkinje cell loss and retinal degeneration appeared in the pcd mice (hereafter also referred to as CCP1−/− mice; Wang et al., 2006; Chakrabarti et al., 2008). A recent study showed that CCP1 specifically removes the penultimate glutamate chains from the detyrosinated α-tubulin (Rogowski et al., 2010). The other two family members, CCP6 and CCP4, display the similar hydrolytic activities to α-tubulin as CCP1. Here we show that CCP6-deficient mice display underdeveloped MKs and abnormal thrombocytosis, which are different from CCP1-deficient mice. Mad2 is a novel substrate for CCP6 in MKs. Mad2 polyglutamylation plays a crucial role in the regulation of megakaryopoiesis.
RESULTS
CCP6 deficiency results in splenomegaly and an increased number of dysfunctional platelets
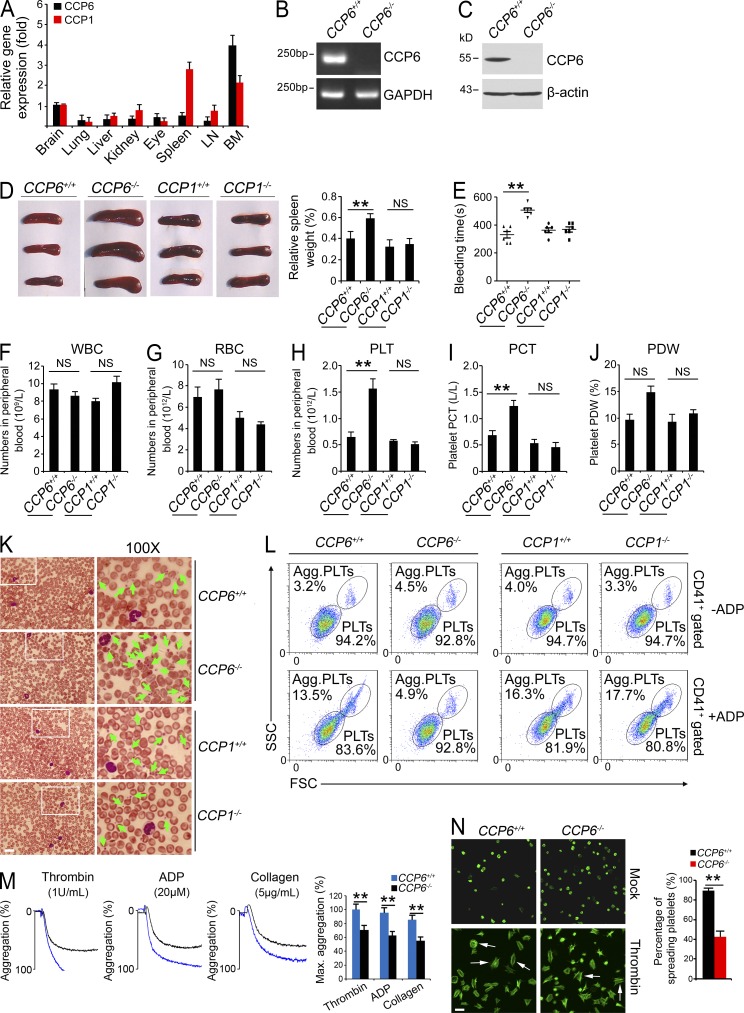
CCP6 was recently defined to hydrolyze the penultimate glutamate chains from detyrosinated α-tubulin, and the gene-coded glutamates of MLCK1 and telokin at their C termini, which is reminiscent of its CCP family members CCP1 and CCP4 (Rogowski et al., 2010). Controlling the length of polyglutamate chains on tubulin mediated by polyglutamylases and deglutamylating enzymes plays an important role in neuronal survival. To examine other roles of CCP6 in addition to the regulation of neuronal survival, we detected expression patterns of CCP6 in different mouse tissues. We found that CCP6 was most highly expressed in BM (Fig. 1 A), whereas CCP6 and CCP1 had distinct expression patterns. Other independent primer pairs for CCP6 or CCP1 obtained similar gene expression trends (unpublished data). The high gene expression of CCP6 in BM implies that CCP6 may play a role in hematopoiesis.
Figure 1.
Deficiency in CCP6 and not in CCP1 results in splenomegaly and increased numbers of dysfunctional platelets. (A) CCP6 and CCP1 total RNA was extracted from different tissues and analyzed by real-time qPCR. Relative fold of gene expression values was normalized to endogenous β-actin. Primer pairs are shown in Materials and methods. Results are shown as means ± SD. n = 5. (B and C) BM CCP6 levels were measured by RT-PCR (B) and immunoblotting (C). GAPDH and β-actin were used as loading controls, respectively. (D) Photographs of spleens (left) and relative weight of spleens (right) were obtained. Results are shown as means ± SD. **, P < 0.01. n = 15 for each group. (E) Bleeding times. Results are shown as means ± SD. n = 6. **, P < 0.01. (F–H) Peripheral blood cell counts were obtained for CCP6-deficient, CCP1-deficient, and littermate control mice. F, WBCs; G, RBCs; H, platelets (PLT). (I and J) PCT and PDW were analyzed. For F–J, results are shown as means ± SD, n = 10. **, P < 0.01. (K) Representative peripheral blood smears with Wright’s staining. Magnified images were visualized using an oil-immersion 100× lens. White boxes in left panels are shown in right panels. Bar, 20 µm. Green arrowheads denote platelets. Data were repeated five times with similar results. (L) Mouse CD41+ platelets were stimulated with or without ADP treatment and analyzed by flow cytometry. n = 4 for each group. (M) ADP, thrombin, and collagen-stimulated platelet aggregations were measured by aggregometry in CCP6+/+ and CCP6−/− mice. Typical results for percentages of aggregation (%) are shown and maximum percentages of aggregation (Max. aggregation %) of each condition were calculated. n = 6 for each group. Results are shown as means ± SD. **, P < 0.01. (N) Platelets were treated with thrombin and placed on poly-l-lysine–coated slides for spreading. Representative confocal immunofluorescence images are shown on the left. Bar, 5 µm. White arrows denote typical spreading platelets. Percentages of spreading platelets in different fields were calculated and shown on right as means ± SD. **, P < 0.01. All data are representative of at least four independent experiments.
To further determine the physiological role of CCP6, CCP6-deficient (CCP6−/−) mice of the FVB strain were generated through a transposon mutagenesis strategy (Ding et al., 2005). The CCP6−/− mice were gene-specific insertion mutations (Sun et al., 2008; Yang et al., 2009). Consistently, CCP6 was successfully deleted in CCP6−/− mice (Fig. 1, B and C). Interestingly, CCP6-deficient mice had no obvious phenotypes in neural degeneration or motor skill disorders (unpublished data), which were totally different from those of CCP1-deficient mice (also known as pcd mice). CCP1 was completely deleted in CCP1−/− mice (unpublished data). Surprisingly, CCP6−/− mice displayed increased weight of spleens (Fig. 1 D), defective hemostasis, and significantly elongated bleeding times compared with the littermate control mice (Fig. 1 E). However, CCP1-deficient mice showed a normal size of spleens and normal bleeding times comparable to the WT CCP1+/+ mice.
Prolonged bleeding times may be due to a decreased platelet count and/or platelet dysfunction. Actually, we found that CCP6−/− mice had similar counts of WBCs and RBCs, comparable to the littermate control mice (Fig. 1, F and G; and Table 1). Similarly, CCP1−/− mice also showed normal counts of WBCs and RBCs compared with the WT mice. However, CCP6−/− mice displayed an about threefold increase in platelet counts compared with the littermate control mice (Fig. 1 H). In contrast, CCP1-deficient mice showed a similar platelet number to WT mice. Moreover, CCP6-deficient mice had significantly elevated plateletcrit (PCT) and platelet distribution width (PDW) compared with the littermate control mice (Fig. 1, I and J; and Table 1). Similar results were observed from peripheral blood smears with Wright’s staining (Fig. 1 K). In contrast, CCP1−/− mice showed similar platelet counts and normal morphology compared with WT mice (Fig. 1, H–K).
Table 1.
Hematopoietic parameters
| Mouse | Parameter | ||||
| WBC | RBC | PLT | PCT | PDW | |
| ×103/µl | ×106/µl | ×103/µl | liter/liter | % | |
| CCP6+/+ (n = 10) | 9.3 ± 0.7 | 7.0 ± 0.9 | 645.0 ± 43.7 | 0.7 ± 0.1 | 9.6 ± 1.1 |
| CCP6−/− (n = 10) | 8.6 ± 0.5 | 7.7 ± 0.9 | 1,567.3 ± 180.4 | 1.2 ± 0.1 | 14.8 ± 1.2 |
| CCP1+/+ (n = 10) | 8.0 ± 0.3 | 4.9 ± 0.6 | 582.8 ± 16.8 | 0.5 ± 0.1 | 9.3 ± 1.4 |
| CCP1−/− (n = 10) | 10.2 ± 0.7 | 4.4 ± 0.3 | 510.5 ± 48.0 | 0.5 ± 0.1 | 10.8 ± 0.7 |
Blood samples were analyzed using an XFA6030 automated hemocytometer (Slpoo). Cell numbers and percentages were counted for each population. Data are shown as means ± SD. PLT, platelet.
In vitro platelet function was assayed by analyzing aggregation of activated platelets in response to adenosine-5′-diphosphate (ADP) stimulation (Smith et al., 2012). In a resting status, CCP6−/− platelets showed normal forward scatter (FSC) and side scatter (SSC) compared with the littermate control mice (Fig. 1 L, top). After ADP stimulation, platelets from CCP6+/+ mice were activated and remarkably aggregated with bigger particle sizes out of the FSC/SSC gate (Fig. 1 L, bottom). However, CCP6−/− platelets did not display apparent aggregation in response to ADP treatment (Fig. 1 L). In contrast, platelet function of CCP1−/− mice appeared to be normal comparable to WT CCP1+/+ mice (Fig. 1 L). Similarly, CCP6−/− platelets exhibited impaired aggregation compared with those of CCP6+/+ mice by assays of several agonists (Fig. 1 M). Additionally, CCP6−/− platelets showed defective spreading compared with those of WT mice (Fig.1 N). To address the morphological change of dysfunctional platelets, we performed ultrastructural analysis of CCP6−/− platelets through electron microscopy. WT platelets showed easily identified dense α-granules, whereas CCP6−/− platelets had significantly declined numbers of α-granules and with many vacuoles (unpublished data). These data indicate that CCP6−/− platelets have abnormal morphology and lack normal hemostatic function.
Furthermore, we examined blood clotting tetrachoric analyses of CCP6-deficient mice and littermate controls. There were no significant defects of coagulation factors and fibrinogens in CCP6-deficient mice (unpublished data). Collectively, CCP6 deficiency causes an increase number of dysfunctional platelets.
CCP6 deficiency causes abnormal megakaryopoiesis
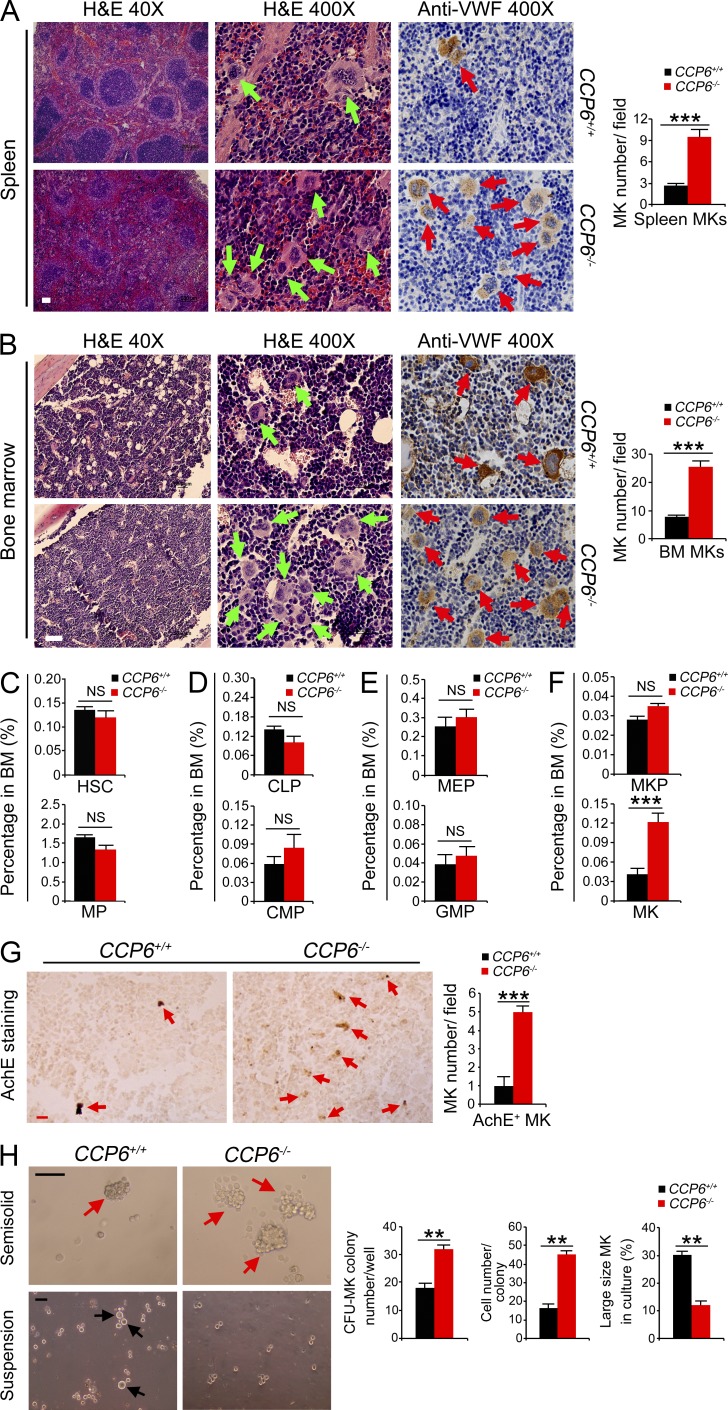
As described above, CCP6-deficient adult mice displayed enlarged spleens. We further observed that CCP6-deficient mouse spleen displayed disordered architecture with apparently extramedullary myelopoiesis (Fig. 2 A). CCP6-deficient mice displayed remarkably increased MKs in spleens. Importantly, MK numbers were also significantly increased in the BM of CCP6-deficient mice (Fig. 2 B). Additionally, CCP6-deficient MKs exhibited smaller sizes with weaker VWF signals than those of the littermate control MKs (Fig. 2, A and B), suggesting that CCP6-deficient mice produce underdeveloped MKs. In contrast, CCP1-deficient mice had similar counts and morphology of MKs comparable to WT mice (unpublished data).
Figure 2.
Deficiency in CCP6 and not in CCP1 causes abnormal megakaryopoiesis. (A and B) Paraffin sections from spleen and femurs of 6-wk-old mice were stained with H&E or anti-VWF antibody (brown)/hematoxylin (blue) by IHC. Bar, 100 µm. Green arrows in left panels and red arrows in right panels denote MKs. MKs in different fields (n = 20 fields) were enumerated (right graphs). Results are shown as means ± SD. ***, P < 0.001. (C) Flow cytometry analysis of HSCs (Lin−Sca-1+c-Kit+/LSK) and myeloid progenitors (Lin−c-Kit+Sca-1−) from BM. (D) Flow cytometry analysis of CLPs (Lin−CD127+Sca-1lowc-Kitlow) and CMPs (Lin−c-Kit+Sca-1−CD34+CD16/32−) from BM. (E) Flow cytometry analysis of MEPs (Lin−c-Kit+Sca-1−CD34−CD16/32−) and GMPs (Lin−c-Kit+Sca-1−CD34+CD16/32+) from BM. (F) Flow cytometry analysis of MKPs (Lin−c-Kit+CD41+) and MKs (Lin−c-Kit−CD41+) from BM. n = 10 for each group. Results are shown as means ± SD. ***, P < 0.001. (G) Paraffin sections of BM-containing MKs were stained for AchE by IHC (left). AchE+ cells represent all MKs. Red arrows denote MKs. Data are representative of at least three separate experiments. Bar, 20 µm. Total numbers of AchE+ cells were enumerated in the right graph (n = 20 fields). Results are shown as means ± SD. ***, P < 0.001. (H) In vitro MK development assays from sorted BM MKPs using MethoCult semisolid media for CFU-MK formation (top left) and liquid suspension media for large size MK maturation (bottom left). Representative microscopy images are shown (left). Bar, 100 µm. Red arrows in top left denote CFU-MK colony and black arrows in bottom left denote large size MKs. 1 × 102 sorted MKPs from BM of the indicated mice were mixed with 1 ml MethoCult media and plated on a 35-mm dish for incubation. MK colonies were enumerated and the colony types of MKs were confirmed by AchE staining. CFU-MK assays from total BM cells (5 × 104) were also performed and displayed similar results (not depicted). Data are representative of at least three separate experiments. n = 5 for each group. Cell numbers were enumerated as specified (right graphs). Results are shown as means ± SD. **, P < 0.01.
Underdeveloped MKs and platelet defects suggest abnormal megakaryopoiesis in CCP6-deficient mice. We found that CCP6-deficient mice showed normal numbers of HSCs, MPs (myeloid progenitors), CLPs (common lymphoid progenitors), and CMPs (common myeloid progenitors) compared with the littermate control mice (Fig. 2, C and D; and not depicted). Moreover, CCP6-deficient mice exhibited similar numbers of GMPs (granulocyte/macrophage progenitors) and MEPs compared with the littermate control mice (Fig. 2 E and not depicted). However, CCP6-deficient mice had significantly increased CD41+ MKs (Fig. 2 F). BM acetylcholinesterase (AchE) staining recognizes all mature and immature MKs (Ichikawa et al., 2004; Smith et al., 2012). As expected, CCP6-deficient mice had around a fivefold increase of AchE+ cells in BM sections (Fig. 2 G). However, CCP1-deficient mice had no apparent abnormality in dispositions of blood progenitors or MKs (unpublished data). We next isolated MKPs from CCP6−/− and CCP1+/+ mice followed by an in vitro MKP development assay. Isolated MKPs were incubated with thrombopoietin (TPO) to induce MK maturation. We found that CCP6−/− MKPs displayed higher MK colony forming ability than those of CCP6+/+ mice (Fig. 2 H). MK colonies from CCP6−/− mice contained much more CFU-MK cells, suggesting enhanced proliferation rates of CCP6−/− MKs. With suspension culture in the presence of TPO, CCP6−/− MKPs developed fewer large-size MKs, indicating immaturity of these cells. Overall, these data suggest that CCP6 plays a critical role in the maturation of MKs in BM.
CCP6 deficiency leads to accumulation of immature MKs in BM
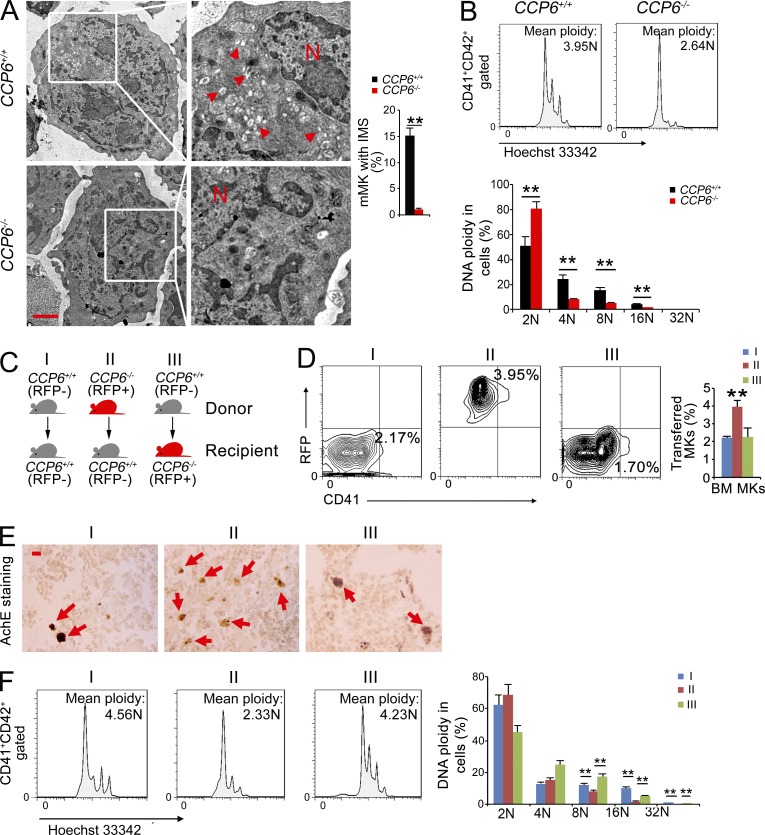
Platelets are generated from their precursor MKs, which reside in the BM of adult mice (Thon and Italiano, 2012; Machlus and Italiano, 2013). Underdeveloped MKs and platelet defect in CCP6-deficient mice suggest abnormal megakaryopoiesis in BM. To further examine defective MKs in BM, MK ultrastructures were analyzed by transmission electron microscopy. CCP6-deficient MKs displayed a poorly developed invaginated membrane system (IMS; also termed DMS [demarcation membrane system]; Fig. 3 A), indicating immaturity of the CCP6−/− MKs. Furthermore, CCP6-deficient MKs displayed much lower DNA ploidy compared with the CCP6+/+ mice (Fig. 3 B). CD42 is a later marker of MKs (Poirault-Chassac et al., 2010). CD42 was significantly declined in CCP6-deficient MKs compared with those of the littermate control mice (unpublished data). Additionally, CCP6-deficient MKs exhibited much smaller sizes than those of the littermate control mice (unpublished data). These results indicate that CCP6 deficiency suppresses the maturation of MKs.
Figure 3.
CCP6 deficiency causes accumulation of immature MKs in the BM. (A) Representative electron micrographs of BM mMKs (left). N: nucleus; arrowheads denote IMS. Bar, 2 µm. mMKs with IMS in different fields were enumerated (right graphs). n ≥ 300 MKs were analyzed for each group. Data were repeated three times with similar results. Results are shown as means ± SD. **, P < 0.01. (B) DNA polyploidy was analyzed by DNA contents of CD41+CD42+ MKs using flow cytometry (top). Percentages of DNA ploidy are shown as means ± SD. **, P < 0.01 (bottom). n = 6 for each group. Data were repeated three times with similar results. (C) Schematic representation of transplantation. BM MKs and peripheral blood platelets of recipient mice were analyzed 4 wk after transplantation. (D) Flow cytometry analysis of CD41+ MKs from the BM of donor mice (left). Percentage of MKs are shown (right graph). Results are shown as means ± SD. n = 6 for each group. **, P < 0.01. (E) Paraffin sections of donor BM containing MKs were stained for AchE. Bar, 20 µm. Red arrows denote AchE+ MKs. Data were repeated three times with similar results. (F) DNA polyploidy of CD41+CD42+ MKs from the BM of donor mice by flow cytometry (left). n = 6 for each group, results are shown as means ± SD. **, P < 0.01 (right). Data are representative of at least four separate experiments.
We next wanted to determine whether CCP6 deficiency-mediated platelet dysfunction is intrinsic or extrinsic. We transplanted BM cells from donor mice (RFP− for control and RFP+ for CCP6-deficient mice) into irradiated recipient mice (Fig. 3 C). BM MKs and peripheral blood platelets of recipient mice were analyzed 4 wk after transplantation. CCP6-deficient cells transplanted into the irradiated recipient mice resulted in an increase number of CD41+ MKs (Fig. 3 D), which was verified by AchE staining (Fig. 3 E). Additionally, CCP6-deficient cell transplantation led to lower DNA ploidy of CD41+ MKs in BM (Fig. 3 F). Moreover, this transplantation caused increased platelet counts and dysfunction of platelets (unpublished data). However, WT cells transplanted into CCP6-deficient mice rescued megakaryopoiesis and platelet function in the recipient CCP6-deficient mice (Fig. 3, D–F). Collectively, CCP6 is an intrinsic factor in the regulation of MKs in BM.
Mad2 is a novel substrate for CCP6 in MKs
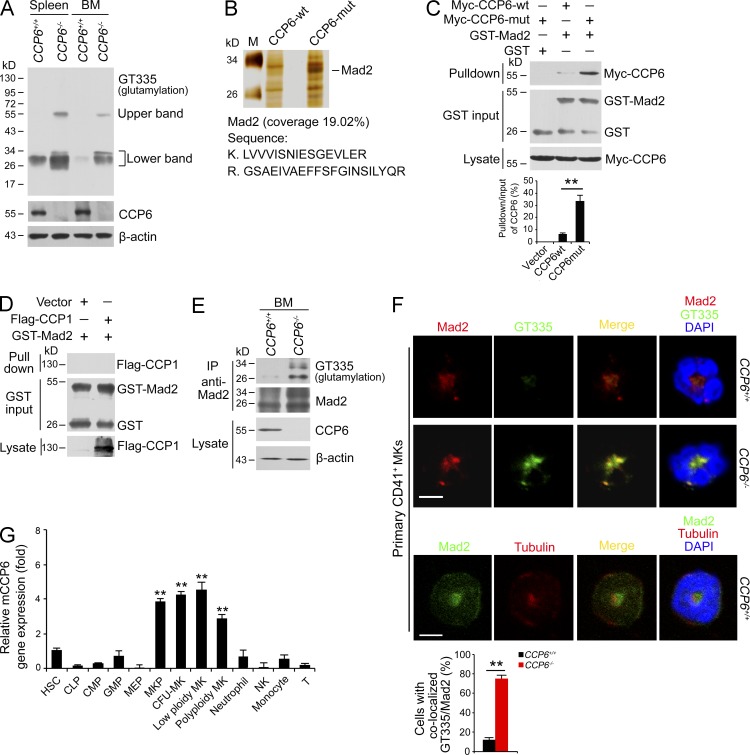
CCP6 was reported to be a CCP that catalyzes the removal of polyglutamate chains of its substrate α-tubulin (Rogowski et al., 2010). To further explore the molecular mechanism of CCP6-mediated immaturity of MKs, we analyzed lysates of BM and spleen from CCP6-deficient mice and littermate controls by immunoblotting with a polyglutamylation-specific antibody GT335. GT335 specifically recognizes the branching point of glutamate side chains and detects all glutamylated forms of target proteins (Wolff et al., 1992). Two blot bands around 55 and 30 kD appeared in the lanes of CCP6-deficient spleen and BM lysates (Fig. 4 A). These two blot bands were undetectable in the corresponding lane locations from the littermate WT control spleen and BM lysates. These observations suggest that the two bands may be potential candidate substrates for CCP6. To identify the candidate substrates of CCP6, we generated an enzymatically inactive mutant of CCP6 (CCP6-mut) through H230S and E233Q mutations as previously described (Rogowski et al., 2010). WT CCP6 (CCP6-wt) and CCP6-mut were immobilized with Affi-gel10 resin to go through mouse BM lysates for affinity chromatography. The eluted fractions were visualized by SDS-PAGE followed by silver staining. These two bands appeared in the CCP6-mut gel and were cut for mass spectrometry. The upper 55 kD band was identified to be α-tubulin, a known substrate for CCP6 as previously described (Rogowski et al., 2010). Interestingly, the lower 30 kD band was Mad2 (Fig. 4 B and not depicted), a novel candidate substrate for CCP6.
Figure 4.
Mad2 is a new substrate for CCP6 in MKs. (A) Protein polyglutamylation was assessed by immunoblotting with GT335 antibody. Spleen and BM lysates from CCP6-deficient and littermate control mice were analyzed. Data were repeated three times with similar results. (B) Recombinant CCP6-wt and inactive CCP6 mutant (CCP6-mut) were immobilized with Affi-gel10 resin, to which mouse BM lysates were added for affinity chromatography. The eluted fractions were visualized by SDS-PAGE, followed by silver staining. M: molecular weight marker. The differential band of 30 kD seen in CCP6-mut gel was cut for mass spectrometry and identified as Mad2. The peptide sequences and coverage analyzed by LC-LTQ MS/MS are shown in the bottom graph. (C) Glutamylated rGST-Mad2 binding to CCP6-mut protein was analyzed by GST pulldown. rGST-Mad2 protein was incubated with lysates from Myc-tagged CCP6-wt or Myc-tagged CCP6-mut expressed 293T cells at 37°C for 2 h, followed by incubation with GST beads. Ratios of pulldown/input of CCP6 were calculated and shown as means ± SD. **, P < 0.01 (bottom graph). (D) Glutamylated rGST-Mad2 binding to CCP1-mut protein was analyzed by GST-pulldown, rGST-Mad2 was incubated with lysates from Flag-tagged CCP1-mut expressed 293T cells as above. (E) Assessment of Mad2 hyper-polyglutamylation in BM. BM lysates were immunoprecipitated with anti-Mad2 antibody followed by immunoblotting with the indicated antibodies. (F) Assessment of Mad2 colocalization with GT335 or tubulin in BM MKs. GT335, top; tubulin, middle. Primary CD41+ MKs were sorted from BM followed by immunofluorescence staining. Representative confocal microscopy images are shown. Mad2 or tubulin, red; GT335 or Mad2, green; nucleus, blue. Bars, 5 µm. More than 50 mitotic MKs were analyzed. GT335-Mad2 colocalization was measured (bottom graph). Pearson’s correlation coefficient (PCC) between Mad2 and GT335 was calculated by ImageJ for each cell and colocalization was identified as PCC rate ≥ 0.5. Percentages of colocalized cells were counted as means ± SD. **, P < 0.01. (G) RNA expression fold of mCCP6 in blood cells was measured by real-time qPCR. Relative gene expression fold change were counted as means ± SD. **, P < 0.01. All data represent at least three independent experiments.
The association of CCP6 and Mad2 was verified in CCP6-mut and Mad2 cotransfected 293T cells by a coimmunoprecipitation assay (unpublished data). Moreover, glutamylated rGST-Mad2 protein could pull down Myc-tagged CCP6 (Fig. 4 C). Importantly, the glutamylated rGST-Mad2 bound to much more enzymatic dead CCP6-mut protein (Fig. 4 C). However, rGST-Mad2 failed to pull down enzymatic dead CCP1-mut (CCP1-H912S/E915Q) protein (Fig. 4 D). These results indicate that Mad2 is a substrate for CCP6 but not for CCP1. Additionally, Mad2 was highly polyglutamylated in the BM of CCP6-deficient mice (Fig. 4 E). With immunofluorescence staining, hyperglutamylation of Mad2 appeared in primary CCP6-deficient MKs (Fig. 4 F). However, Mad2 was not colocalized with tubulin in primary CCP6-deficient MKs. Accordingly, CCP6 exhibited peak expression in all the stages of MKs among the detected blood cells (Fig. 4 G). Altogether, Mad2 is a substrate for CCP6 in MKs.
Mad2 is polyglutamylated at Glu168 by TTLL6 and TTLL4 in MKs
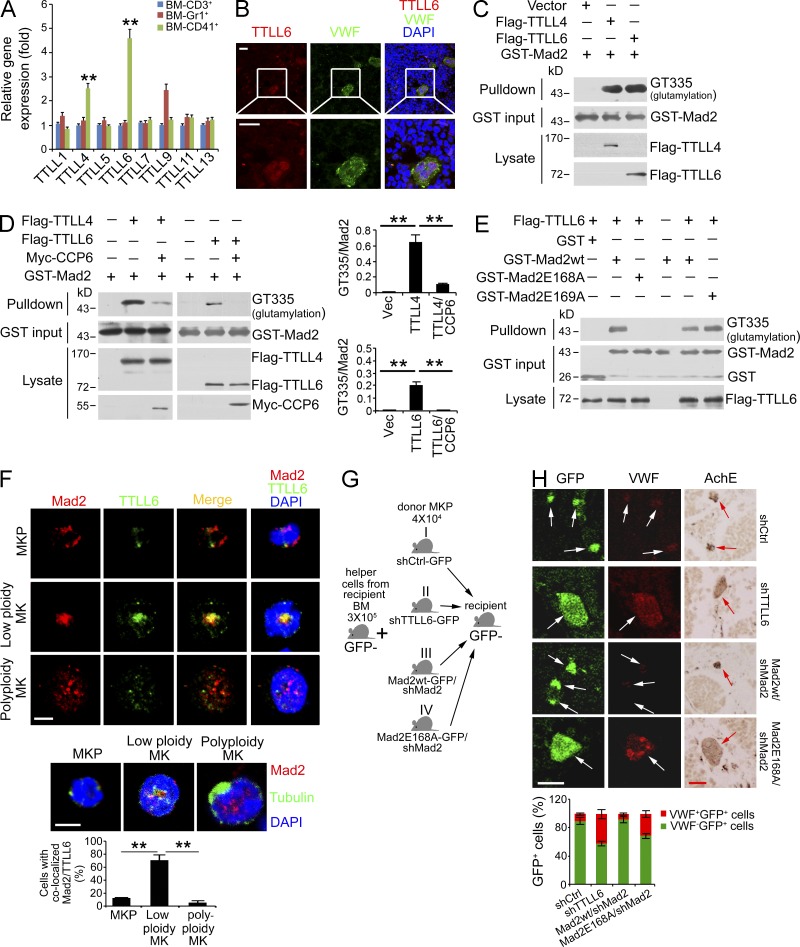
13 polyglutamylases (TTLL1-13) have been identified to date (van Dijk et al., 2007; Rogowski et al., 2010). Among them, only eight TTLLs were reported to glutamylate target proteins (van Dijk et al., 2007; Wasylyk et al., 2010). To determine the physiological polyglutamylases catalyzing Mad2 glutamylation in MKs, we analyzed all eight TTLLs correlated to protein glutamylation in primary BM cells by quantitative real-time PCR. We found that TTLL4 and TTLL6 were highly expressed in primary mouse MKs (Fig. 5 A), whereas TTLL6 was most highly expressed in mouse MKs. These gene expression patterns of TTLLs were confirmed by another separate primer pair (unpublished data). Similar gene expression patterns of TTLLs were also observed in human BM CD41+ MKs (unpublished data). Notably, CCP6 was also highly expressed in human BM CD41+ MKs (unpublished data), displaying a similar trend to that of mice. TTLL6 expression in primary MKs was further confirmed by immunofluorescence staining (Fig. 5 B).
Figure 5.
Mad2 is polyglutamylated by TTLL4 and TTLL6 in MKs. (A) TTLL4 and TTLL6 expression in BM MKs. mRNAs were analyzed by real time qPCR. n = 6. Relative gene expression folds were normalized to endogenous β-actin. Primer pairs are shown in Materials and methods. Relative gene expression fold change was counted as means ± SD. **, P < 0.01. (B) Representative confocal images of immunofluorescence staining of TTLL6 and VWF in BM. Bars, 20 µm. White boxes in top panels are shown in bottom panels. (C) Assessment of rGST-Mad2 polyglutamylation by both TTLL4 and TTLL6 in GST pulldown assays. (D) Assessment of CCP6-mediated rGST-Mad2 deglutamylation by TTLL4 or TTLL6 in GST pulldown assays (left). Ratios of GT335/Mad2 were calculated and shown as means ± SD. **, P < 0.01 (right graphs). (E) Assessment of Mad2 polyglutamylation by TTLL6 at Glu168. 293T cells were transfected with Flag-TTLL6 and lysates were incubated with GST-Mad2 or GST-Mad2 mutant at 37°C for 2 h for in vitro glutamylation followed by GST pulldown assays. (F) Assessment of Mad2 colocalization with TTLL6 or tubulin in low ploidy MKs. Primary mouse MKPs were isolated and cultured for maturation as described in Materials and methods. Representative confocal images are shown (top). Bars, 5 µm. Mad2, red; nucleus, blue; TTLL6, green (top); tubulin, green (middle). For each staining, >50 typical cells were enumerated. Pearson’s correlation coefficient (PCC) between Mad2 and TTLL6 of each cell was calculated by ImageJ and colocalization was identified as PCC rate ≥ 0.5. Percentages of colocalized cells were counted as means ± SD (bottom). **, P < 0.01. (G) Schematic representation of transplantation. (H) Primary mouse MKP cells were sorted. TTLL6 or Mad2 was silenced (shTTLL6 or shMad2). shMad2 silenced MKPs were then rescued with Mad2-wt (Mad2-wt/shMad2) or Mad2E168A (Mad2E168A/shMad2) by retrovirus infection and transplanted into recipients along with helper cells (n = 6). After 7 d, GFP+ and VWF+ MKs derived from donor cells were analyzed by immunofluorescence (top left). AchE staining was also analyzed in separate sections for MK verification (top right). Representative microscopy images are shown. Bars, 20 µm. White and red arrows denote MKs. Percentages of transplanted immature MKs (GFP+ VWF−, small in size) and mature MKs (GFP+ VWF+, large in size) were calculated and shown as means ± SD (bottom). All data are representative of at least four separate experiments.
We next incubated rGST-Mad2 with Flag-tagged TTLL4 or TTLL6 in vitro. We found that rGST-Mad2 was polyglutamylated by both TTLL4 and TTLL6 (Fig. 5 C). Importantly, TTLL6- or TTLL4-mediated Mad2 polyglutamylation was hydrolyzed by CCP6 (Fig. 5 D). Glutamate-rich stretches and acidic environment at the acceptor sites were reported to be important for glutamylation modification (van Dijk et al., 2008). Based on the conservative amino acid sequence analysis, Mad2 has 14 conserved glutamic acid residues, which were potential acceptor site candidates for glutamylation (unpublished data). Among them, only Glu168 and Glu169 were the two identical glutamic residues localized on the flexible loop of Mad2 (unpublished data). From analysis of the Mad2 structural feature, the flexible loop was susceptible to be modified for glutamylation. We then mutated these two glutamic residues and incubated Mad2-E168A or Mad2-E169A with Flag-tagged TTLL6 in vitro. We found that E168A mutation of Mad2 (Mad2-E168A) abolished TTLL6-mediated Mad2 polyglutamylation (Fig. 5 E), whereas Mad2-E169A mutant was still glutamylated. Glu168 was also the receptor residue for TTLL4-mediated Mad2 polyglutamylation (unpublished data). These data indicate that Mad2 is polyglutamylated by TTLL6 and TTLL4 at Glu168.
We next explored the physiological relevance of Mad2 polyglutamylation in MK maturation. We found that TTLL6 was mainly expressed in low ploidy MKs and colocalized with Mad2 (Fig. 5 F). Significant colocalization of Mad2 with TTLL6 was detected in 37 out of 50 low ploidy MKs. However, TTLL6 was weakly expressed in MKPs or large MKs. The anti-TTLL6 antibody we used specifically recognized mouse TTLL6 protein (unpublished data). In contrast, Mad2 was not overlaid with tubulin in the different stages of MKs (Fig. 5 F). TTLL6 and Mad2 were successfully knocked down in freshly isolated mouse MKPs by LMP retrovirus infection (shTTLL6 or shMad2; unpublished data). Additionally, in Mad2 silenced MKPs, we rescued with Mad2-wt (Mad2-wt/shMad2) or Mad2-E168A (Mad2-E168A/shMad2), respectively. Above-treated MKPs were transplanted into irradiated recipient mice together with helper cells (Fig. 5 G). Consequently, TTLL6 depletion significantly promoted MK maturation (Fig. 5 H and not depicted). Rescue of Mad2-wt did not induce MK maturation, whereas restoration of Mad2-E168A mutant was able to enhance MK maturity. In summary, Mad2 polyglutamylation plays a crucial role in the regulation of MK maturation.
CCP6 deficiency causes hyperglutamylation of Mad2 to promote Aurora B activation leading to suppression of MK maturation
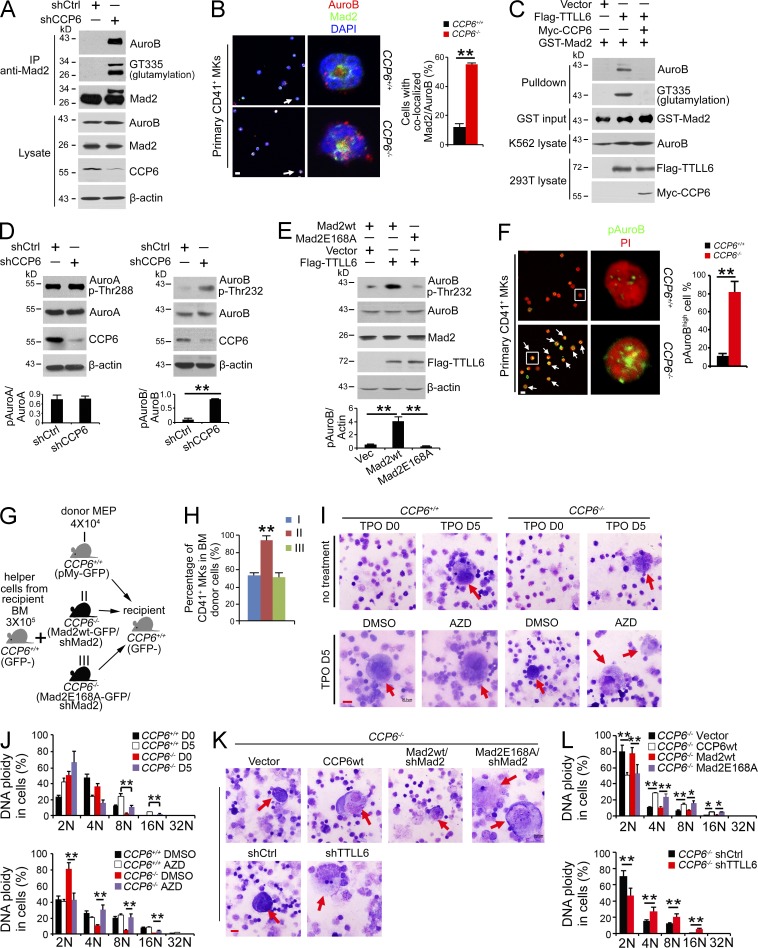
Mad2 was reported to stabilize k-MT attachment and Aurora B activation (Kabeche and Compton, 2012). Aurora B is required for the endomitosis and maturation of MKs. Aurora B transgenic mice display an increased MK number and decreased DNA ploidy (Zhang et al., 2004). The prominent phenotypes of CCP6-deficient mice were reminiscent of those of Aurora B transgenic mice. Interestingly, we observed that CCP6 knockdown dramatically induced Mad2 polyglutamylation (Fig. 6 A), which enhanced its interaction with Aurora B. Moreover, we found that CCP6 deficiency augmented colocalization of Mad2 with Aurora B in primary MKs (Fig. 6 B). Importantly, polyglutamylated Mad2 associated with Aurora B by an in vitro assay (Fig. 6 C). However, CCP6 treatment abrogated the interaction of Mad2 with Aurora B. These results indicate that Mad2 polyglutamylation is involved in its interaction with Aurora B.
Figure 6.
CCP6 deficiency causes Mad2 hyperglutamylation, promoting Aurora B hyperactivation with generation of dysplastic MKs. (A) Assessment of Mad2 polyglutamylation and association with Aurora B upon CCP6 silencing (shCCP6) in an immunoprecipitation assay (shCtrl as control). K562 lysates were immunoprecipitated with anti-Mad2 antibody followed by immunoblotting. (B) Assessment of Mad2 colocalization with Aurora B (AuroB) in primary CD41+ MKs. Representative confocal images are shown (left: Aurora B, red; Mad2, green; nucleus, blue. Bar, 5 µm). 50 primary CD41+ cells were analyzed for each condition. In left panels, arrows point to cells shown in right panels. White arrows denote colocalization of Mad2 with Aurora B. Pearson’s correlation coefficient (PCC) between Mad2 and Aurora B of each cell was calculated by ImageJ and colocalization was identified as PCC rate ≥ 0.5. Percentages of colocalized cells are shown as means ± SD. **, P < 0.01 (right graph). (C) Assessment of polyglutamylated Mad2 association with Aurora B by GST pulldown assays. rGST-Mad2 was incubated with lysates from 293T cells transfected with Flag-TTLL6 and Myc-CCP6 at 37°C for 2 h, followed by GST pulldown in K562 cell lysates. (D) Assessment of Thr232 phosphorylation of Aurora B and Thr288 phosphorylation of Aurora A upon CCP6 silencing (shCCP6; shCtrl as control). K562 lysates were analyzed by immunoblotting with anti-Aurora B, anti–Thr232-Aurora B, anti–Aurora A, and anti–Thr288-Aurora A antibodies (top). Ratios of pThr/Aurora kinase were calculated and shown as means ± SD. **, P < 0.01 (bottom graph). (E) Assessment of Thr232 phosphorylation of Aurora B in the presence of an E168A Mad2 mutant. Flag-TTLL6, pMY-Mad2-wt and pMY-Mad2-E168A vectors were transfected into 293T cells, followed by immunoblotting (top). Ratios of pAurora B/actin were calculated and shown as means ± SD. **, P < 0.01 (bottom graph). (F) Assessment of Thr232 phosphorylation of Aurora B in CCP6-deficient MKs. Representative confocal images are shown (left). Phospho-Thr232 of Aurora B (pAuroB), green; nucleus, red. Bar, 5 µm. The percentage of cells with high phospho-Thr232 of Aurora B (pAuroBhigh) was quantitated in >50 primary CD41+ cells for each group and shown as means ± SD. **, P < 0.01 (right graph). White boxes in left panels are shown in right panels. White arrows denote pAuroBhigh MKs. The fluorescence intensity of pAuroB in each cell was calculated by ImageJ and pAuroBhigh was identified as higher intensity than the mean fluorescence intensity in the CCP6+/+ group. (G) Schematic representation of transplantation. (H) Mad2 silenced BM MEPs from CCP6+/+ or CCP6−/− donor mice were rescued with pMY-Mad2-wt or pMY-Mad2E168A together with BM helper cells, and then transplanted into irradiated recipients. BM MKs of recipients were analyzed 12 d after transplantation. n = 6. Percentages of CD41+ MKs are shown as means ± SD. **, P < 0.01. (I and J) Primary MKPs were treated with the Aurora B inhibitor AZD-1152, and then stimulated with 20 ng/ml TPO for 5 d. Cell morphology was analyzed by Giemsa staining (Bar, 20 µm; red arrows denote mature MKs; n = 6; I). DNA contents were analyzed by flow cytometry. Percentages of DNA ploidy are shown as means ± SD. **, P < 0.01 (J). (K and L) Primary MKPs were infected with the indicated retrovirus then stimulated with 20 ng/ml TPO for 5 d. Cell morphology was analyzed by Giemsa staining (Bar, 20 µm; red arrows denote mature MKs; n = 6; K). DNA contents were analyzed by flow cytometry. Percentages of DNA ploidy are shown as means ± SD. *, P < 0.05; **, P < 0.01 (L). Data represent at least three separate experiments.
Aurora B is phosphorylated at Thr232 for its activation (Lordier et al., 2010). We found that CCP6 depletion induced Aurora B activation (Fig. 6 D). Given that Aurora A has been reported to regulate polyploidization of MKs (Wen et al., 2012), we also examined Aurora A in CCP6 silenced MKs. However, unlike the activation of Aurora B, CCP6 depletion did not affect the activity of Aurora A (Fig. 6 D). More importantly, overexpression of TTLL6 significantly promoted phosphorylation of Aurora B (Fig. 6 E). However, Mad2-E168A mutant did not promote the activation of Aurora B. Accordingly, CCP6-deficient MKs showed remarkably increased phosphorylation of Aurora B (Fig. 6 F). These data indicate that CCP6 deficiency causes hyperactivation of Aurora B in MKs.
To further confirm the physiological role of Mad2 polyglutamylation in MK maturation, primary MEPs from WT or CCP6-deficient mice were silenced for Mad2 and then rescued with GFP-tagged Mad2-wt or Mad2-E168A mutant, followed by transplantation into irradiated recipient mice together with helper cells (Fig. 6 G). Notably, restoration of Mad2-wt still maintained an increased MK number in the recipient mice (Fig. 6 H), accompanied by lower DNA ploidy (not depicted). However, rescue of Mad2-E168A mutant caused a normal MK number in the recipient mice (Fig. 6 H), exhibiting normal DNA ploidy (not depicted). These data suggest that polyglutamylation of Mad2 plays a critical role in the regulation of MK maturation.
After 5 d incubation in liquid media in the presence of TPO, WT MKPs generated morphological maturity and high DNA ploidy of MKs (Fig. 6, I and J). However, CCP6-deficient MKPs produced immature MK morphology and low DNA ploidy. Notably, Aurora B inhibitor (AZD-1152) treatment caused MK maturation and normal DNA ploidy (Fig. 6, I and J). Additionally, TTLL6 silencing in CCP6-deficient MKPs could promote MK maturation and DNA polyploidization (Fig. 6, K and L). Similar observations were obtained for TTLL4 depletion in CCP6-deficient MKPs (unpublished data). Importantly, restoration of CCP6-wt in CCP6-deficient MKPs was able to rescue MK maturity and normal DNA polyploidization (Fig. 6, K and L). As expected, restoration of Mad2-E168A mutant could rescue the maturation of MKs (Fig. 6, K and L). However, restoration of WT Mad2-wt failed to promote the maturation of MKs. In sum, hyperglutamylation of Mad2 promotes Aurora B activation that leads to suppression of MK maturation.
DISCUSSION
MKs generate platelets that play an important role in normal hemostasis and other physiological functions. Herein, we show that CCP6 deficiency leads to dysmaturity of MKs with dysfunction of platelets. We identify Mad2 as a new substrate for CCP6 but not for CCP1. CCP6 deficiency causes hyperglutamylation of Mad2, which promotes activation of Aurora B kinase activity. Aberrantly activated Aurora B suppresses MK maturation and DNA ploidization. Therefore, control of Mad2 polyglutamylation by its polyglutamylases and CCP6 plays a critical role in regulation of megakaryopoiesis.
Recently, CCP family members were reported to catalyze deglutamylation of α-tubulin and display enzymatic specificities (Rogowski et al., 2010). Among them, CCP1, CCP4, and CCP6 deglutamylate the shortening of penultimate polyglutamate chains of α-tubulin, whereas CCP5 specially catalyzes the branching site glutamate. Additionally, CCP1, CCP4, and CCP6 also hydrolyze gene-encoded glutamates of target protein substrates, such as MLCK and telokin, two important regulators of myosin functions (Yin et al., 2006; Rogowski et al., 2010). In this study, we show that CCP6 is most highly expressed in BM and MKs, and exhibits different expression levels in different tissues and cell types. CCP6 expression patterns are different from those of CCP1. Different tissue and cell type distributions of CCPs imply that they exert unique roles in the regulation of different physiological processes.
TTLLs catalyze the formation of glutamate side chains of target proteins (Janke et al., 2005; van Dijk et al., 2007). It has been reported that TTLL4 catalyzes several other non-tubulin substrates (van Dijk et al., 2008), whereas TTLL6 alone has no obvious activity with any of these substrates. We observed that TTLL4 and TTLL6 are most highly expressed in both mouse and human MKs. Both TTLL4 and TTLL6 can catalyze Mad2 polyglutamylation at Glu168. Moreover, TTLL4- or TTLL6-mediated Mad2 polyglutamylation is also hydrolyzed by CCP6. Importantly, depletion of TTLL4 or TTLL6 in CCP6-deficient MKPs is able to restore MK maturation. Altogether, we conclude that both TTLL4 and TTLL6 can catalyze Mad2 polyglutamylation during MK maturation. However, whether TTLL4 and TTLL6 exert a synergistic effect on Mad2 polyglutamylation in the regulation of MK maturation needs to be further investigated.
Mad2 is one of the major components for spindle assembly checkpoint that plays a crucial role in mitosis (Shah and Cleveland, 2000; Chao et al., 2012). Deficiency of Mad2 causes a premature onset of anaphase and abnormal DNA ploidy (Michel et al., 2001). Mad2 has two topologic structural states, the open state (O-Mad2) and the closed state (C-Mad2), which control its association with Cdc20 (Mapelli et al., 2007). Mad2 is reported to be phosphorylated at Ser195 on its C-terminal region. Mad2 phosphorylation at Ser195 is the known posttranslational mechanism to change its conformation (Kim et al., 2010). However, whether Mad2 has other distinct modifications to modulate its function remains unclear. Here, we show that Mad2 is a novel substrate of CCP6 but not CCP1. Mad2 is polyglutamylated by TTLL4 or TTLL6 in MKs. Thus, Mad2 polyglutamylation may be another critical modification for the regulation of cell functions. Moreover, Mad2 is polyglutamylated by TTLL6 and TTLL4 at Glu168, which is a conserved glutamic residue localized on the key flexible loop of the “safety belt” region. Whether this newly identified modification of Mad2 can affect O-Mad2/C-Mad2 transition or phosphorylation status in mitosis needs to be further investigated. Tubulin is also a substrate of CCP6 (Fig. 4 A), which is consistent with a previous study (Rogowski et al., 2010). However, we observed that only weak tubulin polyglutamylation appears in the mitotic MKPs. Moreover, Mad2 is not colocalized with tubulin during megakaryopoiesis. These results suggest that polyglutamylation of Mad2 and tubulin in MKs may play different roles in the regulation of megakaryopoiesis.
Besides the checkpoint function, Mad2 has been defined to be involved in the regulation of k-MT attachment stabilization and Aurora B activity (Kabeche and Compton, 2012). Aurora B is a well-known component of the chromosome passenger complex that regulates both mitosis and endomitosis (Zhang et al., 2004; Lordier et al., 2010; Campbell and Desai, 2013). However, the role of Aurora B in MK maturation is controversial. Aurora B was reported to be expressed in early endomitosis in human MKs (Geddis and Kaushansky, 2004). Aurora B is required for the regulation of endomitosis of MKs but is dispensable for DNA polyploidization. Of note, the early-stage but not late-stage inhibition of Aurora B in cultured MKs decreased the percentage of CD41+ MKs (Lordier et al., 2010). However, the inhibitors of Aurora kinase family proteins could be used as inducers of polyploidization, which induce acute megakaryocytic leukemia (AMKL) cells to undergo terminal differentiation (Wen et al., 2012). Here, we found that Aurora B was highly expressed in small MKs. Glutamylation of Mad2 can promote Aurora B kinase activity to regulate MK maturation.
Although MKs are a fairly rare subpopulation in BM, they generate platelets to play important roles in normal hemostasis and pathogenesis of many hematopoietic diseases such as AMKL, myelofibrosis neoplasms, ET, and PMF (Papadantonakis et al., 2012; Thon and Italiano, 2012). MK hyperproliferation and alterations of platelet counts are implications in ET and PMF (Wen et al., 2011). ET is often diagnosed incidentally through blood counts and has prognosis risk factors including bleeding, thrombosis, leukemic transformation, and fibrotic progression (Tefferi, 2013). The phenotypes of CCP6-deficient mice are reminiscent of the features of ET. Importantly, we further observed that CCP6 was undetectable in many ET patients (unpublished data). However, whether CCP6 and Mad2 polyglutamylation mediate human ET pathogenesis remains to be further investigated.
MATERIALS AND METHODS
Cell culture.
Human K562 cells were cultured with RPMI1640 supplemented with 10% FBS and 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen). Human 293T cells were cultured with DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. Retrovirus infecting primary BM cells and lentivirus infecting K562 cells were produced in 293T cells using the standard protocols. Transfection was performed using lipofectin (Invitrogen). CCP6 silenced K562 stable cell lines were established by lentivirus infection followed by puromycin selection (2 µg/ml).
Antibodies and reagents.
FITC-anti–mouse CD41, FITC-anti–human CD41, anti–mouse CD127, anti–mouse CD34, and anti–mouse CD41, FITC-anti–mouse c-Kit, APC-anti–mouse Sca-1, PE-CY7-anti–mouse CD16/32 antibodies, and eFluor450 lineage detection cocktail were purchased from eBioscience. PercP-CY5.5 goat anti–rat IgG and the antibodies against myc-tag, Mad2, CCP6, and GST-tag were purchased from Santa Cruz Biotechnology, Inc. The antibodies against Flag-tag, β-actin, and GFP-tag were from Sigma-Aldrich. Anti-VWF antibody was purchased from Millipore. GT335 (anti-glutamylation) antibody was from AdipoGen. Antibody against TTLL6 was from Novus Biologicals. The antibodies against Aurora B, Aurora A, pThr288 of Aurora A, tubulin, and CD42 were from Bioss. Anti-pThr232 antibody of Aurora B was from Antibodies Online. Alexa Fluor 405–, Alexa Fluor 488–, Alexa Fluor 594–, and Alexa Fluor 649–labeled secondary antibodies were from Molecular Probes. ADP, Thrombin, collagen, Hoechst 33342, and phorbol-12-myristate-13-acetate (PMA) were purchased from Sigma-Aldrich. TPO was from PeproTech. Aurora B kinase inhibitor AZD-1152 was from Selleck Chemicals. Methocult media were from STEMCELL Technologies.
Vector constructions.
Mouse CCP6 was cloned into pCDNA4-MycHis expression vector. CCP6-H230S/E233Q mutant and CCP1-H912S/E915Q mutant were generated by site-directed mutagenesis method of DpnI digestion (Rogowski et al., 2010). CCP6-wt and CCP6-mut cDNAs were also subcloned into H-MBP-3C vector and purified using Amylose resin (New England BioLabs, Inc.) according to the manufacturer’s instruction. Human CCP1, human TTLL4, and human TTLL6 were cloned into p3×flag-CMV-9 expression vector. Human WT Mad2 (Mad2-wt), Mad2-E168A mutant, and Mad2-E169A mutant were generated and cloned into pGEX-6P-1 vector (GE Healthcare), expressed in E. coli, and purified using Glutathione Sepharose 4B beads. RNA interference sequences were designed according to BLOCK-IT RNAi Designer system instructions (Invitrogen). The shRNA oligos encoding target sequences against hCCP6 (5′-GCTTGACCTCCTGACGATAAC-3′), mTTLL6 (5′-CCTCTTACTCCCACCCTAACC-3′), mMad2 (5′-GGACTCACCTTGCTTACAACT-3′), or scramble sequence (5′-AATTCTCCGAACGTGTCACGT-3′) were cloned into pSIN-EF2 vector or MSCV-LTRmiR30-PIG vector (LMP; GE Healthcare). Human Mad2-wt and Mad2-E168A mutant were also subcloned into pMY-IRES-GFP vector (Cell Biolabs).
Mouse strains.
Mouse experiments were approved by the Institutional Animal Care and Use Committees at the Institute of Biophysics, Chinese Academy of Sciences. CCP1−/− mice (Pcd mice, BALB/cByJ-Agtpbp1pcd-3J-J) were from The Jackson Laboratory. CCP6−/− mice (FVB background) were generated by a transposon piggyBac insertional mutagenesis program (PBMICE) and provided by X. Wu (Fudan University, Shanghai, China; Sun et al., 2008). The breeding strategy was to cross between heterozygotes to generate homozygote knockout mice and littermate control mice. CCP1 and CCP6 deletion was confirmed by RT-PCR and immunoblotting.
Flow cytometry, cell sorting, and in vitro culture.
LSK (Lin−Sca-1+c-Kit+), CLP (Lin−CD127+Sca-1lowc-Kitlow), MP (Lin−c-Kit+Sca-1−), CMP (Lin−c-Kit+Sca-1−CD34+CD16/32−), GMP (Lin−c-Kit+Sca-1−CD34+CD16/32+), MEP (Lin−c-Kit+Sca-1−CD34−CD16/32−), MKP (Lin−c-Kit+CD41+), and MK (Lin−c-Kit−CD41+) populations were analyzed or sorted with a FACSAria II instrument (BD; Tothova et al., 2007). For DNA content analysis, BM cells or sorted MKPs were cultured in suspension media with IMDM containing 10% FBS, 2 mM glutamine, 100 U/ml penicillin and streptomycin, 50 µM β-mercaptoethanol, and 50 ng/ml TPO. 5 d later, Hoechst 33342 was added in culture for 2 h followed by CD41/CD42 staining. DNA ploidy was measured in the CD41+CD42+ cell population by an LSRFortessa flow cytometer (Lordier et al., 2010). For in vitro culture and maturation induction, primary MKPs were sorted from BM, infected with the indicated retrovirus, and plated in suspension IMDM media containing 50 ng/ml TPO for 5 d (Tong and Lodish, 2004). For in vitro semisolid culture, MKPs were sorted from BM and plated in methylcellulose-based media added with 50 ng/ml TPO for 5 d (Boitano et al., 2012).
Mass spectrometry identification of Mad2.
Recombinant MBP-CCP6-wt and MBP-CCP6-mut proteins (Rogowski et al., 2010) were purified using the Amylose resin (New England BioLabs) according to the manufacturer’s instruction. MBP-CCP6-wt or MBP-CCP6-mut was immobilized with Affi-gel10 resin to go through mouse BM lysates for affinity chromatography as previously described (Fan et al., 2003). The eluted fractions were visualized by SDS-PAGE followed by silver staining. Differential bands in SDS-PAGE gels were tryptic digestion for mass spectrometry with nano LC-ESILTQ MS/MS (Thermo Fisher Scientific).
Staining and immunohistochemistry.
Primary BM CD41+ cells were fixed with 1% paraformaldehyde (PFA; Sigma-Aldrich) and permeabilized with 1% Triton X-100 in PBS, followed by 5% donkey serum added with 0.1% BSA blocking. Cells were then incubated with the according primary antibodies at 4°C overnight followed by incubation with corresponding fluorescence-conjugated secondary antibodies. Nuclei were stained with DAPI. Images were obtained with a laser scanning confocal microscopy (FV1000; Olympus; Wang et al., 2013). ImageJ software (National Institutes of Health) was used for colocalization quantitation. Pearson’s correlation coefficient (PCC) of each cell was calculated and colocalization was identified as PCC rate ≥ 0.5. Percentages of colocalized cells were counted as means ± SD. For each experiment, at least 50 typical cells were observed. For BM histology analysis, femurs were fixed in 4% formaldehyde followed by decalcifying in 10% EDTA-PBS buffer. Longitudinal paraffin sections were prepared for hematoxylin and eosin (H&E) staining, VWF immunohistochemical staining, AchE staining, and immunofluorescence staining as the methods previously described (Ichikawa et al., 2004). For immunohistochemistry quantitation, VWF- or AchE-positive MKs in different fields were enumerated in a 40× lens. For MK maturation staining, primary mouse MKPs were isolated and cultured with 20 ng/ml TPO for 5 d for maturation induction. Cultured cells were sorted after Hoechst 33342 and anti–CD41-PerCP Cy5.5 staining into two fractions. Small-sized, CD41+, and 2N to 4N cells represent low ploidy MKs. Large-sized, CD41+, and ≥ 8N cells represent polyploidy MKs (Lordier et al., 2010). More than 50 typical MK cells were observed for each staining.
Analysis of peripheral blood cells.
Peripheral blood was collected from the inferior vena cava of anesthetized mice with EDTA as anticoagulant. Blood was analyzed using an XFA6030 automated hemocytometer (Slpoo). Cell number and percentage of each population were counted. Platelet characters of PCT and PDW were also tested according to the manufacturer’s instruction. For the morphologically classifying, peripheral blood smears were stained with Wright’s and images taken with an oil-immersion 100× lens.
Analysis of platelet function.
Peripheral blood was collected from the inferior vena cava of anesthetized mice with acid citrate dextrose (ACD) as anticoagulant. Blood were stimulated with 1 mM ADP for 5 min and diluted with PBS. Platelets were stained with FITC-anti–mouse CD41 antibody. Platelet aggregation was analyzed by flow cytometry with a FACS Influx instrument (BD; Smith et al., 2012). Bleeding time assay were performed as previously described. In brief, 8–10-wk-old mice were anesthetized and 0.5 cm of tails was removed. Bleeding times were measured as bleeding stopped with the tails held in 37°C PBS (Smith et al., 2012). For platelet aggregation assay, peripheral blood was collected from the inferior vena cava of anesthetized mice with acid citrate dextrose (ACD) as an anticoagulant. Blood was diluted with Hepes-Tyrode’s buffer and spun at 100 g to get platelet-rich plasma (PRP). Platelet-poor plasma (PPP) was obtained by an additional centrifuge of 860 g from PRP as a control. ADP, Thrombin, and collagen-stimulated platelet aggregations were measured by Chrono log 560 aggregometry (Woulfe et al., 2004). For platelet spreading assay, mouse platelets were isolated from PRP. Mock or 0.1 U/ml thrombin was treated for 5 min. Activated platelets were placed on the surface of poly-l-lysine–coated slides at 37°C for 45 min. Platelets were fixed, permeabilized, and probed by phalloidin (Mazharian et al., 2013). For quantitation, spreading platelets in different fields were enumerated with a 100× lens. Percentages of spreading cells were counted as means ± SD. More than 500 platelets per condition were counted.
Electron microscopy.
For transmission electron microscopy, CD41+ cells were sorted from BM and fixed with 2.5% glutaraldehyde at 4°C overnight, followed by post-fixation in 1% osmium tetroxide for 2 h. Cells were dehydrating with sequential concentration of ethanol wash and immersed in SPI-PON812 resin. The ultrathin sections were counterstained. Images were scanned by an FEI Tecnai spirit transmission electron microscope (Xia et al., 2013).
Transplantation assays.
For BM transplantation, BM cells from CCP6−/− (RFP+) mice and littermate controls (RFP−) were injected into the lateral tail veins of lethally irradiated recipient mice. BM MKs and peripheral blood platelets of recipients were analyzed 4 wk after transplantation. For transplantation of MKPs, WT MKPs were sorted and directly infected with LMP-shCtrl, LMP-shTTLL6, LMP-shMad2, pMY-Mad2-wt, and pMY-Mad2-E168A retrovirus. 4 × 104 MKPs (GFP+), together with 3 × 105 helper cells were injected into irradiated recipient mice. 7 d after transplantation, paraffin sections of femurs were performed and GFP+ cells were counted. For transplantation of MEPs, MEPs from CCP6−/− mice and littermate controls were sorted and directly infected with LMP-shMad2, pMY-Mad2-wt, and pMY-Mad2-E168A retrovirus. 4 × 104 MEPs (GFP+), together with 3 × 105 helper cells, were injected into irradiated recipient mice. 12 d after transplantation, the percentage of GFP+ BM MKs was analyzed by flow cytometry (Mercher et al., 2008).
Immunoprecipitation assay.
K562 cells or 293T cells were transfected with the indicated plasmids and harvested. Freshly isolated BM cells were from femurs. Cells were lysed with ice-cold RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% sodium desoxycholate, 0.1% SDS, 5 mM EDTA, 2 mM PMSF, 20 mg/ml aprotinin, 20 mg/ml leupeptin, 10 mg/ml pepstatin A, 150 mM benzamidine, and 1% Nonidet P-40) for 1 h. Lysates were incubated with the indicated antibodies, followed by immunoprecipitation with protein A/G agarose beads and immunoblotting (Wang et al., 2013). Band intensities were calculated by ImageJ software. Normalized ratios were calculated as the percentage of pulldown/input using the equation: band intensity of pulldown/band intensity of input × 100%.
In vitro glutamylation assay.
293T cells were transfected with expression plasmids of CCP6, TTLL4, and TTLL6 for 48 h. Cells were harvested and lysed with PBS-0.2% NP-40. Supernatants were incubated with recombinant GST-Mad2 protein at 37°C for 2 h. rGST-Mad2 was precipitated with Glutathione Sepharose 4B beads, followed by immunoblotting (Rogowski et al., 2010).
Real-time qPCR.
Total RNAs from mouse tissues or FACS-sorted BM cells were extracted with the RNA miniprep kit (LCsciences) according to the manufacturer’s manual. M-MLV reverse transcription (Promega) was used to synthesize cDNA. Quantitative PCR analysis and data collection were performed on the ABI 7300 qPCR system using the primer pairs listed below. mCCP6: forward, 5′-AGGCAGGCAATGATACAGGAA-3′, reverse, 5′-GGTTACCACTTTCAAAGCAAGCA-3′; mCCP1: forward, 5′-GGGGTCGAAGAGCGAGTTTC-3′, reverse, 5′-GAATGGAGTGAGTCTGCACCA-3′; mTTLL1: forward, 5′-GAAGTGGGTCACTGACATTGAG-3′, reverse, 5′-ACGTTGCGAATGGTTTGCAC-3′; mTTLL4: forward, 5′-TGGATGAGAACCTGAAACCCT-3′, reverse, 5′-TGGGGCTGCTGGAACTAGA-3′; mTTLL5: forward, 5′-ACTCCCCAGCTCCCATCTG-3′, reverse, 5′-GGGGCATTGTCAGGAACGG-3′; mTTLL6: forward, 5′-AAGCCCTTCATCATCGACGG-3′, reverse, 5′-TGTCTAGGTTAGGGTGGGAGTAA-3′; mTTLL7: forward, 5′-CTCTGCCTCAAGATGGGGTTA-3′, reverse, 5′-GTTCCGGCAACATTAGCTGTAA-3′; mTTLL9: forward, 5′-TGGAGTGTCGAAAGGAAAAGAGA-3′, reverse, 5′-TGCTCATCCATGTAGGTGTGG-3′; mTTLL11: forward, 5′-CCTGACCAACTACTCCCTGAA-3′, reverse, 5′-GGGATGTCTGACTGGTAGAAAAC-3; mTTLL13: forward, 5′-GGCCTGAAGGAAGTAGGGGA-3′, reverse, 5′-CATGCCAGGGAAGTGGTTGA-3′; and mβ-actin: forward, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′, reverse, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′. Quantitation was normalized to an endogenous GAPDH or β-actin gene.
Statistical analysis.
Student’s t test was used as statistical analysis by using Excel (Microsoft) as previously described (Wang et al., 2013).
Acknowledgments
We thank Dr. Xiaohui Wu (University of Fudan) for providing CCP6−/− mice. We thank Jianhua Wang, Xiang Shi, Zhenwei Yang, Yan Teng, Zhensheng Xie, Chunchun Liu, Junjing Yu, and Junying Jia for technical support. We thank Shu Meng, Xiaoxiao Zhu, Yajie Xu, and Liangming Yao for technical help and assistance with animal procedures. We thank Drs. Jun Peng and Jun Chen for critical suggestions.
This work was supported by the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDA01010407 and XDA01020203), the National Natural Science Foundation of China (81330047, 30830030, and 31372404), the National Key Basic Research Program of China (2010CB911902 and 2014CB964601), the Hundred Talents Program of the Chinese Academy of Sciences (to Y. Tian), and China Postdoctoral Science Foundation (20110490617 and 2012T50145).
The authors declare no competing financial interests.
Author contributions: B. Ye, C. Li, and Z. Yang designed and performed experiments; B. Ye analyzed data and wrote the paper; Y. Wang and L. Sun performed some experiments; Y. Du, L. Hao, B. Liu, and G. Huang constructed vectors; J. Hao performed histology; L. Wang, Y. Li, S. Wang, and P. Xia analyzed data; Y. Tian initiated the study, crossed mice, and analyzed data; Z. Fan initiated the study, organized, designed, and wrote the paper.
Footnotes
Abbreviations used:
- AchE
- acetylcholinesterase
- ADP
- adenosine-5′-diphosphate
- CCP
- cytosolic carboxypeptidase
- ET
- essential thrombocythemia
- HSC
- hematopoietic stem cell
- IMS
- invaginated membrane system
- k-MT
- kinetochore-microtubule
- MEP
- MK/erythroid progenitor
- MK
- megakaryocyte
- pcd
- Purkinje cell degeneration
- PCT
- plateletcrit
- PDW
- platelet distribution width
- PMF
- primary myelofibrosis
- TPO
- thrombopoietin
- TTLL
- tubulin tyrosine ligase-like
References
- Bobinnec Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., and Bornens M.. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano A.E., de Lichtervelde L., Snead J.L., Cooke M.P., and Schultz P.G.. 2012. An image-based screen identifies a small molecule regulator of megakaryopoiesis. Proc. Natl. Acad. Sci. USA. 109:14019–14023 10.1073/pnas.1212545109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch Grau M., Gonzalez Curto G., Rocha C., Magiera M.M., Marques Sousa P., Giordano T., Spassky N., and Janke C.. 2013. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J. Cell Biol. 202:441–451 10.1083/jcb.201305041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.S., and Desai A.. 2013. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 497:118–121 10.1038/nature12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Eng J., Martinez R.A., Jackson S., Huang J., Possin D.E., Sopher B.L., and La Spada A.R.. 2008. The zinc-binding domain of Nna1 is required to prevent retinal photoreceptor loss and cerebellar ataxia in Purkinje cell degeneration (pcd) mice. Vision Res. 48:1999–2005 10.1016/j.visres.2008.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao W.C., Kulkarni K., Zhang Z., Kong E.H., and Barford D.. 2012. Structure of the mitotic checkpoint complex. Nature. 484:208–213 10.1038/nature10896 [DOI] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., and Xu T.. 2005. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 122:473–483 10.1016/j.cell.2005.07.013 [DOI] [PubMed] [Google Scholar]
- Eddé B., Rossier J., Le Caer J.P., Desbruyères E., Gros F., and Denoulet P.. 1990. Posttranslational glutamylation of alpha-tubulin. Science. 247:83–85 10.1126/science.1967194 [DOI] [PubMed] [Google Scholar]
- Fan Z., Beresford P.J., Oh D.Y., Zhang D., and Lieberman J.. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 112:659–672 10.1016/S0092-8674(03)00150-8 [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A., La Spada A.R., Treadaway J., Higdon J.C., Harris B.S., Sidman R.L., Morgan J.I., and Zuo J.. 2002. Purkinje cell degeneration (pcd) phenotypes caused by mutations in the axotomy-induced gene, Nna1. Science. 295:1904–1906 10.1126/science.1068912 [DOI] [PubMed] [Google Scholar]
- Gagnon C., White D., Cosson J., Huitorel P., Eddé B., Desbruyères E., Paturle-Lafanechère L., Multigner L., Job D., and Cibert C.. 1996. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J. Cell Sci. 109:1545–1553. [DOI] [PubMed] [Google Scholar]
- Gao Y., Smith E., Ker E., Campbell P., Cheng E.C., Zou S., Lin S., Wang L., Halene S., and Krause D.S.. 2012. Role of RhoA-specific guanine exchange factors in regulation of endomitosis in megakaryocytes. Dev. Cell. 22:573–584 10.1016/j.devcel.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddis A.E., and Kaushansky K.. 2004. Megakaryocytes express functional Aurora-B kinase in endomitosis. Blood. 104:1017–1024 10.1182/blood-2004-02-0419 [DOI] [PubMed] [Google Scholar]
- Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., and Hirai H.. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299–304 10.1038/nm997 [DOI] [PubMed] [Google Scholar]
- Ikegami K., Mukai M., Tsuchida J., Heier R.L., Macgregor G.R., and Setou M.. 2006. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 281:30707–30716 10.1074/jbc.M603984200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., and Bulinski J.C.. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- Janke C., Rogowski K., Wloga D., Regnard C., Kajava A.V., Strub J.M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., et al. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 308:1758–1762 10.1126/science.1113010 [DOI] [PubMed] [Google Scholar]
- Kabeche L., and Compton D.A.. 2012. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr. Biol. 22:638–644 10.1016/j.cub.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina E., Biswas R., Berezniuk I., Hermoso A., Aviles F.X., and Fricker L.D.. 2007. A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21:836–850 10.1096/fj.06-7329com [DOI] [PubMed] [Google Scholar]
- Kim S., Sun H., Ball H.L., Wassmann K., Luo X., and Yu H.. 2010. Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc. Natl. Acad. Sci. USA. 107:19772–19777 10.1073/pnas.1009000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Yanagisawa H.A., Yagi T., Hirono M., and Kamiya R.. 2010. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr. Biol. 20:441–445 10.1016/j.cub.2009.12.058 [DOI] [PubMed] [Google Scholar]
- Lordier L., Chang Y., Jalil A., Aurade F., Garçon L., Lécluse Y., Larbret F., Kawashima T., Kitamura T., Larghero J., et al. 2010. Aurora B is dispensable for megakaryocyte polyploidization, but contributes to the endomitotic process. Blood. 116:2345–2355 10.1182/blood-2010-01-265785 [DOI] [PubMed] [Google Scholar]
- Machlus K.R., and Italiano J.E. Jr. 2013. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 201:785–796 10.1083/jcb.201304054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M., Massimiliano L., Santaguida S., and Musacchio A.. 2007. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 131:730–743 10.1016/j.cell.2007.08.049 [DOI] [PubMed] [Google Scholar]
- Mazharian A., Mori J., Wang Y.J., Heising S., Neel B.G., Watson S.P., and Senis Y.A.. 2013. Megakaryocyte-specific deletion of the protein-tyrosine phosphatases Shp1 and Shp2 causes abnormal megakaryocyte development, platelet production, and function. Blood. 121:4205–4220 10.1182/blood-2012-08-449272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercher T., Cornejo M.G., Sears C., Kindler T., Moore S.A., Maillard I., Pear W.S., Aster J.C., and Gilliland D.G.. 2008. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 3:314–326 10.1016/j.stem.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., and Benezra R.. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Nakorn T.N., Miyamoto T., and Weissman I.L.. 2003. Characterization of mouse clonogenic megakaryocyte progenitors. Proc. Natl. Acad. Sci. USA. 100:205–210 10.1073/pnas.262655099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadantonakis N., Matsuura S., and Ravid K.. 2012. Megakaryocyte pathology and bone marrow fibrosis: the lysyl oxidase connection. Blood. 120:1774–1781 10.1182/blood-2012-02-402594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirault-Chassac S., Six E., Catelain C., Lavergne M., Villeval J.L., Vainchenker W., and Lauret E.. 2010. Notch/Delta4 signaling inhibits human megakaryocytic terminal differentiation. Blood. 116:5670–5678 10.1182/blood-2010-05-285957 [DOI] [PubMed] [Google Scholar]
- Regnard C., Desbruyères E., Huet J.C., Beauvallet C., Pernollet J.C., and Eddé B.. 2000. Polyglutamylation of nucleosome assembly proteins. J. Biol. Chem. 275:15969–15976 10.1074/jbc.M000045200 [DOI] [PubMed] [Google Scholar]
- Rodriguez de la Vega M., Sevilla R.G., Hermoso A., Lorenzo J., Tanco S., Diez A., Fricker L.D., Bautista J.M., and Avilés F.X.. 2007. Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 21:851–865 10.1096/fj.06-7330com [DOI] [PubMed] [Google Scholar]
- Rogowski K., van Dijk J., Magiera M.M., Bosc C., Deloulme J.C., Bosson A., Peris L., Gold N.D., Lacroix B., Bosch Grau M., et al. 2010. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 143:564–578 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Shah J.V., and Cleveland D.W.. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 103:997–1000 10.1016/S0092-8674(00)00202-6 [DOI] [PubMed] [Google Scholar]
- Sirajuddin M., Rice L.M., and Vale R.D.. 2014. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16:335–344 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Thon J.N., Devine M.T., Lin S., Schulz V.P., Guo Y., Massaro S.A., Halene S., Gallagher P., Italiano J.E. Jr, and Krause D.S.. 2012. MKL1 and MKL2 play redundant and crucial roles in megakaryocyte maturation and platelet formation. Blood. 120:2317–2329 10.1182/blood-2012-04-420828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.V., Jin K., Liu Y., Yang W., Xie X., Ye L., Wang L., Zhu L., Ding S., Su Y., et al. 2008. PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice. Nucleic Acids Res. 36:D729–D734 10.1093/nar/gkm790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S., Eddé B., Fox L.A., Guerrero S., Hard R., Hennessey T., Kabi A., Malison D., Pennock D., Sale W.S., et al. 2010. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr. Biol. 20:435–440 10.1016/j.cub.2009.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A.2013. Polycythemia vera and essential thrombocythemia: 2013 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 88:507–516 10.1002/ajh.23417 [DOI] [PubMed] [Google Scholar]
- Thon J.N., and Italiano J.E.. 2012. Platelets: production, morphology and ultrastructure. Handbook Exp. Pharmacol. 210:3–22 10.1007/978-3-642-29423-5_1 [DOI] [PubMed] [Google Scholar]
- Tong W., and Lodish H.F.. 2004. Lnk inhibits Tpo-mpl signaling and Tpo-mediated megakaryocytopoiesis. J. Exp. Med. 200:569–580 10.1084/jem.20040762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., and Janke C.. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 26:437–448 10.1016/j.molcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- van Dijk J., Miro J., Strub J.M., Lacroix B., van Dorsselaer A., Edde B., and Janke C.. 2008. Polyglutamylation is a post-translational modification with a broad range of substrates. J. Biol. Chem. 283:3915–3922 10.1074/jbc.M705813200 [DOI] [PubMed] [Google Scholar]
- Wang T., Parris J., Li L., and Morgan J.I.. 2006. The carboxypeptidase-like substrate-binding site in Nna1 is essential for the rescue of the Purkinje cell degeneration (pcd) phenotype. Mol. Cell. Neurosci. 33:200–213 10.1016/j.mcn.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Wang S., Xia P., Ye B., Huang G., Liu J., and Fan Z.. 2013. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 13:617–625 10.1016/j.stem.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Zambrano A., Zhao C., Brants J., Abecassis J., Schalken J.A., Rogatsch H., Schaefer G., Pycha A., Klocker H., and Wasylyk B.. 2010. Tubulin tyrosine ligase like 12 links to prostate cancer through tubulin posttranslational modification and chromosome ploidy. Int. J. Cancer. 127:2542–2553 10.1002/ijc.25261 [DOI] [PubMed] [Google Scholar]
- Wen Q., Goldenson B., and Crispino J.D.. 2011. Normal and malignant megakaryopoiesis. Expert Rev. Mol. Med. 13:e32 10.1017/S1462399411002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q., Goldenson B., Silver S.J., Schenone M., Dancik V., Huang Z., Wang L.Z., Lewis T.A., An W.F., Li X., et al. 2012. Identification of regulators of polyploidization presents therapeutic targets for treatment of AMKL. Cell. 150:575–589 10.1016/j.cell.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., de Néchaud B., Chillet D., Mazarguil H., Desbruyères E., Audebert S., Eddé B., Gros F., and Denoulet P.. 1992. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59:425–432. [PubMed] [Google Scholar]
- Wong C.H., Jenne C.N., Petri B., Chrobok N.L., and Kubes P.. 2013. Nucleation of platelets with blood-borne pathogens on Kupffer cells precedes other innate immunity and contributes to bacterial clearance. Nat. Immunol. 14:785–792 10.1038/ni.2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe D., Jiang H., Morgans A., Monks R., Birnbaum M., and Brass L.F.. 2004. Defects in secretion, aggregation, and thrombus formation in platelets from mice lacking Akt2. J. Clin. Invest. 113:441–450 10.1172/JCI200420267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Du Y., Zhao Z., Shi L., Sun L., Huang G., Ye B., Li C., Dai Z., et al. 2013. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32:2685–2696 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Jin K., Xie X., Li D., Yang J., Wang L., Gu N., Zhong Y., and Sun L.V.. 2009. Development of a database system for mapping insertional mutations onto the mouse genome with large-scale experimental data. BMC Genomics. 10:S7 10.1186/1471-2164-10-S3-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Hoggatt A.M., Zhou J., and Herring B.P.. 2006. 130-kDa smooth muscle myosin light chain kinase is transcribed from a CArG-dependent, internal promoter within the mouse mylk gene. Am. J. Physiol. Cell Physiol. 290:C1599–C1609 10.1152/ajpcell.00289.2005 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Nagata Y., Yu G., Nguyen H.G., Jones M.R., Toselli P., Jackson C.W., Tatsuka M., Todokoro K., and Ravid K.. 2004. Aberrant quantity and localization of Aurora-B/AIM-1 and survivin during megakaryocyte polyploidization and the consequences of Aurora-B/AIM-1-deregulated expression. Blood. 103:3717–3726 10.1182/blood-2003-09-3365 [DOI] [PubMed] [Google Scholar]