Yang et al. demonstrate that Hrd1 plays an important role in DC induction of CD4 T cell immunity. The underlying mechanism involves the ability of Hrd1 to ubiquitinate and degrade BLIMP-1, thus releasing CIITA from transcriptional repression and promoting MHCII expression. As a consequence, Hrd1−/− DCs protect mice from MOG-induced experimental autoimmune encephalomyelitis.
Abstract
The ubiquitin pathway plays critical roles in antigen presentation. However, the ubiquitin ligases that regulate MHC gene transcription remain unidentified. We showed that the ubiquitin ligase Hrd1, expression of which is induced by Toll-like receptor (TLR) stimulation, is required for MHC-II but not MHC-I transcription in dendritic cells (DCs). Targeted Hrd1 gene deletion in DCs diminished MHC-II expression. As a consequence, Hrd1-null DCs failed to prime CD4+ T cells without affecting the activation of CD8+ T cells. Hrd1 catalyzed ubiquitination and degradation of the transcriptional suppressor B lymphocyte–induced maturation protein 1 (BLIMP1) to promote MHC-II expression. Genetic suppression of Hrd1 function in DCs protected mice from myelin oligodendrocyte glycoprotein (MOG)–induced experimental autoimmune encephalomyelitis (EAE). We identified Hrd1-mediated BLIMP1 ubiquitination as a previously unknown mechanism in programming DC for CD4+ T cell activation during inflammation.
MHC class I (MHC-I) and class II (MHC-II) are cell surface glycoproteins on APCs involved in the binding and presentation of peptide antigens to the TCRs of T lymphocytes. MHC-I presents antigen from endogenous sources to CD8+ T cells, whereas MHC-II presents peptide from exogenous sources to CD4+ T cells (Rudolph et al., 2006; Adams and Luoma, 2013; Merad et al., 2013). These MHC–peptide–TCR complexes are required to generate antigen-specific immune responses. MHC-II is constitutively expressed in APCs at a relatively low level under naive conditions but can be induced by IFN-γ (Collins et al., 1984; Koeffler et al., 1984). In addition, pathogen-associated molecule patterns, including LPS and bacterial DNA, are capable of inducing MHC-II gene expression through binding to TLRs (Reis e Sousa, 2004). TLR-induced MHC-II expression plays critical roles in host immunity against microbial infections and is associated with inflammatory autoimmune diseases (Friese et al., 2005).
The MHC class II transactivator (CIITA) is critical for both constitutive and inducible MHC-II gene transcription (Steimle et al., 1993). IFN-γ and TLR stimulation induce CIITA to activate MHC-II gene transcription during inflammation (Drozina et al., 2005). CIITA-deficient mice do not express conventional MHC-II molecules on the surface of splenic B cells, macrophages, and DCs (Chang et al., 1994). It has been reported that B lymphocyte–induced maturation protein 1 (BLIMP1), a transcriptional repressor that is capable of triggering plasma cell differentiation, is a repressor of CIITA transcription. BLIMP1-mediated CIITA suppression turns off MHC-II–mediated antigen presentation to induce B cell differentiation into plasma cells (Piskurich et al., 2000). BLIMP1-deficient DCs exhibit elevated expression of MHC-II and facilitate CD4+ T cell immunity (Piskurich et al., 2000; Tooze et al., 2006; Chen et al., 2007; Zhao et al., 2007; Kim et al., 2013).
The ubiquitin pathway has been shown to play a critical role in regulating MHC-II antigen presentation at multiple levels (Moffat et al., 2013). Ubiquitination of CIITA is required for its transcriptional activation of MHC-II gene expression (Greer et al., 2003). Ubiquitination also directly regulates MHC-II degradation, catalyzed by the membrane-associated RING (really interesting new gene)-CH (MARCH) family of E3 ubiquitin ligases (Ishido et al., 2009). Although several MARCH family members have been suggested as regulators of both innate and the adoptive immune responses, MARCH 1, which targets CD86 and MHC-II for ubiquitination-mediated degradation, is the most well characterized member (Matsuki et al., 2007; De Gassart et al., 2008; Young et al., 2008; Walseng et al., 2010; Tze et al., 2011). Given the critical roles of MHC-II in antigen presentation and the activation of the adaptive immune system, it is not surprising that a tight regulatory mechanism is necessary to ensure appropriate MHC-II antigen presentation. However, how the ubiquitin pathway controls MHC-II antigen presentation, in particular the specific E3 ubiquitin ligases that are required in this process, remains largely unidentified.
Hrd1, also known as Synoviolin, is a membrane-spanning protein on the endoplasmic reticulum (ER). It has a RING finger domain followed by a long proline-rich C terminus in its cytoplasmic portion, which is likely involved in recruiting cytoplasmic proteins for ubiquitination. Hrd1 was initially identified as a ubiquitin ligase involved in degrading misfolded proteins (Carvalho et al., 2006; Denic et al., 2006). Because Hrd1 expression is often up-regulated in synovial fibroblasts in patients with rheumatoid arthritis, it was renamed Synoviolin (Amano et al., 2003). We recently reported that proinflammatory cytokines, including TNF and IL-1β, are responsible for inducing Hrd1 expression in synovial fibroblasts (Gao et al., 2006). We further observed that Hrd1 ubiquitinates IRE1α (inositol-requiring enzyme 1α), a critical kinase in regulating the ER stress response (Gao et al., 2008). It has been shown that Hrd1 targets the misfolded MHC-I for degradation in the in vitro cultured cell lines (Burr et al., 2011; Huang et al., 2011). Although the ER stress functions of Hrd1 in misfolded protein degradation have been well studied, its physiological roles in immune regulation are not known.
RESULTS
Hrd1 promotes MHC-II expression by DCs
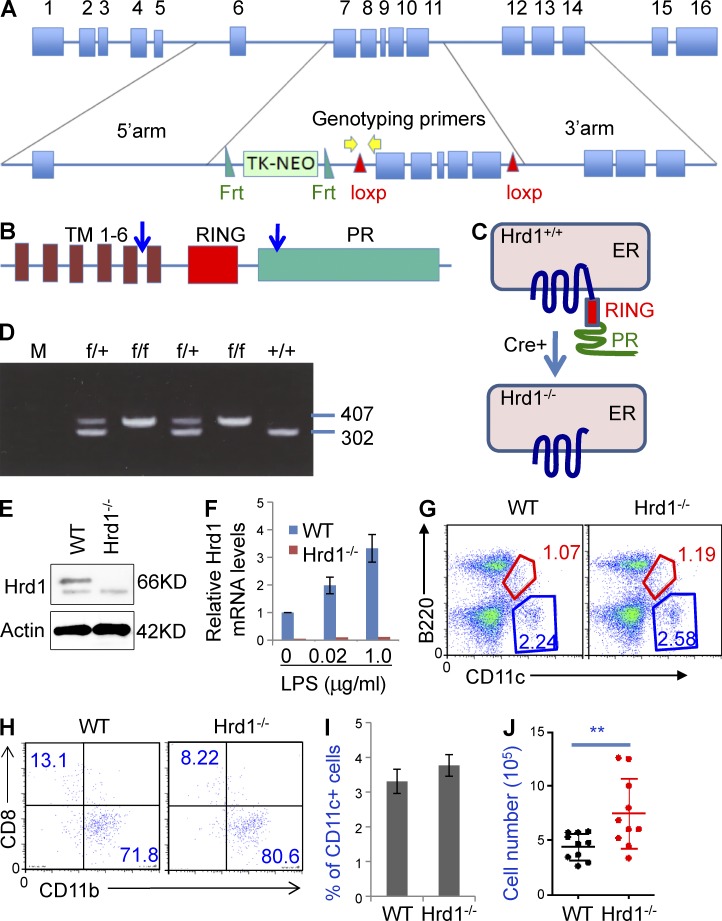
To study the physiological functions of Hrd1 in DCs, we generated Hrd1 floxed mice. The Hrd1 gene contains 16 exons (Fig. 1 A), we floxed exons 8–11 that encode a large region of the Hrd1 protein from its fifth transmembrane domain (TM) to the proline-rich sequence leading to deletion of the functional RING finger (Fig. 1, B and C). To exclude the potential effects of the neomycin selection cassette on Hrd1 expression, this cassette was flanked by two flippase recognition target (FRT) sites, which can be deleted by FLP recombinase. This targeting vector was transfected into an embryonic stem cell line generated from C57/BL6 mice. Neomycin selects were screened by PCR. Seven clones were obtained and confirmed to be correct targeting by Southern blotting. Blastocyst injections resulted in several chimeric mice with the capacity for germline transmission. Breeding of heterozygous mice yielded Hrd1wt/wt, Hrd1wt/f, and Hrd1f/f offspring without phenotypic abnormalities in expected Mendelian ratios (Fig. 1 D and not depicted). DC-specific Hrd1 knockout (Hrd1f/fCD11c-Cre+) mice were generated by breeding Hrd1f/f mice with CD11c-Cre+ transgenic mice. Both Hrd1 protein (Fig. 1 E) and mRNA (Fig. 1 F) were eliminated in purified CD11c+ cells from Hrd1f/fCD11c-Cre+ (Hrd1−/−) mice.
Figure 1.
Targeted disruption of the Hrd1 gene in DCs. (A) Structures of the Hrd1 WT and targeted alleles. Exons and the neomycin phosphotransferase gene (Neo) driven by the thymidine kinase (TK) promoter are shown. The TK-NEO cassette is flanked by 2 FRT sites and exons 7–11 are flanked by 2 LoxP sites. (B and C) Domain structure of Hrd1 protein. The ER membrane-anchoring protein Hrd1 carries 6 transmembrane (TM) domains, one RING finger domain, and a C terminus proline-rich domain. The deletion of floxed Hrd1 gene by Cre recombinase destroys Hrd1 protein expression. (D) Genotyping of Hrd1-floxed mice. Tail snips from a litter of Hrd1flox/wt X Hrd1flox/wt offspring were collected for DNA extraction and PCR analysis. The 302-bp PCR product is the WT allele and the 407-bp product is the mutant allele. (E and F) BM cells were isolated from WT and Hrd1 conditional KO (Hrd1−/−) mice and cultured under DC differentiation conditions. (E) Hrd1 protein expression was analyzed by immunoblotting (top) using β-actin as a loading control (bottom). (F) Hrd1 mRNA levels were determined by real-time quantitative RT-PCR. Hrd1 levels in WT DCs increased with LPS treatment. (G) Cell surface expression of B220 and CD11c in total splenocytes from WT and Hrd1−/− mice was analyzed by flow cytometry. (H) CD11c+B220− cells were gated and the expression of CD8 and CD11b was analyzed. (I and J) The mean percentages (I) and absolute numbers (J) of CD11c+ cells in the spleens of 10 pairs of WT and Hrd1−/− mice are shown (n = 10).
Because Hrd1 has been identified as an anti-apoptotic molecule that protects cells from ER stress-induced apoptosis (Amano et al., 2003), we asked whether Hrd1 gene deletion affects CD11c+ DC survival. Surprisingly, loss of Hrd1 function in DCs did not reduce survival; rather, it led to a slight increase in the percentage and a statistically significant increase in the total numbers of CD11c+ DCs in the spleen. In addition, the percentages of CD11c+B220− conventional DCs and CD11c+B220low plasmacytoid DCs were not altered in the spleens of Hrd1−/− mice compared with WT mice (Fig. 1 G). Moreover, analysis of the gated CD11c+B220− DCs by their expression of CD11b or CD8 did not detect any changes in the percentages of CD11c+CD11b+CD8−B220− myeloid DCs and CD11c+CD11b−CD8+B220− lymphoid DCs with Hrd1 gene deletion (Fig. 1, G and H). In addition, a slight increase in the percentage (Fig. 1 I) and a statistical significant increase in the total numbers (Fig. 1 J) of CD11c+ cells were detected in the spleen of DC-specific Hrd1 knockout mice.
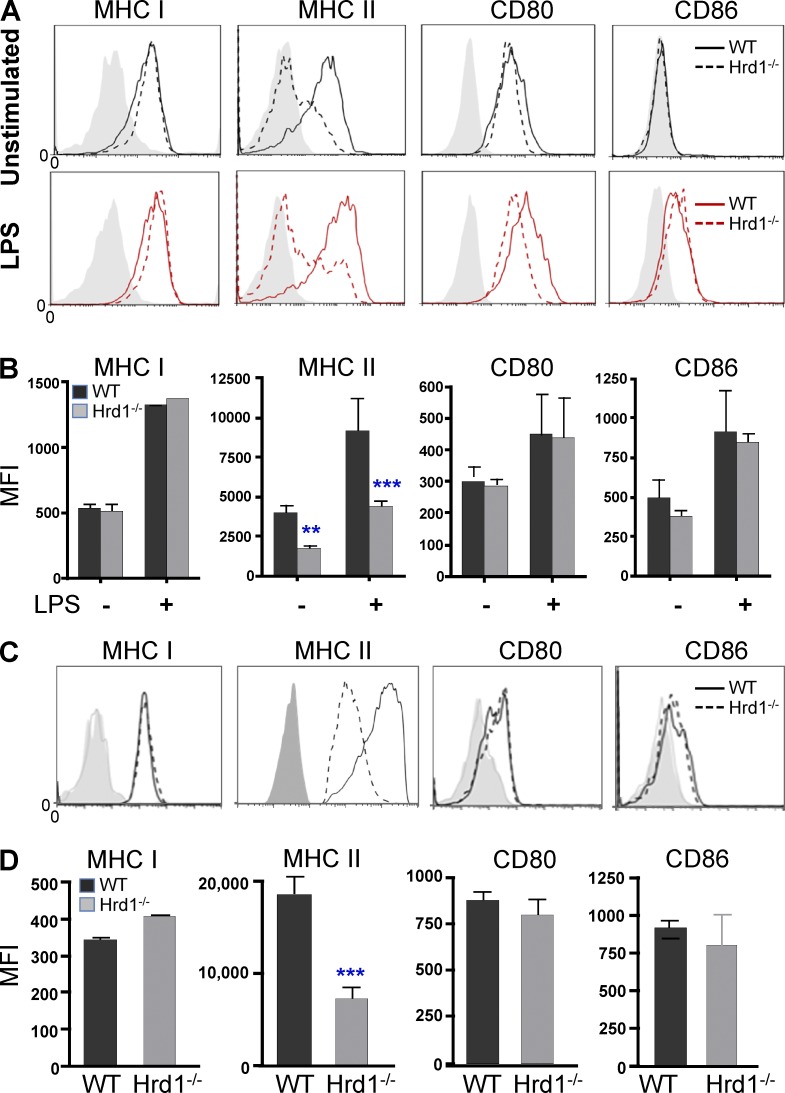
Notably, we detected a significant reduction in MHC-II expression on the surface of immature BM-derived DCs (BMDCs). Stimulation with LPS for 24 h led to a dramatic increase in MHC-II expression in WT DCs but failed to up-regulate MHC-II expression in Hrd1-null DCs. In contrast, the expression levels of MHC-I, CD80, and CD86 were not altered by Hrd1 gene deletion (Fig. 2, A and B). A similar reduction in MHC-II expression in gated CD11c+ cells from the spleens of Hrd1−/− mice was further confirmed (Fig. 2, C and D). These results indicate that Hrd1 gene deletion in mice selectively impairs MHC-II expression by DCs.
Figure 2.
Hrd1 promotes MHC-II expression in DCs. (A and B) WT and Hrd1−/− BMDCs were cultured overnight with or without LPS (200 ng/ml). Expression levels of MHC-I, MHC-II, CD80, and CD86 were analyzed by flow cytometry. (A and B) Representative images (A) and the mean fluorescence identity (MFI; B) from 7 independent experiments are shown (n = 7). (C and D) The expression levels of MHC-I, MHC-II, CD80, and CD86 on gated CD11c+ DCs in WT and Hrd1−/− splenocytes were analyzed. (C and D) Representative images (C) and the average MFI (D) from nine pairs of mice are shown (n = 9). (B and D) Data are reported as mean ± SD and were analyzed by Student’s t test. **, P < 0.01; ***, P < 0.005.
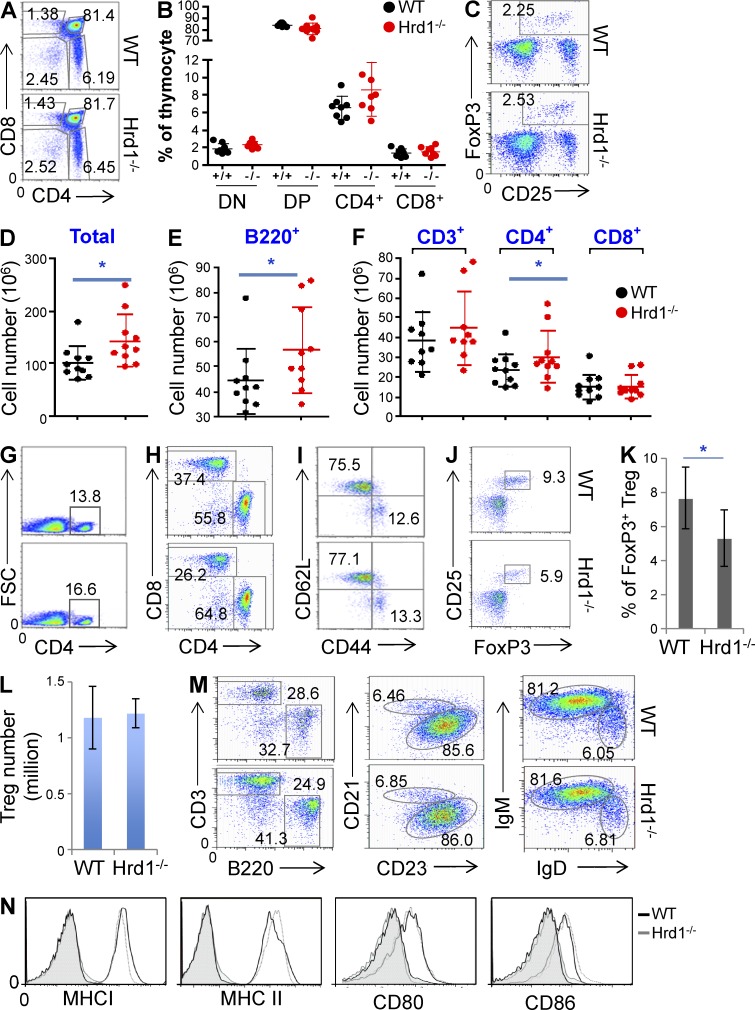
Because MHC-II expression is critical for CD4+ T cell development and homeostatic proliferation, we asked whether the decrease in MHC-II expression in Hrd1-null DCs alters T cell development in mice. Neither the percentages nor the absolute numbers of CD4 and CD8 double-negative, double-positive, and single-positive T cell populations were altered in the thymus of Hrd1−/− mice (Fig. 3, A and B). In contrast, a 20–30% increase in total splenocytes in Hrd1−/− mice was detected (Fig. 3 D). This enlarged spleen size of Hrd1−/− mice appeared to be due to a mean 30% increase in the percentage and total numbers of B220+ B cells (Fig. 3 E) and CD4+ cells (Fig. 3, F–H). Although there is a reduction in the percentage of CD8+ T cells, presumably due to the increase in CD4+ T cell populations in spleen, their absolute numbers were not affected (Fig. 3, F and H). Analysis of CD44 and CD62L expression on the surface of CD4+ T cells showed no changes in CD4+ T cell activation in DC-specific Hrd1-null mice (Fig. 3 I), excluding the possibility that chronic activation caused the observed increase in CD4+ T cells. In addition, there was an ∼30% reduction in the percentage of DC25+FoxP3+ T reg cells in the spleens of Hrd1−/− mice (Fig. 3, J and K), but absolute T reg cell numbers were not altered (Fig. 3 L), suggesting that the reduction in T reg cell percentage was likely a consequence of the CD4+ T cell increase. To support this, FoxP3+ T reg cells were not changed in the thymus of DC-specific Hrd1-null mice (Fig. 3 C). Further characterization of the splenic B cells did not did not detect any changes in the percentages of follicular and marginal zone B cells by their expression of CD21 and CD23, and no changes in the cell surface IgM and IgD expression (Fig. 3 M). Similar to CD4+ T cells, no increase in chronic B cell activation was detected because their expression levels of MHC-I and II, CD80, and CD86 were indistinguishable between WT and DC-specific Hrd1 KO mice (Fig. 3 N).
Figure 3.
Characterization of T and B lymphocytes in Hrd1f/fCD11c-Cre+ (Hrd1−/−) mice. (A–C) Analysis of T cell development in thymus of Hrd1−/− mice. CD4 and CD8 expression in total thymocytes was analyzed by flow cytometry. (A) Representative images are shown from experiments run using seven pairs of WT and Hrd1−/− mice. (B) The percentages of CD4/CD8 double-negative (DN), double-positive (DP), and single-positive (SP) cells from seven pairs of mice are shown. (C) The FoxP3+CD25+ T reg cells in the gated CD4 SP T cells were analyzed; representative images from seven pairs of mice are shown. (D–N) Cellularity analysis in the spleens of Hrd1−/− mice. The absolute numbers of total splenocytes (D), B220+ B cells (E), and T cells (F) from nine pairs of mice are shown. (G) CD4+ T cells in the spleens of WT and Hrd1−/− mice were analyzed by flow cytometry. (H) The ration of CD4 and CD8 T cells in the gated CD3+ population was analyzed. (I–L) Gated CD4+ T cells were used for the analysis of CD44 and CD62L expression (I) and FoxP3+CD25+ T reg cells (J). (K) Mean percentages (K) and total numbers of T reg cells (L) in nine pairs of mice are shown. Data are reported as means ± SEM and Student’s t test was used for the statistical analysis. *, P < 0.05. (M and N) Analysis of B cells in the spleens of Hrd1−/− mice. (M) The percentages of CD3+ T cells and B220+ B cells in the spleen of WT and Hrd1 conditional KO mice are analyzed (left). The gated B220+ B cells were analyzed for their expression of CD21 and CD23 (middle), and IgM and IgD (right). (N) The expression levels of MHC-I, MHC-II, CD80, and CD86 on the surface of gated B220+ B cells were analyzed. Images are representative for at least nine pairs of mice (n = 9).
Hrd1 regulates MHC-II expression at the mRNA level in DCs
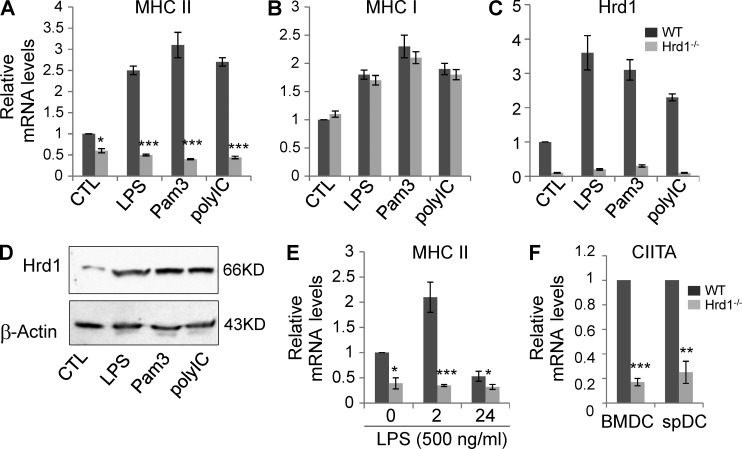
To delineate the mechanism by which Hrd1 regulates MHC-II expression in DCs, we analyzed MHC-II mRNA expression in WT and Hrd1-null BMDCs. Consistent with the observed reduction in MHC-II protein expression in Hrd1-null BMDCs by flow cytometry (Fig. 2), MHC-II mRNA expression was largely diminished in these cells (Fig. 4 A). Importantly, TLR stimulation for 2 h failed to enhance MHC-II transcription in Hrd1-null BMDCs. In contrast, the transcription of MHC-I was not affected by Hrd1 deficiency (Fig. 4 B). Diminished Hrd1 mRNA expression was confirmed in BMDCs from Hrd1−/− mice (Fig. 4 C). In addition, a significant increase in Hrd1 mRNA and protein expression was detected in WT BMDCs upon TLR stimulation (Fig. 4, C and D), indicating that TLR signaling normally induces Hrd1 expression in DCs.
Figure 4.
Hrd1 regulates MHC-II expression at the mRNA level. WT and Hrd1−/− BMDCs were stimulated with the indicated TLR agonists for 2 h. Total mRNA was extracted and levels of MHC-II (A), MHC-I (B), and Hrd1 (C). (D) WT and Hrd1−/− BMDCs were stimulated with each indicated TLR agonist for 24 h. Hrd1 protein expression levels were determined by Western blotting with β-actin as a loading control. (E) WT and Hrd1−/− BMDCs were stimulated with 500 ng/ml LPS for 2 or 24 h. The expression levels of MHC-II were determined by real-time PCR. (F) CIITA expression in BMDCs and sorted CD11c+ splenic DCs without TLR stimulation was quantified by real-time RT-PCR using β-actin as a control. Data are reported as mean ± SD from five independent experiments (n = 5). Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; **, P < 0.01; ***, P < 0.005.
It has been reported that TLR stimulation suppresses MHC-II mRNA transcription in DCs 18–24 h after stimulation (Landmann et al., 2001; Pai et al., 2002; Yao et al., 2006), but with a transient increase during the early stage (0.5–3 h) of stimulation (Landmann et al., 2001; Casals et al., 2007). We dynamically analyzed the effects of Hrd1 deletion on TLR-mediated MHC-II transcription. Indeed, consistent with these previous studies, after a transient increase at 2 h after stimulation, a 40–50% reduction in MHC-II transcription was detected in WT DCs 24 h after LPS stimulation. In contrast, this dynamic transcription of MHC-II in Hrd1-null DCs was dismissed (Fig. 4 E). The transcription factor CIITA has been identified as a critical factor for TLR-induced MHC-II transcription in DCs (Steimle et al., 1993). Interestingly, we detected that CIITA mRNA expression was diminished in BMDCs and in the gated CD11c+ DCs in the spleens of Hrd1−/− mice (Fig. 4 F), indicating that Hrd1 may regulate MHC-II expression by promoting CIITA gene transcription.
DC Hrd1 is a positive regulator for CD4+ T cell priming
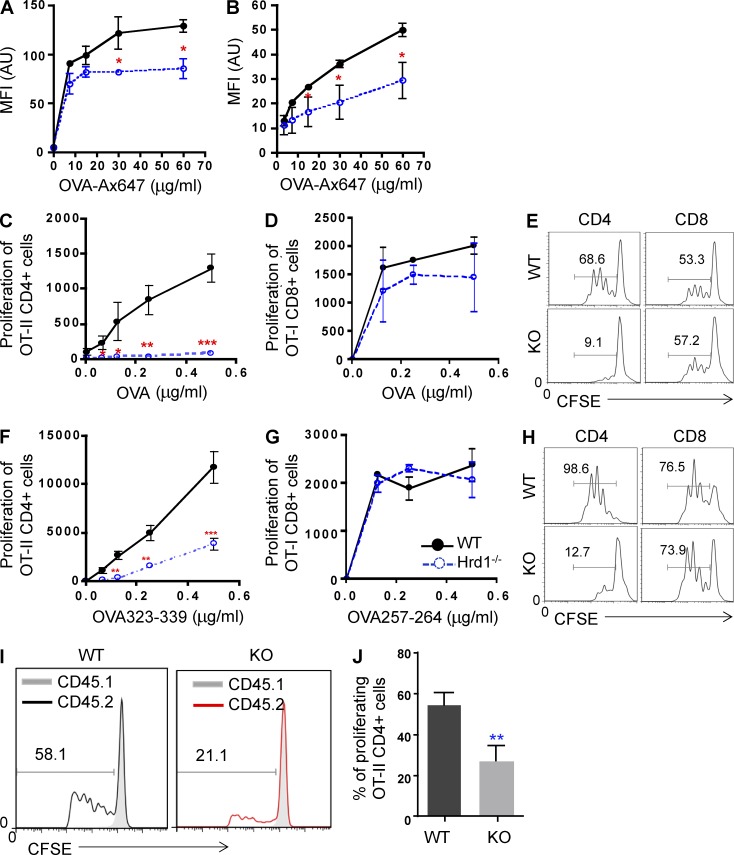
The finding of reduced MHC-II expression on the surface of Hrd1-null DCs led us to test whether Hrd1 positively regulates antigen presentation by DCs. Alexia Fluor 647–conjugated chicken ovalbumin (Ax647-OVA) was developed as a convenient approach to measure antigen uptake and presentation as the processed peptides fluoresce (Mansour et al., 2006). We incubated WT BMDCs with Ax647-OVA and found that mean fluorescence intensity increased in a dose-dependent manner, indicating normal antigen presentation and processing. In contrast, mean fluorescence intensity was significantly reduced in Hrd1-null BMDCs incubated with Ax647-OVA (Fig. 5 A). A similar reduction in fluorescence intensity was observed in freshly sorted CD11c+ splenic DCs (Fig. 5 B), indicating that Hrd1 functions are required for antigen presentation by DCs.
Figure 5.
Hrd1 deficiency impairs DC-mediated CD4+ T cell priming. (A and B) BMDCs (A) and sorted CD11c+ cells (B) from the spleen were cultured with Ax647-OVA at the indicated concentrations for 1 h and washed. Fluorescence intensity was analyzed by flow cytometry and data are reported as mean ± SD from five independent experiments. (C–E) BMDCs were stimulated with LPS in the presence of OVA protein overnight, washed, and co-cultured with either CD4+ OT-II T cells or CD8+ OT-II T cells for 3 d. T cell proliferation was analyzed by 3H-thymidine incorporation (C and D) or CFSE dilution (E). (F–H) BMDCs were stimulated with LPS in the presence of either OVA323-339 (F and H [left]) or OVA257-264 (G and H [right]) peptides overnight, washed, and co-cultured with either CD4+ OT-II T cells or CD8+ OT-II T cells for 3 d. T cell proliferation was analyzed by 3H-thymidine incorporation (F and G) or CFSE dilution (H). (I and J) In vivo OVA-specific CD4+ T cell priming was analyzed as described in Materials and methods. (I and J) Representative images (I) and data reported as mean ± SD (J) from five pairs of WT and Hrd1−/− recipients are shown (n = 5). Student’s t test was used for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
MHC-II expression by DCs is essential for antigen-specific CD4+ T cell priming by directly presenting peptide antigens to their TCRs. Because Hrd1 deficiency in DCs selectively impaired MHC-II expression, we tested whether Hrd1-null DCs were still able to prime CD4+ T cells. WT and Hrd1-null BMDCs were stimulated with LPS in the presence of OVA overnight, washed, and then incubated with naive CD4+ T cells from OT-II TCR transgenic mice. Naive CD8+ T cells from the OT-I mice were used as controls. Antigen-specific T cell proliferation was determined by 3H-thymidine incorporation and CFSE dilution as previously described (Zhang et al., 2009). OT-II CD4+ T cells proliferated vigorously when co-cultured with WT BMDCs. Conversely, OT-II CD4+ T cell proliferation was diminished when co-cultured with Hrd1-null BMDCs and OVA protein as measured by 3H-thymidine incorporation (Fig. 5 C) or CFSE dilution (Fig. 5 E). As expected, the proliferation of CD8+ OT-I T cells was comparable when co-cultured with either WT or Hrd1-null DCs (Fig. 5, D and E). As an E3 ubiquitin ligase that promotes protein degradation, the DC Hrd1 may regulate CD4 T cell activation by controlling the protein antigen processing onto MHC-II complex. To test this notion, we analyzed the T cell proliferation by co-cultivating with OVA peptides. Similar to the results from OVA protein co-culture experiments, loss of Hrd1 in DCs dramatically impaired their ability to prime CD4+ T cells in the presence of the MHC-II–presented OVA323-339 peptide as measured by both 3H-thymidine incorporation assay (Fig. 5 F) and CFSE analysis (Fig. 5 H). In contrast, Hrd1 gene deletion in DCs did not affect CD8+ T cell proliferation when co-cultured with the MHC-I–presented OVA257-264 peptide (Fig. 5, G and H). Therefore, genetic deletion of Hrd1 in DCs impairs CD4 T cell priming is largely due to the reduced MHC-II expression.
Next, we studied the role of DC Hrd1 in regulating CD4+ T cell priming in vivo using an adoptive transfer approach. Naive CD45.2+OT-II CD4+ T cells and control CD45.1+CD4+ T cells were mixed at a 1:1 ratio, stained with CFSE, and adoptively transferred into lethally irradiated WT and DC-specific Hrd1 KO mice. 1 d after the adoptive transfer, recipients were immunized with OVA323-339/CFA. 5 d after immunization, vigorous proliferation of the CD45.2+CD4+ OT-II T cells in the WT recipients was detected. In contrast, proliferation of CD45.2+CD4+ OT-II T cells in the DC-specific Hrd1−/− recipients was dramatically impaired (Fig. 5, I and J). These results indicate that DC Hrd1 plays a critical role in antigen-specific priming of CD4+ T cells in mice.
Hrd1 interacts with BLIMP1 in DCs
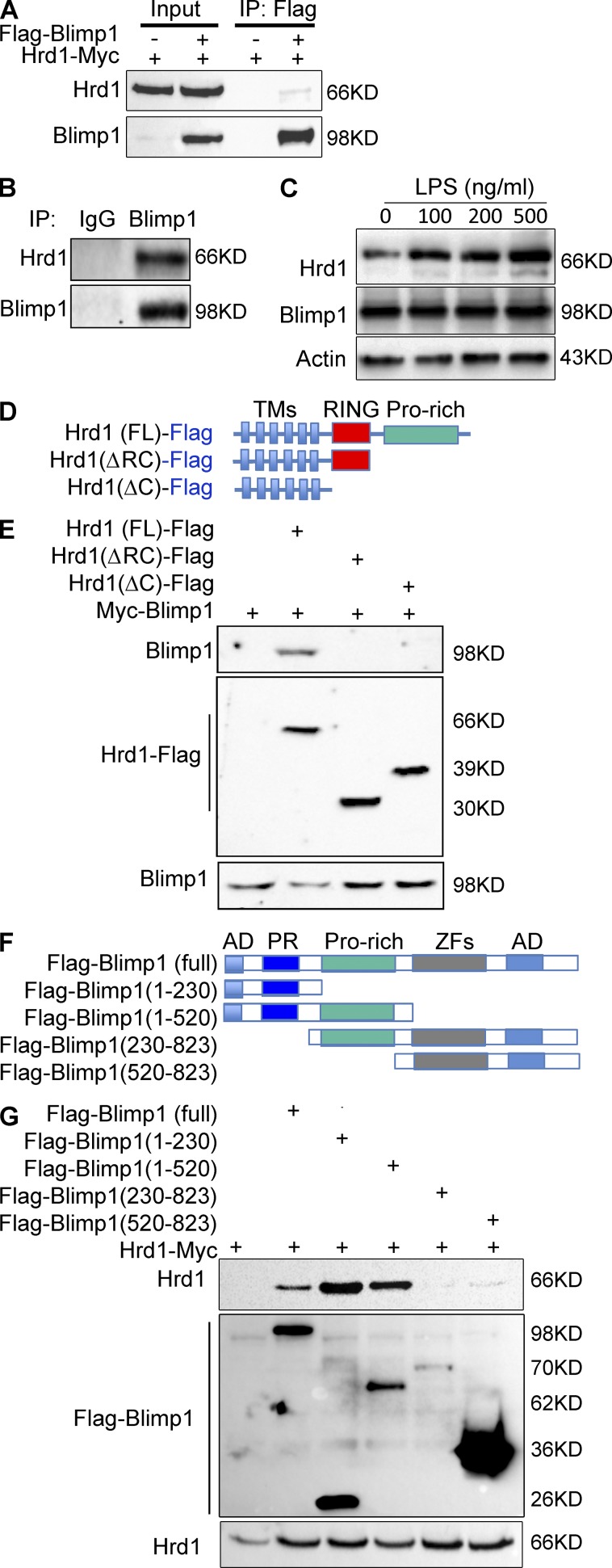
The significant reduction in both MHC-II and CIITA mRNA levels in Hrd1-null DCs led us to ask whether Hrd1 regulates MHC-II expression by targeting upstream regulatory factors. The transcription factor BLIMP1 has been shown to suppress the expression of both CIITA and MHC-II (Piskurich et al., 2000). We speculated that Hrd1 controls MHC-II and CIITA expression through targeting BLIMP1 in DCs. To test this hypothesis, we first determined whether Hrd1 interacts with BLIMP1. Indeed, Hrd1 protein was detected in anti-BLIMP1 immunoprecipitates from HEK293 cells cotransfected with Hrd1 and BLIMP1, but not in cells transfected with Hrd1 alone (Fig. 6 A). Interaction between endogenous Hrd1 and BLIMP1 in mouse primary BMDCs was confirmed by coimmunoprecipitation and immunoblotting because Hrd1 was detected in immunoprecipitates from BMDC lysates using anti-BLIMP1 but not normal rabbit IgG (Fig. 6 B). In addition, the interaction between Hrd1 and BLIMP1 appears to be regulated by TLR signaling as LPS stimulation of BMDCs for 2 h enhanced their interaction in a dose-dependent manner (Fig. 6 C).
Figure 6.
Hrd1 interacts with BLIMP1. (A) A Myc-Hrd1 expression plasmid was transfected with or without Flag-BLIMP1 expression plasmid into HEK293 cells. Expression of Hrd1 and BLIMP1 in whole cell lysates was confirmed by immunoblotting (lanes 1 and 2). Hrd1 interaction with BLIMP1 was determined by immunoprecipitation (IP) with anti-Flag antibody followed by immunoblotting with anti-Myc antibody (top, lanes 3 and 4). The same membrane was reprobed with anti-Flag antibody to confirm BLIMP1 expression (bottom). (B) Interaction between endogenous Hrd1 and BLIMP1 in mouse BMDCs was analyzed by IP with anti-BLIMP1 using normal rabbit IgG as a control, followed by immunoblotting with anti-Hrd1 (top). The same membrane was reprobed with anti-BLIMP1 (bottom). (C) BMDCs were stimulated with different doses of LPS as indicated for 2 h. The interaction between BLIMP1 and Hrd1 was determined as in B. (D and E) Truncation mutants of Hrd1 were generated (D) and their interactions with BLIMP1 in transiently transfected HEK293 cells were determined by IP and immunoblotting (E) as described in A. (F and G) Truncation mutants of BLIMP1 were generated (F) and their interactions with full-length Hrd1 in the transiently transfected HEK293 cells were determined by IP and immunoblotting (G) as described in A. TM, transmembrane; Pro, proline; AD, acidic domain; PR, positive regulatory; ZF, zinc finger.
Hrd1 protein contains six transmembrane (TM) domains and its cytoplasmic tail carries an E3 ligase catalytic RING finger and a long proline-rich C terminus (Fig. 6 D). Analysis of BLIMP1 interaction with Hrd1 truncation mutants identified that the Hrd1 proline-rich region mediates its interaction with BLIMP1, as deletion of this region completely abolished the Hrd1–BLIMP1 interaction (Fig. 6 E). The BLIMP1 protein carries 2 acidic domains (AD), one positive regulatory (PR) domain, a proline-rich region, and 5 DNA-binding zinc finger (ZF) regions (Fig. 6 F). We generated a series of truncated BLIMP1 mutants and mapped the domains that interact with Hrd1. As indicated in Fig. 6 G, the BLIMP1 PR domain appears to be required for its interaction with Hrd1, as the N-terminal portion containing the PR domain interacted with Hrd1 and deletion of the PR domain-containing C terminus completely abolished the Hrd1 interaction.
Hrd1 promotes BLIMP1 protein degradation through ubiquitination in DCs
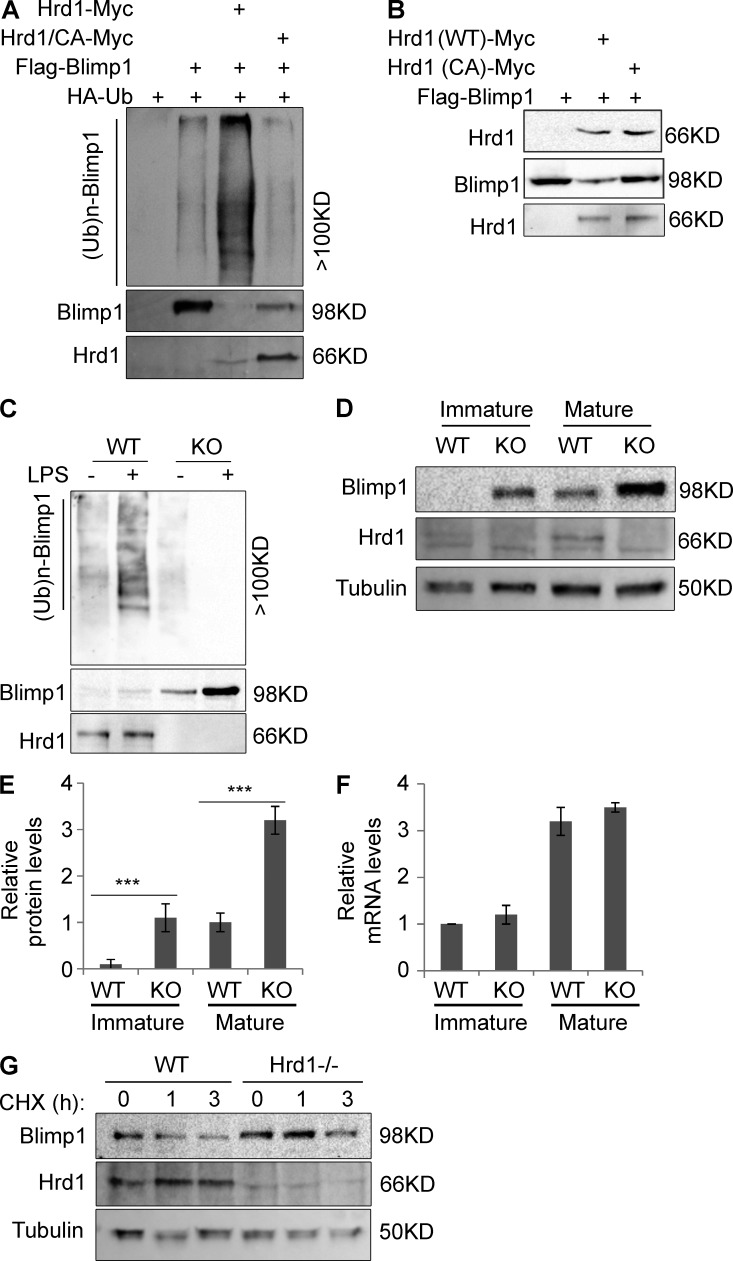
E3 ligases often catalyze ubiquitin conjugation onto interacting proteins to modulate their function. We asked whether Hrd1 catalyzes BLIMP1 ubiquitination. Indeed, transient Hrd1 expression in HEK293 dramatically enhanced BLIMP1 ubiquitination. In contrast, only a low level of BLIMP1 ubiquitination was detected in cells without Hrd1 co-transfection, presumably catalyzed by endogenous Hrd1 (Fig. 7 A). Mutation of a critical cysteine to alanine in the RING finger of Hrd1 (Hrd1/CA) that inactivates its E3 ligase catalytic activity (Gao et al., 2008) completely abolished its ability to catalyze BLIMP1 ubiquitination (Fig. 7 A), although interaction between Hrd1 and BLIMP1 was not affected (Fig. 7 B). These results indicate that Hrd1 is an E3 ubiquitin ligase of BLIMP1 and its functional RING finger is required for Hrd1-mediated BLIMP1 ubiquitination. We then speculated that loss of Hrd1 function would impair or abolish BLIMP1 ubiquitination in DCs. Compared with Hrd1-null DCs, an anti-ubiquitin antibody detected bands with higher molecular weights in anti-BLIMP1 immunoprecipitates from WT BMDCs, suggesting that BLIMP1 protein ubiquitination in WT DCs is lost in Hrd1-null DCs (Fig. 7 C, lanes 1 and 3). Similar to our results that TLR signaling positively regulates Hrd1–BLIMP1 interaction (Fig. 6 C), LPS stimulation further enhanced BLIMP1 ubiquitination in WT DCs (Fig. 7 C, lanes 1 and 2) but failed to enhance BLIMP1 ubiquitination in Hrd1-null BMDCs (Fig. 7 C, lanes 3 and 4). These results indicate that Hrd1 is required for BLIMP1 ubiquitination in mouse DCs and that TLR signaling enhances BLIMP1 ubiquitination.
Figure 7.
Hrd1 is an E3 ubiquitin ligase of BLIMP1. (A) Flag-BLIMP1 and HA-ubiquitin (Ub) plasmids were cotransfected with WT Hrd1 or its CA mutant. BLIMP1 protein in the lysates of transfected cells was immunoprecipitated (IP) with anti-Flag antibody and polyubiquitination ((Ub)n) of BLIMP1 was analyzed by immunoblotting with anti-HA antibody (top). The same membrane was reblotted with anti-BLIMP1 (middle) and again with anti-Hrd1 to confirm expression in cell lysates (bottom). (B) Interaction of BLIMP1 with WT Hrd1 or its CA mutant in transiently transfected HEK293 cells was determined by IP and immunoblotting as described in A. (C) WT and Hrd1−/− BMDCs were treated with or without LPS (200 ng/ml) for 24 h and lysed. BLIMP1 protein in the lysates was immunoprecipitated with anti-BLIMP1, followed by immunoblotting with anti-Ub antibody (top). The same membrane was reprobed with anti-BLIMP1 (middle) and again with anti-Hrd1 to confirm expression in cell lysates (bottom). (D–F) WT and Hrd1−/− BMDCs were stimulated with (mature) or without (immature) LPS (200 ng/ml) for 24 h. (D) Protein expression levels of BLIMP1 (top), Hrd1 (middle), and Tubulin (bottom) in the whole cell lysates were determined by immunoblotting. (E) Relative levels of BLIMP1 protein expression reported as mean ± SD from three independent experiments. Student’s t test was used for statistical analysis. ***, P < 0.005. (F) Total RNA was extracted from immature and mature DCs and the levels of BLIMP1 mRNA were determined by real-time quantitative PCR. Data represent means ± SD. (G) Mature WT and Hrd1−/− BMDCs were treated with cycloheximide (CHX) for the indicated times. Expression levels of BLIMP1 (top), Hrd1 (middle), and Tubulin (bottom) in the whole cell lysates were determined by immunoblotting. Representative results from 3 independent experiments are shown (n = 3).
We noticed a significant reduction in BLIMP1 protein expression in cells when WT Hrd1, but not the Hrd1/CA mutant, was coexpressed (Fig. 7, A and B). This prompted us to ask whether Hrd1 is involved in regulating BLIMP1 protein degradation. We sorted CD11c+ splenic DCs from WT and Hrd1−/− mice and analyzed the expression levels of both BLIMP1 protein and its mRNA. BLIMP1 protein expression was relatively low in the immature mouse splenic DCs, and stimulation with LPS for 24 h significantly induced BLIMP1 protein expression (Fig. 7 D, lane 1 vs. 3). Notably, compared with immature WT DCs, a dramatically higher level of BLIMP1 protein was detected in immature Hrd1-null DCs, which again was further enhanced by LPS stimulation (Fig. 7, D and E). Conversely, BLIMP1 mRNA levels were indistinguishable between WT and Hrd1-null DCs, before or after LPS stimulation (Fig. 7 F), indicating that Hrd1 suppresses BLIMP1 expression posttranscriptionally. To confirm this conclusion, we demonstrated that BLIMP1 protein expression level and half-life are significantly increased in Hrd1-null BMDCs (Fig. 7 G). Therefore, our results collectively indicate that Hrd1 is an E3 ubiquitin ligase of BLIMP1 that controls BLIMP1 protein stability in DCs.
Hrd1 regulates MHC-II expression through suppression of BLIMP1
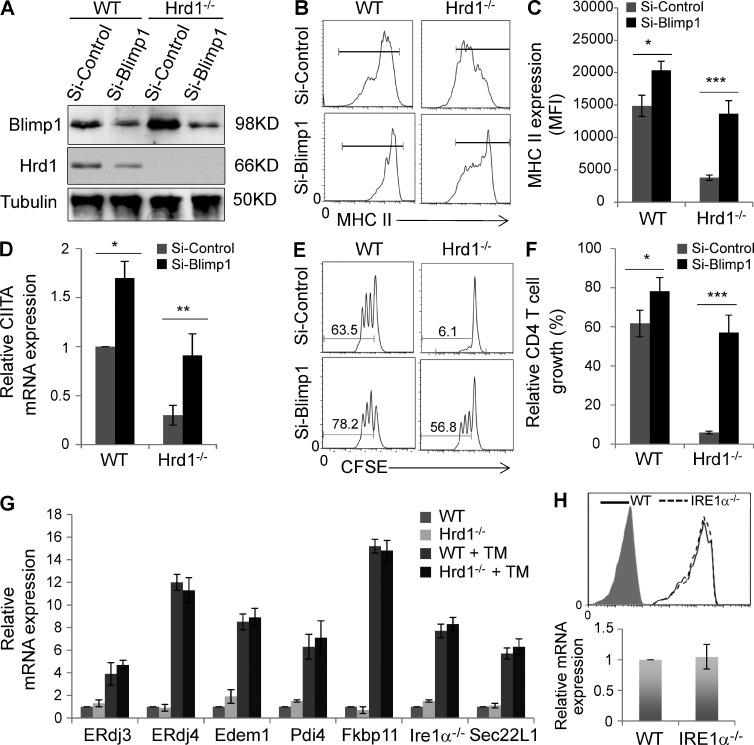
Because BLIMP1 has been shown to suppress CIITA and MHC-II expression (Kim et al., 2013), and we demonstrated that protein expression of BLIMP1 is elevated in Hrd1-null DCs (Fig. 7 D), we hypothesized that Hrd1 promotes MHC-II expression by catalyzing BLIMP1 protein degradation. To test this hypothesis, we used an shRNA-mediated knockdown approach and examined whether BLIMP1 suppression rescues MHC-II expression in Hrd1-null DCs. shRNA-mediated knockdown inhibited >95% of BLIMP1 protein expression in both WT and Hrd1-null DCs (Fig. 8 A). Suppression of BLIMP1 expression rescued MHC-II expression in Hrd1-null BMDCs to levels comparable to those of WT BMDCs (Fig. 8, B and C). We also noticed that the MHC-II expression level in Hrd1-null BMDCs with BLIMP1 knockdown was slightly but statistically significantly lower than that in WT BMDCs with BLIMP1 knockdown; this may be due to incomplete BLIMP1 knockdown in Hrd1-null DCs. We also found that BLIMP1 knockdown rescued CIITA mRNA expression in Hrd1-null DCs (Fig. 8 D). Collectively, these data suggest that Hrd1 diminishes BLIMP1-mediated CIITA suppression to promote MHC-II expression by ubiquitination and degradation of BLIMP1 protein.
Figure 8.
Hrd1 promotes MHC-II expression through degradation of BLIMP1. WT and Hrd1−/− BMDCs were transfected with control or BLIMP1-specific shRNA. (A) 3 d after transfection, protein expression levels of BLIMP1 (top) and Hrd1 (middle) were determined by immunoblotting, using Tubulin as a loading control (bottom). (B and C) MHC-II expression levels were analyzed by flow cytometry. Representative images (B) and mean fluorescence intensities ± SD (C) from 3 independent experiments are shown. (D) CIITA mRNA expression levels were determined by real-time RT-PCR. (E and F) BLIMP1 knockdown and control BMDCs were stimulated with LPS (200 ng/ml) and OVA323-339 peptide overnight, washed, and co-cultured with CD4+ T cells from OT-II mice. Representative images of CD4+ T cell proliferation (E) and mean percentages of dividing CD4+ cells ± SD (F) from three independent experiments are shown. Student’s t test was used for the statistical analysis of the data in (C, D, and F). *, P < 0.05; **, P < 0.01; ***, P < 0.005. (G) WT and Hrd1−/− splenic DCs were stimulated with or without tunicamycin for 2 h and the mRNA expression levels of each indicated UPR genes were analyzed by real-time PRC. (H) The expression levels of MHC-II on the surface of WT and IRE1a−/− splenic DCs (top) and MHC-II mRNA were analyzed. Error bars represent data from five pairs of mice (n = 5).
Next, we determined whether BLIMP1 knockdown could rescue the ability of Hrd1-null BMDCs to prime CD4+ T cells. As shown in Fig. 8 E, CD4+ T cells from OT-II mice failed to proliferate when co-cultured with Hrd1-null DCs and OVA323-339 peptides, confirming that Hrd1 is required for DC function in priming CD4+ T cells. BLIMP1 knockdown in Hrd1-null BMDCs rescued CD4+ OT-II T cell proliferation (Fig. 8, E and F). BLIMP1 knockdown in WT BMDCs also resulted in a slight but statistically significant increase in CD4+ T cell proliferation (Fig. 8, E and F). Therefore, BLIMP1 protein accumulation in Hrd1-null BMDCs appears to interfere with the function of these cells in CD4+ T cell priming, supporting the model that DC Hrd1 regulates CD4+ T cell activation through BLIMP1 degradation.
Hrd1 has been initially identified as an E3 ubiquitin ligase critical for misfolded protein degradation, raising the possibility that loss of Hrd1 expression attenuates MHC-II expression through unfolded protein response (UPR). However, analysis of the downstream UPR genes, including ERdj3, ERdj4, and Edem1 (ER-associated degradation), Pdi4 and Fkbp11 (protein folding), Ire1α (ER stress transducer), and Sec22L1 (ER-Golgi transport; Zhang et al., 2011), did not detect any significant increases in their transcription levels in Hrd1-null BMDCs, even after stimulation with the pharmacological UPR inducer tunicamycin (Fig. 8 G). Together with our observation that Hrd1 deficiency did not facilitate the ER stress–induced DC death (Fig. 2), our results largely exclude the possibility of misfolded protein responses caused by Hrd1 deficiency in DCs. In addition, we have previously reported that Hrd1 promotes the degradation of IRE1α (Gao et al., 2008). Indeed, we did detect that the IRE1α protein expression levels are increased in Hrd1-null DCs (unpublished data), raising a possibility that loss of Hrd1 expression may inhibits MHC-II expression due to the elevated IRE1α functions. However, the MHC-II expression levels, both its protein and mRNA, are not affected in IRE1α-null DCs (Fig. 8 H), excluding the possible role of IRE1α in MHC-II expression in DCs. Collectively, our results indicate that Hrd1 regulates MHC-II expression through targeting BLIMP1 degradation.
Genetic Hrd1 deletion attenuates autoimmune response in mice
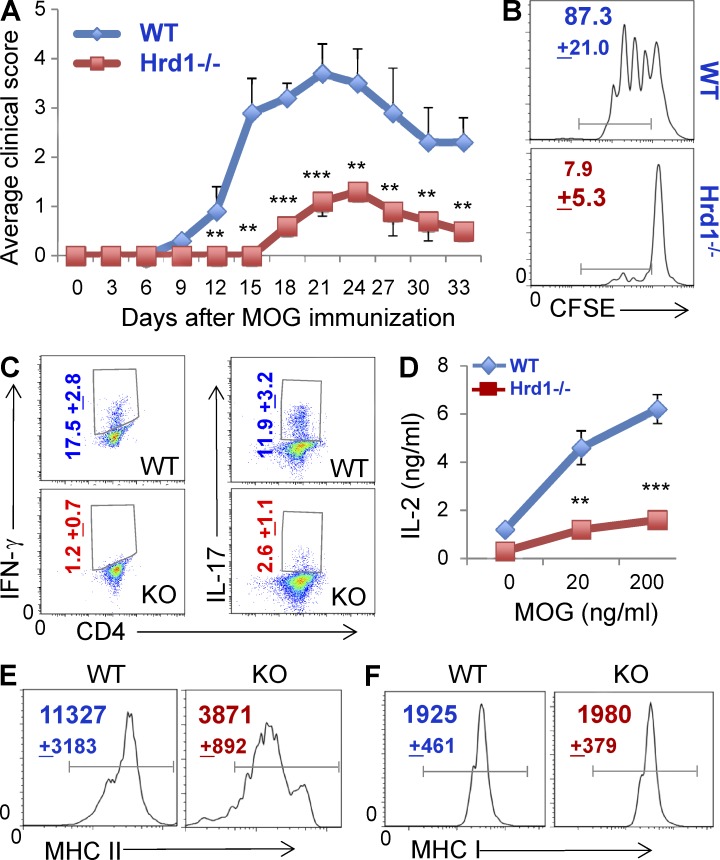
When primed by specific antigens, CD4+ T cells undergo clonal expansion and then differentiate into effector T helper (Th) cells including Th1, Th2, Th17, and T reg cells. Both Th1 and Th17 are critical pathogenic factors in multiple sclerosis in humans and experimental autoimmune encephalomyelitis (EAE) in mice (Pierson et al., 2012). Because Synv-null DCs are incapable of priming CD4+ T cells, we reasoned that genetic deletion of Hrd1 in DCs would attenuate the CD4+ T cell–mediated autoimmune inflammatory response and disease progression. We used a myelin oligodendrocyte glycoprotein (MOG)–induced EAE mouse model to test this hypothesis. Because both CD4+ T cells and B cells are increased in DC-specific Hrd1−/− mice (Fig. 3), we generated Hrd1f/fCD11c-Cre+/RAG1−/− double KO (Hrd1−/−/RAG1−/−) mice and used an adoptive transfer approach. CD11c+ cell–depleted splenocytes from WT C57/B6 mice were adoptively transferred into Hrd1−/−/RAG1−/− double KO and Hrd1+/+/RAG1−/− mice. EAE induction with MOG was initiated 1 d after adoptive transfer and the clinical symptoms were scored daily. As shown in Fig. 9 A, in contrast to the severe disease that developed in Hrd1+/+RAG1−/− recipients after MOG immunization, only modest symptoms with a dramatic delay in onset were observed in Hrd1−/−/RAG1−/− mice, indicating that Hrd1 suppression in DCs protects mice from MOG-induced EAE. MOG-specific CD4+ T cell proliferation (Fig. 9 B) and IL-2 production (Fig. 9 C) were largely diminished in the draining lymph nodes of Hrd1−/−/RAG1−/− recipients. As a consequence of impaired CD4+ T cell priming during disease progression in Hrd1-null mice, the differentiation of MOG-specific Th1 and Th17 cells was significantly inhibited (Fig. 9 D). In addition, we confirmed a significant reduction in MHC-II, but not MHC-I, expression on CD11c+ DCs in the draining lymph nodes of Hrd1−/−/RAG1−/− mice (Fig. 9, E and F). Therefore, our studies collectively indicate that DC-specific Hrd1 suppression attenuates autoimmune EAE through down-regulation of MHC-II expression, which inhibits MOG-specific CD4+ T cell priming.
Figure 9.
Genetic deletion of Hrd1 gene in DCs partially protects mice from MOG-induced EAE. (A) CD11c-depleted splenocytes from WT C57/B6 mice were adoptively transferred into 8-wk-old RAG1−/− and Hrd1−/−/RAG1−/− double KO mice. 1 d after transfer, recipients were immunized with MOG35-55 peptide (100 µg per mouse, emulsified with CFA). Mice were also given pertussis toxin (200 ng per mouse) on days 0 and 2 via tail vein injection. All mice were weighed and examined for clinical symptoms. Error bars represent data from six pairs of mice (mean ± SD; n = 6). **, P < 0.01. (B–F) Splenocytes from MOG-immunized mice were isolated, stained with CFSE, and co-cultured with MOG peptide for 3 d. (B) Proliferation of CD4+ T cells was analyzed by flow cytometry. (C) Percentages of IFN-γ–producing Th1 and IL-17–producing Th17 cells were analyzed by intracellular staining. (D) IL-2 production in the culture supernatant was examined by ELISA. Data represent means ± SD from 6 pairs of mice. **, P < 0.01. (E and F) CD11c+ conventional DCs in the draining lymph nodes from mice during disease were analyzed for their expression of MHC-II (E) and MHC-I (F) by flow cytometry. The mean fluorescence identity is indicated (mean + SD).
DISCUSSION
Our study shows that the ER membrane-spanning E3 ubiquitin ligase Hrd11 is required for DC-controlled CD4+ T cell priming in the autoimmune inflammatory response. This conclusion is supported by the following observations: targeted deletion of the Hrd1 gene in DCs impaired MHC-II expression at the transcriptional level; Hrd1-null DCs failed to prime CD4+ T cells without affecting CD8+ T cell activation; Hrd1 targets BLIMP1, a nuclear transcriptional repressor, for ubiquitin-mediated degradation in DCs; Hrd1 promotes MHC-II gene transcription and CD4+ T cell priming through BLIMP1 degradation; and genetic deletion of Hrd1 gene in DCs partially protects mice from MOG-induced EAE.
Hrd1 appeared to specifically regulate MHC-II expression without affecting the expression of either MHC-I or costimulation molecules, including CD80 and CD86. As a consequence, Hrd1-null DCs failed to prime CD4+ T cells, but antigen-specific CD8+ T cell activation was not affected. Recent studies showed that Hrd1 catalyzes the degradation of misfolded MHC-I and is possibly involved in MHC-I–restricted antigen presentation (Burr et al., 2011; Huang et al., 2011). However, we found that in mouse primary DCs, Hrd1 is not required for MHC-I protein and mRNA expression. Therefore, it is likely that Hrd1 selectively regulates MHC II gene transcription but degrades the misfolded MHC I molecules in DCs.
We identified Hrd1 as a specific E3 ubiquitin ligase that catalyzes BLIMP1 ubiquitination and degradation in mouse DCs. Loss of Hrd1 led to an accumulation of BLIMP1 in DCs, which reduced MHC-II expression; BLIMP1 knockdown rescued CIITA and MHC-II expression as well as CD4+ T cell priming. Therefore, Hrd1-mediated BLIMP1 degradation promotes the transcription of both CIITA and MHC-II genes. To our knowledge, Hrd1 is the first identified E3 ubiquitin ligase that targets BLIMP1. In addition to its important physiological functions in DCs, BLIMP1 has been shown as a critical regulator in B cell differentiation of plasma cells (Kikuchi et al., 1995; Simard et al., 2011; Maseda et al., 2012), follicular T helper cell differentiation (Crotty, 2011), CD8+ memory T cell development (Martins and Calame, 2008), and myeloid cell functions (Chang et al., 2000). It will be interesting to determine whether Hrd1-mediated BLIMP1 ubiquitination and degradation regulate the functions of these immune cell types. In addition to BLIMP1, this ER membrane-anchoring E3 ubiquitin ligase Hrd1 has been shown to target transcription factors including Nrf1 (Steffen et al., 2010; Tsuchiya et al., 2011) and p53 (Yamasaki et al., 2007). More recently, we have shown that Hrd1 catalyzes Nrf2 degradation to suppress Nrf2-mediated cellular protection during liver cirrhosis (Wu et al., 2014). As expected, similar to BLIMP1, Hrd1 recognizes these target proteins through its cytoplasmic C terminus domain. More importantly, the regulation of these transcription factors is unlikely triggered by misfolded protein response. Therefore, the accumulated evidences suggest that Hrd1 is involved in a variety of pathobiological functions by degrading cellular signaling molecules through its cytoplasmic domain.
MHC-II expression is critical for CD4+ T cell development. Mice with targeted gene deletion of MHC-II, or factors required for MHC-II gene transcription such as CIITA, lack CD4+ T cells (Chang et al., 1994; Madsen et al., 1999). Although the DC-specific Hrd1 knockout mice showed a significant reduction in MHC-II expression, the development of CD4+ and CD8+ T cells in their thymus was not affected. On the contrary, CD4+ T cells and B cells, but not CD8+ T cells, in the peripheral lymphoid organs were slightly increased in the DC-specific Hrd1 knockout mice compared with WT mice. The mechanisms underlying the increase in CD4+ T cells and B cells are not clear. It is possible that the relatively lower percentage of FoxP3+ T reg cells, which normally suppress the homeostatic proliferation of both T cells and B cells, permits an increase in the CD4+ T cells and B cell populations in the Hrd1 KO mice. However, T reg cell reduction cannot completely explain why the increase is specific to CD4+ T cells and B cells without affecting CD8+ T cells. The reduced percentage of T reg cells is probably not related to a developmental defect because their absolute numbers in the peripheral lymphoid organs of Hrd1-null mice were not different than in WT controls. Rather, it is likely that the reduction in T reg cell percentage is a consequence of the increase in CD4+ T cells. However, because MHC-II–restricted expression of self-antigen is critical for FoxP3+ T reg cell development (Sakaguchi, 2005), the possibility that Hrd1 regulates DC MHC-II expression to modulate T reg cell development cannot be fully excluded. Further, in addition to DCs, CD11c expression can be detected in other cell types (Miller et al., 2012). Therefore, further studies are needed to delineate how Hrd1 expression in DCs, as well as other types of CD11c+ cells, affects CD4+ and B cell homeostatic proliferation in mice.
Hrd1 has been identified as an anti-apoptotic factor that protects cells from ER stress-induced cell death (Carvalho et al., 2006; Denic et al., 2006), raising the possibility that loss of Hrd1 might cause DC apoptosis. However, the percentages of CD11+ cells in the spleens of Hrd1−/− mice were not altered and the absolute numbers of CD11+ cells in the spleen were increased. It has been shown that the ER stress responsive transcription factor XBP-1 (X-box binding protein 1) is required for the development and survival of CD11c+ DCs (Iwakoshi et al., 2007). We have recently reported that Hrd1 can promote degradation of IRE1α (Gao et al., 2008), the only known enzyme that activates XBP-1 (Shen et al., 2001), and our unpublished data show that the IRE1α protein, but not its mRNA, is increased in Hrd1-null DCs. Together with our observations that Ire1α gene deletion did not alter MHC-II expression on DCs, these studies suggest that loss of Hrd1 may promote DC survival due to an elevation in IRE1α-mediated XBP-1 activation, but not for the impaired MHC-II expression. We are currently generating DC-specific IRE1α−/−/Hrd1−/− double knockout mice to study the role of the Hrd1–IRE1α pathway in DC survival.
In summary, our studies reveal a previously unappreciated molecular mechanism that regulates MHC-II gene transcription, involving the ER membrane-spanning E3 ubiquitin ligase Hrd1. In this model, TLR signaling induces Hrd1 expression to promote BLIMP1 ubiquitination and degradation. Because BLIMP1 is a critical transcriptional suppressor of MHC-II expression, TLR-induced Hrd1 expression enhances MHC-II expression to facilitate CD4+ T cell response.
MATERIALS AND METHODS
Generation of Hrd1 floxed mice.
The Hrd1-targeting vector was generated as in Fig. 1 A, and then transfected into an embryonic stem cell line generated from C57BL/6 mice. Neomycin selects were screened by PCR. Seven clones were obtained and confirmed by Southern blotting. Blastocyst injections resulted in several chimeric mice with the capacity for germline transmission. Breeding of heterozygous mice yielded Hrd1wt/wt, Hrd1wt/f, and Hrd1f/f mice without phenotypic abnormalities in expected Mendelian ratios (Fig. 1 C). The DC-specific Hrd1-null mice were generated by breeding Hrd1 floxed mice with CD11c-Cre transgenic mice.
CD11c-Cre transgenic mice, OT-I and OT-II TCR transgenic mice, and RAG1 knockout mice, all of which are at the C57BL/6 genetic background, were purchased from The Jackson Laboratory. IRE1α floxed mice were used as previously described (Qiu et al., 2013). All mice used in this study were maintained and used at the Northwestern University mouse facility under pathogen-free conditions according to institutional guidelines. All animal study proposals have been approved by the institutional animal care and use committees (IACUC) at Northwestern University.
Cell lines, antibodies, and plasmids.
Human embryonic kidney (HEK) 293 cells were maintained in DMEM (Invitrogen). The media was supplemented with 10% FBS, 100 U/ml penicillin, 200 µg/ml streptomycin, and 0.25 µg/ml amphotericin B. Polyclonal antibodies against the epitope tags (Flag, HA, and Myc), Ubiquitin, BLIMP1, and β-actin were obtained from Santa Cruz Biotechnology, Inc. Anti-Hrd1 and anti-Tubulin were purchased from Sigma-Aldrich. Fluorescence-labeled antibodies, including CD11c, CD11b, CD4, CD8, CD45.1, CD45.2, MHC-I, MHC-II, CD80, and CD86, were used for flow cytometry analysis (eBioscience). Hrd1 and the ubiquitin expression plasmids were obtained as reported previously (Gao et al., 2008). Flag-BLIPM1 expression plasmids were purchased from Addgene. The truncation mutants of both Hrd1 and BLIMP1 were generated by PCR and subcloned into pCMV-Flag (Sigma-Aldrich) or pCMV-Myc vectors (Invitrogen).
BMDC cultivation and activation.
BMDCs were generated as previously described (Yang et al., 2013). BM cells were isolated from leg bones of 8–10-wk-old WT and Hrd1f/fCD11c-Cre+ (Hrd1−/−) mice and were cultured in RPMI medium containing 10% FCS and GM-CSF (20 ng/ml; BioLegend). Cell cultures were fed on days 3, 6, and 8 and used on day 9 or 10. To select pure DCs, cells were purified by CD11c microbeads (Miltenyi Biotec) and stimulated with TLR agonists LPS (Sigma-Aldrich), Pam3 (Sigma-Aldrich), and polyIC (InvivoGen).
Antigen presentation and the antigen-specific T cell proliferation assay.
For the analysis of antigen presentation, WT and Hrd1-null DCs were activated with 200 µg/ml LPS overnight and then incubated with different doses of OVA–Alexia Fluor 647 (Invitrogen) for 1 h at 37°C. Fluorescence intensity was analyzed by flow cytometry. For the study of antigen-specific T cell proliferation, WT and Hrd1-null DCs were cultivated with LPS in the presence of either OVA protein or with OVA peptides (OT-I: OVA257-264; OT-II: OVA323-339). The DCs were then washed and co-cultured with naive CD4+ T cells from OT-II TCR transgenic mice or CD8+ T cells from OT-I TCR transgenic mice for 3 d. Proliferation of co-cultured T cells was determined by either CFSE dilution or 3H-thymidine incorporation.
For in vivo antigen-specific CD4+ T cell activation, naive CD4+ T cells were isolated from OT-II TCR transgenic mice with CD4+ T cells from B6/Sjl mice as controls and stained with CFSE. The CFSE-stained cells were adoptively transferred into lethally irradiated recipient mice (RAG1−/− or RAG1−/−/Hrd1−/− double KO mice) by intravenous injection. 1 d after transfer, mice were immunized with OVA/CFA and CD4+ T cell growth was analyzed by flow cytometry.
Cell surface receptor expression by flow cytometry and IL-2 production analysis.
Single cell suspensions of thymocytes, splenocytes, or BMDCs were used for staining using specific antibodies against CD11c, CD11b, MHC-I, MHC-II, CD80, CD86, CD4, CD8, CD25, B220, CD44, and CD62L for 30 min on ice. The stained cells were washed with ice-cold PBS, fixed, and analyzed by flow cytometry. The levels of IL-2 in the culture supernatants were determined by ELISA as previously described (Zhang et al., 2009).
Real-time quantitative RT PCR.
WT and Hrd1-null CD11c+ BMDCs were activated with or without TLR agonists for 4 h. Total RNA was extracted from DCs with TRIzol reagent according to the manufacturer’s instructions (Invitrogen). cDNA was synthesized with qScript cDNA Synthesis kit (Quanta Biosciences). iQ5 and SYBRGreen Detection system (Bio-Rad Laboratories) was used for qPCR as previously described (Lee et al., 2008, 2009). Data were normalized to the expression of β-actin in each sample. Primers used to detect the expression of MHC-I, MHC-II, CIITA, BLIMP1, and Hrd1 are shown in Table S1.
Cell transfection, immunoblotting, and coimmunoprecipitation assay.
Transient transfection was performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions, in 60-mm dishes and using 2–3 µg of total DNA per transfection. 2 d after transfection, cells were lysed in 1× Nonidet P-40 lysis buffer and freshly added protease inhibitor cocktail. The cell lysates were mixed with antibodies (1 µg) for 2 h, followed by the addition of 30 µl of fast flow protein G–Sepharose beads (GE Healthcare) for an additional 2 h at 4°C. Immunoprecipitates were washed four times with Nonidet P-40 lysis buffer and boiled in 20 µl of 2× Laemmli’s buffer. Samples were subjected to 8–12% SDS-PAGE analysis and electro-transferred onto polyvinylidene difluoride membranes (Millipore). Membranes were probed with the indicated primary antibodies against Hrd1 (Sigma-Aldrich), BLIMP1 (Santa Cruz Biotechnology, Inc.), and Tubulin (Santa Cruz Biotechnology, Inc.), followed by horseradish peroxidase–conjugated secondary antibodies. Membranes were then washed and visualized with an enhanced chemiluminescence detection system (ECL; GE Healthcare). When necessary, membranes were stripped by incubation in stripping buffer (Bio-Rad Laboratories), washed, and then reprobed with other antibodies as indicated.
EAE induction, immunohistochemistry, and isolation of infiltrated lymphocytes from brain and spinal cord.
6–8-wk-old C57/BL6 mice were immunized with MOG35-55 peptide (emulsified with CFA [200 µg per mouse]). Mice were also given pertussis toxin (200 ng per mouse) on days 0 and 2 via tail vein injection. All mice were weighed and examined daily for clinical symptoms and assigned scores on a scale of 0–5 as follows: 0, no overt signs of disease; 1, limp tail; 2, limp tail and partial hindlimb paralysis; 3, complete hindlimb paralysis; 4, complete hindlimb and partial forelimb paralysis; 5, moribund state or death.
Online supplemental material.
Table S1 shows the sequence details of primers used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140283/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Chyung-Ru Wang for critical reading of the manuscript and constructive suggestions during our research.
This work was supported by National Institutes of Health (NIH) RO1 grants (AI079056 and AI108634) to D. Fang.
The authors declare no competing financial interests.
Author contributions: H. Yang, S. Kong, Q. Qiu, Z. Lin, and B. Gao performed the experiments and analyzed the data. D. Fang analyzed the data and wrote the manuscript.
Footnotes
Abbreviations used:
- BLIMP1
- B lymphocyte–induced maturation protein 1
- BMDC
- BM-derived DC
- CIITA
- MHC class II transactivator
- EAE
- experimental autoimmune encephalomyelitis
- ER
- endoplasmic reticulum
- MOG
- myelin oligodendrocyte glycoprotein
- UPR
- unfolded protein response
References
- Adams E.J., and Luoma A.M.. 2013. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol. 31:529–561 10.1146/annurev-immunol-032712-095912 [DOI] [PubMed] [Google Scholar]
- Amano T., Yamasaki S., Yagishita N., Tsuchimochi K., Shin H., Kawahara K., Aratani S., Fujita H., Zhang L., Ikeda R., et al. 2003. Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel pathogenic factor for arthropathy. Genes Dev. 17:2436–2449 10.1101/gad.1096603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr M.L., Cano F., Svobodova S., Boyle L.H., Boname J.M., and Lehner P.J.. 2011. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. USA. 108:2034–2039 10.1073/pnas.1016229108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Casals C., Barrachina M., Serra M., Lloberas J., and Celada A.. 2007. Lipopolysaccharide up-regulates MHC class II expression on dendritic cells through an AP-1 enhancer without affecting the levels of CIITA. J. Immunol. 178:6307–6315 10.4049/jimmunol.178.10.6307 [DOI] [PubMed] [Google Scholar]
- Chang C.H., Fontes J.D., Peterlin M., and Flavell R.A.. 1994. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J. Exp. Med. 180:1367–1374 10.1084/jem.180.4.1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.H., Angelin-Duclos C., and Calame K.. 2000. BLIMP-1: trigger for differentiation of myeloid lineage. Nat. Immunol. 1:169–176 10.1038/77861 [DOI] [PubMed] [Google Scholar]
- Chen H., Gilbert C.A., Hudson J.A., Bolick S.C., Wright K.L., and Piskurich J.F.. 2007. Positive regulatory domain I-binding factor 1 mediates repression of the MHC class II transactivator (CIITA) type IV promoter. Mol. Immunol. 44:1461–1470 10.1016/j.molimm.2006.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins T., Korman A.J., Wake C.T., Boss J.M., Kappes D.J., Fiers W., Ault K.A., Gimbrone M.A. Jr, Strominger J.L., and Pober J.S.. 1984. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 81:4917–4921 10.1073/pnas.81.15.4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S.2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., and Gatti E.. 2008. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. USA. 105:3491–3496 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Quan E.M., and Weissman J.S.. 2006. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 126:349–359 10.1016/j.cell.2006.05.045 [DOI] [PubMed] [Google Scholar]
- Drozina G., Kohoutek J., Jabrane-Ferrat N., and Peterlin B.M.. 2005. Expression of MHC II genes. Curr. Top. Microbiol. Immunol. 290:147–170. [DOI] [PubMed] [Google Scholar]
- Friese M.A., Jones E.Y., and Fugger L.. 2005. MHC II molecules in inflammatory diseases: interplay of qualities and quantities. Trends Immunol. 26:559–561 10.1016/j.it.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Gao B., Calhoun K., and Fang D.. 2006. The proinflammatory cytokines IL-1β and TNF-α induce the expression of Synoviolin, an E3 ubiquitin ligase, in mouse synovial fibroblasts via the Erk1/2-ETS1 pathway. Arthritis Res. Ther. 8:R172 10.1186/ar2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B., Lee S.M., Chen A., Zhang J., Zhang D.D., Kannan K., Ortmann R.A., and Fang D.. 2008. Synoviolin promotes IRE1 ubiquitination and degradation in synovial fibroblasts from mice with collagen-induced arthritis. EMBO Rep. 9:480–485 10.1038/embor.2008.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S.F., Zika E., Conti B., Zhu X.S., and Ting J.P.. 2003. Enhancement of CIITA transcriptional function by ubiquitin. Nat. Immunol. 4:1074–1082 10.1038/ni985 [DOI] [PubMed] [Google Scholar]
- Huang L., Marvin J.M., Tatsis N., and Eisenlohr L.C.. 2011. Cutting Edge: Selective role of ubiquitin in MHC class I antigen presentation. J. Immunol. 186:1904–1908 10.4049/jimmunol.1003411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido S., Goto E., Matsuki Y., and Ohmura-Hoshino M.. 2009. E3 ubiquitin ligases for MHC molecules. Curr. Opin. Immunol. 21:78–83 10.1016/j.coi.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Iwakoshi N.N., Pypaert M., and Glimcher L.H.. 2007. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J. Exp. Med. 204:2267–2275 10.1084/jem.20070525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Yasue T., Miyake K., Kimoto M., and Takatsu K.. 1995. CD38 ligation induces tyrosine phosphorylation of Bruton tyrosine kinase and enhanced expression of interleukin 5-receptor α chain: synergistic effects with interleukin 5. Proc. Natl. Acad. Sci. USA. 92:11814–11818 10.1073/pnas.92.25.11814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.J., Gregersen P.K., and Diamond B.. 2013. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J. Clin. Invest. 123:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeffler H.P., Ranyard J., Yelton L., Billing R., and Bohman R.. 1984. γ-interferon induces expression of the HLA-D antigens on normal and leukemic human myeloid cells. Proc. Natl. Acad. Sci. USA. 81:4080–4084 10.1073/pnas.81.13.4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmann S., Mühlethaler-Mottet A., Bernasconi L., Suter T., Waldburger J.M., Masternak K., Arrighi J.F., Hauser C., Fontana A., and Reith W.. 2001. Maturation of dendritic cells is accompanied by rapid transcriptional silencing of class II transactivator (CIITA) expression. J. Exp. Med. 194:379–392 10.1084/jem.194.4.379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Gao B., and Fang D.. 2008. FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood. 111:3599–3606 10.1182/blood-2007-09-115014 [DOI] [PubMed] [Google Scholar]
- Lee S.M., Gao B., Dahl M., Calhoun K., and Fang D.. 2009. Decreased FoxP3 gene expression in the nasal secretions from patients with allergic rhinitis. Otolaryngol. Head Neck Surg. 140:197–201 10.1016/j.otohns.2008.08.016 [DOI] [PubMed] [Google Scholar]
- Madsen L., Labrecque N., Engberg J., Dierich A., Svejgaard A., Benoist C., Mathis D., and Fugger L.. 1999. Mice lacking all conventional MHC class II genes. Proc. Natl. Acad. Sci. USA. 96:10338–10343 10.1073/pnas.96.18.10338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour M.K., Latz E., and Levitz S.M.. 2006. Cryptococcus neoformans glycoantigens are captured by multiple lectin receptors and presented by dendritic cells. J. Immunol. 176:3053–3061 10.4049/jimmunol.176.5.3053 [DOI] [PubMed] [Google Scholar]
- Martins G., and Calame K.. 2008. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26:133–169 10.1146/annurev.immunol.26.021607.090241 [DOI] [PubMed] [Google Scholar]
- Maseda D., Smith S.H., DiLillo D.J., Bryant J.M., Candando K.M., Weaver C.T., and Tedder T.F.. 2012. Regulatory B10 cells differentiate into antibody-secreting cells after transient IL-10 production in vivo. J. Immunol. 188:1036–1048 10.4049/jimmunol.1102500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., et al. 2007. Novel regulation of MHC class II function in B cells. EMBO J. 26:846–854 10.1038/sj.emboj.7601556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium. 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.M., Mintern J.D., and Villadangos J.A.. 2013. Control of MHC II antigen presentation by ubiquitination. Curr. Opin. Immunol. 25:109–114 10.1016/j.coi.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Pai R.K., Askew D., Boom W.H., and Harding C.V.. 2002. Regulation of class II MHC expression in APCs: roles of types I, III, and IV class II transactivator. J. Immunol. 169:1326–1333 10.4049/jimmunol.169.3.1326 [DOI] [PubMed] [Google Scholar]
- Pierson E., Simmons S.B., Castelli L., and Goverman J.M.. 2012. Mechanisms regulating regional localization of inflammation during CNS autoimmunity. Immunol. Rev. 248:205–215 10.1111/j.1600-065X.2012.01126.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurich J.F., Lin K.I., Lin Y., Wang Y., Ting J.P., and Calame K.. 2000. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat. Immunol. 1:526–532 10.1038/82788 [DOI] [PubMed] [Google Scholar]
- Qiu Q., Zheng Z., Chang L., Zhao Y.S., Tan C., Dandekar A., Zhang Z., Lin Z., Gui M., Li X., et al. 2013. Toll-like receptor-mediated IRE1α activation as a therapeutic target for inflammatory arthritis. EMBO J. 32:2477–2490 10.1038/emboj.2013.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis e Sousa C.2004. Activation of dendritic cells: translating innate into adaptive immunity. Curr. Opin. Immunol. 16:21–25 10.1016/j.coi.2003.11.007 [DOI] [PubMed] [Google Scholar]
- Rudolph M.G., Stanfield R.L., and Wilson I.A.. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466 10.1146/annurev.immunol.23.021704.115658 [DOI] [PubMed] [Google Scholar]
- Sakaguchi S.2005. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat. Immunol. 6:345–352 10.1038/ni1178 [DOI] [PubMed] [Google Scholar]
- Shen X., Ellis R.E., Lee K., Liu C.Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D.M., Mori K., and Kaufman R.J.. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 107:893–903 10.1016/S0092-8674(01)00612-2 [DOI] [PubMed] [Google Scholar]
- Simard N., Konforte D., Tran A.H., Esufali J., Leonard W.J., and Paige C.J.. 2011. Analysis of the role of IL-21 in development of murine B cell progenitors in the bone marrow. J. Immunol. 186:5244–5253 10.4049/jimmunol.1004040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen J., Seeger M., Koch A., and Krüger E.. 2010. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell. 40:147–158 10.1016/j.molcel.2010.09.012 [DOI] [PubMed] [Google Scholar]
- Steimle V., Otten L.A., Zufferey M., and Mach B.. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell. 75:135–146 10.1016/0092-8674(93)90685-J [DOI] [PubMed] [Google Scholar]
- Tooze R.M., Stephenson S., and Doody G.M.. 2006. Repression of IFN-γ induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling. J. Immunol. 177:4584–4593 10.4049/jimmunol.177.7.4584 [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Morita T., Kim M., Iemura S., Natsume T., Yamamoto M., and Kobayashi A.. 2011. Dual regulation of the transcriptional activity of Nrf1 by β-TrCP- and Hrd1-dependent degradation mechanisms. Mol. Cell. Biol. 31:4500–4512 10.1128/MCB.05663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tze L.E., Horikawa K., Domaschenz H., Howard D.R., Roots C.M., Rigby R.J., Way D.A., Ohmura-Hoshino M., Ishido S., Andoniou C.E., et al. 2011. CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10-driven MARCH1-mediated ubiquitination and degradation. J. Exp. Med. 208:149–165 10.1084/jem.20092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walseng E., Furuta K., Bosch B., Weih K.A., Matsuki Y., Bakke O., Ishido S., and Roche P.A.. 2010. Ubiquitination regulates MHC class II-peptide complex retention and degradation in dendritic cells. Proc. Natl. Acad. Sci. USA. 107:20465–20470 10.1073/pnas.1010990107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Zhao F., Gao B., Tan C., Yagishita N., Nakajima T., Wong P.K., Chapman E., Fang D., and Zhang D.D.. 2014. Hrd1 suppresses Nrf2-mediated cellular protection during liver cirrhosis. Genes Dev. 28:708–722 10.1101/gad.238246.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S., Yagishita N., Sasaki T., Nakazawa M., Kato Y., Yamadera T., Bae E., Toriyama S., Ikeda R., Zhang L., et al. 2007. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 26:113–122 10.1038/sj.emboj.7601490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lee S.M., Gao B., Zhang J., and Fang D.. 2013. Histone deacetylase sirtuin 1 deacetylates IRF1 protein and programs dendritic cells to control Th17 protein differentiation during autoimmune inflammation. J. Biol. Chem. 288:37256–37266 10.1074/jbc.M113.527531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Xu Q., Kwon M.J., Matta R., Liu Y., Hong S.C., and Chang C.H.. 2006. ERK and p38 MAPK signaling pathways negatively regulate CIITA gene expression in dendritic cells and macrophages. J. Immunol. 177:70–76 10.4049/jimmunol.177.1.70 [DOI] [PubMed] [Google Scholar]
- Young L.J., Wilson N.S., Schnorrer P., Proietto A., ten Broeke T., Matsuki Y., Mount A.M., Belz G.T., O’Keeffe M., Ohmura-Hoshino M., et al. 2008. Differential MHC class II synthesis and ubiquitination confers distinct antigen-presenting properties on conventional and plasmacytoid dendritic cells. Nat. Immunol. 9:1244–1252 10.1038/ni.1665 [DOI] [PubMed] [Google Scholar]
- Zhang J., Lee S.M., Shannon S., Gao B., Chen W., Chen A., Divekar R., McBurney M.W., Braley-Mullen H., Zaghouani H., and Fang D.. 2009. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 119:3048–3058 10.1172/JCI38902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Wang S., Malhotra J., Hassler J.R., Back S.H., Wang G., Chang L., Xu W., Miao H., Leonardi R., et al. 2011. The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J. 30:1357–1375 10.1038/emboj.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Flynt F.L., Hong M., Chen H., Gilbert C.A., Briley N.T., Bolick S.C., Wright K.L., and Piskurich J.F.. 2007. MHC class II transactivator (CIITA) expression is upregulated in multiple myeloma cells by IFN-γ. Mol. Immunol. 44:2923–2932 10.1016/j.molimm.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.