Wang et al. examine how influenza A virus causes GI symptoms. Intranasal infection in mice causes intestinal pathology via virally activated CD4 T cells in the lung up-regulating CCR9 and migrating to the intestine where they secrete IFN-γ that alters homeostasis of the microbiota. Subsequent induction of IL-15 aids differentiation into pathogenic Th17 cells in the gut.
Abstract
Influenza in humans is often accompanied by gastroenteritis-like symptoms such as diarrhea, but the underlying mechanism is not yet understood. We explored the occurrence of gastroenteritis-like symptoms using a mouse model of respiratory influenza infection. We found that respiratory influenza infection caused intestinal injury when lung injury occurred, which was not due to direct intestinal viral infection. Influenza infection altered the intestinal microbiota composition, which was mediated by IFN-γ produced by lung-derived CCR9+CD4+ T cells recruited into the small intestine. Th17 cells markedly increased in the small intestine after PR8 infection, and neutralizing IL-17A reduced intestinal injury. Moreover, antibiotic depletion of intestinal microbiota reduced IL-17A production and attenuated influenza-caused intestinal injury. Further study showed that the alteration of intestinal microbiota significantly stimulated IL-15 production from intestinal epithelial cells, which subsequently promoted Th17 cell polarization in the small intestine in situ. Thus, our findings provide new insights into an undescribed mechanism by which respiratory influenza infection causes intestinal disease.
Influenza is an infectious respiratory disease affecting many bird and mammal species (Laver and Webster, 1979; Reid et al., 1999). Clinically, the most common symptoms include cough, fever, headache, and weakness (Monto et al., 2000). These symptoms are often accompanied by gastroenteritis-like symptoms in many influenza patients, such as abdominal pain, nausea, vomiting, and diarrhea, especially in young children (Baden et al., 2009; Shinde et al., 2009; Dilantika et al., 2010). However, the immune mechanisms underlying these clinical manifestations in the intestine during a lung-tropic viral influenza infection remain unclear.
The intestinal tracts in humans and other animals are inhabited by hundreds of diverse species of commensal bacteria, which are essential in shaping intestinal immune responses during both health and disease (Hooper and Gordon, 2001; Chervonsky, 2009). Distinct components of commensal bacteria were associated with special status of the immune system. Although most commensal bacteria are beneficial (Ichinohe et al., 2011), a few can be potentially harmful in some conditions; for example, some commensal bacteria have been suggested to influence susceptibility to inflammatory bowel disease (IBD; Garrett et al., 2007; Mazmanian et al., 2008). Thus, when conditions in the host are unfavorable, such as during infection, the resulting changes within the intestinal tract environment may promote growth of the harmful bacteria that induce intestinal disease.
It is well known that the respiratory and intestinal tracts are both mucosal tissues. Over 30 years ago, John Bienenstock hypothesized that the immune cells and structures contained in mucosal tissues were universally connected within the whole body. This “common mucosal immune system” concept speculated that the mucosal immune system was itself an “organ” in which the mucosal immune cells distributed throughout the body could interplay between or among different mucosal tissues or organs (McDermott and Bienenstock, 1979; McDermott et al., 1980). Although this term was coined three decades ago, appreciation of its importance is only just beginning. Much was learned from the numerous studies conducted on the mucosal immune system during this time, which mainly focused on understanding its individual components (Holmgren and Czerkinsky, 2005; Sato and Kiyono, 2012). Although a few studies have suggested that the mucosal immune system is a system-wide organ (Gallichan et al., 2001; Sobko et al., 2010), some questions still need to be clarified. For example, how do the different components affect each other, and how is cross talk achieved among the various mucosal sites (Gill et al., 2010)?
In this study, we found that lymphocytes derived from the respiratory mucosa specifically migrated into the intestinal mucosa during respiratory influenza infection by the CCL25–CCR9 chemokine axis and destroyed the intestinal microbiota homeostasis in the small intestine, finally leading to intestinal immune injury. Our findings may provide new insights into not only the mechanisms underlying intestinal immune injury induced by influenza infection of the lung but also the interplay of immune cells between or among different mucosal sites.
RESULTS
Intranasal (i.n.), but not intragastric (i.g.), infection with influenza virus causes intestinal immune injury
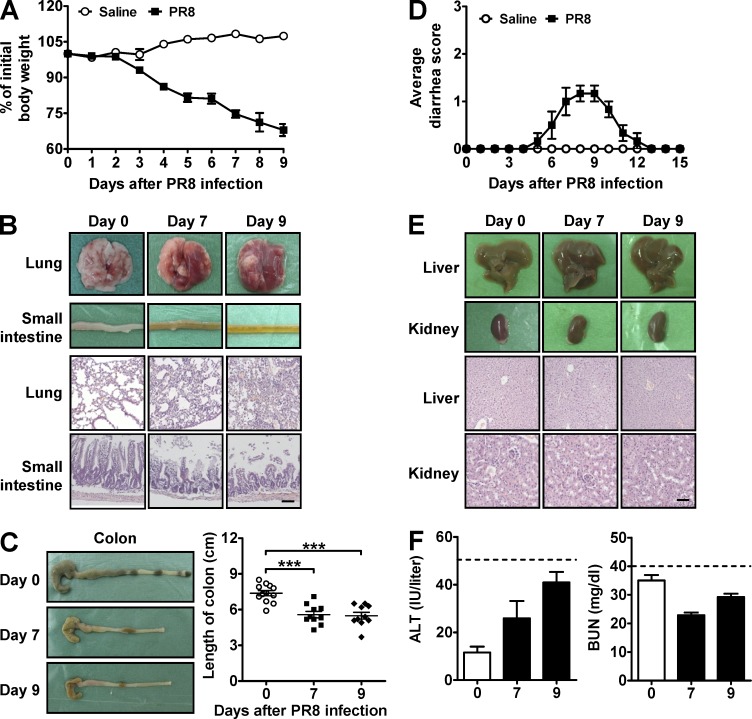
To test whether intestinal injury was also a feature in a mouse model of influenza, we infected mice i.n. with the A/PR/8/34 (PR8) influenza virus strain. Indeed, their body weight gradually decreased from days 2 to 9 as compared with saline-treated controls, which maintained their body weight over the same period (Fig. 1 A). Furthermore, both the lung and small intestine had severe injury after PR8 infection (Fig. 1 B). Colon length was shortened (Fig. 1 C) and mild diarrhea occurred (Fig. 1 D), further indicating intestinal injury (Zaki et al., 2010; Murray and Rubio-Tapia, 2012). In contrast, nonmucosal liver and kidney tissues appeared normal after PR8 infection (Fig. 1 E), which was also supported by ALT and BUN analysis (Fig. 1 F). Together, these data indicate that respiratory influenza infection causes severe immune injury not only in the lung but also in the intestine.
Figure 1.
Respiratory influenza virus infection causes lung and intestinal immune injury. C57BL/6 mice were i.n. infected with saline or 0.1 HA of PR8. (A) Body weight was monitored after PR8 infection. (B) The pathology of lung and small intestine was assayed after PR8 infection. (C) The length of colon was recorded after PR8 infection. (D) The severity of the diarrhea was scored after PR8 infection (0, normal stool or absent; 1, slightly wet and soft stool; 2, wet and unformed stool with moderate perianal staining of the coat; and 3, watery stool with severe perianal staining of the coat). (E) The pathology of liver and kidney was assayed after PR8 infection. (F) Serum ALT and BUN levels were measured after PR8 infection (dashed lines represent damage threshold). All tissue sections were stained with H&E. Bars, 100 µm. Data represent three independent experiments with at least five mice/group in A, C, and D or three mice/group in B, E, and F. Data are expressed as mean ± SEM by a Student’s t test. ***, P < 0.001.
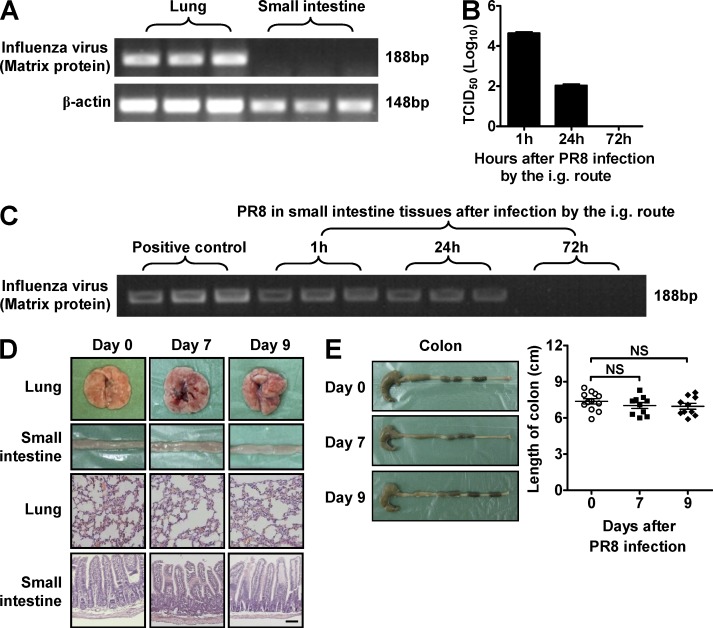
To rule out the possibility that the influenza virus entered the gastrointestinal tract and directly caused immune injury at this site, we tested for the presence of virus within the small intestine after i.n. infection and found that the influenza virus could not be detected at this site (Fig. 2 A). To test this possibility in a more rigorous way, we i.g. infected mice with PR8 and found that live virus could be detected in the intestinal contents and intestinal tissues in a short time after infection, and virus was completely cleared from these sites 3 d after infection (Fig. 2, B and C). However, pathological injury was not found in any of the examined tissues (Fig. 2, D and E). These results collectively suggest that influenza infection does not directly cause immune injury in the small intestine. Thus, we unexpectedly observed that influenza infection induced severe immune injury within the intestine only when the virus infected the respiratory tract and immune injury occurred in the lung.
Figure 2.
Influenza virus does not infect the small intestine directly. (A) C57BL/6 mice were i.n. infected with 0.1 HA of PR8. The levels of the influenza virus–derived matrix protein gene in lung and small intestine were detected by PCR. (B–E) C57BL/6 mice were i.g. infected with saline or 0.1 HA of PR8. Viral titer in intestinal contents was determined by 50% tissue culture infective dose (TCID50) assay after PR8 infection (B). The levels of the influenza virus–derived matrix protein gene in small intestine were detected by PCR after PR8 infection (C). The pathology of lung and small intestine was assayed after PR8 infection, and tissue sections were stained with H&E. Bar, 100 µm (D). The length of colon was recorded after PR8 infection (E). Data represent three independent experiments with at least three mice/group in A–E. Data are expressed as mean ± SEM by a Student’s t test. NS: not significant.
Intestinal microbiota is required for influenza-induced intestinal immune injury
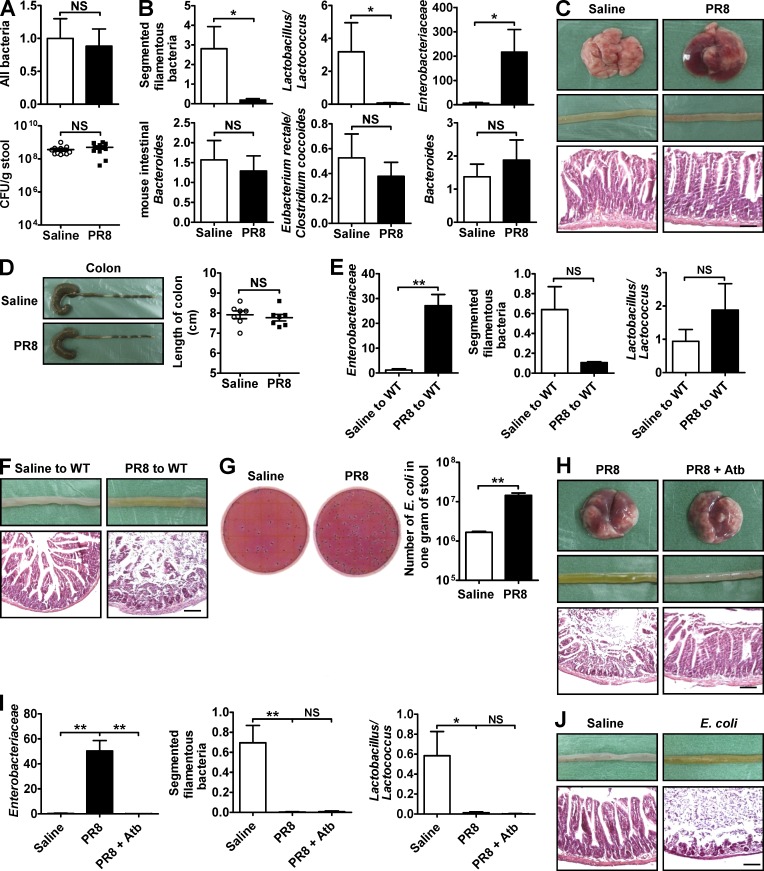
Changes in intestinal microbiota are often involved in the occurrence of intestinal inflammation in many mouse models (Lupp et al., 2007; Maslowski et al., 2009). To determine whether intestinal microbiota was involved in influenza–induced intestinal immune injury, we first assayed whether viral infection affected the relative composition of several major bacterial groups within the intestinal microbiota. Although the number of total bacteria remained the same after infection as quantified by both real-time PCR and selective culture (Fig. 3 A), the numbers of segmented filamentous bacteria (SFB) and Lactobacillus/Lactococcus decreased after PR8 infection, whereas the number of Enterobacteriaceae increased; moreover, the numbers of mouse intestinal Bacteroides, Eubacterium rectale/Clostridium coccoides, and Bacteroides were unchanged (Fig. 3 B). We next administered combinatorial antibiotics to the mice via their drinking water to deplete intestinal microbiota (Ichinohe et al., 2011) 4 wk before infecting them with PR8. In antibiotic-treated mice, the lungs still sustained severe immune-mediated injury after PR8 infection, but the small intestine and colon were protected (Fig. 3, C and D). In another way, transferring intestinal microbiota from PR8-infected mice into healthy WT mice increased the number of Enterobacteriaceae and caused intestinal immune injury in recipient mice even in the absence of viral infection as compared with the intestinal microbiota from saline-treated mice (Fig. 3, E and F). Thus, these data suggest that respiratory influenza infection induces intestinal immune injury by altering the composition of intestinal microbiota.
Figure 3.
Antibiotic treatment reduces influenza-induced intestinal immune injury. (A) Bacteria in the small intestine were assayed by real-time PCR and selective culture in blood plate 7 d after PR8 infection. (B) Several major bacterial groups in intestinal microbiota were assayed by real-time PCR 7 d after PR8 infection. (C and D) C57BL/6 mice were subjected to a 4-wk oral treatment of combinatorial antibiotics in drinking water, followed by i.n. infection with saline or 0.1 HA of PR8. The pathology of lung and small intestine was assayed 7 d after PR8 infection (C). The length of colon was recorded 7 d after PR8 infection (D). (E and F) Transfer of intestinal microbiota from saline-treated or PR8-infected mice into healthy WT mice by the i.g. route. Major bacterial groups in the intestinal microbiota (E) and the pathology of small intestine were assayed 6 d later (F). (G) The number of E. coli in stool was detected by E. coli/Coliform Count Plates 6 d after PR8 infection. (H and I) C57BL/6 mice were subjected to a 1-wk oral treatment of streptomycin in their drinking water and then were i.n. infected with 0.1 HA of PR8. The pathology of lung and small intestine (H) and major bacterial groups in intestinal microbiota (I) were assayed 6 d after PR8 infection. (J) C57BL/6 mice were i.g. infected with saline or 5 × 108 E. coli, and the pathology of small intestine was assayed 3 d later. All tissue sections were stained with H&E. Bars, 100 µm. Data represent two independent experiments with three mice/group in I and J or three independent experiments with at least three mice/group in A–H. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; NS: not significant.
Escherichia coli is an important component of Enterobacteriaceae, and pathogenic E. coli infection often causes vomiting and diarrhea in humans (Ochoa and Contreras, 2011). The number of E. coli in the intestinal tract significantly increased after PR8 infection (Fig. 3 G). Treating mice with streptomycin––an antibiotic to which E. coli is sensitive––protected mice against PR8 infection-induced immune injury to the small intestine by inhibiting the increase of Enterobacteriaceae (Fig. 3, H and I). Furthermore, directly infecting mice i.g. with E. coli caused immune injury in the small intestine (Fig. 3 J). Thus, these data suggest that the increase of E. coli may be the primary cause for intestinal immune injury during influenza infection.
Th17 cells mediate influenza-induced intestinal immune injury
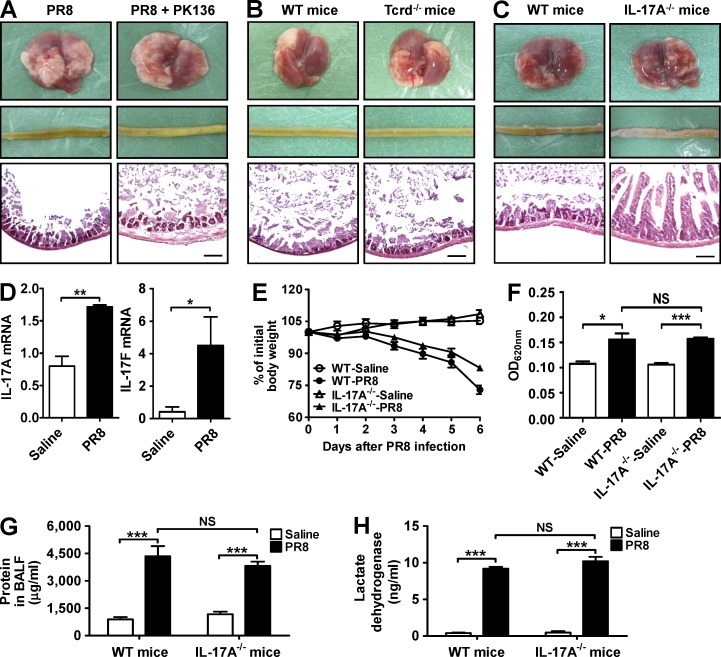
To explore the mechanism by which intestinal bacteria caused intestinal immune injury during influenza infection, many different types of proinflammatory cells involved in intestinal inflammation (Zhou et al., 2007a; Kleinschek et al., 2009; Leppkes et al., 2009) were examined. Depletion of NK1.1+ by specific antibodies or γδ T cell deficiency could not reduce the PR8 infection-induced intestinal immune injury in our study (Fig. 4, A and B). However, no intestinal injury was observed in IL-17A−/− mice after PR8 infection (Fig. 4 C), suggesting that Th17 cells might be involved in influenza-induced intestinal immune injury.
Figure 4.
IL-17A deficiency reduces influenza-induced immune injury in small intestine but not in lung. (A) The pathology of lung and small intestine from control and PK136-treated mice was assayed 6 d after PR8 infection. (B) The pathology of lung and small intestine from WT and Tcrd−/− mice was assayed 6 d after PR8 infection. (C) The pathology of lung and small intestine from WT and IL-17A−/− mice was assayed 6 d after PR8 infection. (D) IL-17A and IL-17F expressions in the lung from WT mice were detected by real-time PCR 6 d after PR8 infection. (E) Body weight of WT and IL-17A−/− mice was monitored after PR8 infection. (F) Evans blue dye concentration in BALF from WT and IL-17A−/− mice was determined by spectrophotometer 6 d after PR8 infection. (G and H) Total protein (G) and lactate dehydrogenase (H) levels in BALF from WT and IL-17A−/− mice were determined by ELISA 6 d after PR8 infection. All tissue sections were stained with H&E. Bars, 100 µm. Data represent two independent experiments with five mice/group in E–H or three mice/group in A–D. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS: not significant.
To rule out the possibility that lung injury might also be reduced in IL-17A−/− mice after influenza infection, which subsequently resulted in reducing the small intestinal injury indirectly, we compared the degree of the lung injury after influenza infection between WT and IL-17A−/− mice. The results showed that both IL-17F and IL-17A expressions in lung from WT mice were increased after PR8 infection (Fig. 4 D). Compared with WT mice, IL-17A−/− mice exhibited reduced body weight loss during PR8 infection (Fig. 4 E). However, the degree of lung leak and the levels of total protein and lactate dehydrogenase in bronchoalveolar lavage fluid (BALF) were not significantly different between WT and IL-17A−/− mice (Fig. 4, F–H), suggesting that the lung injury did not reduce in IL-17A−/− mice after influenza infection when compared with WT mice. Thus, these data suggest that the decrease of immune injury in the small intestine from IL-17A−/− mice after influenza infection is independent of the decrease of lung injury.
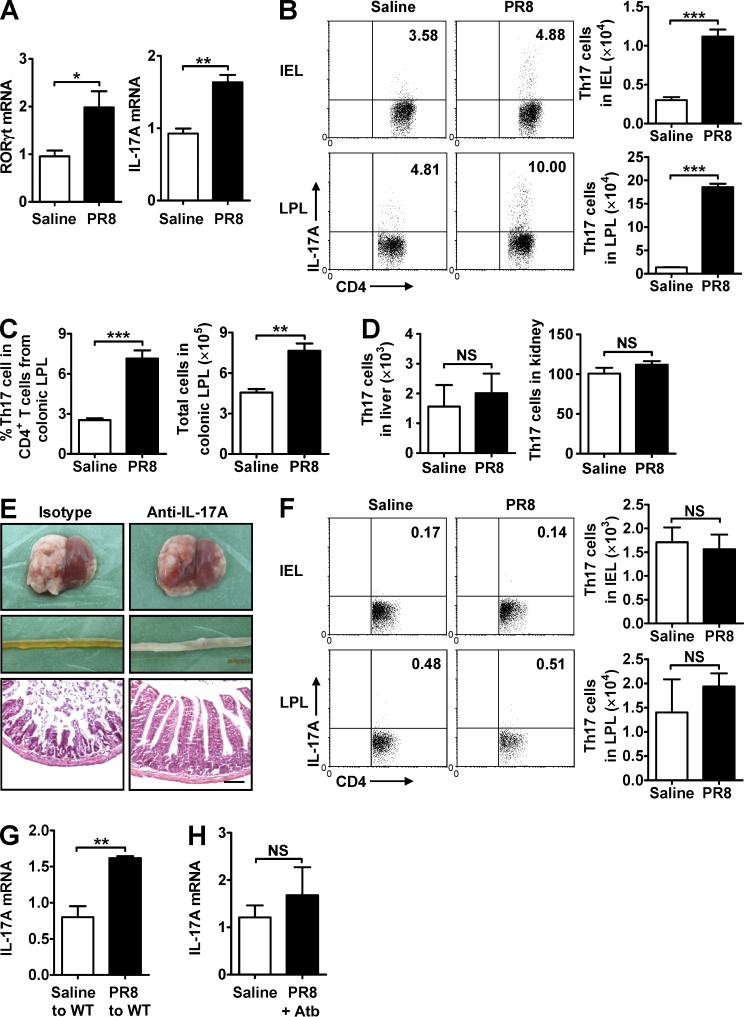
To further determine that Th17 cells were responsible for influenza-induced intestinal immune injury, we detected the expression of Th17-specific transcription factor RORγt and IL-17A and found that their expressions increased in the small intestine after PR8 infection (Fig. 5 A). The percentage and number of Th17 cells increased in the small intestine and colon after PR8 infection (Fig. 5, B and C), but not in the liver or kidney (Fig. 5 D), consistent with previous observations (Esplugues et al., 2011). Furthermore, treating mice i.p. with a neutralizing anti–IL-17A antibody during PR8 infection effectively reduced intestinal injury (Fig. 5 E). Together, these data suggest that influenza infection–induced intestinal immune injury is dependent on Th17 cells.
Figure 5.
Increased Th17 cells occur in the small intestine during influenza virus infection. (A) RORγt and IL-17A expressions in the small intestine were detected by real-time PCR 7 d after PR8 infection. (B) The percentage and number of Th17 cells in intestinal IEL and LPL were detected 7 d after PR8 infection. (C) The percentage and number of Th17 cells in colonic LPL were detected 7 d after PR8 infection. (D) The number of Th17 cells in liver and kidney was detected 7 d after PR8 infection. (E) C57BL/6 mice were i.p. treated with a neutralizing anti–IL-17A antibody during PR8 infection. The pathology of lung and small intestine was assayed 6 d after PR8 infection, and tissue sections were stained with H&E. Bars, 100 µm. (F) The percentage and number of Th17 cells in IEL and LPL were detected 7 d after PR8 infection in antibiotic-treated mice. (G) Transfer of intestinal microbiota from saline-treated or PR8-infected mice into healthy WT mice by the i.g. route. IL-17A expression in the small intestine was detected by real-time PCR 6 d later. (H) IL-17A expression in the small intestine was detected by real-time PCR at day 6 after PR8 infection in streptomycin-treated mice. Data represent two independent experiments with three mice/group in A, D, E, G, and H or three independent experiments with three mice/group in B, C, and F. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS: not significant.
Because we observed that influenza–induced intestinal immune injury is dependent on both intestinal microbiota and Th17 cells, we wondered whether there was an association between intestinal bacteria and Th17 cells. The results showed that the percentage and number of Th17 cells in the small intestine were unchanged in antibiotic-treated mice after PR8 infection as compared with uninfected control mice (Fig. 5 F); transferring intestinal microbiota from PR8-infected mice into healthy WT mice promoted IL-17A expression in the small intestine of recipient mice (Fig. 5 G); and streptomycin treatment inhibited IL-17A expression in the small intestine during PR8 infection (Fig. 5 H). Collectively, these data suggest that changes in intestinal microbiota induced by influenza infection promote Th17 cell production, which subsequently causes intestinal immune injury.
CCL25/CCR9 mediates the recruitment of lung-derived CD4+ T cells into the small intestine
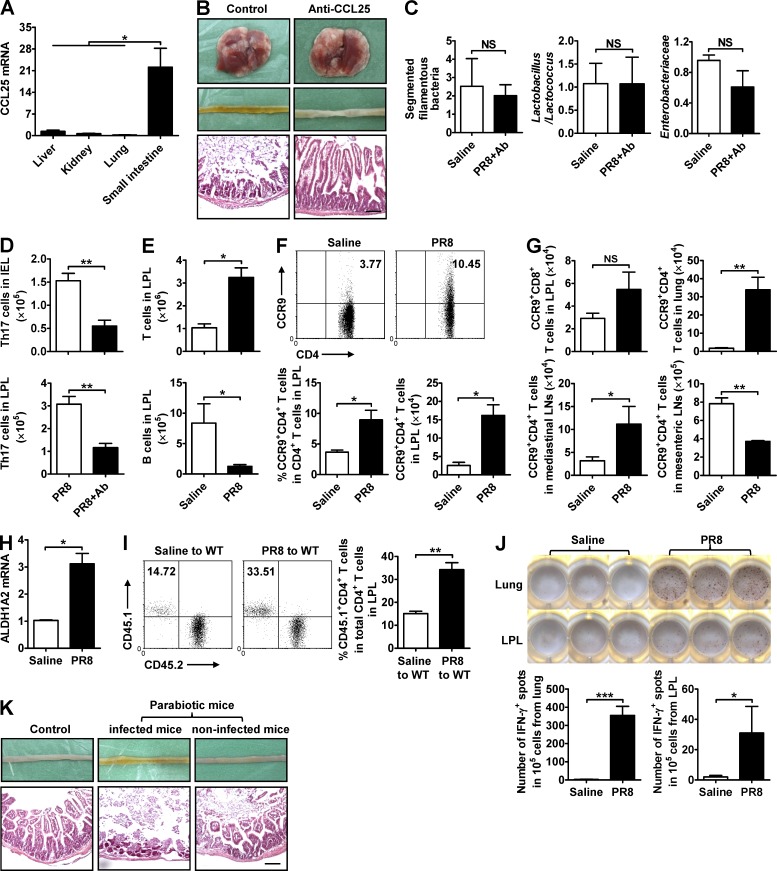
Because respiratory influenza infection influences the composition of intestinal microbiota, which subsequently promotes Th17 cell production and causes intestinal immune injury, we wanted to know how respiratory influenza infection destroyed the microecological homeostasis of the intestinal microbiota. Given that influenza infection specifically caused immune injury in the respiratory and intestinal mucosal tissues, but not in the nonmucosal liver or kidney in our study, an interconnected relationship existed between them was intriguing according to the “common mucosal immune system” theory (McDermott and Bienenstock, 1979; McDermott et al., 1980). The CCL25 chemokine is expressed by intestinal epithelial cells (IECs) and functions to specifically guide CCR9-expressing effector lymphocytes into the small intestine as a homing mechanism (Campbell and Butcher, 2002). Consistent with previous observations, CCL25 expression in the small intestine tissue was much higher than any other tissues, including liver, kidney, and lung (Fig. 6 A). Treating mice i.v. with a neutralizing anti-CCL25 antibody during PR8 infection reduced intestinal immune injury (Fig. 6 B), inhibited the changes in intestinal microbiota (Fig. 6 C), and reduced the number of Th17 cells in the small intestine (Fig. 6 D). These results suggest that the CCL25–CCR9 axis contributes to altering the composition of the intestinal microbiota after influenza infection and the subsequent development of intestinal inflammation via recruiting effector lymphocytes into the intestinal mucosa.
Figure 6.
Anti-CCL25 antibody treatment reduces influenza–induced intestinal immune injury. (A) CCL25 expression in various tissues was detected by real-time PCR 4 d after PR8 infection. (B–D) C57BL/6 mice were i.v. treated with a neutralizing anti-CCL25 antibody during PR8 infection. The pathology of lung and small intestine (B), major bacterial groups in intestinal microbiota (C), and the number of Th17 cells in IEL and LPL were assayed 7 d after PR8 infection (D). (E–G) C57BL/6 mice were i.n. infected with saline or 0.1 HA of PR8. The number of T and B cells in LPL (E), the percentage and number of CCR9+CD4+ T cells in small intestine (F), and the number of CCR9+CD8+ T cells in LPL and CCR9+CD4+ T cells in lung, mediastinal LNs, and mesenteric LNs were assayed 7 d after PR8 infection (G). (H) ALDH1A2 expression in lung was detected by real-time PCR 6 d after PR8 infection. (I) CD4+ T cells from the lungs of saline- or PR8-infected CD45.1+ mice were adoptively transferred into WT CD45.2+ mice, and the percentage of CD45.1+CD4+ T cells in total CD4+ T cells in LPL from recipient CD45.2+ mice was detected by flow cytometry 48 h later. (J) C57BL/6 mice were i.n. infected with saline or 0.1 HA of PR8. CD4+ T cells in the lung and LPL were purified 6 d later by MACS and then co-cultured with antigen-presenting cells and heat-killed PR8 in an IFN-γ ELISPOT plate. The number of positive spots was counted 20 h later. (K) Parabiotic pairs of WT mice were established first, and the left partner was i.n. infected with PR8 2 wk later. The pathology of small intestine was assayed 6 d after PR8 infection. All tissue sections were stained with H&E. Bars, 100 µm. Data represent three independent experiments with three mice/group in A–H and K or three wells/treatment in J. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS: not significant.
Next, we explored which lymphocyte subsets were recruited by CCL25 in our influenza model. Although the total number of T cells increased in the LPL after PR8 infection, the total number of B cells decreased (Fig. 6 E). Within the T cell population, the CCR9+CD4+ T cell subset increased (Fig. 6 F), whereas the CCR9+CD8+ T cell subset remained unchanged (Fig. 6 G), indicating that CCR9+CD4+ T cells might play a key role in altering the intestinal microbiota. Evaluating this subpopulation in other tissues revealed that the number of CCR9+CD4+ T cells was significantly increased in the lung and in the mediastinal LNs after PR8 infection, but not in the mesenteric LNs (Fig. 6 G), suggesting that the lung and mediastinal LNs might be the main sources of CCR9+CD4+ T cells recruited to the small intestine during PR8 infection. Retinoic acid is reported to promote the expression of CCR9 on T cells (Ohoka et al., 2011), and the production of retinoic acid is regulated by the aldehyde dehydrogenase (ALDH) 1A2 (Yokota et al., 2009). In our study, the expression of ALDH1A2 in lung increased after influenza infection (Fig. 6 H), suggesting that the increase of retinoic acid in lung after influenza infection might be responsible for promoting the CCR9 expression on lung CD4+ T cells.
To determine whether influenza infection–activated lung CD4+ T cells tended to migrate into the small intestine, we adoptively transferred lung CD4+ T cells from saline- or PR8-infected CD45.1+ mice into recipient WT CD45.2+ mice and found that LPL in recipient mice contained a higher frequency of CD45.1+CD4+ T cells from PR8-infected CD45.1+ mice (Fig. 6 I). Moreover, PR8-specific CD4+ T cells were detected not only in lung but also in the small intestine after PR8 infection, as assessed by the IFN-γ ELISPOT plate (Fig. 6 J), and, in a parabiotic mice model, PR8 infection in one partner caused small intestinal injury to occur in a noninfected partner (Fig. 6 K). Thus, these data suggest that the CCL25–CCR9 axis mediates the recruitment of lung-derived effector CD4+ T cells into the small intestine as well as the alterations to the intestinal microbiota composition during influenza infection.
Lung-derived CD4+ T cells destroy microbiota homeostasis and promote resident Th17 cell polarization
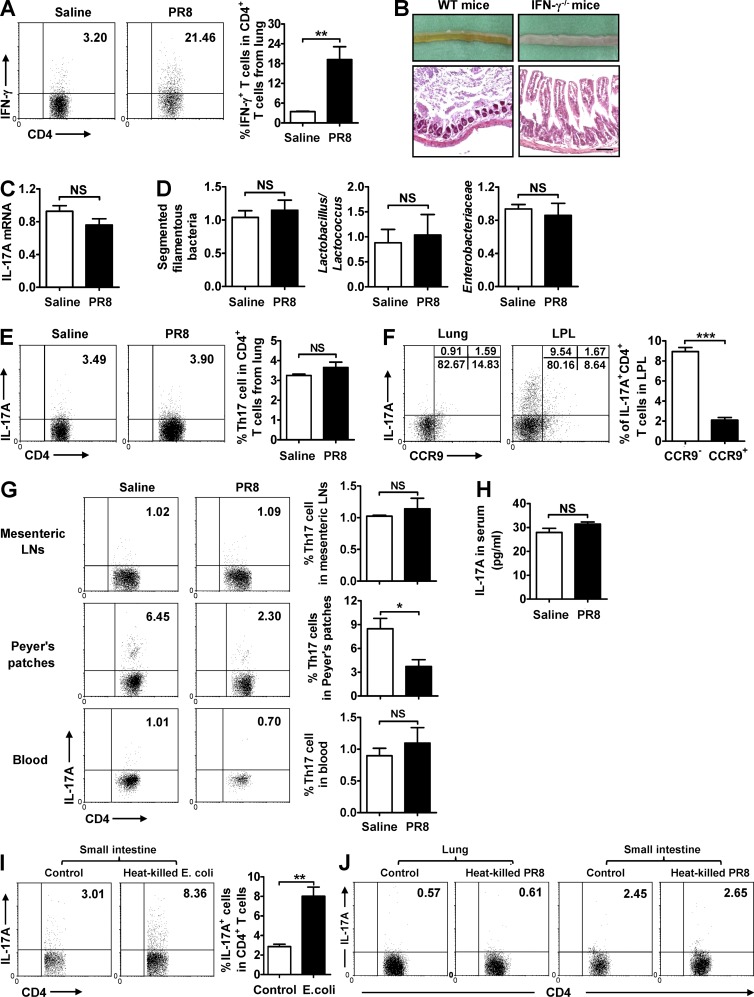
As we found that lung-derived effector CD4+ T cells are recruited into the small intestine and alter the intestinal microbiota during influenza infection, we wondered how they influenced the intestinal microbiota composition and whether Th17 cells in the small intestine originated from the polarization of them. For the first question, IFN-γ expression was found to be significantly increased in lung CD4+ T cells after PR8 infection (Fig. 7 A). When IFN-γ was deficient, the mice exhibited reduced intestinal immune injury, normal IL-17A expression, and unchanged intestinal microbiota in the small intestine after PR8 infection (Fig. 7, B–D). Thus, these data suggest that lung-derived effector CD4+ T cells destroy the homeostasis of intestinal microbiota by secreting IFN-γ. For the second question, Th17 cells were not found to be increased in lung after PR8 infection (Fig. 7 E) and, although some CCR9+ Th17 cells were present in the small intestine, most Th17 cells (∼90%) exhibited a CCR9− phenotype (Fig. 7 F). Meanwhile, Th17 cells were also not found increased in the mesenteric LNs, Peyer’s patches, and blood (Fig. 7 G), and IL-17A levels in blood did not increased after PR8 infection (Fig. 7 H). More convincing evidences showed that E. coli–specific Th17 cells could be detected in the small intestine (Fig. 7 I), but PR8-specific Th17 cells could not be detected both in the lung and small intestine (Fig. 7 J). Thus, these data suggest that Th17 cell polarization, but not recruitment, occurs in the small intestine in situ during influenza infection.
Figure 7.
Lung-derived CD4+ T cells influence microbiota and intestine injury by secreting IFN-γ. (A) IFN-γ expression in CD4+ T cells from lung was detected by flow cytometry 6 d after PR8 infection in WT mice. (B) The pathology of small intestine was assayed at day 7 after PR8 infection in WT and IFN-γ−/− mice, and tissue sections were stained with H&E. Bars, 100 µm. (C and D) IL-17A expression in the small intestine (C) and major bacterial groups in intestinal microbiota (D) were assayed at day 7 after PR8 infection in IFN-γ−/− mice. (E and F) IL-17A expression in CD4+ T cells from lung (E) and the percentages of CCR9− Th17 cells and CCR9+ Th17 cells in lung and LPL (F) were detected 6 d after PR8 infection in WT mice. (G) IL-17A expression in CD4+ T cells from mesenteric LNs, Peyer’s patches, and blood was detected by flow cytometry 6 d after PR8 infection in WT mice. (H) IL-17A level in serum was detected by ELISA 6 d after PR8 infection in WT mice. (I) LPL from WT mice at day 6 after PR8 infection was stimulated by heat-killed E. coli in vitro; 24 h later, the expression of IL-17A in CD4+ T cells was detected by flow cytometry. (J) Lung lymphocytes and LPL from WT mice at day 6 after PR8 infection were stimulated by heat-killed PR8 in vitro; 24 h later, the expression of IL-17A in CD4+ T cells was detected by flow cytometry. Data represent three independent experiments with three mice/group in A–H or three wells/treatment in I and J. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS: not significant.
Intestinal microbiota–induced IL-15 production promotes intestinal Th17 cell polarization
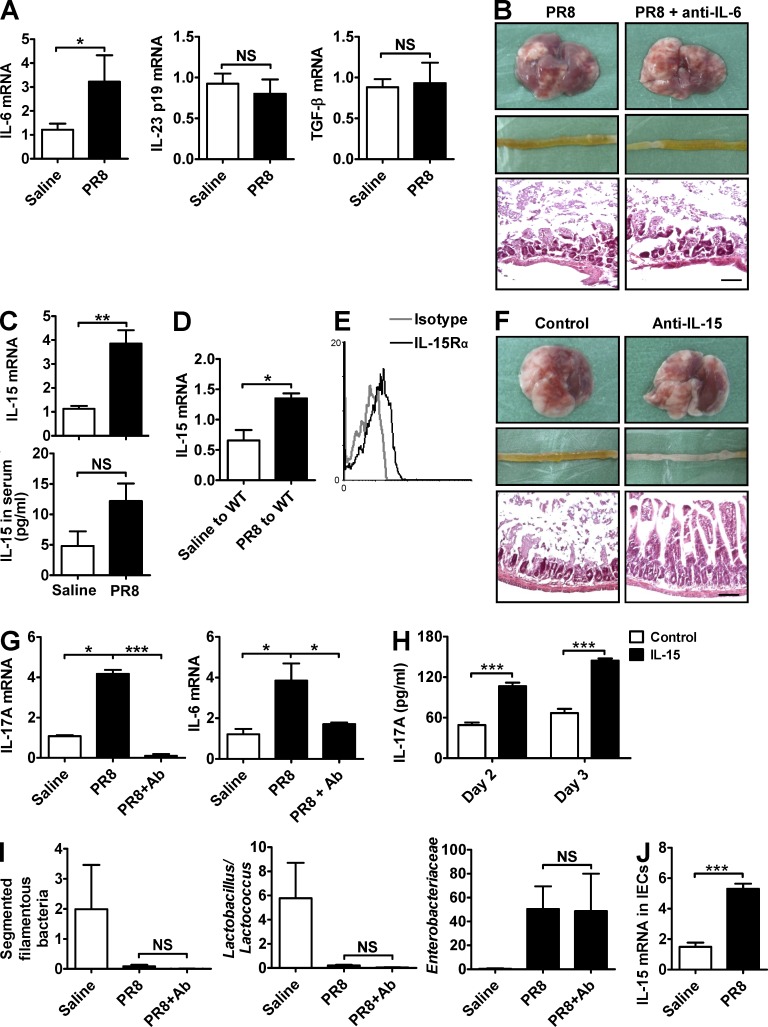
Because Th17 cell polarization occurs in the small intestine in situ during influenza infection, we next explore what kind of factors mediated this process. IL-6 expression in the small intestine was increased after PR8 infection, but IL-23 and TGF-β expressions were unchanged (Fig. 8 A). However, treating mice i.v. with a neutralizing anti–IL-6 antibody during PR8 infection could not reduce intestinal immune injury (Fig. 8 B). Thus, the increase of IL-6 is not the main reason for Th17 cell polarization in our study. IL-15 has been reported to contribute to intestinal inflammation in various mouse models (Zhou et al., 2007b; Schulthess et al., 2012) and, importantly, it has been shown to induce IL-17A expression in both mice and human CD4+ T lymphocytes (Ziolkowska et al., 2000; Ferretti et al., 2003). In our study, IL-15 expression in the small intestine, but not in serum, was up-regulated after PR8 infection (Fig. 8 C). Transferring intestinal microbiota from PR8-infected mice also increased IL-15 expression in the small intestine of recipient mice (Fig. 8 D). To explore whether IL-15 contributed to Th17 cell polarization in our study, we first assayed the expression of IL-15 receptor and found that intestinal CD4+ T cells expressed the IL-15 receptor after PR8 infection (Fig. 8 E). Next, treating mice with a neutralizing anti–IL-15 antibody during PR8 infection effectively reduced intestinal immune injury (Fig. 8 F). Thus, IL-15, which was induced by intestinal bacteria, contributes to intestinal immune injury during influenza infection. Further experiments showed that IL-15 neutralization inhibited IL-17A and IL-6 expression in the small intestine after PR8 infection (Fig. 8 G) and, consistent with the previous observations (Ziolkowska et al., 2000; Ferretti et al., 2003), exogenous IL-15 promoted IL-17A secretion in purified CD4+ T cells from LPL in vitro (Fig. 8 H), suggesting that intestinal bacteria–induced IL-15 might promote Th17 cell polarization in the small intestine in situ by a direct and/or indirect way. However, IL-15 neutralization did not influence the changes of the intestinal microbiota (Fig. 8 I), suggesting that IL-15 functioned upstream of IL-17A production but downstream of the change in microbiota after PR8 infection. Exploring the in vivo cellular sources of IL-15, high IL-15 expression was detected in IECs after PR8 infection (Fig. 8 J), suggesting that IECs might be an important source of IL-15 in the small intestine during influenza infection.
Figure 8.
Intestinal microbiota induces Th17 cell polarization in situ via triggering IL-15 production. (A) IL-6, IL-23, and TGF-β expressions in the small intestine were detected by real-time PCR 6 d after PR8 infection. (B) The pathology of lung and small intestine from control and anti–IL-6–treated mice was assayed 6 d after PR8 infection. (C) IL-15 expression in the small intestine and serum was detected 6 d after PR8 infection. (D) Transfer of intestinal microbiota from saline-treated or PR8-infected mice into healthy WT mice by the i.g. route. IL-15 expression in the small intestine was detected 6 d later. (E) IL-15Rα expression on CD4+ T cells in LPL from WT mice was detected at day 6 after PR8 infection. (F and G) C57BL/6 mice were i.p. treated with a neutralizing anti–IL-15 antibody during PR8 infection. The pathology of lung and small intestine (F) as well as IL-17A and IL-6 expressions in the small intestine were assayed 6 d after PR8 infection (G). (H) MACS-purified CD4+ T cells from LPL were stimulated by IL-15 in vitro, and IL-17A levels in supernatant were measured at days 2 and 3 by ELISA. (I) Major bacterial groups in the intestinal microbiota from control and anti–IL-15–treated mice were assayed by real-time PCR 6 d after PR8 infection. (J) IL-15 expression in IECs was detected 6 d after PR8 infection in WT mice. All tissue sections were stained with H&E. Bars, 100 µm. Data represent three independent experiments with three mice/group in A–G, I, and J or three wells/treatment in H. Data are expressed as mean ± SEM by a Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS: not significant.
DISCUSSION
Mucosal tissues, including the gastrointestinal, respiratory, and urogenital tracts, etc., are the first line of host defense against external invaders. Although much has been learned from studying each of these components individually, the mucosal immune system has not yet been examined from a holistic point of view as a system-wide organ (Gill et al., 2010), as conceptualized by the common mucosal immune system hypothesis. Unexpectedly, we observed that respiratory influenza infection in mice caused immune injury not only in the lung but also specifically in the intestine, as it had no influence on the pathology in nonmucosal organs such as the liver or kidney. Because this resembles the symptoms exhibited by humans after influenza infection, these influenza virus–infected mice provide a good model in which to study the mechanisms underlying how respiratory influenza infection causes intestinal immune injury; furthermore, these observations provide further evidence to support the existence of a common mucosal immune system.
Pathogens extensively disseminate beyond the limits of the primary infection site in almost all cases of infectious diseases, particularly at the early stage of infection. Although some studies suggest that influenza virus disseminates into extrapulmonary tissues or organs during infection (Korteweg and Gu, 2008), others contradict this finding (Mauad et al., 2010), and direct evidence for viral replication in extrapulmonary tissues or organs has not yet been shown (Kuiken and Taubenberger, 2008). It therefore remains a mystery how influenza infection can be associated with immune injury to extrapulmonary tissues or organs if these injuries are not induced by direct virus infection of these tissues or organs (Polakos et al., 2006; Mauad et al., 2010). In our mouse model of respiratory influenza infection, no influenza virus was detected in the small intestine, and i.g. administration of the influenza virus directly into the intestine did not lead to intestinal immune injury. Thus, the intestinal immune injury observed in our study was not directly caused by influenza infection of the intestine.
An explosive increase in neutrophils is responsible for influenza-induced acute lung injury and death. IL-17 is a potent regulator for the neutrophils recruitment. Previous studies have shown that respiratory influenza infection increases the IL-17A and IL-17F expressions in lung, and IL-17RA−/− mice exhibit reduced lung injury and higher survival rates after influenza infection (Crowe et al., 2009; Li et al., 2012). In our mouse model of respiratory influenza infection, IL-17A and IL-17F expressions in lung also increased after infection, but IL-17A deficiency could not reduce influenza-induced lung injury. Thus, based on above results, we speculated that IL-17A and IL-17F played the same function during influenza infection, and IL-17F alone might be enough to function to activate IL-17RA and recruit neutrophils when IL-17A was deficiency.
Recruitment and infiltration of inflammatory cells into the gastrointestinal mucosa critically regulates the development as well as progression of IBD (Wurbel et al., 2011). Differential expression of chemokine receptors and adhesion molecules on lymphocytes not only determine their migration into different tissues but also their localization within these tissues. CCL25 is constitutively expressed by the epithelium of the small intestine (Papadakis et al., 2000), and the CCL25–CCR9 chemokine axis is considered to be one of the few non-promiscuous chemokine/receptor pairs involved in gut-specific migration of lymphocytes (Stenstad et al., 2006). In our study, the percentage and number of CCR9+CD4+ T cells in both lung and small intestine increased after influenza infection, and neutralizing CCL25 with antibody treatment reduced CCR9+CD4+ T cell recruitment and intestinal immune injury. Thus, these data might explain why influenza infection specifically caused immune injury in the intestine, but not the liver or kidney, in our study.
IBD is a common disease characterized by severe inflammation of the intestine (Hooper and Macpherson, 2010). However, the exact causes of this disease remain unclear. Some studies suggest that IBD arises from dysregulated control of host–microorganism interactions. For example, patients with this disease have an increased number of epithelial cell surface–associated bacteria (Swidsinski et al., 2005), suggesting the failure of a mechanism designed to limit direct contact between the epithelium and the microbiota. Similarly, in our study, we also found that CCR9+CD4+ T cell recruitment correlated to intestinal inflammation by the following mechanism: the CCR9+CD4+ T cells destroyed the homeostasis of the intestinal microbiota, the altered microbiota then promoted Th17 cell polarization in the small intestine in situ, and the resulting IL-17A secretion finally mediated the intestinal immune injury.
This concept of the common mucosal immune system proposed by John Bienenstock suggests that the mucosal immune system may be considered as one large interconnected network (McDermott and Bienenstock, 1979; McDermott et al., 1980), which is supported by some recent researches. For example, i.n. immunization results in vaginal protection against genital HSV-2 infection (Neutra and Kozlowski, 2006), and antibiotics used in neonates increases the risk to develop asthma (Sobko et al., 2010). These findings suggest that potential exists for an undetermined link between mucosal immune components and that each component is efficient at sharing information distally (Gill et al., 2010). In our study, we found that lung-derived virus-specific effector CCR9+CD4+ T cells were recruited into the small intestine and destroyed the homeostasis of intestinal microbiota by secreting IFN-γ after influenza infection. Thus, we speculated that the effector CCR9+CD4+ T cells might enter into the small intestine by a special way as described above and remained in the active state to secrete IFN-γ even in the absence of antigen stimulation.
The intestinal microbiota is extensively accepted in the field as a virtual metabolic organ in and of itself (O’Hara and Shanahan, 2006). Beyond this role in metabolism, the intestinal microbiota has a conspicuous effect on host immune functions, as indicated by comparing immune responses between germ-free and conventional animals. A previous study showed that commensal SFBs induce IL-6 and IL-23 production to stimulate Th17 cell polarization (Ivanov et al., 2009). However, in our mouse model of respiratory influenza infection, the number of SFB decreased while the number of E. coli increased in the intestinal tract after influenza infection; E. coli promoted IL-15 expression in IECs, and this IL-15 then promoted Th17 cell polarization. Moreover, overgrowth of existing strain and/or acquisition of new pathogenic strain are involved in E. coli–caused gastrointestinal symptoms (Nguyen et al., 2006; Ochoa and Contreras, 2011). In our study, considering that the mice live in an SPF environment and that intestinal inflammation occurs in different kinds of mice, we think that the overgrowth of existing E. coli in the gut may be the primary cause for intestinal immune injury during influenza infection.
The function of IL-15 in regulation of Th17 responses has been studied extensively, but there are still some controversies. Some studies found that IL-15 induces IL-17A expression in both mice and human CD4+ T lymphocytes directly (Ziolkowska et al., 2000; Ferretti et al., 2003), which was also demonstrated in our study. However, another study showed that IL-15 inhibits Th17 cell polarization in a mouse model of EAE (Pandiyan et al., 2012). We thought that there were two main reasons to explain why IL-15 played the opposite effect in different mouse model: (1) the immuno-microenvironment in different mouse model is different; (2) IL-15 is reported to activate both STAT3 and STAT5 (Johnston et al., 1995), which is inferred either to inhibit or to promote Th17 cells.
MATERIALS AND METHODS
Mice, virus, and bacteria.
Male C57BL/6 mice were purchased from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences. IFN-γ−/−, Tcrd−/−, and IL-17A−/− mice were purchased from The Jackson Laboratory. All mice were housed under specific pathogen-free conditions at the School of Life Sciences, University of Science and Technology of China (USTC), and were used at 6–10 wk of age. Animal care and experimental procedures were followed in accordance with the experimental animal guidelines at USTC. Mouse-adapted influenza A/PR/8/34 strain (H1N1) was a gift from H. Meng (Institute of Basic Medicine, Shandong Academy of Medical Sciences, Shandong, China). For influenza infection studies, mice were anesthetized and infected i.n. with 0.1 HA of PR8 in 50 µl sterile saline. E. coli strain was isolated from stool of PR8-infected mice by the 3M Petrifilm E. coli/Coliform Count Plate and was cultured in broth medium for amplification. For E. coli infection, mice were anesthetized and infected i.g. with 5 × 108 E.coli in 500 µl sterile saline.
Histopathology.
Lung, small intestine, liver, and kidney tissues were removed and fixed immediately in 10% neutral-buffered formalin in PBS for >24 h, embedded in paraffin, and cut into 5–7-µm sections. The sections were deparaffinized and stained with hematoxylin and eosin (H&E) to determine histological changes.
Analysis of lung injury.
Lung leakage: 1 h before sacrificing mice, 20 mg/kg Evans blue dye was administered i.v. The lung was instilled with 1 ml of saline, and the BALF was collected. After centrifugation, Evans blue dye concentration in supernatant was determined by spectrophotometer at 620 nm. Total protein and lactic dehydrogenase in BALF: The lung was instilled with 1 ml saline, and the BALF was collected. After centrifugation, the level of total protein in supernatant was assayed by the BCA protein assay kit, and the level of lactic dehydrogenase in supernatant was assayed by ELISA kit (Cloud-Clone Corp).
Analysis of liver and kidney function.
Serum from infected mice or control mice were collected and stored at −80°C until analysis. Liver function was determined by measuring serum ALT (alanine aminotransferase) using a commercially available kit (Rong Sheng). Kidney function was assessed by measuring serum BUN (blood urea nitrogen) using a commercially available kit (Jiancheng Bioengineering Institute).
Determination of virus and bacteria.
Influenza virus in the lung and small intestine were detected by PCR. The primer sequences to detect the gene encoding the matrix protein within the influenza virus were as follows: 5′-GGACTGCAGCGTAGACGCTT-3′ (forward) and 5′-CATCCTGTTGTATATGAGGCCCAT-3′ (reverse). Intestinal bacterial genomic DNA was extracted from the stool using a stool kit (QIAGEN) according to the manufacturer’s instruction (the optional high-temperature step was performed). The abundance of total and specific intestinal bacterial groups was measured by real-time PCR with corresponding 16S rDNA gene primers (Sangon Biotech; Table S1). The number of E. coli in stool was detected by the 3M Petrifilm E. coli/Coliform Count Plate according to the manufacturer’s instructions.
Isolation of IEC, IEL, and LPL.
IECs were isolated as described in a previous study (Zhou et al., 2007a). IELs and LPLs were isolated as previously described with minor modifications (Das et al., 2003; Kamanaka et al., 2006; Esplugues et al., 2011). In brief, small intestines were harvested and washed with PBS, and mesentery and Peyer’s patches were carefully dissected out. Intestines were opened longitudinally and then cut into 1-cm pieces. Intestinal pieces were incubated in 10 ml of extraction buffer (5% FCS, 1 mM DTT, and 5 mM EDTA in PBS) at 37°C for 30 min. The released cells were loaded onto a Percoll gradient and centrifuged. The cells at the interface of a 40/70% Percoll solution were collected and used as IELs. The remaining segments were incubated twice in extraction buffer to remove IELs and isolate LPLs. The tissue was digested with prewarmed complete RPMI1640 containing 2 mg/ml collagenase IV at 37°C for 60 min, loaded onto a Percoll gradient, and centrifuged. The cells at the interface of a 40/70% Percoll solution were collected and used as LPLs.
Flow cytometry.
After blocking the Fc receptor with anti-CD16/CD32, single-cell suspensions were incubated with the fluorescently labeled mAbs at 4°C for 30 min in PBS and then washed twice. For intracellular cytokine staining, cells were first stimulated for 4 h at 37°C with 50 ng/ml PMA, 1 µg/ml ionomycin, and 10 µg/ml monensin (all from Sigma-Aldrich); cells were then stained for extracellular markers, fixed, permeabilized, and stained with the fluorescently labeled mAbs against the indicated intracellular cytokines or isotype control Abs. Samples were collected by a flow cytometer (LSR II; BD) and analyzed by FlowJo and WinMDI 2.9 software.
Real-time PCR.
Total RNA was extracted from tissues using TRIzol (Invitrogen), and cDNA was then synthesized. Real-time PCR was performed according to the manufacturer’s instructions using a SYBR Premix Ex Taq (Takara Bio Inc.). For analysis, target gene expression was normalized to the housekeeping gene β-actin. Gene expression values were then calculated using the mean from the control samples as a calibrator. Real-time PCR primers were synthesized by Sangon Biotech (Table S1).
Neutralizing antibodies and antibiotic treatment.
For in vivo neutralization, the following neutralizing antibodies were administered: 100 µg/mouse anti–IL-17A (TC11-18H10.1), 100 µg/mouse anti-CCL25 (89818), 100 µg/mouse anti–IL-6 (MP5-20F3), or 100 µg/mouse anti–IL-15 (AIO.3) were administered into mice at days 0, 2, and 4 after PR8 infection. For in vivo cell depletion, 200 µg/mouse anti-NK1.1 was administered i.v. into mice 2 d before PR8 infection. For intestinal microbiota depletion, mice were treated with a mixture of antibiotics (1 mg/ml ampicillin, 0.5 mg/ml vancomycin, 1 mg/ml neomycin sulfate, and 1 mg/ml metronidazole [Sangon Biotech]) added to their drinking water beginning 4 wk before PR8 infection and continuing until sacrifice, as previously described (Ichinohe et al., 2011). For intestinal E. coli depletion, mice were treated with 1 mg/ml streptomycin (Sangon Biotech) added to their drinking water beginning 1 wk before PR8 infection and continuing until sacrifice. Antibiotic-containing water was changed twice a week.
Microbiota transplantation.
Cecal contents from saline- or PR8-infected mice were suspended in 1 ml saline and were administered (0.5 ml per mouse) immediately to WT mice by the i.g. route. Transplanted mice were maintained in sterile cages and detected intestinal immune injury 7 d later.
Transfer of T cells and PR8-specific CD4+ T cells assay.
For T cells transfer, 5 × 105 CD4+ T cells from the lungs of saline- or PR8-infected CD45.1+ mice were adoptively transferred i.v. into WT CD45.2+ mice, and the percentage of CD45.1+CD4+ T cells in total CD4+ T cells in LPL from recipient CD45.2+ mice was detected 48 h later by flow cytometry. For PR8-specfic CD4+ T cells assay, CD4+ T cells in the lung and LPL from saline-treated and PR8-infected mice were purified 6 d later by MACS and then co-cultured with antigen-presenting cells and heat-killed PR8 in an IFN-γ ELISPOT plate. The number of positive spots was counted 20 h later according to the manufacturer’s instructions.
Statistical analysis.
A two-tailed Student’s t test was used for statistical analyses. Data were expressed as the mean ± SEM, and the data were considered statistically significant when differences achieved values of P < 0.05.
Online supplemental material.
Table S1 shows primers used for real-time PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140625/DC1.
Supplementary Material
Acknowledgments
We thank H. Meng (Shandong Academy of Medical Sciences) for the influenza A/PR/8/34 strain.
This work was supported by Ministry of Science & Technology of China (#2010CB911901 and #2013CB530506), Natural Science Foundation of China (#31300753 and #31400783), Fundamental Research Funds for the Central Universities (#WK2070000039), and China Postdoctoral Science Foundation (#2013M531532 and #2014T70599).
Author contributions: J. Wang and F. Li performed experiments. J. Wang, F. Li, H. Wei, Z.-X. Lian, R. Sun, and Z. Tian designed the research. J. Wang, F. Li, and Z. Tian wrote the manuscript. Z. Tian supervised the project. J. Wang and F. Li contributed equally to this paper.
Footnotes
Abbreviations used:
- BALF
- bronchoalveolar lavage fluid
- IBD
- inflammatory bowel disease
- IEC
- intestinal epithelial cell
- i.g.
- intragastrical(ly)
- i.n.
- intranasal(ly)
- SFB
- segmented filamentous bacteria
References
- Baden L.R., Drazen J.M., Kritek P.A., Curfman G.D., Morrissey S., and Campion E.W.. 2009. H1N1 influenza A disease—information for health professionals. N. Engl. J. Med. 360:2666–2667 10.1056/NEJMe0903992 [DOI] [PubMed] [Google Scholar]
- Campbell D.J., and Butcher E.C.. 2002. Intestinal attraction: CCL25 functions in effector lymphocyte recruitment to the small intestine. J. Clin. Invest. 110:1079–1081 10.1172/JCI0216946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervonsky A.2009. Innate receptors and microbes in induction of autoimmunity. Curr. Opin. Immunol. 21:641–647 10.1016/j.coi.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe C.R., Chen K., Pociask D.A., Alcorn J.F., Krivich C., Enelow R.I., Ross T.M., Witztum J.L., and Kolls J.K.. 2009. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol. 183:5301–5310 10.4049/jimmunol.0900995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Augustine M.M., Das J., Bottomly K., Ray P., and Ray A.. 2003. An important regulatory role for CD4+CD8αα T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 100:5324–5329 10.1073/pnas.0831037100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilantika C., Sedyaningsih E.R., Kasper M.R., Agtini M., Listiyaningsih E., Uyeki T.M., Burgess T.H., Blair P.J., and Putnam S.D.. 2010. Influenza virus infection among pediatric patients reporting diarrhea and influenza-like illness. BMC Infect. Dis. 10:3 10.1186/1471-2334-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esplugues E., Huber S., Gagliani N., Hauser A.E., Town T., Wan Y.Y., O’Connor W. Jr, Rongvaux A., Van Rooijen N., Haberman A.M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature. 475:514–518 10.1038/nature10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti S., Bonneau O., Dubois G.R., Jones C.E., and Trifilieff A.. 2003. IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger. J. Immunol. 170:2106–2112 10.4049/jimmunol.170.4.2106 [DOI] [PubMed] [Google Scholar]
- Gallichan W.S., Woolstencroft R.N., Guarasci T., McCluskie M.J., Davis H.L., and Rosenthal K.L.. 2001. Intranasal immunization with CpG oligodeoxynucleotides as an adjuvant dramatically increases IgA and protection against herpes simplex virus-2 in the genital tract. J. Immunol. 166:3451–3457 10.4049/jimmunol.166.5.3451 [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., and Glimcher L.H.. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131:33–45 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill N., Wlodarska M., and Finlay B.B.. 2010. The future of mucosal immunology: studying an integrated system-wide organ. Nat. Immunol. 11:558–560 10.1038/ni0710-558 [DOI] [PubMed] [Google Scholar]
- Holmgren J., and Czerkinsky C.. 2005. Mucosal immunity and vaccines. Nat. Med. 11:S45–S53 10.1038/nm1213 [DOI] [PubMed] [Google Scholar]
- Hooper L.V., and Gordon J.I.. 2001. Commensal host-bacterial relationships in the gut. Science. 292:1115–1118 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Hooper L.V., and Macpherson A.J.. 2010. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10:159–169 10.1038/nri2710 [DOI] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., and Iwasaki A.. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 108:5354–5359 10.1073/pnas.1019378108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Bacon C.M., Finbloom D.S., Rees R.C., Kaplan D., Shibuya K., Ortaldo J.R., Gupta S., Chen Y.Q., Giri J.D., et al. 1995. Tyrosine phosphorylation and activation of STAT5, STAT3, and Janus kinases by interleukins 2 and 15. Proc. Natl. Acad. Sci. USA. 92:8705–8709 10.1073/pnas.92.19.8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanaka M., Kim S.T., Wan Y.Y., Sutterwala F.S., Lara-Tejero M., Galán J.E., Harhaj E., and Flavell R.A.. 2006. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 25:941–952 10.1016/j.immuni.2006.09.013 [DOI] [PubMed] [Google Scholar]
- Kleinschek M.A., Boniface K., Sadekova S., Grein J., Murphy E.E., Turner S.P., Raskin L., Desai B., Faubion W.A., de Waal Malefyt R., et al. 2009. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 206:525–534 10.1084/jem.20081712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korteweg C., and Gu J.. 2008. Pathology, molecular biology, and pathogenesis of avian influenza A (H5N1) infection in humans. Am. J. Pathol. 172:1155–1170 10.2353/ajpath.2008.070791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., and Taubenberger J.K.. 2008. Pathology of human influenza revisited. Vaccine. 26:D59–D66 10.1016/j.vaccine.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver W.G., and Webster R.G.. 1979. Ecology of influenza viruses in lower mammals and birds. Br. Med. Bull. 35:29–33. [DOI] [PubMed] [Google Scholar]
- Leppkes M., Becker C., Ivanov I.I., Hirth S., Wirtz S., Neufert C., Pouly S., Murphy A.J., Valenzuela D.M., Yancopoulos G.D., et al. 2009. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 136:257–267 10.1053/j.gastro.2008.10.018 [DOI] [PubMed] [Google Scholar]
- Li C., Yang P., Sun Y., Li T., Wang C., Wang Z., Zou Z., Yan Y., Wang W., Wang C., et al. 2012. IL-17 response mediates acute lung injury induced by the 2009 pandemic influenza A (H1N1) virus. Cell Res. 22:528–538 10.1038/cr.2011.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C., Robertson M.L., Wickham M.E., Sekirov I., Champion O.L., Gaynor E.C., and Finlay B.B.. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2:204 10.1016/j.chom.2007.08.002 [DOI] [PubMed] [Google Scholar]
- Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D., Schilter H.C., Rolph M.S., Mackay F., Artis D., et al. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 461:1282–1286 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauad T., Hajjar L.A., Callegari G.D., da Silva L.F., Schout D., Galas F.R., Alves V.A., Malheiros D.M., Auler J.O. Jr, Ferreira A.F., et al. 2010. Lung pathology in fatal novel human influenza A (H1N1) infection. Am. J. Respir. Crit. Care Med. 181:72–79 10.1164/rccm.200909-1420OC [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., and Kasper D.L.. 2008. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 453:620–625 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- McDermott M.R., and Bienenstock J.. 1979. Evidence for a common mucosal immunologic system. I. Migration of B immunoblasts into intestinal, respiratory, and genital tissues. J. Immunol. 122:1892–1898. [PubMed] [Google Scholar]
- McDermott M.R., Clark D.A., and Bienenstock J.. 1980. Evidence for a common mucosal immunologic system. II. Influence of the estrous cycle on B immunoblast migration into genital and intestinal tissues. J. Immunol. 124:2536–2539. [PubMed] [Google Scholar]
- Monto A.S., Gravenstein S., Elliott M., Colopy M., and Schweinle J.. 2000. Clinical signs and symptoms predicting influenza infection. Arch. Intern. Med. 160:3243–3247 10.1001/archinte.160.21.3243 [DOI] [PubMed] [Google Scholar]
- Murray J.A., and Rubio-Tapia A.. 2012. Diarrhoea due to small bowel diseases. Best Pract. Res. Clin. Gastroenterol. 26:581–600 10.1016/j.bpg.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutra M.R., and Kozlowski P.A.. 2006. Mucosal vaccines: the promise and the challenge. Nat. Rev. Immunol. 6:148–158 10.1038/nri1777 [DOI] [PubMed] [Google Scholar]
- Nguyen R.N., Taylor L.S., Tauschek M., and Robins-Browne R.M.. 2006. Atypical enteropathogenic Escherichia coli infection and prolonged diarrhea in children. Emerg. Infect. Dis. 12:597–603 10.3201/eid1204.051112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara A.M., and Shanahan F.. 2006. The gut flora as a forgotten organ. EMBO Rep. 7:688–693 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa T.J., and Contreras C.A.. 2011. Enteropathogenic Escherichia coli infection in children. Curr. Opin. Infect. Dis. 24:478–483 10.1097/QCO.0b013e32834a8b8b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka Y., Yokota A., Takeuchi H., Maeda N., and Iwata M.. 2011. Retinoic acid-induced CCR9 expression requires transient TCR stimulation and cooperativity between NFATc2 and the retinoic acid receptor/retinoid X receptor complex. J. Immunol. 186:733–744 10.4049/jimmunol.1000913 [DOI] [PubMed] [Google Scholar]
- Pandiyan P., Yang X.P., Saravanamuthu S.S., Zheng L., Ishihara S., O’Shea J.J., and Lenardo M.J.. 2012. The role of IL-15 in activating STAT5 and fine-tuning IL-17A production in CD4 T lymphocytes. J. Immunol. 189:4237–4246 10.4049/jimmunol.1201476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis K.A., Prehn J., Nelson V., Cheng L., Binder S.W., Ponath P.D., Andrew D.P., and Targan S.R.. 2000. The role of thymus-expressed chemokine and its receptor CCR9 on lymphocytes in the regional specialization of the mucosal immune system. J. Immunol. 165:5069–5076 10.4049/jimmunol.165.9.5069 [DOI] [PubMed] [Google Scholar]
- Polakos N.K., Cornejo J.C., Murray D.A., Wright K.O., Treanor J.J., Crispe I.N., Topham D.J., and Pierce R.H.. 2006. Kupffer cell-dependent hepatitis occurs during influenza infection. Am. J. Pathol. 168:1169–1178, quiz :1404–1405 10.2353/ajpath.2006.050875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A.H., Fanning T.G., Hultin J.V., and Taubenberger J.K.. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA. 96:1651–1656 10.1073/pnas.96.4.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., and Kiyono H.. 2012. The mucosal immune system of the respiratory tract. Curr. Opin. Virol. 2:225–232 10.1016/j.coviro.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Schulthess J., Meresse B., Ramiro-Puig E., Montcuquet N., Darche S., Bègue B., Ruemmele F., Combadière C., Di Santo J.P., Buzoni-Gatel D., and Cerf-Bensussan N.. 2012. Interleukin-15-dependent NKp46+ innate lymphoid cells control intestinal inflammation by recruiting inflammatory monocytes. Immunity. 37:108–121 10.1016/j.immuni.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Shinde V., Bridges C.B., Uyeki T.M., Shu B., Balish A., Xu X., Lindstrom S., Gubareva L.V., Deyde V., Garten R.J., et al. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616–2625 10.1056/NEJMoa0903812 [DOI] [PubMed] [Google Scholar]
- Sobko T., Schiött J., Ehlin A., Lundberg J., Montgomery S., and Norman M.. 2010. Neonatal sepsis, antibiotic therapy and later risk of asthma and allergy. Paediatr. Perinat. Epidemiol. 24:88–92 10.1111/j.1365-3016.2009.01080.x [DOI] [PubMed] [Google Scholar]
- Stenstad H., Ericsson A., Johansson-Lindbom B., Svensson M., Marsal J., Mack M., Picarella D., Soler D., Marquez G., Briskin M., and Agace W.W.. 2006. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood. 107:3447–3454 10.1182/blood-2005-07-2860 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Weber J., Loening-Baucke V., Hale L.P., and Lochs H.. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380–3389 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbel M.A., McIntire M.G., Dwyer P., and Fiebiger E.. 2011. CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PLoS ONE. 6:e16442 10.1371/journal.pone.0016442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota A., Takeuchi H., Maeda N., Ohoka Y., Kato C., Song S.Y., and Iwata M.. 2009. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int. Immunol. 21:361–377 10.1093/intimm/dxp003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Boyd K.L., Vogel P., Kastan M.B., Lamkanfi M., and Kanneganti T.D.. 2010. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 32:379–391 10.1016/j.immuni.2010.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Wei H., Sun R., and Tian Z.. 2007a. Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J. Immunol. 178:4548–4556 10.4049/jimmunol.178.7.4548 [DOI] [PubMed] [Google Scholar]
- Zhou R., Wei H., Sun R., Zhang J., and Tian Z.. 2007b. NKG2D recognition mediates Toll-like receptor 3 signaling-induced breakdown of epithelial homeostasis in the small intestines of mice. Proc. Natl. Acad. Sci. USA. 104:7512–7515 10.1073/pnas.0700822104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowska M., Koc A., Luszczykiewicz G., Ksiezopolska-Pietrzak K., Klimczak E., Chwalinska-Sadowska H., and Maslinski W.. 2000. High levels of IL-17 in rheumatoid arthritis patients: IL-15 triggers in vitro IL-17 production via cyclosporin A-sensitive mechanism. J. Immunol. 164:2832–2838 10.4049/jimmunol.164.5.2832 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.