Akiyama et al. show that transcription factor Spi-B is up-regulated by RANKL to trigger mTEC differentiation. Osteoprotegerin is also induced by this signaling pathway and acts as a negative feedback loop to attenuate mTEC development and thymic T reg cells.
Abstract
Medullary thymic epithelial cells (mTECs) expressing the autoimmune regulator AIRE and various tissue-specific antigens (TSAs) are critical for preventing the onset of autoimmunity and may attenuate tumor immunity. However, molecular mechanisms controlling mTEC development remain elusive. Here, we describe the roles of the transcription factor Spi-B in mTEC development. Spi-B is rapidly up-regulated by receptor activator of NF-κB ligand (RANKL) cytokine signaling, which triggers mTEC differentiation, and in turn up-regulates CD80, CD86, some TSAs, and the natural inhibitor of RANKL signaling, osteoprotegerin (OPG). Spi-B–mediated OPG expression limits mTEC development in neonates but not in embryos, suggesting developmental stage–specific negative feedback regulation. OPG-mediated negative regulation attenuates cellularity of thymic regulatory T cells and tumor development in vivo. Hence, these data suggest that this negative RANKL–Spi-B–OPG feedback mechanism finely tunes mTEC development and function and may optimize the trade-off between prevention of autoimmunity and induction of antitumor immunity.
Of the various T cell types that develop in the thymus, medullary thymic epithelial cells (TECs [mTECs]) are crucial to the induction of T cell tolerance to endogenous tissues during thymic T cell development (Anderson and Takahama, 2012). Mature mTECs highly express MHC class II (MHC II) and co-stimulatory molecules, such as CD80 and CD86, and function as self-antigen–presenting cells in the thymus (Kyewski and Klein, 2006; Klein et al., 2009; Hinterberger et al., 2010). Uniquely, mature mTECs promiscuously express a wide variety of endogenous tissue-specific antigens (TSAs), including insulin, C-reactive protein, and caseins (Kyewski and Klein, 2006; Klein et al., 2009). The autoimmune regulator Aire, mutations in which cause human autoimmune diseases, is a transcription factor that is highly expressed in mature mTECs and that enhances TSA diversity (Abramson et al., 2010). Consequently, mature mTECs promote clonal deletion and regulatory T cell (T reg cell) conversion of potentially TSA-reactive T cells; these are critical for preventing the onset of autoimmunity. Furthermore, recent studies have shown that Aire deficiency inhibits tumor growth and T reg cell accumulation in tumors (Träger et al., 2012; Malchow et al., 2013; Zhu et al., 2013), suggesting that mTECs induce immunological tolerance in tumor and normal tissues. This implies that precise regulation of mTEC-mediated tolerance may be critical for balancing prevention of autoimmunity with induction of tumor immunity, but the molecular mechanisms underlying development and function of mTECs are poorly understood.
We and others previously reported that the receptor activator of NF-κB (RANK) ligand (RANKL) promotes development of mature mTECs (Rossi et al., 2007; Akiyama et al., 2008, 2012b; Hikosaka et al., 2008). Moreover, several signal transducers regulating NF-κB activation pathways, such as TNF receptor–activated factor 6 (TRAF6), NF-κB–inducing kinase (NIK), and the NF-κB family member RelB, are required for mTEC development (Burkly et al., 1995; Weih et al., 1995; Kajiura et al., 2004; Akiyama et al., 2005). Thus, RANKL probably triggers mTEC differentiation by activating NF-κB pathways (Akiyama et al., 2012b), but the molecular events involved remain unknown.
The Ets transcription factor family member Spi-B (Ray et al., 1992) regulates plasmacytoid dendritic cell development and function, B cell antigen receptor signaling, early T cell lineage decisions, and intestinal M cell development (Garrett-Sinha et al., 1999; Schotte et al., 2004; Dontje et al., 2006; Kanaya et al., 2012; Sasaki et al., 2012). The locus of human SPIB has also been associated with autoimmune primary biliary cirrhosis (Liu et al., 2010), implicating it in prevention of autoimmunity.
Here, we demonstrate that Spi-B links RANKL–NF-κB signaling with up-regulation of several molecules expressed in mature mTECs, including CD80, CD86, some TSAs, and osteoprotegerin (OPG), the natural inhibitor of RANKL. Moreover, we show that Spi-B–mediated OPG expression in the thymus limits the development of mature mTECs via a negative feedback regulatory circuit that may facilitate immune responses to tumors.
RESULTS
RANKL signaling up-regulates Spi-B expression in mTECs through an NIK-dependent pathway
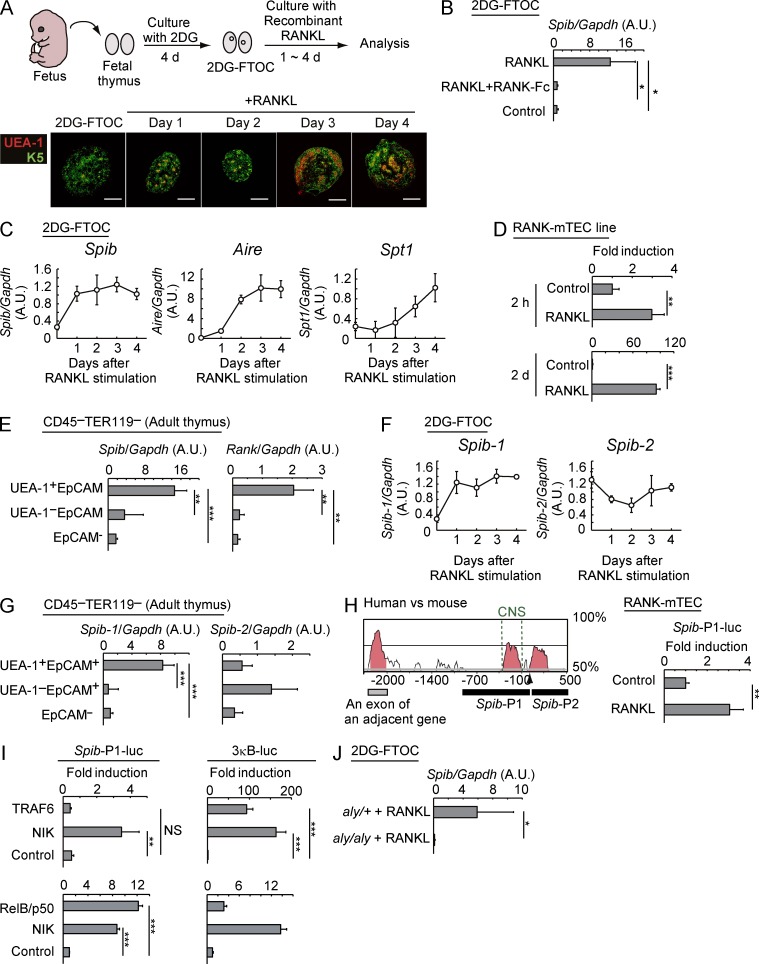
We recently identified candidate transcriptional regulators of mTEC development by microarray analysis (Ohshima et al., 2011). Spi-B was selected for further analysis because of its possible involvement in autoimmune disease (Liu et al., 2010). We first investigated whether RANKL signaling induces the expression of Spi-B in mTECs. RANKL stimulation is known to induce differentiation of mature mTECs expressing Aire, TSAs, and an mTEC marker, UEA-1 lectin ligand (Fig. 1 A) in in vitro organ culture of fetal thymic stroma (2′-deoxyguanosine [2DG]–fetal thymus organ culture [FTOC]; Rossi et al., 2007; Akiyama et al., 2008), which is prepared by depleting cells of hematopoietic origin from fetal thymus (Aichinger et al., 2012). Quantitative RT-PCR (qPCR) revealed that Spib mRNA was significantly up-regulated by RANKL stimulation in 2DG-FTOCs (Fig. 1 B). Addition of RANK-Fc blocked RANKL-dependent Spib expression (Fig. 1 B), confirming the requirement of RANKL–RANK interactions. RANKL-dependent up-regulation of Spib preceded that of Aire, the TSA Salivary protein 1 (Spt1), and UEA-1 (Figs. 1, A and C), suggesting that Spib is an early gene that responds to RANKL signaling.
Figure 1.
RANK signaling up-regulates Spi-B expression through an NIK-dependent pathway in mTECs. (A) Fetal thymic stromal organ cultures (2DG-FTOCs) were prepared and stimulated with recombinant RANKL protein. Thymic lobes isolated from an E15 mouse fetus were cultured in the presence of 2DG to eliminate hematopoietic cells, yielding 2DG-FTOCs. Addition of recombinant RANKL induced mTECs expressing Aire and TSAs in 2DG-FTOCs. Bottom panels show immunohistochemical staining of 2DG-FTOC cryosections with UEA-1, a mature mTEC marker, and keratin-5 (K5) before and at days 1, 2, 3, and 4 after RANKL stimulation. Bars, 200 µm. (B) qPCR analysis of Spib mRNA expression in 2DG-FTOCs at 4 d after incubation without (control) or with 1 µg/ml recombinant RANKL. For blocking experiments (RANKL + RANK-Fc), 5 µg/ml RANK-Fc was added to the culture medium. The values are arbitrary units (A.U.) normalized to Gapdh expression. (C) Expression of Spib, Aire, and Spt1 in 2DG-FTOC at 1–4 d after incubation with 1 µg/ml recombinant RANKL or before RANKL stimulation (day 0) were analyzed by qPCR. The values are normalized to Gapdh expression. (D) RANKL stimulation induces Spib expression in an established mTEC line. Expression of Spib in the RANK-mTEC line at 2 h and at 2 d after RANKL stimulation was determined by qPCR. Values are normalized to Gapdh expression and further normalized to the control values. (E) The UEA-1+EpCAM+ CD45−TER119− (mTEC fraction), UEA-1−EpCAM+ CD45−TER119− (cTEC-rich fraction), and EpCAM−CD45−TER119− (non-TEC fraction) fractions were sorted from 6-wk-old C57BL/6 mice. The expression levels of Spib and Rank in each fraction were determined by qPCR. Values are arbitrary units normalized to Gapdh expression. (F) qPCR analysis of expression of Spib1 and Spib2, transcripts from two different promoters of Spib, in 2DG-FTOCs after 1–4 d of incubation with 1 µg/ml recombinant RANKL or before stimulation (day 0). Values are normalized to Gapdh expression and further normalized to the control values. (G) Expression of Spib1 and Spib2 in thymic stromal cells of adult mice. mTEC, cTEC-rich, and non-TEC fractions were sorted from 6-wk-old C57BL/6 mice. Expression levels of Spib1 and Spib2 in each fraction were determined by qPCR. Values are arbitrary units normalized to Gapdh expression. (H) Human and mouse Spib genes were compared using the VISTA algorithm. The arrowhead indicates the transcriptional start site for Spib1 (transcribed from Spib-P1). The CNS shows a conserved noncoding sequence upstream of the Spib1 initiation site. The black bars (Spib-P1 and Spib-P2) indicate the promoter regions isolated for the reporter assay. After RANK-mTECs were transfected with Spib-P1 luciferase reporter plasmid and a β-galactosidase plasmid and stimulated with 1 µg/ml recombinant RANKL, luciferase activities were determined. The fold induction was calculated on the basis of the luciferase activity of unstimulated cells. (I) Overexpression of NIK or RelB complex, but not TRAF6, activates Spib-P1. HEK293T cells were transfected with expression vectors encoding NIK, TRAF6, or a combination of RelB and p50 together with a Spib-P1 luciferase reporter plasmid or artificial NF-κB promoter (3κB) and a β-galactosidase plasmid as an internal control for transfection efficiency. The 3κB promoter contains three typical NF-κB–binding sites. The fold induction was calculated on the basis of the vector control sample. Representative data from three independent triplicate experiments are shown in the graphs. (J) RANKL-mediated Spib expression in 2DG-FTOCs is dependent on functional NIK. qPCR analysis of Spib expression in aly/+ and aly/aly 2DG-FTOCs at 4 d after incubation with 1 µg/ml recombinant RANKL was performed. Error bars represent mean ± one SD for three independent experiments (B–H and J) or triplicate experiments (I). *, P < 0.05; **, P < 0.01; and ***, P < 0.001 (Student’s t test).
We next investigated whether RANKL signaling induces Spib expression in an established mTEC line because 2DG-FTOCs contain several types of stromal cells, including TECs. This mTEC line was retrovirally transduced with RANK (RANK-TEC) because endogenous RANK expression was undetectable in the currently available mTEC lines. Stimulation with RANKL rapidly and continuously up-regulated Spib expression in the RANK-TECs (Fig. 1 D). These findings suggest that Spib expression is rapidly induced by RANKL signaling in mTECs.
Subsequently, we determined whether Spi-B is expressed in mTECs in vivo. Adult mouse thymic stroma cells (CD45−TER119−) were sorted by staining with anti-EpCAM (an epithelial cell marker) and UEA-1 and were subsequently processed for qPCR gene expression assays. The mTEC fraction (UEA-1+EpCAM+), which exhibits high RANK expression levels (Fig. 1 E), showed significantly higher levels of Spi-B expression than did the cTEC-rich fraction (UEA-1−EpCAM+) and the nonepithelial stromal cell fraction (EpCAM−; Fig. 1 E). These data were consistent with increased Spi-B expression after RANKL signaling in thymic RANK-expressing mTECs.
Next, we investigated the mechanisms regulating Spi-B expression that occur downstream of RANKL signaling. Previous studies showed that murine Spib is transcribed from two different promoters (Chen et al., 1998), which can be discriminated using qPCR (Bartholdy et al., 2006). RANKL stimulation selectively induced 1 of the Spib transcripts (Spib1) in 2DG-FTOC (Fig. 1 F). Moreover, Spib1 expression was higher in mTECs than in other stromal cell fractions (Fig. 1 G). Thus, increased Spib expression in mTECs (Fig. 1 E) follows expression of Spib1 but not that of Spib2. Because RANKL signaling selectively activates the promoter that drives Spib1 transcription, the genomic region containing the conserved noncoding sequence upstream of the Spib1 initiation site (Fig. 1 H) was linked to a luciferase gene for reporter assays (Spib-P1-luc). As expected, Spib-P1-luc was responsive to RANKL stimulation in the RANK-mTEC line (Fig. 1 H), suggesting that this region contains the promoter that drives Spib transcription.
Previous studies showed that the signal transducers TRAF6 and NIK, which mediate classical and nonclassical NF-κB activation pathways, respectively, are required during RANKL-mediated mTEC differentiation (Kajiura et al., 2004; Akiyama et al., 2005, 2008). Thus, we investigated whether overexpression of these two signal transducers activated Spib-P1 in cells. Interestingly, Spib-P1 was activated only in NIK-overexpressing cells (Fig. 1 I), whereas an artificial NF-κB promoter (3κB-luc) was effectively activated by NIK or TRAF6 overexpression (Fig. 1 I). Induction of NIK activates the RelB complex (Thu and Richmond, 2010; Sun, 2012). Overexpression of the latter effectively activated the Spib-P1 promoter (Fig. 1 I). Moreover, a dysfunctional point mutation in Nik (aly/aly mutation) markedly diminished RANKL-mediated Spib expression in 2DG-FTOCs (Fig. 1 J; Shinkura et al., 1999). Thus, these data suggested that Spib is a target gene of RANKL–NIK–NF-κB signaling that promotes mTEC development (Kajiura et al., 2004; Akiyama et al., 2008, 2012b). Therefore, we investigated the roles of Spi-B in mature mTEC development in Spi-B–deficient (Spib−/−) mice.
Spi-B limits development of mature mTECs but promotes CD80 expression in mature mTECs
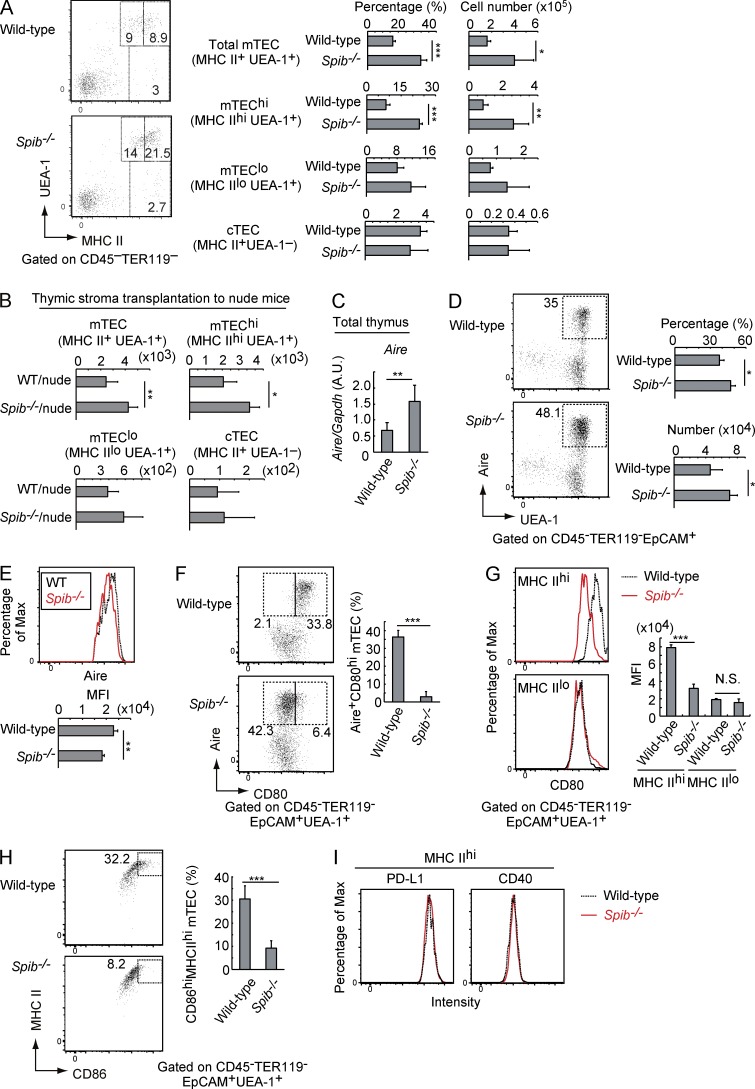
Adult murine mTECs are classified into subsets according to the expression levels of MHC II. mTECs expressing high MHC II levels (mTEChi) express a wide variety of TSAs (Kyewski and Klein, 2006; Anderson and Takahama, 2012) and are considered more mature than those expressing low MHC II (mTEClo) levels (Gray et al., 2007; Anderson et al., 2009). Flow cytometry revealed that the numbers and frequencies of total mTECs and mTEChi were significantly increased in the absence of Spi-B (Fig. 2 A), indicating that Spi-B is a negative regulator of mature mTEC development. To confirm that expression of Spi-B in TECs, but not in thymocytes, is responsible for this phenotype, we transplanted 2DG-FTOCs from Spib−/− and WT fetal thymus into the kidney capsules of athymic nude mice. In these chimeric mice, the thymus comprises Spi-B–deficient thymic stroma and thymocytes derived from nude mice. mTEChi numbers were significantly increased in the developed thymus of mice receiving Spib−/− 2DG-FTOCs (Fig. 2 B), indicating that Spi-B in thymic stromal cells limits the development of mTEChi cells.
Figure 2.
Spi-B limits development of mature mTECs and up-regulates CD80 and CD86 expression in mature mTECs. (A) Thymic stromal cells (CD45−TER119−) among total thymic cells of 3-wk-old WT and Spib−/− mice were analyzed by staining with MHC II (I-A/I-E) antibody and UEA-1 lectin, an mTEC marker (left). Flow cytometric data are summarized in the graphs. The percentages of MHC IIhiUEA-1+ (mTEChi), MHC IIloUEA-1+ (mTEClo), and UEA-1−MHC II+ (mainly cTEC) cells among thymic stroma cells and their numbers in the thymus are shown. (B) 2DG-FTOC prepared from a Spib−/− or WT fetus (six lobes each) were transplanted into the kidneys of nude mice. The grafted thymuses were analyzed by flow cytometry at 6 wk after transplantation. The numbers of MHC II+UEA-1+, MHC IIhiUEA-1+ (mTEChi), MHC IIloUEA-1+ (mTEClo), and UEA-1−MHC II+ (mainly cTEC) cells in the transplanted thymuses are shown. (C) Expression levels of Aire in WT and Spib−/− thymuses of 3-wk-old mice were determined by qPCR. Values are arbitrary units (A.U.) normalized to Gapdh expression. (D) Flow cytometric profiles of Aire-expressing mTECs in WT and Spib−/− TECs (left) in 3-wk-old mice. TECs (EpCAM+CD45−TER119−) were analyzed by staining with a combination of UEA-1 lectin and Aire antibody. The percentage of Aire+ mTECs among TECs and cell number of Aire+ mTECs in the thymus are shown in the graphs on the right. (E) Flow cytometric profiles of Aire expression in Aire-expressing mTECs of 3-wk-old WT and Spib−/− mice. Expression of Aire in Aire+UEA-1+EpCAM+CD45−TER119− of WT and Spib−/− mice was analyzed. Expression levels of Aire as means of fluorescence intensity (MFI) are summarized in the graph below. (F) Flow cytometric profiles of Aire-expressing mTECs in WT and Spib−/− TECs (left). TECs (EpCAM+CD45−TER119−) of 3-wk-old mice were analyzed by staining with a combination of CD80 and Aire antibodies. The percentage of Aire+CD80hi mTECs among TECs is shown in the graph on the right. (G) Flow cytometric profiles of CD80 expression in mTECs of 3-wk-old WT and Spib−/− mice. Expression of CD80 in mTECs expressing high levels of MHC II (MHC IIhi) and mTECs expressing low levels of MHC II (MHC IIlo) of WT and Spib−/− mice were analyzed. Expression levels of CD80 as means of fluorescence intensity are summarized in the graph on the right. (H) Flow cytometric profiles of MHC II+CD86+ cells in mTECs (UEA-1+EpCAM+CD45−TER119−) of WT and Spib−/− mice (left). The percentage of MHC IIhiCD86hi cells (left, rectangles) among mTECs is shown in the graph on the right. (I) Flow cytometric profiles of PD-L1 and CD40 expression in mTECs expressing high levels of MHC II (MHC IIhi) of WT and Spib−/− mice. Error bars represent mean ± one SD for four (A and D–H) or three (B and C) independent experiments. *, P < 0.05, **, P < 0.01; and ***, P < 0.001 (Student’s t test).
We next investigated the expression of other mTEC marker proteins in Spib−/− thymuses using flow cytometry. The mTEChi subset includes Aire-expressing mTECs (Gray et al., 2007). Moreover, mTEChi cells express high levels of co-stimulatory molecules, such as CD80, CD86, PD-L1, and CD40 (Kyewski and Klein, 2006; Klein et al., 2009; Hinterberger et al., 2010). As expected, Aire expression levels in whole thymus were increased by the lack of Spi-B (Fig. 2 C). Consistently, the number and frequency of Aire-expressing mTECs were increased in Spib−/− thymuses (Fig. 2 D), whereas Aire expression levels in Aire-expressing mTECs were slightly reduced by the absence of Spi-B (Fig. 2 E). Remarkably, CD80 expression was diminished in Spib−/− mTEChi, including Aire-expressing mTECs (Fig. 2, F and G). Moreover, mTECs expressing high levels of CD86 were significantly reduced in Spib−/− mice (Fig. 2 H). In contrast, expression of the other co-stimulatory molecules, PD-L1 and CD40, was not affected by the absence of Spi-B (Fig. 2 I), suggesting that Spi-B selectively up-regulates CD80 and CD86 among co-stimulatory molecules in mTEChi cells. Altogether, these data demonstrated two distinct novel functions of Spi-B during mTEC development: Spi-B limits mature mTEC development and promotes expression of co-stimulatory molecules CD80 and CD86 in mTECs.
Spi-B facilitates expression of OPG and some TSAs in mTECs
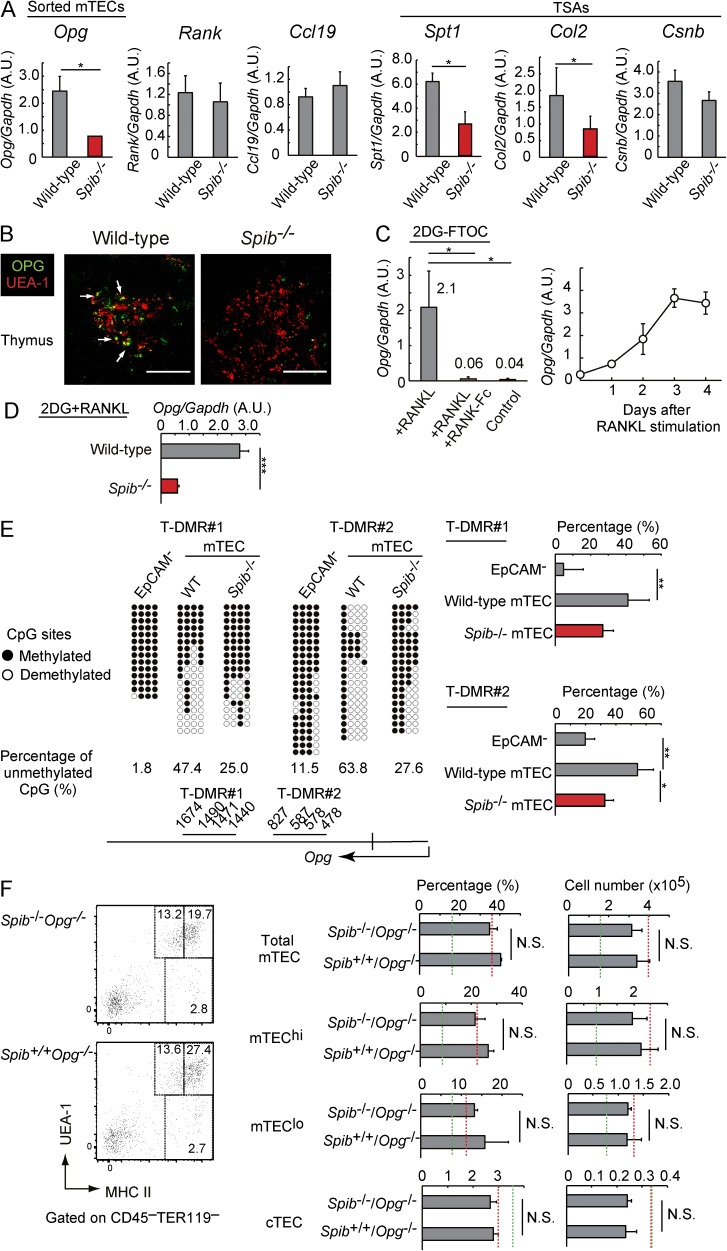
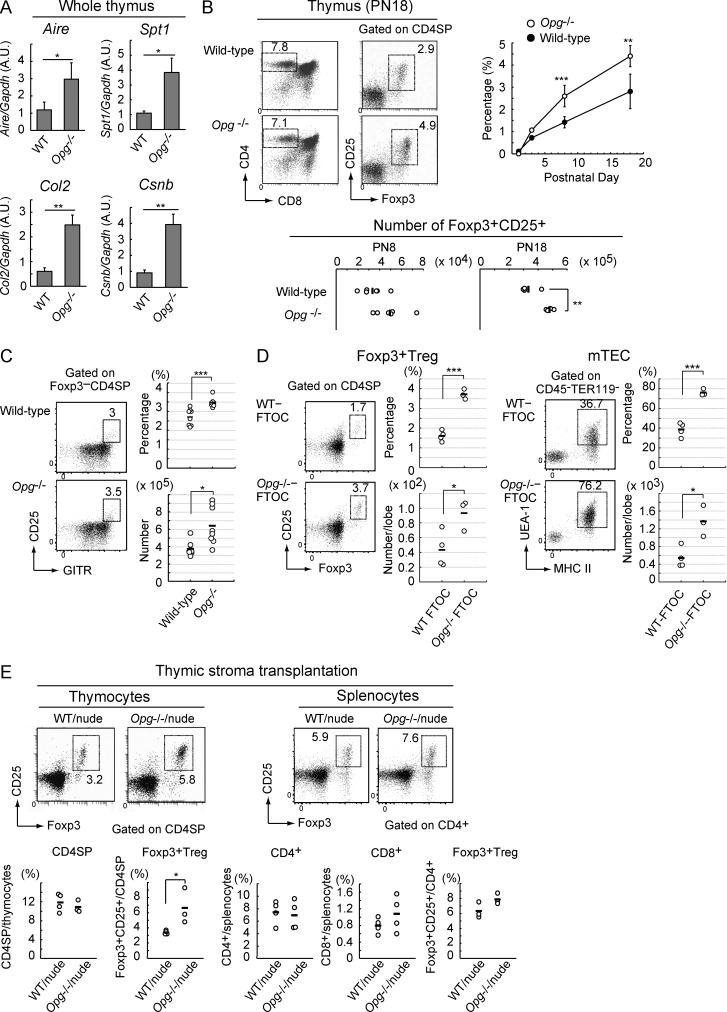
Because Spi-B may selectively regulate gene expression in mTECs, we subsequently identified other genes that are regulated by Spi-B in mTECs. We sorted mTEC fractions (EpCAM+UEA-1+) from Spib−/− and WT mice and investigated gene expression using qPCR. Indeed, expression of the TSAs Spt1 and Col2 was significantly reduced in Spib−/− mTEC fractions compared with those in WT mTEC fractions (Fig. 3 A). Thus, Spi-B may facilitate the expression of some TSA members by an Aire-independent mechanism. Notably, the expression of OPG was significantly reduced in Spib−/− mTECs (Fig. 3 A), and reduced OPG protein expression was confirmed by immunohistochemical staining of thymic sections (Fig. 3 B). OPG competitively inhibits RANKL–RANK interactions as a decoy receptor of RANKL (Leibbrandt and Penninger, 2008), thereby inhibiting mTEC development (Hikosaka et al., 2008). Because this inhibitory effect (Fig. 2) may relate to reduced OPG expression, we further characterized Spi-B–dependent OPG expression in mTECs.
Figure 3.
Spi-B promotes expressions of decoy receptor OPG and thereby limits mTEC development. (A) Expression of Opg, Rank, Ccl19, Spt1, Col2, and Csnb in mTECs sorted from the thymuses of Spib−/− and WT mice was analyzed by qPCR. Salivary protein 1, collagen 2, and casein-β are TSAs promiscuously expressed in mTECs. Values are arbitrary units (A.U.) normalized to Gapdh expression. Per experiment, the mTECs were isolated and pooled from three WT or three Spib−/− mice (3–4 wk old). (B) Thymic cryosections of 3-wk-old WT and Spib−/− mice were immunostained with anti-OPG and UEA-1 antibodies. White arrows indicate OPG expression. Figures are representatives of three independent experiments. Bars, 200 µm. (C, left) qPCR analysis of Opg expression in 2DG-FTOCs at 4 d after incubation with 1 µg/ml recombinant RANKL (+RANKL) or without RANKL (control). For blocking experiments (+RANKL + RANKL-Fc), 5 µg/ml RANK-Fc was added to the culture medium. Values are arbitrary units normalized to Gapdh expression. (right) Expression of Opg in 2DG-FTOC at 1–4 d after incubation with 1 µg/ml recombinant RANKL (+RANKL) or before RANKL stimulation (day 0) was analyzed by qPCR. Values are normalized to Gapdh expression and further normalized to the control values. (D) qPCR analysis was performed to determine Opg expression levels in WT and Spib−/− 2DG-FTOCs at 4 d after incubation with 1 µg/ml recombinant RANKL. Values are arbitrary units normalized to Gapdh expression. (E) DNA methylation analysis of Opg in the mTEC and EpCAM− stromal cell fractions sorted from the thymuses of 3-wk-old Spib−/− and WT mice. Each T-DMR contains four CpG sites. The DNA methylation status of CpGs in T-DMRs is indicated as open (unmethylated) and closed (methylated) circles. Percentages of unmethylated CpGs in T-DMR are indicated under each panel. T-DMRs and the transcription start site of Opg are schematically shown at the bottom of the panels. Data of methylated CpGs were obtained from three independent pairs of Spib−/− and WT mice and are summarized in the graphs on the right. (F) Typical flow cytometric profiles of TECs prepared from 3-wk-old Opg−/− and Spib−/−Opg−/− mice (C57BL/6 background). Flow cytometric analyses of Opg−/− and Spib−/−Opg−/− thymic stromal cells are summarized in the graphs. The percentage of MHC II+UEA-1+, MHC IIhiUEA-1+ (mTEChi), MHC IIloUEA-1+ (mTEClo), and UEA-1−MHC II+ (mainly cTEC) cells among thymic cells and their cell numbers are shown. Red and green dotted lines indicate data from 3-wk-old Spib−/− mice and WT mice, respectively. Error bars represent mean ± one SD for four (A and F) or three (C–E) independent experiments. *, P < 0.05, **, P < 0.01; and ***, P < 0.001 (Student’s t test).
As expected, Opg mRNA expression was induced by RANKL signaling in 2DG-FTOCs (Fig. 3 C). RANKL stimulation consecutively induced Spib and Opg expression (compare Fig. 3 C with Fig. 1 C). RANKL-dependent Opg expression was significantly reduced in the absence of Spi-B in 2DG-FTOCs (Fig. 3 D). These data suggested that RANKL signaling up-regulates Spib expression and promotes expression of its own inhibitor, OPG, in mTECs.
Subsequently, we investigated the mechanism regulating Opg expression via Spi-B in mTECs. Because Spi-B overexpression did not activate the putative proximal promoter of Opg in reporter assays (not depicted), Spi-B was expected to control Opg transcription without directly binding to the proximal promoter of the gene. Thus, we analyzed CpG methylation of Opg in sorted mTEC fractions. Initially, we observed two CpG-rich regions that were hypomethylated in mTECs compared with EpCAM− thymic stromal cells (Fig. 3 E), suggesting that Opg is tissue-dependently and differentially methylated (Yagi et al., 2008) in these regions (referred to as tissue-dependently and differentially methylated region [T-DMR] #1 and T-DMR#2). T-DMRs were hypermethylated in Spib−/− mTECs compared with control mTECs (Fig. 3 E). Thus, Spi-B expression in mTECs leads to either demethylation of CpG sites or maintenance of their hypomethylated states in Opg, which may enhance Opg expression.
Moreover, we genetically verified that enhancement of mTEC development in Spib−/− mice was dependent on OPG expression. As reported earlier (Hikosaka et al., 2008), OPG-deficient (Opg−/−) mice showed enhanced development of mTECs (Fig. 3 F) compared with WT controls (Figs. 2 A and 3 F [green lines]). Indeed, mTEC development was similar between Opg−/− mice and Spi-B and OPG double-deficient (Spib−/−Opg−/−) mice (Fig. 3 F). Thus, Spi-B does not attenuate mTEC development in the absence of OPG, strongly suggesting that the difference in mTEC development between WT and Spib−/− mice was caused by differential expression of OPG.
Spi-B–mediated negative feedback regulation of RANKL signaling initiates perinatally, limiting mTEC development
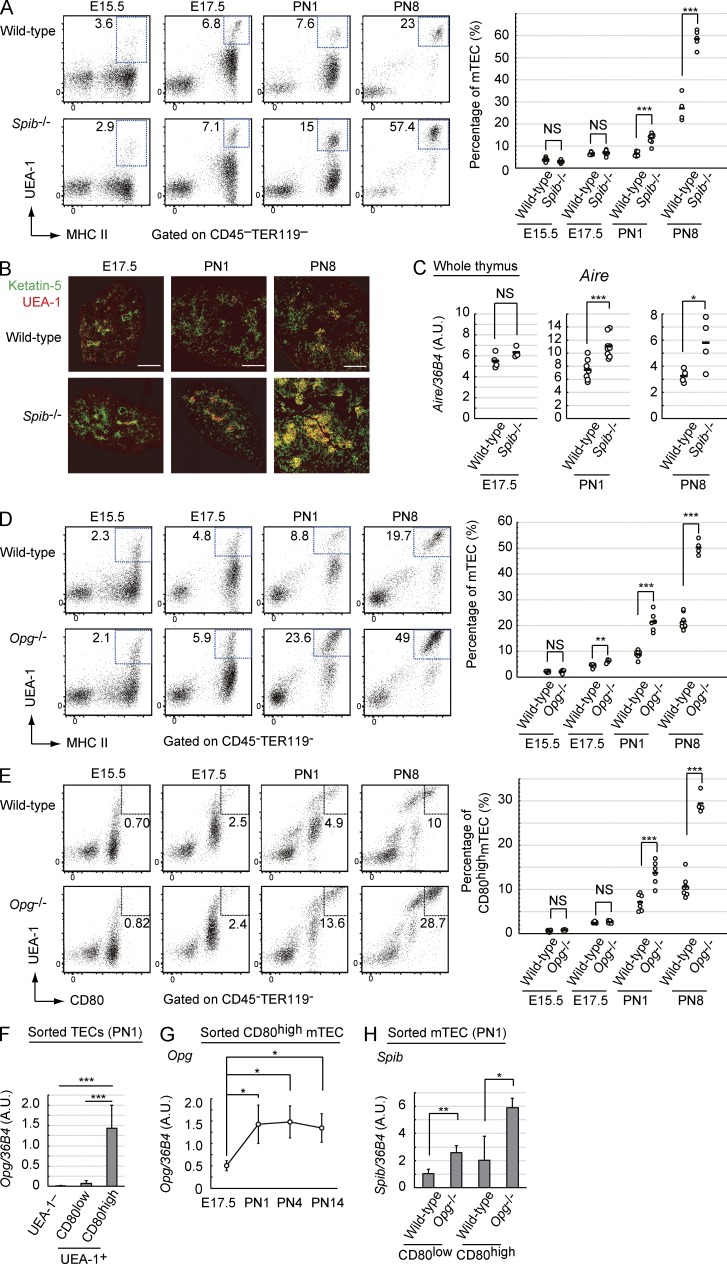
Aire-expressing mTECs in the perinatal period are essential and sufficient for establishment of long-lasting self-tolerance (Guerau-de-Arellano et al., 2009). Therefore, we investigated mTEC development in embryonic and neonatal Spib−/− mice. Flow cytometric analyses of embryonic day (E) 15.5, E17.5, postnatal day (PN) 1, and PN8 thymuses revealed incremental development of mTECs (MHC II+UEA-1+) in Spib−/− PN1 and PN8 mice, but not in E15.5 and E17.5 mice (Fig. 4 A). Immunohistochemical analysis confirmed enhanced development of mTECs in postnatal Spib−/− thymuses (Fig. 4 B). Moreover, Aire expression levels in mature mTECs were significantly increased only in postnatal Spib−/− thymuses (Fig. 4 C). Thus, negative regulation of mTEC development by Spi-B characteristically starts perinatally. Although an increment in mTEC number in response to a lack of OPG has been reported only in adult mice (Hikosaka et al., 2008), OPG function in embryonic and neonatal mTEC development remained unknown. We therefore investigated whether OPG attenuates mTEC development in a developmental stage–dependent manner, similar to Spi-B. Indeed, mTEC development was not influenced by OPG deficiency in E15.5 thymuses and minimally enhanced in E17.5 Opg−/− thymuses (Fig. 4 D). In contrast, mTEC development was markedly enhanced in PN1 and PN8 Opg−/− mice (Fig. 4 D). In embryonic and neonatal thymus, CD80 is a more expedient marker for mature mTECs than MHC II. Consistently, CD80hi mature mTECs were increased in Opg−/− PN1 and PN8, but not in E15.5 and E17.5 mice (Fig. 4 E). Overall, these data suggested that the negative regulation of mature mTEC development by the Spi-B–OPG system is initiated perinatally. We subsequently identified the stromal cell type that expresses Opg using qPCR. We found that Opg is expressed in CD80hi mature mTECs in PN1 mice (Fig. 4 F). Interestingly, Opg expression levels in mature mTECs were significantly up-regulated perinatally and plateaued in neonatal thymuses (Fig. 4 G). This may explain the stage-dependent negative regulation of mTEC development by OPG and implies a qualitative change in mature mTECs during the perinatal period.
Figure 4.
Spi-B–OPG system fine-tunes mTEC development from neonates via negative feedback regulation. (A) Flow cytometric analysis of mTECs prepared from WT and Spib−/− mice at E15.5 (WT, n = 9; and Spib−/−, n = 6; P > 0.05), at E17.5 (WT, n = 8; and Spib−/−, n = 8; P > 0.05), PN1 (WT, n = 7; and Spib−/− mice, n = 9; P = 1.52 × 10−6), and PN8 (WT, n = 4; and Spib−/−, n = 5; P = 3.03 × 10−5). Typical flow cytometric profiles of MHC II (I-A/I-E) and UEA-1 lectin–positive cells, which are also EpCAM+, among thymic stroma cells (CD45+TER119−) are shown. Percentages of mTECs among thymic stroma cells are summarized in the graph on the right. (B) Immunostaining of thymic sections from WT and Spib−/− mice at E17.5, PN1, and PN8. Thymic cryosections were immunostained with keratin-5 and UEA-1 lectin antibodies. Figures are representative of three independent experiments. Bars, 200 µm. (C) Expression levels of Aire in whole thymus from WT and Spib−/− mice at E17.5 (WT, n = 4; and Spib−/−, n = 3; P > 0.1), PN1 (WT, n = 7; and Spib−/−, n = 9; P = 7.4 × 10−4), and PN8 (WT, n = 4; and Spib−/−, n = 4; P = 0.045) were determined by qPCR. The values are arbitrary units (A.U.) normalized to 36B4 expression. (D) Flow cytometric profiles of thymic cells prepared at E15.5 (WT, n = 5; and Opg−/−, n = 4; P > 0.5), E17.5 (WT, n = 8; and Opg−/−, n = 4; P = 0.0022), PN1 (WT, n = 8; and Opg−/−, n = 6; P = 4.91 × 10−8), and PN8 (WT, n = 8; and Opg−/−, n = 4; P = 1.94 × 10−8). Typical flow cytometric profiles of MHC II (I-A/I-E)– and UEA-1 lectin–positive cells, which are also EpCAM+ (not depicted), among thymic stroma cells (CD45+TER119−) are shown on the left. Percentages of mTECs among thymic stroma cells are summarized in the graph on the right. (E) Flow cytometric profiles of thymic cells prepared at E15.5 (WT, n = 5; and Opg−/−, n = 4; P > 0.2), E17.5 (WT, n = 8; and Opg−/−, n = 4; P > 0.8), PN1 (WT, n = 8; and Opg−/−, n = 6; P = 1.19 × 10−5), and PN8 (WT, n = 8; and Opg−/−, n = 4; P = 1.21 × 10−7). Typical flow cytometric profiles of CD80- and UEA-1 lectin-positive cells, which are also EpCAM+ (not depicted), among thymic stroma cells (CD45+TER119−) are shown on the left. Percentages of CD80hi mature mTECs in thymic stroma cells are summarized in the graph on the right. (A and C–E) Black bars indicate mean values. (F) qPCR analysis of Opg in mature mTECs (CD80hiUEA-1+EpCAM+CD45−TER119−), immature mTECs (CD80loUEA-1+EpCAM+CD45−TER119−), and cTECs (CD80loUEA-1−EpCAM+CD45−TER119−) sorted from thymuses of WT mice at PN1 (cTECs, n = 6; immature mTECs, n = 7; and mature mTECs, n = 5). Values are arbitrary units normalized to 36B4 expression. Per experiment, the mTECs were isolated from three neonates and pooled for RNA preparation. (G) qPCR analysis of Opg in mature mTECs (CD80hiUEA-1+EpCAM+CD45−TER119−) sorted from thymuses of WT mice at E17.5 (n = 3), PN1 (n = 5), PN4 (n = 4), and PN14 (n = 4). Values are arbitrary units normalized to 36B4 expression. Per experiment, the mTECs were isolated from several embryos and neonates and pooled for RNA preparation. Error bars represent mean ± one SD. (H) qPCR analysis of Spib in immature mTECs (CD80loUEA-1+EpCAM+CD45−TER119−) and mature mTECs (CD80hiUEA-1+EpCAM+CD45−TER119−) sorted from thymuses of Opg−/− and WT mice at PN1 (n = 3). Per experiment, mTECs were isolated from several neonates and pooled for RNA preparation. Values are arbitrary units normalized to 36B4 expression. (F and H) Error bars represent mean ± one SD for four independent experiments. *, P < 0.05, **, P < 0.01; and ***, P < 0.001 (Student’s t test).
We confirmed the RANKL–RANK–Spi-B–OPG–RANKL feedback loop in the perinatal period. Given that such a negative feedback loop exists, the absence of OPG would enhance Spi-B expression in mTECs by increasing RANKL signaling. Indeed, Spi-B expression was significantly enhanced in Opg−/− mTECs at PN1 (Fig. 4 H). Interestingly, Spi-B was expressed in both CD80lo immature mTECs and CD80hi mature mTECs (Fig. 4 H) and was also up-regulated in the absence of OPG in both mTEC types, suggesting that RANKL signaling rapidly induces Spi-B in immature mTECs before promoting differentiation into mature mTECs. Cumulatively, these data strongly suggested that Spi-B–mediated Opg expression attenuates the development of mTECs by a negative feedback regulation of RANKL signaling.
Thymic stromal OPG attenuates cellularity of T reg cells in the thymus
We subsequently determined whether this negative regulation via RANKL–Spi-B–OPG influences mTEC-mediated thymic functions. We investigated the effects of abolishing this negative feedback loop in Opg−/− mice rather than Spib−/− mice because Spi-B reduces expression of CD80, CD86, and some TSAs in mTECs (Figs. 3 and 4). As reported earlier (Khan et al., 2014), expression levels of TSAs and Aire were significantly increased in Opg−/− thymuses (Fig. 5 A). Because mature mTECs promote T reg cell development as self-antigen–presenting cells (Hinterberger et al., 2010; Lei et al., 2011; Cowan et al., 2013; Malchow et al., 2013) and an increase in mTEC cellularity enhances thymic T reg cell development (Hauri-Hohl et al., 2014), we investigated whether OPG deficiency facilitates T reg cell development in the thymus. Indeed, numbers and frequency of Foxp3+ T reg cells were significantly increased in thymuses of neonatal and young Opg−/− mice (Fig. 5 B). Moreover, the frequency of thymic T reg cell precursors (CD4+CD8−Foxp3−GITR+CD25+; Lio and Hsieh, 2008) was increased in Opg−/− thymuses (Fig. 5 C). In vitro FTOC experiments in which mTECs and T reg cells develop by endogenous signaling suggested that the increase in T reg cells by OPG deficiency is caused in a thymus-intrinsic manner (Fig. 5 D). Fetal thymic stroma transplantation experiments indicated that OPG deficiency in thymic stromal cells caused an increment of T reg cell frequency in thymuses, whereas the increase in splenic T reg cells was not significant (Fig. 5 E). Overall, these data supported the idea that negative feedback regulation via RANKL–Spi-B–OPG attenuates mTEC-mediated thymic generation of T reg cells, although it is possible that more T reg cells are retained in the Opg−/− thymus.
Figure 5.
OPG attenuates TSA expressions and frequency of T reg cells in the thymus. (A) Expression of Aire, Spt1, Col2, and Csnb in the whole thymus of Opg−/− and WT mice was analyzed by qPCR. *, P < 0.05; and **, P < 0.01 (Student’s t test). Values are arbitrary units (A.U.) normalized to Gapdh expression. Error bars represent mean ± one SD for three independent experiments. (B) Flow cytometric analysis of thymocytes of WT and Opg−/− mice at PN1 (WT, n = 3; and Opg−/−, n = 3), PN3 (WT, n = 8; and Opg−/−, n = 5), PN8 (WT, n = 6; and Opg−/−, n = 6), and PN18 (WT, n = 5; and Opg−/−, n = 5). Typical flow cytometric profiles at PN18 are shown on the left. Percentages of Foxp3+CD25+ in CD4SP are summarized in the graph on the right. P-values were determined by Student’s t test between Opg−/− and WT mice: **, P < 0.01; ***, P < 0.001 (P = 3.6 × 10−4 at PN8 and P = 4.6 × 10−3 at PN18). Error bars represent mean ± one SD. T reg cell numbers are shown in the graphs below. P-values were determined by Student’s t test between Opg−/− and WT mice: **, P < 0.01 (P = 0.052 at PN8 and P = 4.2 × 10−3 at PN18). (C) Flow cytometric analysis of thymocytes of WT and Opg−/− mice at PN20 (WT, n = 8; and Opg−/−, n = 8). The percentages of GITR+CD25+ in Foxp3−CD4SP and numbers of GITR+CD25+Foxp3−CD4SP are summarized in the graphs. P-values were determined by Student’s t test between Opg−/− and WT mice: *, P = 0.036; ***, P = 8.1 × 10−4. (D) mTECs and T reg cells of in vitro FTOC from WT and Opg−/− mice were analyzed by flow cytometry. Percentages of mTECs in stroma cells (CD45−TER119−) and T reg cells in CD4SP cells and numbers of mTECs and T reg cells per thymic lobe in WT and Opg−/− FTOC are summarized in the graphs (WT FTOC, n = 4; and Opg−/− FTOC, n = 3). The fetal thymuses of WT or Opg−/− mice were cultured for 7 d. Per experiment, FTOCs from three to four embryos were pooled. Cell numbers per thymic lobe were exhibited in graphs. P-values were determined by Student’s t test between Opg−/− and WT mice: *, P = 0.036 for Foxp3+ Treg; *, P = 0.013 for mTEC; ***, P = 1.3 × 10−4 for Foxp3+ Treg; and ***, P = 4.1 × 10−4 for mTEC. (E) 2DG-FTOCs prepared from Opg−/− or WT fetuses were transplanted into the kidney of nude mice (Opg−/−/nude or WT/nude). The grafted thymuses and spleen were analyzed by flow cytometry 8 wk after transplantation. Typical flow cytometric profiles are shown at the top. The ratios of CD4SP cells to total thymocytes, Foxp3+CD25+ cells to CD4SP cells, CD4+ and CD8+ T cells to splenocytes, and Foxp3+CD4+ T cells to splenocytes are summarized in the graphs (WT/nude, n = 4; and Opg−/−/nude, n = 3). P-values were determined by Student’s t test between Opg−/− and WT mice: *, P < 0.05. (B–E) Black bars indicate mean values.
Thymic stromal OPG attenuates tumor incidence and growth and facilitates humoral immune responses to tumors
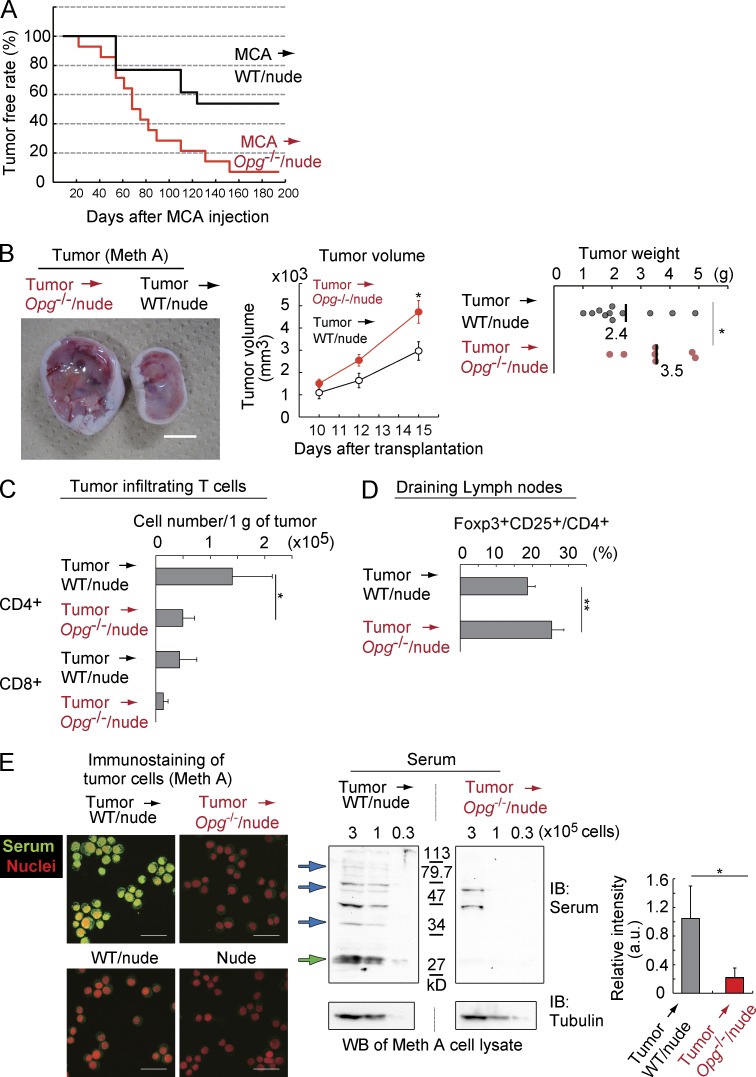
Because negative feedback regulation of mTEC development via RANKL–Spi-B–OPG is initiated perinatally (Figs. 4 and 5), establishing long-term self-tolerance by Aire-expressing mTECs (Guerau-de-Arellano et al., 2009), we investigated the role of this negative regulation in modulation of immunological tolerance in vivo. Recent studies suggested that Aire-expressing mTECs impede antitumor immunity (Träger et al., 2012; Malchow et al., 2013; Zhu et al., 2013). Therefore, we speculated that this feedback regulation may contribute to antitumor immunity by attenuating mTEC-dependent tolerance to tumor-derived antigens. We tested this hypothesis in mouse carcinogen-induced tumor and tumor transplantation models. To exclude any other effects caused by increased RANKL activity, we initially adoptively transplanted Opg−/− 2DG-FTOCs (BALB/c) into kidney capsules of BALB/c nude mice (Leibbrandt and Penninger, 2008; Akiyama et al., 2012a). A limited dose of the carcinogenic polycyclic aromatic hydrocarbon, methylcholanthrene (MCA), was injected into chimeric nude mice that had received Opg−/− 2DG-FTOCs (Opg−/−/nude) and control mice that had received WT 2DG-FTOCs. Tumor incidence was significantly increased in Opg−/−/nude mice compared with control mice (Fig. 6 A), indicating that thymic stromal OPG limits MCA-dependent tumor incidence in vivo.
Figure 6.
OPG expressed in thymic stromal cells attenuates tumor development and growth. (A) Chimeric nude mice receiving Opg−/− or WT 2DG-FTOCs were examined once a week for tumor development after MCA injection. The red line indicates the tumor-free rate of the chimeric nude mice receiving Opg−/− 2DG-FTOCs (n = 14), and the black line indicates that of chimeric mice receiving WT 2DG-FTOCs (n = 13). P = 0.033: log-rank test between Opg−/− and WT chimeric mice sets regarding the tumor-free rate. (B) Tumor cells (Meth A; female, BALB/c background) were subcutaneously transferred to nude mice receiving Opg−/− (tumor → Opg−/−/nude; n = 7) or WT 2DG-FTOCs (tumor → WT/nude; n = 11). The left panel shows a typical image of tumors that developed in these mice. Tumor volumes were measured on days 10, 12, and 15 (middle graph) after tumor transfer. Asterisks indicate statistical significance: *, P < 0.05 (Student’s t test). Error bars represent one SEM. Tumor weight was measured after sacrificing the mice (right graph; asterisk indicates significant difference by Mann–Whitney U test: *, P < 0.05). (C) Tumors developed from Meth A cells were analyzed. Tumor-infiltrating lymphocytes in nude mice receiving Opg−/− (tumor → Opg−/−/nude) or WT 2DG-FTOC (tumor → WT/nude) were analyzed by flow cytometry. The number of CD4+ T and CD8+ T cells in 1 g of tumor from nude mice receiving Opg−/− 2DG-FTOCs (tumor → Opg−/−/nude) and WT 2DG-FTOCs (tumor → WT/nude) was determined on day 15 after tumor cell (Meth A) transfer. Asterisks indicate statistical significance: *, P < 0.05 (Student’s t test). Error bars represent mean ± one SD for the indicated number of independent experiments (WT, n = 5; and KO, n = 4). (D) The ratio of Foxp3+CD25+ cells to CD4+ cells in the lymph nodes of nude mice receiving Opg−/− (tumor → Opg−/−/nude) or WT 2DG-FTOCs (tumor → WT/nude) was determined on day 15 after tumor cell (Meth A) transfer. Asterisks indicate statistical significance: **, P < 0.01 (Student’s t test). Error bars represent mean ± one SD for the indicated number of independent experiments (WT, n = 11; and KO, n = 7). (E) Serum antibodies obtained from the nude mice receiving Opg−/− (tumor → Opg−/−/nude) or WT 2DG-FTOCs (tumor → WT/nude) were analyzed for tumor reactivity after tumor cell (Meth A) transfer. Meth A cells were immunostained with mouse sera (left: green, sera; red, propidium iodide for nuclear staining). Immunostaining data of Meth A with sera from tumor-transplanted mice are shown in the two top panels. Immunostaining data with WT/nude sera (without tumor transplantation) and nude sera (without transplantation of 2DG-FTOCs and tumor) are shown in bottom panels. Data are representative of three independent experiments. Western blot analysis of Meth A cell lysate using the serum from tumor → Opg−/−/nude or WT tumor → WT/nude mice (middle). Cell numbers of Meth A loaded as cell lysate are indicated at the top of the panels. Data are representative of three independent experiments. Arrows indicate protein bands that were reduced or disappeared in Western blots using sera of Opg−/−/nude mice receiving tumors. Molecular masses (kilodaltons) are indicated between the two panels. The intensity of each band indicated by a green arrow was quantified and normalized to tubulin levels (right graph). Error bars represent mean ± one SD for three independent experiments. *, P < 0.05 (Student’s t test). Bars: (B) 1 cm; (E) 50 µm.
After congenic mouse tumor cells were subcutaneously transferred into nude recipients, tumor growth was significantly increased in Opg−/−/nude compared with WT/nude mice (Fig. 6 B). Tumor-infiltrating T cells were reduced in tumors of Opg−/−/nude mice (Fig. 6 C). In addition, T reg cell numbers were significantly increased in the draining lymph nodes of these mice (Fig. 6 D). Titers of antitumor cell antibodies evoked by tumor cell transplantation (determined by immunochemistry) were decreased in the serum of tumor-transplanted Opg−/−/nude mice compared with those in WT/nude control mice (Fig. 6 E), suggesting weaker humoral immune responses to tumors in the former. Notably, the kinds of tumor-derived proteins recognized by serum antibodies upon Western blotting were also reduced in tumor-transplanted Opg−/−/nude mice (Fig. 6 E, arrows), implying that TCR repertoires against tumor-derived proteins were decreased by the absence of OPG in thymic stroma. Thus, OPG expression in thymic stromal cells enhances immune responses to and limits the growth of tumors. These data further indicated that negative regulation via RANKL–Spi-B–OPG fine-tunes induction of mTEC-mediated tolerance and may ensure immune responses to tumors.
DISCUSSION
Although RANKL signaling is known to trigger differentiation of mature mTECs, molecular events linking RANKL signaling to the expression of molecules regulating mTEC functions have been largely unexplored. Here, we demonstrated that Spi-B expression links RANKL–NF-κB signaling with the expression of CD80, CD86, some TSAs, and OPG, implying a key regulatory role for Spi-B in mTECs. Interestingly, Spi-B deficiency did not affect the expression of Aire, MHC II, PDL1, or CD40, which are up-regulated in mature mTECs. This suggests that other unidentified mechanisms must regulate the expression of these molecules downstream of RANKL–NF-κB signaling.
Spi-B regulates high expression of CD80 and CD86 in mature mTECs, suggesting its positive role in mTEC-mediated function. Interestingly, Spi-B, CD80, and NF-κB1 have been associated with primary biliary cirrhosis (Liu et al., 2010; Mells et al., 2011), implying that Spi-B–mediated CD80 expression is correlated to the onset of autoimmunity.
Regulation of gene expression by Spi-B in mTECs remains unclear. However, the nearest homologue of Spi-B, PU.1, reportedly induces nucleosome remodeling and H3K4 monomethylation (Heinz et al., 2010). These epigenetic events are usually associated with changes in DNA methylation (Jones and Baylin, 2007). Given that Spi-B promotes demethylation of T-DMRs in Opg, Spi-B may exert epigenetic controls over gene expression in mTECs. Genome-wide epigenetic and expression studies of Spib−/− mTECs may verify this hypothesis in future.
The mechanisms that activate self-tolerance by mTECs require stochastic encounters between self-reactive T cells and mature mTECs or dendritic cells that receive TSAs from mTECs. This mechanism may be inherently imperfect (Bouneaud et al., 2000) because of its stochasticity. The negative feedback regulation described herein ensures inadequacy of mTEC-dependent tolerance and could mediate detrimental effects by increasing the prevalence of autoreactive T cells. However, our data suggest that this negative regulation may contribute to tumor immunity and may optimize the trade-off between prevention of autoimmunity and induction of tumor immunity. Future analyses of this regulatory mechanism will promote understanding of tumor immunosurveillance and may lead to the development of new therapeutic approaches for cancer.
MATERIALS AND METHODS
Mice.
Spib−/− mice were established on a C57BL/6 background (Sasaki et al., 2012). The C57BL/6, BALB/cA, BALB/cA nude/nude, aly/+, and aly/aly mice were obtained from CLEA Japan. The Opg−/− mice have been described previously (Mizuno et al., 1998). The Opg−/− mice on a BALB/cA background were prepared by backcrossing with BALB/cA mice for 10 generations. Littermates or age-matched WT mice from the same colonies as the mutant mice were used as controls. All mice were maintained under specific pathogen–free conditions and were handled in accordance with the Guidelines for Animal Experiments of the Institute of Medical Science, The University of Tokyo (Tokyo, Japan). E0.5 was defined as the first morning a vaginal plug was observed. Pregnant female mice were checked every morning and evening, and discovery of new pups was marked as day 1.
Antibodies and reagents.
Rabbit anti–mouse keratin-5 antibody was purchased from Covance. Biotinylated UEA-1 was obtained from Vector Laboratories. The APC-Cy7–conjugated rat anti–mouse CD45, PE-Cy7–conjugated TER119, and PE-conjugated anti–mouse EpCAM (G8.8) antibodies were obtained from BD or BioLegend. The PE-conjugated rat anti-CD45, PE-conjugated mouse TER-119, FITC-conjugated mouse MHC II (M5/114.15.1), and APC-conjugated Aire antibodies were obtained from eBioscience. PE-conjugated CD80, CD86, CD40, and PD-L1 antibodies, purified anti–mouse CD16/32, FITC-conjugated ant–mouse EpCAM (G8.8), APC-Cy7–conjugated CD4, PE–Cy-7–conjugated CD8, and APC-conjugated GITR antibodies were obtained from BioLegend. PE-conjugated CD8, FITC-conjugated CD4, and FITC-conjugated CD3e antibodies were obtained from BD. PE-conjugated anti-Foxp3 (FJK-16s) was purchased from eBioscience. 7-aminoactinomycin D and recombinant mouse RANKL were purchased from Wako Pure Chemical Industries. Anti–mouse OPG antibody was purchased from R&D Systems. A RANK-Fc chimera was obtained from Sigma-Aldrich. Alexa Fluor 546–conjugated streptavidin and Alexa Fluor 488–conjugated anti–rabbit IgG were obtained from Invitrogen.
Flow cytometric analysis and cell sorting.
Whole thymus was used to prepare a single-cell suspension by digestion in RPMI 1640 medium containing collagenase/disperse (Roche) and DNase I (Sigma-Aldrich). Dead cells were excluded from the analysis after 7-aminoactinomycin D staining. After the Fc-receptor was blocked with CD16/32 antibody, the cells were stained with the indicated antibodies. For the analysis of T reg cells, thymocytes were prepared and used for nuclear staining according to the supplier’s protocol (Foxp3 staining buffer set; eBioscience) and analyzed using a fluorescence-activated cell sorter (Epics XL [Beckmann Coulter] or Canto II [BD]). The single-cell suspension was separated using a Percoll gradient to obtain a TEC-rich fraction, which was stained with anti-CD45, anti–TER-119, anti-EpCAM, and UEA-1 antibodies and sorted using a cell sorter (Aria; BD).
FTOC.
Thymic lobes were isolated from E15 embryos and were cultured on Nucleopore filters (Whatman) placed in R10 medium containing RPMI 1640 (Invitrogen) supplemented with 10% FBS (Equitech-Bio), 2 mM l-glutamine (Wako Pure Chemical Industries), 100 U/ml penicillin (Banyu Pharmaceutical), 100 µg/ml streptomycin (Meiji Seika Kaisha), and 50 µM 2-mercaptoethanol (Wako Pure Chemical Industries). FTOCs proceeded for 7 d and were subsequently analyzed by flow cytometry to investigate mTEC and T reg cell development in vitro. To isolate fetal thymic stromal (2DG-FTOC) cells, thymic lobes were cultured in the presence of 1.35 mM 2DG (Sigma-Aldrich) for 4 d. In some experiments, the 2DG-FTOC cells were further cultured in R10 with 1 µg/ml recombinant RANKL. For the blocking experiments, a RANK-Fc chimera (5 µg/ml) was added to the medium. For each day indicated after ligation, 2DG-FTOC was recovered and used for the analyses. For the transplantation experiments, the 2DG-FTOC was further cultured for 1 d without 2DG to eliminate 2DG before transplantation.
Transplantation of 2DG-FTOC into nude mice and tumor development.
2DG-FTOCs (six lobes each) were transplanted to the kidney capsule of 6-wk-old female nude mice. For tumor transfer experiments, Meth A (5 × 106 cells) was injected subcutaneously into the nude mice receiving 2DG-FTOC at 6 wk after transplantation. Tumor sizes were measured on days 10, 12, and 15 after transplantation. At day 15, the recipient mice were killed, and the inguinal lymph nodes were prepared for flow cytometric analysis. For the carcinogen-induced model, 50 µg MCA was injected subcutaneously into nude mice receiving 2DG-FTOC.
Histopathology and immunohistochemistry.
For immunohistochemistry, tissues were embedded in OCT compound (Sakura) and frozen in liquid nitrogen. Cryostat sections (6-µm thick) were fixed with ice-cold acetone and incubated with primary antibody for 1 h at room temperature. The slides were subsequently incubated with secondary antibody for 40 min at room temperature. For immunostaining of Meth A cells, cells were fixed with methanol and incubated with serum (100× dilution) for 1 h at room temperature and then incubated with secondary antibody (anti–mouse IgG–Alexa Fluor 488) and 1 µg/ml propidium iodide for 40 min at room temperature. Confocal color images were obtained using an FV1000D (Olympus) at 20 or 60 (Fig. 6 E) magnification and Fluoview software (Olympus).
qRT-PCR.
Total RNA was extracted using an RNeasy micro kit (QIAGEN) or TRIzol (Invitrogen) and was subjected to random-primed reverse transcription using a Primescript II first strand cDNA synthesis kit (Takara Bio Inc.). Real-time qPCR was performed using an ABI PRISM 7300 Sequence Detection System (Applied Biosystems) and SYBR Green Master Mix (Roche or TOYOBO). The primers used were 36B4: 5′-TCCAGGCTTTGGGCATCA-3′ and 5′-CTTTATCAGCTGCACATCACT-3′; Gapdh: 5′-ACCATGTAGTTGAGGTCAATGAAGG-3′ and 5′-GGTGAAGGTCGGTGTGAACG-3′; Spib: 5′-CTGCAAGCCCTTCAGTTACC-3′ and 5′-AAAGGCAGCAGTAGCAGGAT-3′; Aire: 5′-GGTTCTGTTGGACTCTGCCCTG-3′ and 5′-TGTGCCACGACGGAGGTGAG-3′; Spt1: 5′-GTGTTGCTTGGTGTTTCCAC-3′ and 5′-GTGTTGCTTGGTGTTTCCAC-3′; Rank: 5′-GCTGGCTACCACTGGAACTC-3′ and 5′-GTGCAGTTGGTCCAAGGTTT-3′; Spib1: 5′-CTCTGAACCACCATGCTTGCT-3′ and 5′-TCCTTCTGGGTACAAACAGCTTAA-3′; Spib2: 5′-AGGGCGGCCCTGACAT-3′ and 5′-TCCTTCTGGGTACAAACAGCTTAA-3′; Opg: 5′-GGGCGTTACCTGGAGATCG-3′ and 5′-GAGAAGAACCCATCTGGACATTT-3′; Ccl19: 5′-GCTAATGATGCGGAAGACTG-3′ and 5′-ACTCACATCGACTCTCTAGG-3′; Col2: 5′-AGAACAGCATCGCCTACCTG-3′ and 5′-CTTGCCCCACTTACCAGTGT-3′; and Csnb: 5′-GGCACAGGTTGTTCAGGCTT-3′ and 5′-AAGGAAGGGTGCTACTTGCTG-3′.
CpG methylation analysis.
Cell pellets were subjected to bisulfite treatment using the EZ DNA methylation-Direct kit (Zymo Research) according to the manufacturer’s instructions. PCR was performed using treated DNA obtained from 200–300 cells. For bisulfite sequencing, PCR products were cloned into a pGEM-T easy vector (Promega), and >16 clones were sequenced using the ABI 3130 sequencer and Big-Dye terminator kit (Applied Biosystems).
Preparation of tumor-infiltrating lymphocytes.
Tumors were minced with razor blades and digested in RPMI 1640 (Wako Pure Chemical Industries) containing 1 mg/ml collagenase D (Roche), 0.1 mg/ml DNase I, and 10% FBS for 1 h, with shaking. The remaining undigested fragments were passed through a needle. After addition of EDTA (final concentration: 10 mM), the cell suspension was filtered through a nylon mesh. The cells were collected and resuspended in 5 ml RPMI 1640 containing 45% Percoll and 10% FBS, and the mixture was layered on top of the same volume of 67.5% Percoll. After the cells were centrifuged at 800 g for 20 min at room temperature, the layer between 45% and 67.5% Percoll was collected.
Statistical analysis.
P-values were calculated using Student’s t test, Mann–Whitney U test with two-tailed distribution, and two-sample equivalent variance parameters. For statistical analysis of the tumor-free rate, the log-rank test was performed.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research from the Precursory Research for Embryonic Science and Technology (PRESTO) program of the Japan Science and Technology Agency (to T. Akiyama), grants from the Japanese Society for the Promotion of Science (to T. Akiyama), Takeda Science Foundation (to T. Akiyama), Princess Takamatsu Cancer Research Fund (to T. Akiyama), grants for Priority Area Research (to T. Akiyama and J.-i. Inoue), and a Grant-in-Aid for Challenging Exploratory Research (to T. Akiyama).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 2DG
- 2′-deoxyguanosine
- FTOC
- fetal thymus organ culture
- MCA
- methylcholanthrene
- mTEC
- medullary TEC
- NIK
- NF-κB–inducing kinase
- OPG
- osteoprotegerin
- qPCR
- quantitative RT-PCR
- RANK
- receptor activator of NF-κB
- RANKL
- RANK ligand
- T-DMR
- tissue-dependently and differentially methylated region
- TEC
- thymic epithelial cell
- TSA
- tissue-specific antigen
References
- Abramson J., Giraud M., Benoist C., and Mathis D.. 2010. Aire’s partners in the molecular control of immunological tolerance. Cell. 140:123–135 10.1016/j.cell.2009.12.030 [DOI] [PubMed] [Google Scholar]
- Aichinger M., Hinterberger M., and Klein L.. 2012. Probing gene function in thymic epithelial cells. Eur. J. Cell Biol. 91:24–30 10.1016/j.ejcb.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Maeda S., Yamane S., Ogino K., Kasai M., Kajiura F., Matsumoto M., and Inoue J.. 2005. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 308:248–251 10.1126/science.1105677 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shimo Y., Yanai H., Qin J., Ohshima D., Maruyama Y., Asaumi Y., Kitazawa J., Takayanagi H., Penninger J.M., et al. 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 29:423–437 10.1016/j.immuni.2008.06.015 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Shinzawa M., and Akiyama N.. 2012a. RANKL-RANK interaction in immune regulatory systems. World J. Orthop. 3:142–150 10.5312/wjo.v3.i9.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama T., Shinzawa M., and Akiyama N.. 2012b. TNF receptor family signaling in the development and functions of medullary thymic epithelial cells. Front. Immunol. 3:278 10.3389/fimmu.2012.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G., and Takahama Y.. 2012. Thymic epithelial cells: working class heroes for T cell development and repertoire selection. Trends Immunol. 33:256–263 10.1016/j.it.2012.03.005 [DOI] [PubMed] [Google Scholar]
- Anderson G., Jenkinson E.J., and Rodewald H.R.. 2009. A roadmap for thymic epithelial cell development. Eur. J. Immunol. 39:1694–1699 10.1002/eji.200939379 [DOI] [PubMed] [Google Scholar]
- Bartholdy B., Du Roure C., Bordon A., Emslie D., Corcoran L.M., and Matthias P.. 2006. The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc. Natl. Acad. Sci. USA. 103:11665–11670 10.1073/pnas.0509430103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouneaud C., Kourilsky P., and Bousso P.. 2000. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 13:829–840 10.1016/S1074-7613(00)00080-7 [DOI] [PubMed] [Google Scholar]
- Burkly L., Hession C., Ogata L., Reilly C., Marconi L.A., Olson D., Tizard R., Cate R., and Lo D.. 1995. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 373:531–536 10.1038/373531a0 [DOI] [PubMed] [Google Scholar]
- Chen H.M., Gonzalez D.A., Radomska H.S., Voso M.T., Sun Z., Zhang P., Zhang D.E., and Tenen D.G.. 1998. Two promoters direct expression of the murine Spi-B gene, an Ets family transcription factor. Gene. 207:209–218 10.1016/S0378-1119(97)00629-X [DOI] [PubMed] [Google Scholar]
- Cowan J.E., Parnell S.M., Nakamura K., Caamano J.H., Lane P.J., Jenkinson E.J., Jenkinson W.E., and Anderson G.. 2013. The thymic medulla is required for Foxp3+ regulatory but not conventional CD4+ thymocyte development. J. Exp. Med. 210:675–681 10.1084/jem.20122070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontje W., Schotte R., Cupedo T., Nagasawa M., Scheeren F., Gimeno R., Spits H., and Blom B.. 2006. Delta-like1-induced Notch1 signaling regulates the human plasmacytoid dendritic cell versus T-cell lineage decision through control of GATA-3 and Spi-B. Blood. 107:2446–2452 10.1182/blood-2005-05-2090 [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha L.A., Su G.H., Rao S., Kabak S., Hao Z., Clark M.R., and Simon M.C.. 1999. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity. 10:399–408 10.1016/S1074-7613(00)80040-0 [DOI] [PubMed] [Google Scholar]
- Gray D., Abramson J., Benoist C., and Mathis D.. 2007. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J. Exp. Med. 204:2521–2528 10.1084/jem.20070795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerau-de-Arellano M., Martinic M., Benoist C., and Mathis D.. 2009. Neonatal tolerance revisited: a perinatal window for Aire control of autoimmunity. J. Exp. Med. 206:1245–1252 10.1084/jem.20090300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri-Hohl M., Zuklys S., Holländer G.A., and Ziegler S.F.. 2014. A regulatory role for TGF-β signaling in the establishment and function of the thymic medulla. Nat. Immunol. 15:554–561 10.1038/ni.2869 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka Y., Nitta T., Ohigashi I., Yano K., Ishimaru N., Hayashi Y., Matsumoto M., Matsuo K., Penninger J.M., Takayanagi H., et al. 2008. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 29:438–450 10.1016/j.immuni.2008.06.018 [DOI] [PubMed] [Google Scholar]
- Hinterberger M., Aichinger M., Prazeres da Costa O., Voehringer D., Hoffmann R., and Klein L.. 2010. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nat. Immunol. 11:512–519 10.1038/ni.1874 [DOI] [PubMed] [Google Scholar]
- Jones P.A., and Baylin S.B.. 2007. The epigenomics of cancer. Cell. 128:683–692 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiura F., Sun S., Nomura T., Izumi K., Ueno T., Bando Y., Kuroda N., Han H., Li Y., Matsushima A., et al. 2004. NF-κB-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J. Immunol. 172:2067–2075 10.4049/jimmunol.172.4.2067 [DOI] [PubMed] [Google Scholar]
- Kanaya T., Hase K., Takahashi D., Fukuda S., Hoshino K., Sasaki I., Hemmi H., Knoop K.A., Kumar N., Sato M., et al. 2012. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat. Immunol. 13:729–736 10.1038/ni.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I.S., Mouchess M.L., Zhu M.L., Conley B., Fasano K.J., Hou Y., Fong L., Su M.A., and Anderson M.S.. 2014. Enhancement of an anti-tumor immune response by transient blockade of central T cell tolerance. J. Exp. Med. 211:761–768 10.1084/jem.20131889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., Wirnsberger G., and Kyewski B.. 2009. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9:833–844 10.1038/nri2669 [DOI] [PubMed] [Google Scholar]
- Kyewski B., and Klein L.. 2006. A central role for central tolerance. Annu. Rev. Immunol. 24:571–606 10.1146/annurev.immunol.23.021704.115601 [DOI] [PubMed] [Google Scholar]
- Lei Y., Ripen A.M., Ishimaru N., Ohigashi I., Nagasawa T., Jeker L.T., Bösl M.R., Holländer G.A., Hayashi Y., Malefyt R.W., et al. 2011. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J. Exp. Med. 208:383–394 10.1084/jem.20102327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibbrandt A., and Penninger J.M.. 2008. RANK/RANKL: regulators of immune responses and bone physiology. Ann. N. Y. Acad. Sci. 1143:123–150 10.1196/annals.1443.016 [DOI] [PubMed] [Google Scholar]
- Lio C.W., and Hsieh C.S.. 2008. A two-step process for thymic regulatory T cell development. Immunity. 28:100–111 10.1016/j.immuni.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Invernizzi P., Lu Y., Kosoy R., Lu Y., Bianchi I., Podda M., Xu C., Xie G., Macciardi F., et al. 2010. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat. Genet. 42:658–660 10.1038/ng.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malchow S., Leventhal D.S., Nishi S., Fischer B.I., Shen L., Paner G.P., Amit A.S., Kang C., Geddes J.E., Allison J.P., et al. 2013. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 339:1219–1224 10.1126/science.1233913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mells G.F., Floyd J.A., Morley K.I., Cordell H.J., Franklin C.S., Shin S.Y., Heneghan M.A., Neuberger J.M., Donaldson P.T., Day D.B., et al. Wellcome Trust Case Control Consortium 3. 2011. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat. Genet. 43:329–332 (published erratum appears in Nat. Genet. 2011. 43:1164) 10.1038/ng.789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno A., Amizuka N., Irie K., Murakami A., Fujise N., Kanno T., Sato Y., Nakagawa N., Yasuda H., Mochizuki S., et al. 1998. Severe osteoporosis in mice lacking osteoclastogenesis inhibitory factor/osteoprotegerin. Biochem. Biophys. Res. Commun. 247:610–615 10.1006/bbrc.1998.8697 [DOI] [PubMed] [Google Scholar]
- Ohshima D., Qin J., Konno H., Hirosawa A., Shiraishi T., Yanai H., Shimo Y., Shinzawa M., Akiyama N., Yamashita R., et al. 2011. RANK signaling induces interferon-stimulated genes in the fetal thymic stroma. Biochem. Biophys. Res. Commun. 408:530–536 10.1016/j.bbrc.2011.04.049 [DOI] [PubMed] [Google Scholar]
- Ray D., Bosselut R., Ghysdael J., Mattei M.G., Tavitian A., and Moreau-Gachelin F.. 1992. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol. Cell. Biol. 12:4297–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S.W., Kim M.Y., Leibbrandt A., Parnell S.M., Jenkinson W.E., Glanville S.H., McConnell F.M., Scott H.S., Penninger J.M., Jenkinson E.J., et al. 2007. RANK signals from CD4+3− inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J. Exp. Med. 204:1267–1272 10.1084/jem.20062497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Hoshino K., Sugiyama T., Yamazaki C., Yano T., Iizuka A., Hemmi H., Tanaka T., Saito M., Sugiyama M., et al. 2012. Spi-B is critical for plasmacytoid dendritic cell function and development. Blood. 120:4733–4743 10.1182/blood-2012-06-436527 [DOI] [PubMed] [Google Scholar]
- Schotte R., Nagasawa M., Weijer K., Spits H., and Blom B.. 2004. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J. Exp. Med. 200:1503–1509 10.1084/jem.20041231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R., Kitada K., Matsuda F., Tashiro K., Ikuta K., Suzuki M., Kogishi K., Serikawa T., and Honjo T.. 1999. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-κb-inducing kinase. Nat. Genet. 22:74–77 10.1038/8780 [DOI] [PubMed] [Google Scholar]
- Sun S.C.2012. The noncanonical NF-κB pathway. Immunol. Rev. 246:125–140 10.1111/j.1600-065X.2011.01088.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu Y.M., and Richmond A.. 2010. NF-κB inducing kinase: a key regulator in the immune system and in cancer. Cytokine Growth Factor Rev. 21:213–226 10.1016/j.cytogfr.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träger U., Sierro S., Djordjevic G., Bouzo B., Khandwala S., Meloni A., Mortensen M., and Simon A.K.. 2012. The immune response to melanoma is limited by thymic selection of self-antigens. PLoS ONE. 7:e35005 10.1371/journal.pone.0035005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F., Carrasco D., Durham S.K., Barton D.S., Rizzo C.A., Ryseck R.P., Lira S.A., and Bravo R.. 1995. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-κB/Rel family. Cell. 80:331–340 10.1016/0092-8674(95)90416-6 [DOI] [PubMed] [Google Scholar]
- Yagi S., Hirabayashi K., Sato S., Li W., Takahashi Y., Hirakawa T., Wu G., Hattori N., Hattori N., Ohgane J., et al. 2008. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 18:1969–1978 10.1101/gr.074070.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M.L., Nagavalli A., and Su M.A.. 2013. Aire deficiency promotes TRP-1-specific immune rejection of melanoma. Cancer Res. 73:2104–2116 10.1158/0008-5472.CAN-12-3781 [DOI] [PMC free article] [PubMed] [Google Scholar]