Summary
Degeneration of the knee joint during osteoarthritis often begins with meniscal lesions. Meniscectomy, previously performed extensively after meniscal injury, is now obsolete because of the inevitable osteoarthritis that occurs following this procedure. Clinically, meniscus self-renewal is well documented as long as the outer, vascularized meniscal ring remains intact. In contrast, regeneration of the inner, avascular meniscus does not occur. Here, we show that cartilage tissue harvested from the avascular inner human meniscus during the late stages of osteoarthritis harbors a unique progenitor cell population. These meniscus progenitor cells (MPCs) are clonogenic and multipotent and exhibit migratory activity. We also determined that MPCs are likely to be controlled by canonical transforming growth factor β (TGF-β) signaling that leads to an increase in SOX9 and a decrease in RUNX2, thereby enhancing the chondrogenic potential of MPC. Therefore, our work is relevant for the development of novel cell biological, regenerative therapies for meniscus repair.
Graphical Abstract
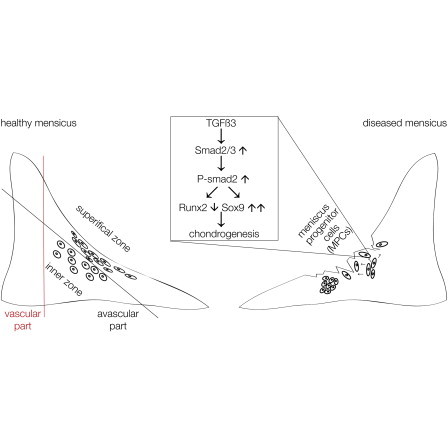
Highlights
-
•
Progenitor cells are found in the inner avascular part of human osteoarthritic menisci
-
•
These meniscus progenitor cells (MPCs) are clonogenic, migratory, and multipotent
-
•
MPCs are governed via the canonical TGF-β pathway
-
•
TGF-β3 via Smad2 reduces Runx2 to enhance the chondrogenic potential of MPCs
Degeneration of knee joints during osteoarthritis often begins with meniscal lesions and regeneration of the inner, avascular meniscus does not occur. In this article, Miosge and colleagues show that cartilage tissue harvested from the inner human meniscus harbors unique meniscus progenitor cells (MPCs). These cells are controlled by canonical TGF-β signaling that leads to an increase in their chondrogenic potential.
Introduction
In the elderly, osteoarthritis (OA) is the most common musculoskeletal disease (Reginster, 2002) and will be the fourth leading cause of disability by the year 2020 (Woolf and Pfleger, 2003). The knee is particularly prone to meniscal lesions that lead to OA (Englund et al., 2012), and a high interdependency of OA and meniscus lesions has been described (Brophy et al., 2012). In fact, meniscal injuries are the most common knee injury and account for more than 50% of the 1.5 million knee arthroscopies performed annually (Englund et al., 2008; Lohmander et al., 2007). The prevalence of meniscal tears increases with age (Loeser, 2013) and may be as high as 56% in men aged 70–90 years old (Englund et al., 2008). Lack of robust meniscal repair in adults with or without surgical intervention has led to the development of allografts or bioengineered meniscal substitutes (Haddad et al., 2013; Steinert et al., 2007), and, whereas these fill the space void created following removal of the meniscus, clinical, radiological, and MRI evaluations show no protection against the development of OA (Hommen et al., 2007). The specific reasons for this lack of effect are unknown; however, a failure to successfully remodel the allograft into living tissue is one likely factor (Steadman and Rodkey, 2005). Almost all patients eventually require joint replacement (Lohmander and Roos, 2007).
The meniscus is best described as a fibrocartilage (Benjamin and Evans, 1990) comprising an outer, “red vascularized” part and an inner “white, unvascularized” part harboring round fibrochondrocytes (Hellio Le Graverand et al., 2001). Additionally, the outer surface of the meniscus is enclosed by a superficial layer with flattened, elongated fibroblast-like cells that predominantly synthesize collagen type I (McDevitt and Webber, 1990), whereas the round fibrochondrocytes from the inner part also produce collagen type II (Chevrier et al., 2009).
It has long been known that the outer, vascularized part of the meniscus is often able to successfully repair itself after injury allowing for normal meniscus function, whereas the inner meniscus produces a repair tissue in response to injury but cannot functionally regenerate. Experimental animal studies support the idea that the meniscus possesses regeneration activity. For example, fibrochondrocytes have been isolated from bovine meniscus tissue (Mauck et al., 2007) and migrating cells are found in healthy adult rabbit meniscus (Webber et al., 1989). Regeneration of the vascular, outer part of the meniscus might be due to the presence of CD34-positive cells (Osawa et al., 2013) or to mesenchymal cells that can be released by collagenase digestion of human meniscus (Segawa et al., 2009). Because application of growth factors and fibrin clots have elicited responses by cells resident in the inner meniscus, several authors have speculated about the importance of activation of meniscal repair cells (Petersen et al., 2005). Up until today, the question remains, as to whether the inner, avascular part of the human meniscus harbors multipotent cells capable of regeneration.
Here, we describe isolation of human meniscus progenitor cells (MPCs) from the inner meniscus harvested from late-stage OA patients prior to knee replacement and show that the regenerative potential of these cells is governed by transforming growth factor β (TGF-β) signaling.
Results
Meniscus Tissue Histology and Molecular Composition
By combining and modifying available classification systems for OA specimens (Pauli et al., 2011; Zhang et al., 2011), we developed a means to discriminate healthier human menisci from their diseased counterparts. Intact meniscus tissue is composed of a superficial zone with flattened cells that primarily synthesize collagen type I (McDevitt and Webber, 1990). This architecture remains in healthier human menisci obtained from patients suffering from OA but is always absent in more disease samples (Figures 1A and 1B). The inner zone, containing more rounded cells that secrete collagen type I and type II, is present regardless of disease severity (Figures 1C and 1F). In diseased human menisci (score greater 4), 23% of the samples exhibit abnormal calcifications (Figure 1D) and 53% display anomalous clusters of cells (Figure 1E).
Figure 1.
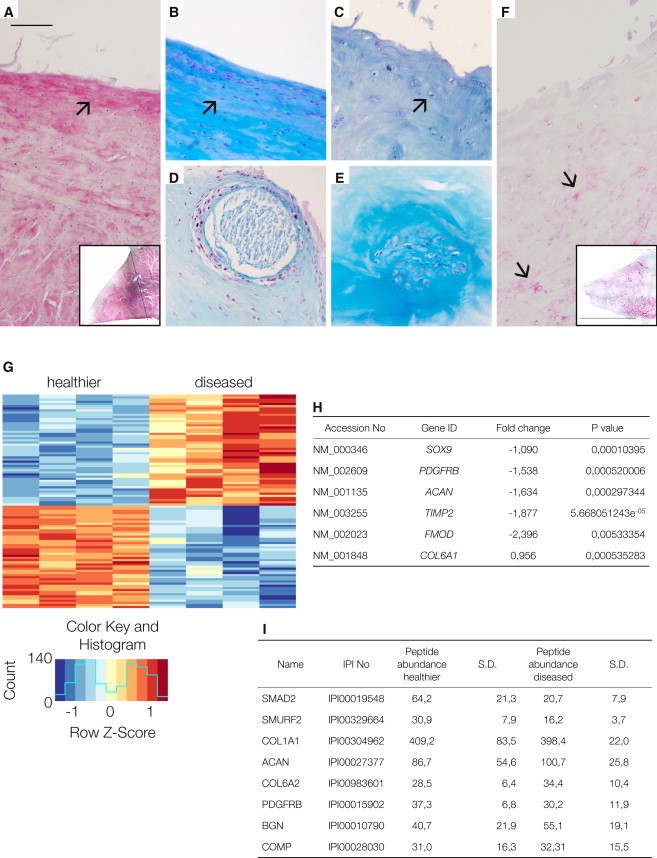
Characterization of Human Healthy and Diseased Meniscus Tissue
(A) Immunohistochemistry of collagen type I in healthier meniscus (arrow); inset: low magnification of a human healthier meniscus stained for collagen type I; the black line indicates the border between the vascular part on the right side and the avascular part on the left side.
(B) Healthier human meniscus with an intact superficial zone and flattened cells (arrow).
(C) Diseased meniscus with a completely degenerated superficial zone where only the round cells of the inner zone remain (arrow).
(D and E) Calcifications (D) and cell clusters (E) are signs of OA.
(F) Immunohistochemistry of collagen type II in diseased meniscus (arrows); inset: low magnification of a human diseased meniscus stained for collagen type II. Magnification in (A)–(F), scale bar, 150 μm; inset bar, 1.7 cm.
(G) Heatmap of the top 100 differentially expressed genes with p values < 0.001. The red color indicates upregulated genes, and the blue color indicates downregulated genes. The upregulated genes in diseased meniscus compared with healthier tissue are ACTR2, KRT, HSPD1, CALU, HSPA9, BAX, CCDC80, C17orf59, HSP90AB1, CDC42, HNRNPH1, FKBP9, MT1DP, G3BP1, CTTN, COL6A1, TAF15, ELAVL1, MCL1, AL, HSPA4, CXorf40A CXorf40B, TCP1, RP5, VHL, PRPF4, BAG5, RIOK3, FCF1, SPTLC1, APH1A, RNF170, RCN3, DENR, CAPZB, MBNL1, CASC4, ASB1, MRPS10, RAB23, PAAF1, FARSB, NASP, NUDC, ZNF346, RIOK1, and GAR1. The downregulated genes in diseased meniscus compared with healthier tissue are RNF103, SLC41A3, SNX19, GTFIP1, ERF, NBPF10 NBPF3, OSBPL10, ADAM15, KIAA0930, ZMAT3, AL845464.3, ANTXR1, ENDOD1, ZNFX1, GLT8D2, RP5-1022P6.4, ENG, POMZP3, C11orf95, LPCAT1, EPN2, POMZP3, RAB11FIP3, TBC1D2B, PCDHGC3, MMP14, TRPM4, HDAC5, ABL1, SOX9, Antxr1, BCAN, ZP3, ABR, SNX33, RPS6KA4, CXCR7, NCOR2, PLEC, GPC1, SEMA3C, PDGFRB, LUM, ACAN, CSPG4, OAS1, TIMP2, VCAM1, LAM5, PRSS54, PCDHB1, and FMOD.
(H) Selected microarray data of genes that are known from the literature to be highly expressed in healthy hyaline cartilage tissue (SOX9, PDFGFRB, ACAN, TIMP2, and FMOD). These genes were significantly downregulated in cells derived from diseased tissue compared with cells from healthier meniscus tissue. Only COL6, as indicator gene of an active pericellular matrix of chondrocytes (Poole et al., 1992), is upregulated.
(I) Peptide abundance of selected genes shown as the mean of four experiments that analyzed cells that migrated out from meniscus from healthier and diseased samples in vitro (SD).
Using microarrays, we then analyzed cells obtained by culturing tissue explants from the inner zone of diseased and healthier human menisci. We focused on the top 100 most abundantly expressed genes, and among these, we identified 48 as being upregulated, i.e., exhibiting a positive fold change, and 52 as being downregulated, i.e., exhibiting a negative fold change (p value <0.001, as listed in the figure legend). Among these top 100 genes, we found only four of the 15 arbitrarily defined potential marker genes: TIMP2, SOX9, ACAN, and MMP14, stressing the importance of these particular genes for discriminating healthier meniscal samples from diseased ones. When we analyzed eight human meniscus tissue samples, the 100 top genes clustered into two groups, healthier and diseased, as shown in the heatmap (Figure 1G). A selection is listed (Figure 1H), and the complete results can be found under GEO (accession number GSE52042).
To complement our microarray analyses, we performed proteomics analyses on cells grown from the healthier and diseased meniscus samples and could identify approximately 4,000 proteins as expressed by meniscal explant cells. However, only a small number of proteins produced by meniscal cells are known to be relevant for OA and meniscus pathology (as listed in Figure 1I), suggesting that for the meniscus, like other tissues studied so far, the correlation between transcriptome and proteome data is weak (Haider and Pal, 2013). Upon close examination, several signaling pathway mediators stood out as being differentially expressed between cells from healthier and diseased meniscal samples. In particular, SMAD2, a mediator of the canonical TGF-β/activin signaling pathway was more abundant in healthier meniscus cells (Figure 1I; for a full listing, visit http://www.miosge.med.uni-goettingen.de/de/?id=17). To follow up on this finding, we performed immunohistochemical (IHC) staining and found that meniscus samples that received a high disease score exhibited a reduced IHC staining for TGF-β and SMAD2. Consistent with these findings, proteome analysis and western blotting of diseased specimens also showed a reduction in SMAD2 protein and an upregulation of RUNX2 compared to healthier specimens. These results, together with existing literature, indicated that the TGF-β/BMP pathway, with its dual osteogenic and chondrogenic actions (Massagué, 2012), was a good candidate to investigate in greater detail.
TGF-β/BMP Signaling in Human Osteoarthritic Meniscus Tissue
Because the deregulation of TGF-β family proteins has been described to be important for meniscus pathology (de Mulder et al., 2013), we examined the TGF-β status of our samples. We observed greater staining for TGF-β3 in healthier (Figure 2A) tissue compared with diseased (Figure 2B) human menisci and also found more SMAD2/3 staining in healthier tissue (Figure 2D). Reduced staining for SMAD1 was also observed in diseased tissue (Figure 2C) compared with healthier specimens (data not shown). SMAD4 staining appeared to be similar between the two groups of human menisci (Figure 2F; data not shown). When we examined the tissues for the presence of TGF-β receptors, we did not find ALK1 expression in healthier meniscus tissues (Figure 2G) but did observe staining in blood vessels of the outer vascularized part of the same specimen, consistent with reported expression of ALK1 in endothelial cells (Figure 2H; Goumans et al., 2003). However, staining for ALK5 was positive in the healthier meniscus (Figure 2I). Some of our IHC findings were verified by western blotting using tissue extracts from the inner zone of the menisci. All blots were quantified using ImageJ64 with α-tubulin used as a control (Figure 2J). A significantly higher level of SMAD2/3 was detected in healthier than in diseased tissue extracts (Figure 2K). Interestingly, we found greater level of RUNX2 and SMAD4 in diseased specimens (Figures 2L and 2N), whereas SOX9 levels were not altered significantly (Figure 2M). Real-time RT-PCR (for primer sequences, see Table 1) revealed results consistent with western blotting for the expression of TGF-β3 (Figure 2O) and RUNX2 (Figure 2P). Expression of SOX9 mRNA was reduced in diseased specimens (Figure 2Q), as was the expression of collagen type II (Figure 2R). Why the SOX9 mRNA reduction is not seen on the protein level remains unclear but may be due to the half-life of the protein. The simultaneous reduction in TGF-β3 and SMAD2/3 and greater levels of RUNX2 seen in diseased meniscus have been identified as a hallmark of osteochondroprogenitor cells that are present in human tissue at late stages of OA (Koelling et al., 2009; Seol et al., 2012) and suggested to us that diseased meniscus may also contain an increased number of progenitor cells.
Figure 2.
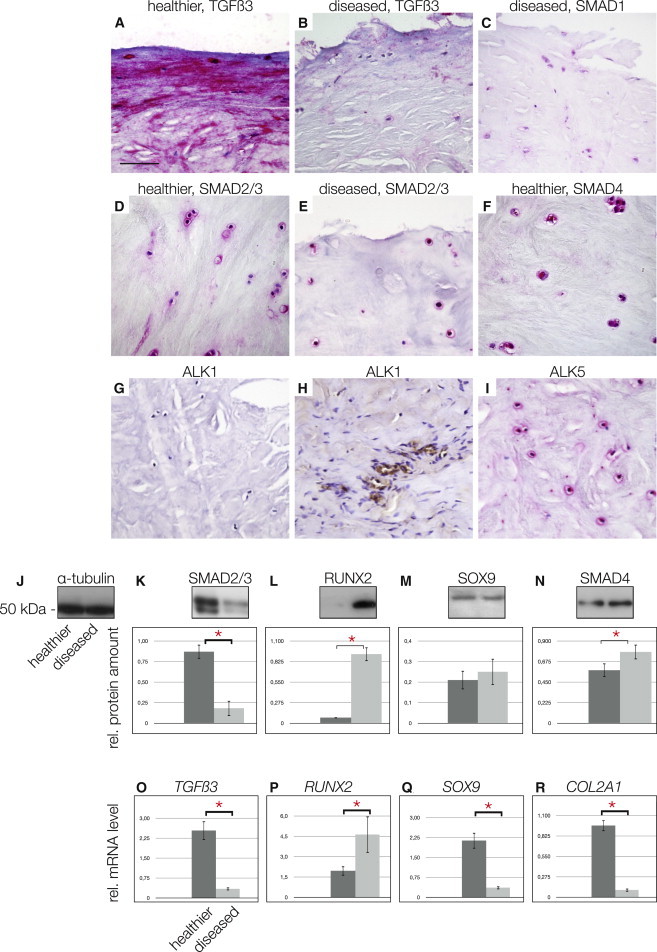
The TGF-β Pathway Players in Healthy and Diseased Meniscus Tissue
(A and B) Healthier meniscus stained for TGF-β3 (A) compared with the sparse staining in diseased meniscus (B).
(C–E) Sparse staining of SMAD1 in diseased meniscus tissue (C). SMAD2/3 in healthier (D) compared to diseased meniscus tissue (E).
(F–H) SMAD4 staining in healthier meniscus tissue (F). No ALK1 staining was detectable in healthier meniscus tissue (G), whereas blood vessels from the outer, vascularized part of the same specimen served as positive control (H).
(I) However, ALK5 was detected in healthier meniscus tissue. Magnification in (A)–(I); scale bar, 150 μm.
(J–N) Western blotting and quantification using ImageJ64: (J) α-tubulin shows equal loading; the first lanes of the blots corresponding to the dark gray bars in the graphs always show the healthier specimens. The diseased specimens are always found in the second lanes of the blots, which correspond to the light gray bars in the graphs. A significantly greater level of SMAD2/3 in healthier meniscus tissues is found (K). RUNX2 is almost undetectable in healthier tissue, whereas it is significantly elevated in diseased meniscus (L). SOX9 showed no significant differences between tissue types (M). SMAD4 was increased significantly in diseased meniscus (N).
(O–R) Quantitative real-time RT-PCR results: (O) TGF-β3 mRNA is significantly increased in healthier meniscus tissue; however, RUNX2 mRNA levels are significantly reduced in healthier meniscus (P). (Q and R) (Q) SOX9 mRNA is significantly reduced in diseased meniscus, which is in line with the significant reduction in collagen type II mRNA (R).
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three different healthier and three different diseased samples of OA patients for mRNA and western blotting experiments.
Table 1.
Primer Sequences
| GeneID | Forward Primer | Reverse Primer | Accession Number |
|---|---|---|---|
| SMAD1 | 5′-TCTTCAGAGCCACCATGAACTAA-3′ | 5′-AACCAGCACAGGAGGAAGTACAG-3′ | NM_005900 |
| SMAD2 | 5′-GTCTCTTGATGGTCGTCTCCA-3′ | 5′-TTCTGTTAGGATCTCGGTGTGTC-3′ | NG_029946 |
| SMAD3 | 5′-CCATCCTGCCTTTCACTC-3′ | 5′-TGGTGATGCACTTGGTGT-3′ | AB004922 |
| SMAD4 | 5′-GCACAAGGTTGGTTGCTAAGA-3′ | 5′-GCAGAACAGTGAGACATTAGGTAGAG-3′ | NG_013013 |
| TGF-β3 | 5′-CTTTGGACACCAATTACTGCTTC-3′ | 5′-GGGTTCAGAGTGTTGTACAGTCC-3′ | NM_003239.2 |
| RUNX2 | 5′-TTCCAGACCAGCAGCACTC-3′ | 5′-CAGCGTCAACACCATCATT-3′ | NM_004348 |
| COL1A1 | 5′-TTCCCCCAGCCACAAAGAGTC-3′ | 5′-CGTCATCGCACAACACCT-3′ | NM_000088 |
| COL2A1 | 5′-CTCCTGGAGCATCTGGAGAC-3′ | 5′-ACCACGATCACCCTTGACTC-3′ | NM_033150 |
| SOX9 | 5′-CAGGCTTTGCGATTTAAGGA-3′ | 5′-CCGTTTTAAGGCTCAAGGTG-3′ | Z46629 |
| β2M | 5′-TGCTGTCTCCATGTTTGATGTATCT-3′ | 5′-TCTCTGCTCCCCACCTCTAA-3′ | NM_004048 |
Cells from Diseased Human Meniscus In Vitro Are Similar to the Cells Found In Vivo
We used explant cultures of diseased human meniscus from late-stage OA to further examine the resident osteochondroprogenitor cell population. Because most OA cases are identified at late disease stages when the superficial zone of the meniscus has been completely lost, we concentrated on the cells from the inner, “white,” avascular zone. In culture, these cells grow clonally and exhibit the spindle shape typical of mesenchymal stem cells (MSCs) (Figure 3A). Consistent with this observation, fluorescence-activated cell sorting (FACS) analysis for the stem cell markers STRO1, CD29, CD90, CD105, and CD106 showed significantly higher levels of these antigens on cells migrating out of the inner zone of diseased meniscus than in cells released after collagenase digestion of healthier specimens where the superficial zone remained intact (Figure 3B). Both populations were negative for the hematopoietic markers CD45 and CD34 (data not shown). When driven toward the chondrogenic lineage (3D alginate culture with standard chondrocyte differentiation medium) cells from damaged menisci showed 60% more SOX9 (Figure 3D), but RUNX2 was no longer detected (Figure 3E). Collagen type II appeared in chondrogenic culture conditions; however, it was undetectable in undifferentiated cells in 2D culture (Figure 3F). The levels of SOX9 (Figure 3G) and collagen type II mRNA (Figure 3H) were also significantly increased when cells were grown in 3D and chondrocyte differentiation medium. In contrast, the levels of collagen type I mRNA decreased (Figure 3I). Following osteogenic differentiation, a proportion of cells became positive for alkaline phosphatase (Figure 3J) and osteocalcin (Figure 3K), as well as alizarin red (data not shown). Oil red O-positive (Figure 3L) and PPARγ-positive (data not shown) adipocytes were identified following adipogenic differentiation.
Figure 3.

Characterization of Cells from the Diseased Meniscus Tissue as MPCs
(A) Explant cell culture of cells migrating out of a diseased meniscus specimen. Magnification in (A), scale bar, 150 μm.
(B) FACS data showed that migratory cells from diseased meniscus explant cultures obtained without collagenase digestion expressed significantly more stem cell markers, including STRO1, CD29, CD105, CD106 (bars on left side), than cells obtained via collagenase digestion from a healthier meniscus (bars on right side). CD90 was expressed equally in the two cell populations.
(C) α-tubulin loading control; the first lanes always represent the 2D control experiments, the second lanes always represent the 3D differentiation, and the third lanes always represent the 3D differentiation together with the chondrogenic differentiation medium (3D + chon. diff.).
(D) Production of SOX9 is significantly increased compared with the control in 2D and 3D cultures.
(E) RUNX2 was undetectable following 3D + chon. diff. culture.
(F) Collagen type II was present after 3D culture and 3D + chon. diff. culturing.
(G and H) Western blotting data were corroborated by mRNA results that revealed significantly higher levels of SOX9 mRNA (G) and collagen type II (H) following 3D + chon. diff. culture compared with control cells in 2D culture.
(I) Collagen type I mRNA decreased significantly following 3D + chon. diff. culture.
(J–L) After osteogenic differentiation, cells become positive for alkaline phosphatase (J) and (K) osteocalcin. Adipogenic differentiation is indicated by positive oil red O staining (L). Scale bars, 150 μm in (J)–(L).
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three individual experiments.
To examine the cell migration potential ex vivo (Figure 4A), a sample of diseased meniscus tissue was placed on the scraped side of a tissue culture flask that had been 80% confluent with GFP-labeled primary meniscus cells. After 3 days, green cells were found inside the tissue sample (Figure 4B), which was confirmed by placing the sample in a new flask (Figure 4C). After an additional 7–10 days, cells, both green and unlabeled, residing in the damaged meniscus sample, were again found outside the tissue. Strikingly, cells migrated only into diseased meniscus tissues and not into the ones containing an intact superficial zone. Furthermore, a standard Boyden chamber assay revealed that the meniscus-derived cells displayed enhanced migration toward a gradient of human recombinant platelet-derived growth factor (PDGF) (Figure 4D), much like the behavior recorded for chondrogenic progenitor cells (Koelling et al., 2009). Taken together, the multilineage differentiation potential and the migration results enabled us to name the cells “meniscus progenitor cells” (MPCs).
Figure 4.
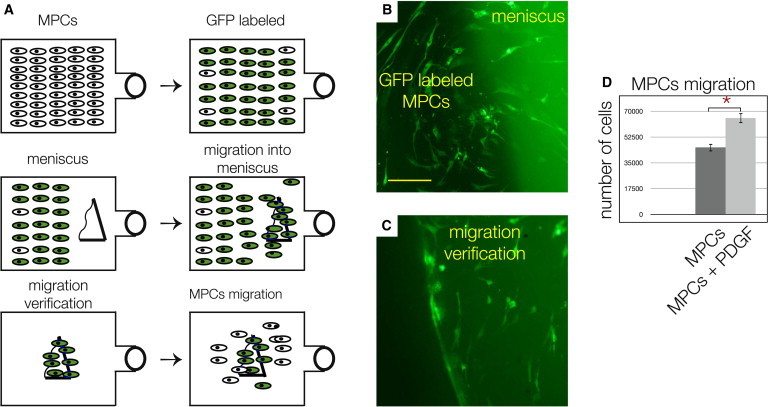
Migration Potential of MPCs
(A, top flasks) MPCs were GFP labeled, enriched via FACS to 97%, and cultured. (Middle flasks) Upon reaching 80% confluence, one side was scraped, and a sample of diseased meniscus tissue was placed on the cell-free side. After 3–4 days, MPCs repopulated and migrated into the tissue sample. (Bottom flasks) The tissues were washed with 1 × PBS solution and transferred into a new flask, and, after 7–10 days, some labeled and unlabeled MPCs migrated out again.
(B) The labeled cells adjacent to the diseased meniscus tissue.
(C) Labeled MPCs in the sample of the diseased tissue after washing and transferal to a new flask. Scale bar, 150 μm in (C) and (D).
(D) Boyden chamber assay results showing that significantly more MPCs were attracted by human recombinant PDGF compared with controls.
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three individual experiments.
The Influence of TGF-β3, RUNX2, and SMADs on the Chondrogenic Potential of MPCs
Using real-time RT-PCR, we observed significantly higher levels of mRNA for TGF-β3 (Figure 5A), SMAD2 (Figure 5B), and SMAD3 (Figure 5C) in the cells derived from healthier menisci, whereas SOX9 was generally unchanged (Figure 5D). When examined by IHC, SMAD1 perinuclear staining (Figure 5E), SMAD2/3 (Figure 5F), and SMAD4 (Figure 5G) were all detected in cells from diseased menisci. To further elucidate the possible connection of the disease stage with the TGF-β/BMP pathways, we performed a human TGF-β/BMP signaling pathway array and found significantly higher levels of mRNA for SMAD7 (Figure 5H), noggin (Figure 5I), and follistatin (Figure 5J) in meniscus cells from diseased tissues.
Figure 5.
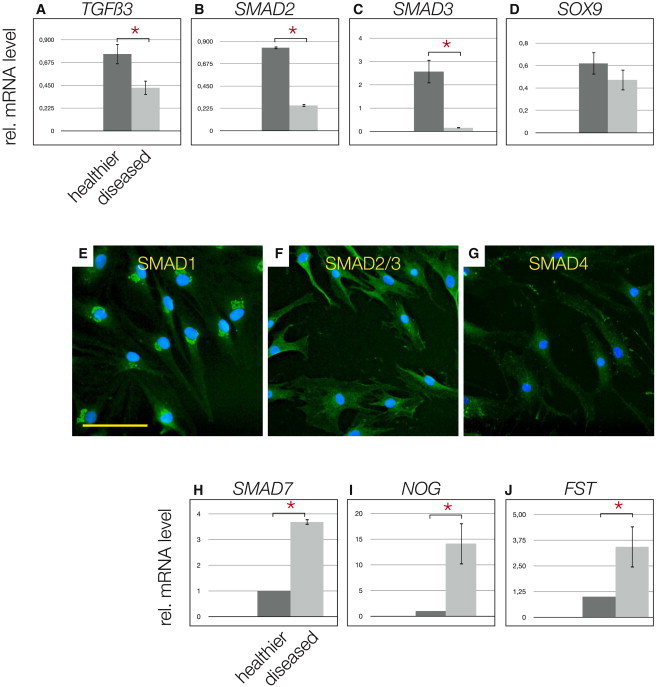
TGF-β Pathway Players in MPCs
(A–C) TGF-β3 mRNA levels are significantly higher in cells from healthier samples (A), which is consistent with SMAD2 (B) and SMAD3 (C) mRNA.
(D) No significant difference was observed for SOX9 mRNA; however, a tendency toward higher levels in cells derived from healthier tissue was noted.
(E–G) Immunocytochemistry of inner diseased meniscal cells positive for SMAD1 (E); note the punctuate perinuclear pattern, SMAD2/3 (F), and SMAD4 (G).
(H–J) The human PCR array for the TGF-β/BMP pathway revealed high levels of mRNA of inhibitors of the TGF-β/BMP axis, for example, SMAD7 (H) noggin (I), follistatin (J), in cells obtained from inner diseased meniscus.
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three different healthier and three different diseased samples of OA patients. Scale bar, 150 μm in (E)–(G).
Because TGF-β enhances chondrogenesis of OA chondrocytes (van der Kraan et al., 2012), we treated MPCs with 10 ng/ml TGF-β3 (Figure 6A). MPCs exposed to TGF-β3 exhibited significantly greater levels of SMAD2/3 (Figure 6B) and also significantly greater levels of p-SMAD2 (Figure 6C). Treatment with TGF-β3 also increased the levels of SOX9 in the MPCs compared with controls (Figure 6D) while also decreasing RUNX2 levels significantly (Figure 6E). In contrast, MPCs treated with 10 ng/ml BMP2 (an activator of the SMAD1/5/7 axis) showed significantly reduced mRNA levels of SMAD2 (Figure 6F) and SOX9 (Figure 6G). When RUNX2 was knocked down in MPCs by small interfering RNA (siRNA) interference (Figures 7A and 7B), SMAD2/3 levels increased (Figure 7C); more importantly, p-SMAD2 became detectable only after the RUNX2 knockdown (Figure 7D).
Figure 6.
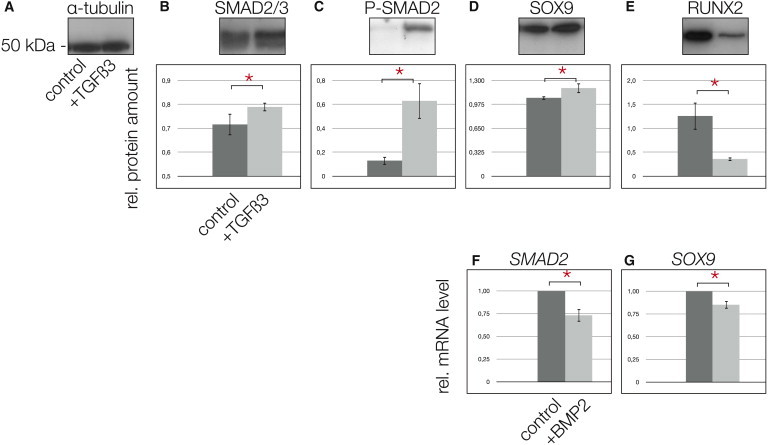
The Effect of TGF-β on MPCs
(A) α-Tubulin indicates equal loading of the gel lanes; also in this figure, the first lanes in the blots and the dark gray bars in the graphs represent control experiments, whereas the second lanes and the light gray bars represent the TGF-β3 stimulation experiments.
(B and C) Following stimulation of MPCs with 10 ng/ml TGF-β3 for 24 hr, total SMAD2/3 (B) was significantly increased and active p-SMAD2 was significantly upregulated (C).
(D and E) In turn, SOX9 increased significantly (D) and RUNX2 was reduced significantly (E).
(F and G) In contrast, following stimulation of MPCs with 10 ng/ml BMP2 for 24 hr, total SMAD2 was reduced significantly (F), as well as SOX9 (G), shown by real-time RT-PCR.
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three individual experiments.
Figure 7.
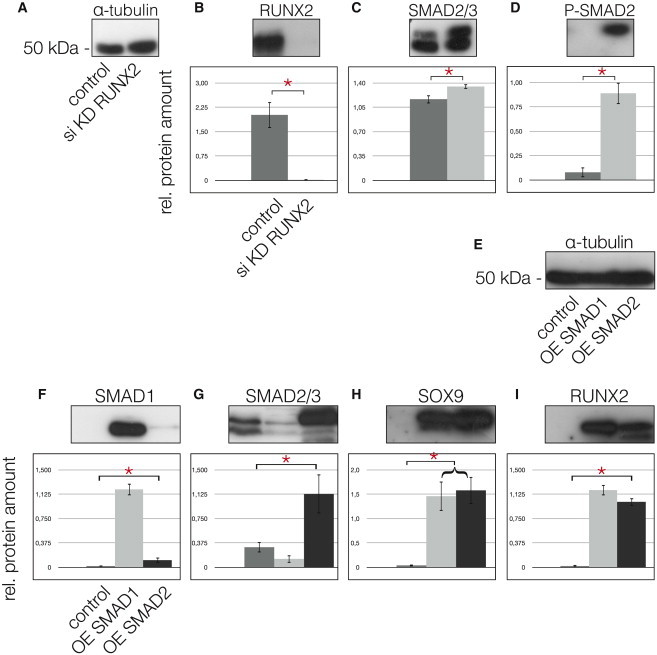
Knockdown of RUNX2 and Overexpression of SMADs
(A) α-tubulin was used for the quantification with ImageJ64; the first lanes of the blots always represent the controls as do the gray bars in the graphs, whereas the second lanes of the blots and the light gray bars of the graphs always represent the RUNX2 KD.
(B) Following RUNX2 KD, the protein was undetectable (lane 2) compared with the control (lane 1).
(C–E) In turn, total SMAD2/3 increased significantly (C) and enabled the detection of p-SMAD2 (D). Overexpression (OE) of SMAD1 and SMAD2. (E) α-tubulin was applied for quantification, the first lanes of the blots always represent the controls as do the dark gray bars in the graphs. The second lanes and the light gray bars represent SMAD1 OE. Finally, the third lanes of the blots and the black bars of the graphs represent the OE of SMAD2.
(F) SMAD1 OE is seen in lane 2.
(G) SMAD2 OE is seen in lane 3.
(H) OE of SMAD1 and SMAD2 results in a significant upregulation of SOX9.
(I) OE of SMAD2 results in a greater reduction of RUNX2 as does OE of SMAD1.
∗Significant differences (p ≤ 0.05); error bars denote the means ± SD of three individual experiments.
Moreover, although the overexpression of SMAD1 or SMAD2 in MPCs (Figures 7E–7G) significantly increased SOX9 expression (Figure 7H), SMAD2 overexpression was more effective in reducing RUNX2 levels then SMAD1 overexpression (Figure 7I). These results demonstrate that the chondrogenic potential of MPCs is likely to be controlled by TGF-β3-mediated phosphorylation of SMAD2, which, in turn, upregulates SOX9 and downregulates RUNX2.
Discussion
By employing a simple histopathological grading system, we have been able to separate OA tissue from patients undergoing knee replacement into samples of early OA (healthier specimens) and severe degenerative disease (diseased specimens). The discriminative power of our grading system was validated by transcriptome analysis, where the molecular pattern found in diseased samples was distinct from that found in early stage OA meniscus. Differential expression patterns have also been shown before in menisci of different age (Rai et al., 2013). Previous results (Koelling et al., 2009; Seol et al., 2012) indicated the existence of progenitor cells in cartilage tissues from late stage OA, and, in fact, when we performed explant cultures using diseased meniscus tissue, we found cells migrating out of these specimens after a few days, which was not observed for healthier menisci. The intact superficial zone, with its flattened cells positive for collagen type I (Chevrier et al., 2009; Hellio Le Graverand et al., 2001) appear to be a barrier for cell migration. When removed, cells are able to migrate out of the less diseased samples. However, cell migration is more effective in diseased meniscus, indicating that inflammation and matrix degrading mediators are needed for migration. FACS analysis of these migratory meniscal cells showed that they were positive for well-known stem cell markers including STRO-1, CD105, or CD106. We also determined that these highly migratory meniscal-derived cells were multipotent; they synthesized collagen type II and suppressed RUNX2 in 3D culture when grown in chondrogenic medium and they can be differentiated into the osteoblastic and adipogenic lineage. Interestingly, a significant difference in marker expression was observed when we compared cells migrating out of diseased meniscal explants with cells derived from meniscal tissue digestions, which indicates that the migratory progenitor cells are a subpopulation of the cells found in diseased human meniscus tissue in vivo. When the protein expression characteristics of the MSC markers are considered together with their clonicity, multipotency, and migratory potential, we named these cells meniscus progenitor cells (MPCs). The cells show signs of senescence (data not shown); therefore, we prefer the term progenitor rather than stem cell.
MPCs are decidedly different from the recently described CPCs (Koelling et al., 2009; Koelling and Miosge, 2010) not only due to their tissue origin and stem cell marker pattern, but also due to their ability to produce collagen type I and type II. However, they are similar as they also appear to be modulated via a balance between RUNX2 and SOX9 (Muhammad et al., 2013). Despite the presence of these multipotent cells in diseased human meniscus, they are not able to contribute substantially to regeneration efforts. Although the cell clusters found might be interpreted as signs of proliferation of the progenitor cells, it is likely that the numerous mediators of inflammation and tissue degeneration present, especially, in the late stages of OA investigated here, inhibit the regenerative capacity of any stem cell present in the diseased tissue.
In articular cartilage, deletion of the type 2 TGF-β receptor results in the upregulation of RUNX2, MMP13, and ADAMTS5, which correlates with the progression of OA disease (Shen et al., 2013), and stimulation of SMAD2/3 signaling in articular chondrocytes has been suggested as a potential therapeutic manipulation for the treatment of OA (van der Kraan et al., 2012). Here, we demonstrate that the treatment of MPCs with TGF-β3 results in an upregulation of SOX9 and a downregulation of RUNX2 similar to findings in articular cartilage. The shift in articular chondrocytes from SMADS2/3 to SMADS1/5 appears to be important for the development of OA, and, recently, a balancing role of endoglin (CD 105) for this shift has been highlighted (Finnson et al., 2010). This change in pathway mediators may also be the case in the diseased human meniscus, because we found lower levels of SMAD2/3 and higher levels of RUNX2 in diseased tissue compared with healthier meniscus tissue.
The complex regulatory role of the signaling balance between TGF-β/BMP in OA pathology is widely acknowledged (Finnson et al., 2010; van der Kraan et al., 2010), and our data suggest this interaction also governs meniscal cell behavior. We observe that MPCs from less diseased meniscus respond to TGF-β via the ALK5 receptor, and that this promotes chondrogenesis, whereas MPCs from more diseased meniscus are prone toward osteogenesis. Furthermore, our findings suggest that specific components of the TGF-β pathway may be suitable targets for regenerative therapies directed at diseased human meniscus in late stages of OA. As OA is a very complex disease, and multiple tissues that are integral to joint function are impacted in different ways (Loeser et al., 2012), we find it unlikely that direct TGF-β treatment of the meniscus will have significant therapeutic benefit, and undesirable side effects of TGF-β therapy have been reported, including synovial hyperplasia, inflammation, or even osteophyte formation (Blaney Davidson et al., 2006; van Beuningen et al., 1998). Our current finding supports the notion that a more nuanced understanding of the regulatory events governing MPCs biology will be necessary to identify a cell-based therapy and the modulatory factors that support using these cells for meniscal regeneration. Identification and characterization of MPCs provides direct evidence that meniscal repair cells are present in highly diseased tissue and also allows for detailed study of the signaling pathways that govern their proliferation and differentiation, a first step in identifying drug targets for the regeneration of diseased meniscus.
Experimental Procedures
Tissues and Preparation
Adult osteoarthritic lateral and medial menisci were obtained from more than 150 patients (ages 62–75 years) suffering from late-stage OA after total knee replacement operations. The patients met the American College of Rheumatology classification criteria and provided their written informed consent. Histopathological classification of OA cartilage confirmed the presence of late-stage OA (Pritzker et al., 2006) of the tibial plateau and femoral condyles in all samples.
Histology and Meniscus Grading
For light microscopy, meniscus specimens (n = 80), including the white and red areas (Figure 1A, inset), were processed as described previously (Koelling and Miosge, 2010), and a combined Alcian blue/nuclear fast-red staining was performed. Based on existing grading systems (Pauli et al., 2011; Zhang et al., 2011), we developed a simple score for meniscus degradation. The presence (1 point) or absence (2 points) of the superficial zone and the intensity of the Alcian blue staining (high = 1 point or low = 2 points) were used for evaluations. The presence of fatty degeneration and/or cell clusters (2 points) or the presence of calcifications (3 points) was also included. A minimum of 2 and a maximum score of 9 points can be reached. The threshold was set to 4 points. Three independent histologists evaluated more than 40 samples and identified 31 with a score of above 4 (diseased) and 16 with scores of below 4 (healthier). The remaining samples were not evaluated unequivocally.
Cell Isolation and Culture
Tissue pieces measuring 7–10 mm3 from the central inner zone (white area) of the healthier (n = 12) and diseased (n = 12) human menisci were excised, and care was taken not to include the outer red zone. Samples of healthier menisci with an intact superficial zone (grading score: 2, n = 12) were also excised. After 7–10 days of incubation, only samples that lacked a superficial zone showed outgrowth of cells, which were harvested, and 103 cells/cm2 were transferred to a monolayer culture under standard conditions. 3D culturing was performed using alginate beads. Samples of 2–3 mm3 in size from the superficial area of healthier menisci (n = 7) and the central area of diseased menisci (n = 7) were harvested and digested with collagenase I (152 U/ml; Invitrogen), collagenase II (280 U/ml; Biochrom), and dispase (15 U/ml; Invitrogen) for 6 hr at 37°C.
Microarray Analysis and Bioinformatic Methods
Quality control and the quantification of total RNA samples was performed, and data from ten human samples were measured using the microarray (Microarrays) ReadyArray (HS1100), which contains 48,958 probes per microarray slide from the Stanford/Illumina collaboration on the HEEBO (Human Exonic Evidence Based Oligonucleotide) set of long oligos. This chip was used in a one-color assay with a reference measurement of internal housekeeping genes. An internal QC was run and data were curated, primarily from nonzero flag values that indicate absent or poor-quality spots, resulting in 13,585 probes. Next, quantile-normalization and logarithmizing followed. We then arbitrarily selected 15 potential marker genes, TIMP1, TIMP2, TIMP3, COL3A1, RUNX2, SOX9, ACAN, PRG4, DCN, MMP9, MMP3, MMP14, ADAMTS20, ADAMTS1, and NID2 (all known to be involved in the pathogenesis of OA), to determine whether each of the ten samples belonged to the “diseased” or “healthier” group. A Pearson correlation-based hierarchical clustering approach including row scaling and Ward’s minimum variance method was chosen. This analysis revealed two groups of four samples each. For the selected eight samples, a differential expression analysis including all 13,585 probes with an empirical Bayes statistic of the limma package, moderated gene-by-gene t tests and p value adjustments via the Benjamini-Hochberg method was performed.
Proteome Analysis
In-gel digestion and peptide extraction was performed using aqueous acetonitrile, and mass spectrometric analysis was performed as described (Christian et al., 2014). The eluent was analyzed using a Top10 method in the Data Dependent Acquisition mode on the Q Exactive high-resolution mass spectrometry system (Thermo Scientific) operated under Tune 2.2 using HCD fragmentation, with normalized collision energy of 25%. Peak lists were generated using the Raw2MSM v1.10 software (MPI for Biochemistry, Martinsried, Germany). All MS/MS samples were analyzed using Mascot (Matrix Science, v.2.4.1) set to search the NCBInr_20130805 database (selected for Homo sapiens, 251,430 entries). Further details, especially about normalization procedures, have already been published (Christian et al., 2014).
Antibodies
Monoclonal anti-type II collagen (CIIC1, 1:500) and anti-STRO1 (STRO1, 1:100) antibodies were obtained from the Developmental Studies Hybridoma Bank, University of Iowa. For western blotting, monoclonal α-tubulin (T6199, 1:2,000, Sigma-Aldrich), anti-SMAD2/3 (c-8, sc-133098, 1:1,000), anti-SMAD1 (A-4, sc-7965, 1:1,000), anti-SOX9 (H-90, sc-2095, 1:1,000), anti-RUNX2 (M-70, sc10758, 1:1,000), anti-SMAD4 (B-8, sc-7966, 1:1,000), anti-TGF-β3 (III, sc-83, 1:100), anti-collagen type I (C-18, sc-8784, 1:100), and anti-ALK1 (c20, sc-19547, 1:100) antibodies were from SC Biotechnology. Polyclonal anti-phospho-SMAD2 (Ser465/467, #3101, 1:1,000) purchased from Cell Signaling Technology, and anti-ALK5 antibodies (Ab-165, E11-1126B, 1:100) from EnoGene Biotech. FACS analysis was performed with PE-coupled anti-CD29 (#554543, 1:500), CD34 (#345802, 1:200), CD106 (#551146, 1:500), and fluorescein isothiocyanate (FITC)-coupled anti- CD45 (#345808, 1:200), CD105 (#312404, 1:500) antibodies from BD Pharmingen. Anti-CD90-PE (#328110, 1:200) was from BioLegend. All secondary antibodies were the ones used previously (Koelling et al., 2009).
Immunohistochemistry
Ten diseased and ten healthier samples were processed for immunohistochemistry with the HiDefDetection alkaline phosphatase mouse/rabbit system (Cell Marque). Immunoreactions were also performed with swine serum as a negative control, and figures show representatives of three different healthier and three different more diseased samples of OA patients.
Immunoblotting
Immunoblotting was performed as described previously (Koelling et al., 2009) and quantified using α-tubulin as a loading control using the ImageJ64 program.
RNA extraction and cDNA Synthesis
Cells from samples of healthier and diseased menisci in P1 monolayer cultures were lysed directly in RLT buffer and subjected to RNA isolation (RNeasy Mini Kit, QIAGEN) as described (Koelling et al., 2009).
Quantitative Real-Time RT-PCR
A 10 μl volume per PCR comprising 1 ng of cDNA, 5 μl of Platinum Sybr Green qPCR SuperMix (Invitrogen) and 20 pmol of each primer was chosen. The program primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/) was used to design the primers (Table 1).
PCRs were performed as described previously, and data were normalized to a highly consistent housekeeping gene (β2M). PCR products were sequenced (Seqlab), and relative ratios were calculated (Pfaffl, 2001). Every PCR was run three times in triplicate. The intratest and intertest variations were <1%. Primer efficiencies ranged from 1.9 to 2.1.
Cell Differentiation
Chondrogenic differentiation was performed using 3D cell culture as described previously (Koelling et al., 2009). For osteogenic differentiation, a total of 1,000 MPCs/cm2 in 75 cm2 flasks were differentiated into cells of the osteoblastic lineage under the influence of NH OsteoDiffMedium or adipogenic differentiation was performed using NH AdipoDiffMedium (Miltenyi Biotec).
Immunofluorescence Microscopy
P1 cells were grown on coverslips, fixed, and incubated with 100 μl of primary antibody (1:50 dilution in PBS) for 1 hr at room temperature. In the case of uncoupled primary antibodies, we then added secondary fluorescence-coupled antibodies (1:500 dilution in PBS) for 20 min at room temperature and stained with DAPI.
FACS Analysis
To test cells for stem cell markers, 106 cells were suspended in 100 μl of PBS containing 1 μl of fluorescence-coupled antibody at room temperature for 1 hr in the dark. Anti-human-PE/FITC immunoglobulin isotype antibodies (BD Pharmingen) served as negative controls for gating. The cells were then washed twice, centrifuged, and subjected to FACS analysis using a FACScan machine (Becton Dickinson), and 10,000 living cells were analyzed (Koelling et al., 2009). For data evaluation, we used the WinMDlv2.9 program (Scripps Research Institute). The FACS Vantage SE (Becton Dickinson) was applied for cell selection and the Cell Quest Pro 2000 software package was used for analysis.
PCR Array
The human TGF-β/BMP signaling pathway array was used (PAHS-035ZA_0123, SAbiosciences), according to the manufacturer’s instructions, and data were normalized using the standard program of the company’s online platform.
GFP Transfection
The cells (5 × 105) were transfected with 2.5 μg of the pmaxGFP vector (Lonza, Koeln, Germany) in 100 μl of nucleofector solution using the Amaxa program U-23 as described before (Koelling et al., 2009).
Migration and Integration Assay
In an ex vivo assay, a sample of diseased meniscus tissue was placed on the scraped side of a flask that had been 80% confluent with GFP-labeled cells. After 5–7 days, the specimens were washed with PBS, transferred to a new flask and examined using a fluorescence microscope. For the in vitro migration assay, we used a commercial two-chamber system (CytoSelect, Cell Biolabs) and extinctions were measured at 560 nm. Human recombinant PDGF (R&D Systems, 10 ng/ml) was applied as chemoattractant.
Cell Induction Experiment
Cells in 3D alginate were stimulated using recombinant human BMP2 (10 ng/ml, lot# MSA3612112) and TGF-β3 (10 ng/ml, lot# MSA3612112), both of which were obtained from R&D Systems. After 24 hr, the cells were harvested and subjected to total protein and RNA extraction.
siRNA Transfections
The iLenti-GFP siRNA expression vector (ABM) was used for RUNX2 knockdown. The probe (CAGCACGCTATTAAATCCAAATT) that targeted Runx2 was placed under the control of H1/H6 and the GFP sequence under the CMV promoter. Control experiments were performed using a vector containing a scrambled siRNA sequence or without any vector. For the transfection of cells, see above.
Overexpression
SMAD1 and SMAD2 cloned into the pCMV5-Flag and pCMV5B-HA vectors were purchased from Addgene, Cambridge, USA and sequenced to verify the inserts. The cells were transfected either with the expression vector or the vector without the insert, as described above.
Statistical Analyses
Statistical product and service solutions (SPSS) software version 13.0 was used. The observed data were tested statistically, and the representative data shown are the means and standard deviation of at least three different healthier and three different more diseased samples of OA patients, if not stated otherwise. After testing for normality of distribution and homogeneity of variances, we performed ANOVAs and post hoc pairwise comparisons of the mean values. The Pearson correlation coefficients were calculated to examine the relationships between parameters. A p value < 0.05 was considered significant.
Author Contributions
Contributions were as follows: H.M., study conception, study design, acquisition of data, analyses and interpretation of data, manuscript preparation, and statistical analyses; B.S., study conception, study design, acquisition of data, analyses and interpretation of data, manuscript and figure preparation, and statistical analyses; C.B., acquisition of data; M.R., acquisition of data; J.A., acquisition of data; S.v.d.H., microarray analysis and bioinformatic methods; V.R., study conception, study design, interpretation of data, and manuscript preparation; and N.M., study conception, study design, interpretation of data, and manuscript preparation.
Acknowledgments
The authors would like to thank Professor H. Urlaub and Dr. C. Lenz from the joint proteome facility at the MPI for Biophysical Chemistry, Goettingen and the Medical Faculty of the University of Goettingen. We would also like to thank Dr. K. Jung from the Institute of Medical Statistics, Medical Faculty Goettingen.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
Data were submitted to Gene Expression Omnibus under accession number GSE52042.
References
- Benjamin M., Evans E.J. Fibrocartilage. J. Anat. 1990;171:1–15. [PMC free article] [PubMed] [Google Scholar]
- Blaney Davidson E.N., Vitters E.L., van der Kraan P.M., van den Berg W.B. Expression of transforming growth factor-beta (TGFbeta) and the TGFbeta signalling molecule SMAD-2P in spontaneous and instability-induced osteoarthritis: role in cartilage degradation, chondrogenesis and osteophyte formation. Ann. Rheum. Dis. 2006;65:1414–1421. doi: 10.1136/ard.2005.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy R.H., Rai M.F., Zhang Z., Torgomyan A., Sandell L.J. Molecular analysis of age and sex-related gene expression in meniscal tears with and without a concomitant anterior cruciate ligament tear. J. Bone Joint Surg. Am. 2012;94:385–393. doi: 10.2106/JBJS.K.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier A., Nelea M., Hurtig M.B., Hoemann C.D., Buschmann M.D. Meniscus structure in human, sheep, and rabbit for animal models of meniscus repair. J. Orthop. Res. 2009;27:1197–1203. doi: 10.1002/jor.20869. [DOI] [PubMed] [Google Scholar]
- Christian H., Hofele R.V., Urlaub H., Ficner R. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014;42:1162–1179. doi: 10.1093/nar/gkt985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mulder E.L., Hannink G., Giele M., Verdonschot N., Buma P. Proliferation of meniscal fibrochondrocytes cultured on a new polyurethane scaffold is stimulated by TGF-β. J. Biomater. Appl. 2013;27:617–626. doi: 10.1177/0885328211417317. [DOI] [PubMed] [Google Scholar]
- Englund M., Guermazi A., Gale D., Hunter D.J., Aliabadi P., Clancy M., Felson D.T. Incidental meniscal findings on knee MRI in middle-aged and elderly persons. N. Engl. J. Med. 2008;359:1108–1115. doi: 10.1056/NEJMoa0800777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund M., Roemer F.W., Hayashi D., Crema M.D., Guermazi A. Meniscus pathology, osteoarthritis and the treatment controversy. Nat Rev Rheumatol. 2012;8:412–419. doi: 10.1038/nrrheum.2012.69. [DOI] [PubMed] [Google Scholar]
- Finnson K.W., Parker W.L., Chi Y., Hoemann C.D., Goldring M.B., Antoniou J., Philip A. Endoglin differentially regulates TGF-beta-induced Smad2/3 and Smad1/5 signalling and its expression correlates with extracellular matrix production and cellular differentiation state in human chondrocytes. Osteoarthritis Cartilage. 2010;18:1518–1527. doi: 10.1016/j.joca.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Goumans M.J., Valdimarsdottir G., Itoh S., Lebrin F., Larsson J., Mummery C., Karlsson S., ten Dijke P. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGFbeta/ALK5 signaling. Mol. Cell. 2003;12:817–828. doi: 10.1016/s1097-2765(03)00386-1. [DOI] [PubMed] [Google Scholar]
- Haddad B., Pakravan A.H., Konan S., Adesida A., Khan W. A systematic review of tissue engineered meniscus: cell-based preclinical models. Curr. Stem Cell Res. Ther. 2013;8:222–231. doi: 10.2174/1574888x11308030007. [DOI] [PubMed] [Google Scholar]
- Haider S., Pal R. Integrated analysis of transcriptomic and proteomic data. Curr. Genomics. 2013;14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellio Le Graverand M.P., Vignon E., Otterness I.G., Hart D.A. Early changes in lapine menisci during osteoarthritis development: part I: cellular and matrix alterations. Osteoarthritis Cartilage. 2001;9:56–64. doi: 10.1053/joca.2000.0350. [DOI] [PubMed] [Google Scholar]
- Hommen J.P., Applegate G.R., Del Pizzo W. Meniscus allograft transplantation: ten-year results of cryopreserved allografts. Arthroscopy. 2007;23:388–393. doi: 10.1016/j.arthro.2006.11.032. [DOI] [PubMed] [Google Scholar]
- Koelling S., Miosge N. Sex differences of chondrogenic progenitor cells in late stages of osteoarthritis. Arthritis Rheum. 2010;62:1077–1087. doi: 10.1002/art.27311. [DOI] [PubMed] [Google Scholar]
- Koelling S., Kruegel J., Irmer M., Path J.R., Sadowski B., Miro X., Miosge N. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4:324–335. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Loeser R.F. Aging processes and the development of osteoarthritis. Curr. Opin. Rheumatol. 2013;25:108–113. doi: 10.1097/BOR.0b013e32835a9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeser R.F., Goldring S.R., Scanzello C.R., Goldring M.B. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmander L.S., Roos E.M. Clinical update: treating osteoarthritis. Lancet. 2007;370:2082–2084. doi: 10.1016/S0140-6736(07)61879-0. [DOI] [PubMed] [Google Scholar]
- Lohmander L.S., Englund P.M., Dahl L.L., Roos E.M. The long-term consequence of anterior cruciate ligament and meniscus injuries: osteoarthritis. Am. J. Sports Med. 2007;35:1756–1769. doi: 10.1177/0363546507307396. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauck R.L., Martinez-Diaz G.J., Yuan X., Tuan R.S. Regional multilineage differentiation potential of meniscal fibrochondrocytes: implications for meniscus repair. Anat. Rec. (Hoboken) 2007;290:48–58. doi: 10.1002/ar.20419. [DOI] [PubMed] [Google Scholar]
- McDevitt C.A., Webber R.J. The ultrastructure and biochemistry of meniscal cartilage. Clin. Orthop. Relat. Res. 1990;(252):8–18. [PubMed] [Google Scholar]
- Muhammad H., Schminke B., Miosge N. Current concepts in stem cell therapy for articular cartilage repair. Expert Opin. Biol. Ther. 2013;13:541–548. doi: 10.1517/14712598.2013.758707. [DOI] [PubMed] [Google Scholar]
- Osawa A., Harner C.D., Gharaibeh B., Matsumoto T., Mifune Y., Kopf S., Ingham S.J., Schreiber V., Usas A., Huard J. The use of blood vessel-derived stem cells for meniscal regeneration and repair. Med. Sci. Sports Exerc. 2013;45:813–823. doi: 10.1249/MSS.0b013e31827d1e06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C., Grogan S.P., Patil S., Otsuki S., Hasegawa A., Koziol J., Lotz M.K., D’Lima D.D. Macroscopic and histopathologic analysis of human knee menisci in aging and osteoarthritis. Osteoarthritis Cartilage. 2011;19:1132–1141. doi: 10.1016/j.joca.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen W., Pufe T., Stärke C., Fuchs T., Kopf S., Raschke M., Becker R., Tillmann B. Locally applied angiogenic factors—a new therapeutic tool for meniscal repair. Ann. Anat. 2005;187:509–519. doi: 10.1016/j.aanat.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole C.A., Ayad S., Gilbert R.T. Chondrons from articular cartilage. V. Immunohistochemical evaluation of type VI collagen organisation in isolated chondrons by light, confocal and electron microscopy. J. Cell Sci. 1992;103:1101–1110. doi: 10.1242/jcs.103.4.1101. [DOI] [PubMed] [Google Scholar]
- Pritzker K.P., Gay S., Jimenez S.A., Ostergaard K., Pelletier J.P., Revell P.A., Salter D., van den Berg W.B. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Rai M.F., Patra D., Sandell L.J., Brophy R.H. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis Rheum. 2013;65:2090–2101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginster J.Y. The prevalence and burden of arthritis. Rheumatology (Oxford) 2002;41(Supp 1):3–6. [PubMed] [Google Scholar]
- Segawa Y., Muneta T., Makino H., Nimura A., Mochizuki T., Ju Y.J., Ezura Y., Umezawa A., Sekiya I. Mesenchymal stem cells derived from synovium, meniscus, anterior cruciate ligament, and articular chondrocytes share similar gene expression profiles. J. Orthop. Res. 2009;27:435–441. doi: 10.1002/jor.20786. [DOI] [PubMed] [Google Scholar]
- Seol D., McCabe D.J., Choe H., Zheng H., Yu Y., Jang K., Walter M.W., Lehman A.D., Ding L., Buckwalter J.A., Martin J.A. Chondrogenic progenitor cells respond to cartilage injury. Arthritis Rheum. 2012;64:3626–3637. doi: 10.1002/art.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Li J., Wang B., Jin H., Wang M., Zhang Y., Yang Y., Im H.J., O’Keefe R., Chen D. Deletion of the transforming growth factor β receptor type II gene in articular chondrocytes leads to a progressive osteoarthritis-like phenotype in mice. Arthritis Rheum. 2013;65:3107–3119. doi: 10.1002/art.38122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman J.R., Rodkey W.G. Tissue-engineered collagen meniscus implants: 5- to 6-year feasibility study results. Arthroscopy. 2005;21:515–525. doi: 10.1016/j.arthro.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Steinert A.F., Palmer G.D., Capito R., Hofstaetter J.G., Pilapil C., Ghivizzani S.C., Spector M., Evans C.H. Genetically enhanced engineering of meniscus tissue using ex vivo delivery of transforming growth factor-beta 1 complementary deoxyribonucleic acid. Tissue Eng. 2007;13:2227–2237. doi: 10.1089/ten.2006.0270. [DOI] [PubMed] [Google Scholar]
- van Beuningen H.M., Glansbeek H.L., van der Kraan P.M., van den Berg W.B. Differential effects of local application of BMP-2 or TGF-beta 1 on both articular cartilage composition and osteophyte formation. Osteoarthritis Cartilage. 1998;6:306–317. doi: 10.1053/joca.1998.0129. [DOI] [PubMed] [Google Scholar]
- van der Kraan P.M., Blaney Davidson E.N., van den Berg W.B. Bone morphogenetic proteins and articular cartilage: to serve and protect or a wolf in sheep clothing’s? Osteoarthritis Cartilage. 2010;18:735–741. doi: 10.1016/j.joca.2010.03.001. [DOI] [PubMed] [Google Scholar]
- van der Kraan P.M., Goumans M.J., Blaney Davidson E., ten Dijke P. Age-dependent alteration of TGF-β signalling in osteoarthritis. Cell Tissue Res. 2012;347:257–265. doi: 10.1007/s00441-011-1194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber R.J., York J.L., Vanderschilden J.L., Hough A.J., Jr. An organ culture model for assaying wound repair of the fibrocartilaginous knee joint meniscus. Am. J. Sports Med. 1989;17:393–400. doi: 10.1177/036354658901700314. [DOI] [PubMed] [Google Scholar]
- Woolf A.D., Pfleger B. Burden of major musculoskeletal conditions. Bull. World Health Organ. 2003;81:646–656. [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Cheriyan T., Martin S.D., Gomoll A.H., Schmid T.M., Spector M. Lubricin distribution in the torn human anterior cruciate ligament and meniscus. J. Orthop. Res. 2011;29:1916–1922. doi: 10.1002/jor.21473. [DOI] [PubMed] [Google Scholar]


