Abstract
The effect of biomaterial topography on healing in vivo and monocyte/macrophage stimulation in vitro was assessed. A series of expanded polytetrafluoroethylene (ePTFE) materials were characterized by increasing average intranodal distance of 1.2 µm (1.2-ePTFE), 3.0 µm (3.0-ePTFE) and 4.4 µm (4.4-ePTFE), but presented consistent surface chemistry with nonporous PTFE (np-PTFE). Subcutaneous implantation of 4.4-ePTFE into mice resulted in a statistically thinner capsule that appeared less organized and less dense than the np-PTFE response. In vitro, isolated monocytes/macrophages cultured on np-PTFE produced low levels of interleukin 1-beta (IL-1β), 1.2-ePTFE and 3.0-ePTFE stimulated intermediate levels, and 4.4-ePTFE stimulated a 15-fold increase over np-PTFE. Analysis of cDNA microarrays demonstrated that additional proinflammatory cytokines and chemokines, including IL-1β, interleukin 6, tumor necrosis factor alpha, monocyte chemotactic protein 1 and macrophage inflammatory protein 1-beta, were expressed at higher levels by monocytes/macrophages cultured on 4.4-ePTFE at four and twenty-four hours. Expression ratios for several genes were quantified by RT-PCR and were consistent with those from the cDNA array results. These results demonstrate the effect of biomaterial topography on early proinflammatory cytokine production and gene transcription by monocytes/macrophages in vitro as well as decreased fibrous capsule thickness in vivo.
Keywords: human monocytes / macrophages, topography, surface dependent behavior, foreign body reaction, microarray
INTRODUCTION
Monocytes are integrally involved in the host inflammatory and foreign body response to biomaterials.1,2 Monocytes initially move from the vasculature into the implant site where they mature to the macrophage phenotype and can further fuse into multinucleated foreign body giant cells at the biomaterial interface. Adherent macrophages are powerful inflammatory mediators and release a large number of molecules, including pro-inflammatory cytokines that can contribute to cell activation, chronic inflammation, and fibrous encapsulation.2 However, macrophages are also involved in orchestrating wound healing and resolution, for which controlled macrophage activation is necessary.3 A better understanding of how biomaterials influence key parameters of the inflammatory response is central to the development of engineered constructs that integrate with host tissues to promote healing and regeneration of function.
There are numerous mechanical and surface properties of materials that can play important roles in macrophage activation. Adherence of monocytes induces an activated phenotype.4 Surface chemistry affects monocyte/macrophage (MC/MO) secretion of cytokines such as interleukin 1-beta (IL-1β), interleukin 6 (IL-6) and tumor necrosis factor-alpha (TNF-α)5,6 and is the subject of proteomic studies.7,8
Biomaterial topography affects cellular and healing responses as well. Micro-range surface roughness modifies cultured cell response in vitro and biocompatibility and tissue attachment in vivo.9 Nanometer and micrometer surface alterations induce changes in macrophage adhesion, orientation, spreading, and cytoskeleton formation.10,11 Capsule thickness varies around filters made of the same material but with different pore sizes.12 Reduced fibrous-capsule formation, increased vascularity, and closer proximity of vessels to the implant surface have been demonstrated with pillar-textured implants and with porous polyvinyl alcohol implants in comparison with the nontextured or smooth control of the same material.13,14
Expanded PTFE (ePTFE) has been used historically in medical device applications such as vascular grafts, to which macrophages have been shown to adhere within 24 hours.15 The influence of ePTFE materials on MC/MO activation and the secretion of proinflammatory cytokines in vitro, as well as fibrous capsule formation in vivo, has been explored in comparison with other surface chemistries.5,16–19 Expanded PTFE materials with varied pore dimensions have displayed a relationship between pore type and healing.20–22 Previous studies have demonstrated MC/MO activation and IL-1β production when these cells were seeded onto a uniaxially-expanded ePTFE material of a single expansion ratio and were stimulated with LPS.5,16,19
To study the effect of topography, we investigated MC/MO activation in vitro and fibrous capsule formation in vivo in response to biaxially-expanded ePTFE materials in a range of pore sizes, and in comparison with nonporous PTFE, in the absence of additional LPS stimulation.
MATERIALS AND METHODS
PTFE materials
Expanded polytetrafluoroethylene (ePTFE) was obtained in a range of pore sizes with nominal filtration sizes of 0.2 µm, 1 µm and 3 µm (Fluoropore filters, Millipore, Bedford, MA). Nonporous PTFE (np-PTFE, Berghof/America, Coral Springs, FL) was included for comparison. The PTFE was cut into disks with diameters of 21 mm for in vitro studies and 8 mm for in vivo studies by a solvent-washed punch or scalpel, sonicated 3 times in 100% ethanol for 20 minutes, and dried aseptically. The disks were tested with the Limulus amebocyte lysate assay (Associates of Cape Cod, Falmouth, MA) according to the protocol developed by the Food and Drug Administration and found to be free of endotoxin with a sensitivity of 0.03 EU/ml.
Scanning electron microscopy
PTFE disks were mounted on studs with colloidal silver paste and sputter coated with a Au/Pd source. The topography of the samples was analyzed using a JEOL JSM-6300F scanning electron microscope with an accelerating voltage of 15 kV. Images were taken either flat or at a tilt of approximately 40°, recorded onto Polaroid film, and scanned into a digital format. Internodal, intranodal and interfiber distances were quantified with NIH Image.
Electron spectroscopy for chemical analysis
PTFE disks were analyzed on a Surface Science Instruments (SSI) X-Probe ESCA instrument at the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO, University of Washington). An aluminum Kα1,2 monochromatized X-ray source was used for the generation of photoelectrons from a sample surface. The samples were analyzed at a take-off angle of 55° with respect to the surface, allowing for analysis of the outermost 80 Å of the sample. The elemental composition was calculated with the SSI data analysis software using the F(1s) and C(1s) peak areas normalized with a fluorine photoemission cross-section of 4.43 and a carbon cross-section of 1.0.
Subcutaneous implantation of PTFE materials in mice
PTFE disks were implanted subcutaneously into the backs of five male Swiss Webster mice (B&K Universal Limited, England). All work with mice was conducted according to the University of Washington Animal Guide for the Care and Use of Laboratory Animals. Each mouse received two disks, placed bilaterally. Four weeks after implantation, the disks and surrounding tissue were excised, fixed in formalin, sectioned, mounted on slides, and stained with hematoxylin and eosin for general morphological analysis or Masson’s trichrome for analysis of collagen organization. Fibrous capsule thickness was measured with NIH Image and was confined to the dermis above the implant which was not disrupted during sectioning.
Monocyte isolation
Primary human monocytes were isolated from peripheral blood drawn from anonymous donors who had signed an informed consent form approved by the University of Washington’s Human Subjects Review Committee. Blood was collected into 10 ml Vacutainer collection tubes containing sodium heparin and 15 ml Vacutainer tubes without anticoagulant (Becton Dickinson, Franklin Lakes, NJ). The tubes without anticoagulant were held at room temperature for 1 hour and at 4°C for 1 hour. Serum was removed by pipette, centrifuged at 2000 rpm for 10 minutes, and filtered through a 0.2 µm filter to produce autologous human serum. The whole blood/anticoagulant mixture in the heparin tubes was diluted with an equal volume of Dulbecco’s phosphate-buffered saline (DPBS, Gibco, Carlsbad, CA) containing 1mM EDTA (Gibco) and 60u/ml heparin sodium salt (Sigma, St. Louis, MO), layered on top of a Histopaque 1077 gradient (Sigma), and centrifuged. The interface layer containing peripheral blood mononuclear cells was separated and washed. Monocytes were isolated by magnetic bead negative selection using the Monocyte Isolation Kit II with MACS Column type LS (Miltenyi Biotec, Auburn, CA) according to the kit protocol. Cells were resuspended in RPMI 1640 with glutamine containing 25 mM HEPES buffer, 1mM sodium pyruvate, 1mM non-essential amino acids, 100 units/ml penicillin, 100mg/ml streptomycin (Gibco), and 15% filtered autologous human serum. Isolated monocytes were incubated for one hour at 37°C prior to use.
Monocyte/macrophage culture on PTFE materials and cytokine production
PTFE disks were placed in 12-well cell culture plates and secured to the bottom of each well with glass rings that had been stringently cleaned to remove endotoxin. The disks were baked overnight at 180°C, autoclaved, and kept sterile until use. Negative controls of tissue culture polystyrene (TCPS) wells without materials were included in each experiment. Isolated monocytes from a single donor were plated at a concentration of 2 × 106 cells/ml and volume of 1 ml/well and were incubated for 2 hours at 37°C and 5% CO2 to allow for adherence. Subsequently, the wells were washed with pre-warmed media. One ml of fresh media was added per well and the plates were incubated an additional 2 or 22 hours to the endpoints of 4 or 24 hours from the original plating. Supernatants from MC/MO cultures were removed 24 hours after seeding, centrifuged to remove cells and cell debris, aliquotted into sterile tubes, and frozen at −80°C. The supernatants were analyzed for IL-1β or basic fibroblast growth factor (bFGF) by ELISA according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN).
The number of adherent and nonadherent cells in each well was determined by a modified lactate dehydrogenase (LDH) method (Roche, Indianapolis, IN). The cells were lysed by addition of 1 ml of cold 2% Triton X-100 in DPBS buffer and vigorous pipetting. A standard curve was generated with nonadherent monocytes. A 100 µl aliquot of lysate solution was added to an equal volume of LDH reagent prepared according to the manufacturer’s suggestion. The optical density was measured at 490 nm minus 650 nm and was compared to the standard curve.
RNA isolation and amplification
Total RNA was isolated with the RNeasy Mini Kit according to the manufacturer’s instructions (Qiagen, Valencia, CA). Briefly, cells were lysed in the wells using the kit lysis buffer. Cell lysates were centrifuged through the QIAshredder homogenization column, and total RNA was isolated. The optional on-column DNase I digestion was completed with the RNase-Free DNase Set (Qiagen). Total RNA was stored at −80°C. Total RNA was amplified by use of the RiboAmp RNA Amplification Kit (Arcturus, Mountain View, CA) according to the manufacturer’s instructions. One round of amplification was completed from 2–5 µg of total RNA, representing 3–4 cell culture wells. Amplified RNA concentration was measured on a UV spectrophotometer.
cDNA labeling and array hybridization
Labeling and hybridization were completed by the Genomics Resource at the Fred Hutchinson Cancer Research Center (Seattle, WA). For each array sample, 5 µg of amplified RNA was reverse-transcribed and labeled by an amino-allyl labeling protocol as previously described, except that random hexamers were used instead of oligo dT primers for the reverse transcription.23 Spotted human cDNA microarrays contained over 17,600 spots representing named genes, ESTs, and control sequences. Certain genes were represented on the array by multiple spots, with either identical sequences or sequences from different portions of the gene.
Array analysis
Each of the arrays from the independent donors was scanned and analyzed with the GenePix 4000 Microarray Scanner and GenePix Pro 3.0× software (Axon Instruments, Inc. Foster, CA). Raw data were acquired by determination of a diameter for each spot on the array. Fluorescence intensity was measured at 532 nm and 635 nm for each pixel within the spot and the local background. For each spot, median fluorescence intensity was calculated and corrected for the local background fluorescence levels. These background-corrected fluorescence intensities were then used to calculate an expression ratio for each spot equal to (F635, Median − B635, Median)/(F532, Median − B532, Median), where FX, Median is the median fluorescence intensity at wavelength X for each spot and BX, Median is the median fluorescence intensity of the local background at wavelength X. The expression ratios were then normalized such that the overall ratio for the entire chip was 1.0, with the assumption that there was similar expression of most genes between the two samples, and corrects for differences between the two dyes. This ratio was verified by the use of several control spots on the chips. Normalized expression ratios were used in the following analysis. Ratios greater than 2.0 represent a higher expression in the 4.4-ePTFE sample, whereas ratios less than 0.5 represent a higher expression in the np-PTFE sample.
RT-PCR analysis
An aliquot of the total RNA was reserved for quantitative reverse transcription polymerase chain reaction (RT-PCR). Total RNA isolated from the first donor was used to verify the array ratios for TNF-α, IL-1β, IL-6, interleukin 8 (IL-8), monocyte chemotactic protein 1 (MCP1), and interferon-gamma–inducible protein (IP10) by a two-step RT-PCR reaction. Primers were selected with Primer324 such that either the left or right primer overlapped an exon/intron boundary (Table I). A reverse transcription (RT) reaction was performed on 0.3 or 0.5 µg of total RNA with Oligo dT20 primers and C. therm polymerase (Roche). The reaction was placed at 60°C for 30 min, heated to 95°C for 5 min, diluted, and used in a quantitative PCR reaction with the Quantitect SYBR Green PCR kit (Qiagen) and the LightCycler instrument (Roche). The cycler program had an initial activation step of 95°C for 15 minutes and a PCR cycle of 94°C for 15s, 55°C for 20s, 72°C for 15s for 50 cycles. Samples were run in triplicate at one or two dilutions. A standard curve consisting of known concentrations of plasmids (American Type Culture Collection, Manassas, VA) containing full-length clones of the chosen gene was run simultaneously (Table I). The LightCycler Data Analysis software was used to determine the threshold cycle for each sample and standard. The standard curve was subsequently used to determine the concentration of each sample, which was normalized based on total RNA concentration as quantified on an Agilent Bioanalyzer. Expression ratios and standard deviations for the ratios were calculated.
Table I.
Primers for RT-PCR
| Gene | Accession Number | Primer Sequence |
|---|---|---|
| TNF | BC028148 | F: tgtagcccatgttgtagcaaac R: cagcttgagggtttgctaca |
| IL-1β | BC008678 | F: cttcgacacatgggataacg R: gacatggagaacaccacttgt |
| IL-6 | BC015511 | F: tggattcaatgaggagacttg R: gcaggaactggatcaggac |
| IL-8 | BC013615 | F: gtttttgaagagggctgagaa R: tcttgtattgcatctggcaac |
| MCP1 | BC009716 | F: ctcagccagatgcaatcaa R: aatcctgaacccacttctgc |
| IP-10 | BC010954 | F: actctaagtggcattcaaggagta R: catctcttctcacccttctttttc |
F indicates forward primer, R indicates reverse primer. Accession numbers are for plasmids used in development of the standard curve for RT-PCR.
Statistical analysis
Statistical significance for fibrous capsule thickness and MC/MO cytokine production was determined using the unpaired t-test in comparison with np-PTFE.
RESULTS
Scanning electron microscopy of PTFE surfaces
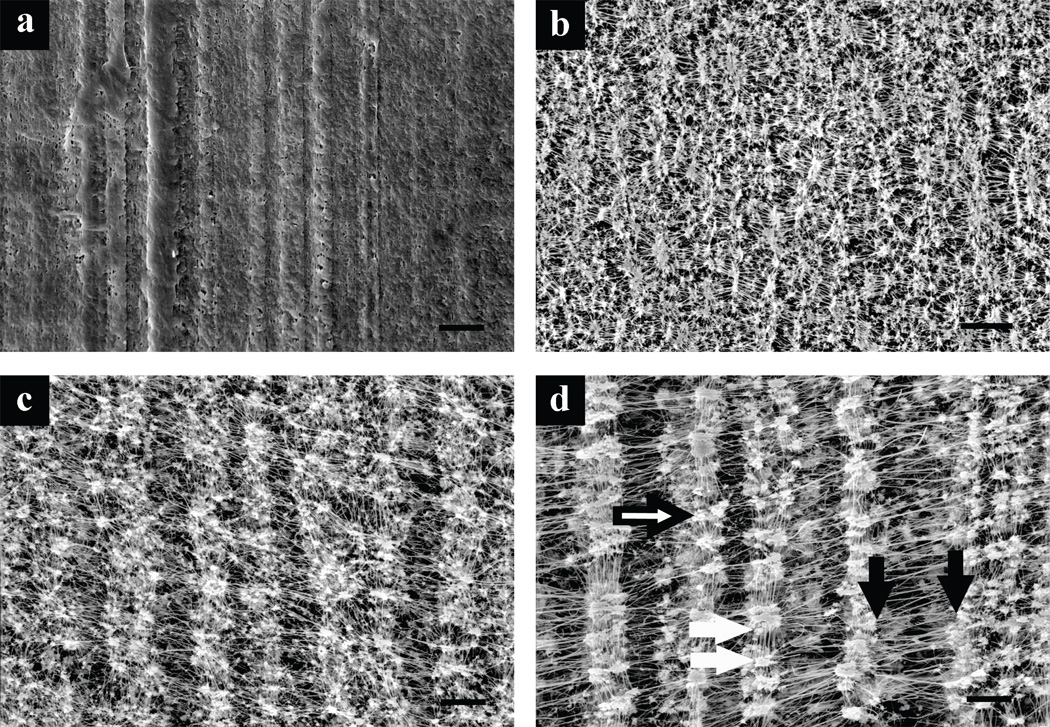
Scanning electron microscopy of the PTFE materials revealed significant differences in surface topographies as shown in Figure 1. The np-PTFE exhibited extrusion or molding lines but did not contain pores visible at the micron scale. The ePTFE contained strands of polymer that were stretched biaxially between nodes, creating a mesh of fibers running perpendicular to each other. The biaxial expansion was non-uniform and displayed a preferential expansion in one direction designated as the major axis of expansion. The pore sizes were quantified by measurement of the distances between the fibers by use of the parameters specified (Figure 1). Internodal distances were measured along the major axis of expansion between nodes of material, and intranodal distances were measured along the minor axis. Interfiber distances were measured between fibers within the nodes of material. The results, summarized in Table II, demonstrate increasing internodal and intranodal distances with increasing nominal filtration sizes. As the topographical distance is more relevant for this discussion than the nominal filtration size, the materials will be referred to by abbreviations (outlined in Table II) representing the average intranodal distances.
Figure 1.
Scanning electron micrographs demonstrating topography of PTFE materials. (a) Nonporous PTFE (np-PTFE), and ePTFE materials with average intranodal distances of (b) 1.2 µm (1.2-ePTFE), (c) 3.0 µm (3.0-ePTFE), and (d) 4.4 µm (4.4-ePTFE). Distance measurements outlined in Table II are indicated in (d), internodal distance indicated between black arrows (measured at 1000×), intranodal distance between white arrows (measured at 1000×), and interfiber distance at point of black and white arrow head (measured at 3000×). Magnification 1000×, scale bar = 10 µm.
Table II.
Pore Dimensions in ePTFE Materials
| Manufacturer’s Designated Nominal Filtration Size (µm) |
Internodal Distance (µm) ± SD |
Intranodal Distance (µm) ± SD |
Interfiber Distance (µm) ± SD |
Abbreviation |
|---|---|---|---|---|
| 0.2 | 3.1 ± 0.4 | 1.2 ± 0.4 | 0.4 ± 0.2 | 1.2-ePTFE |
| 1 | 7.1 ± 0.4 | 3.0 ± 1.3 | 0.9 ± 0.4 | 3.0-ePTFE |
| 3 | 13.2 ± 0.4 | 4.4 ± 2.2 | 0.9 ± 0.4 | 4.4-ePTFE |
Five separate images for each ePTFE material were analyzed for internodal, intranodal and interfiber distances as shown in Figure 1. Internodal distance measured at 1000×, between 200 and 280 measurements per material. Intranodal distance measured at 1000×, between 300 and 400 measurements per material. Interfiber distance measured at 3000×, between 61 and 74 measurements per material. Distances represent average ± standard deviation.
Electron spectroscopy for chemical analysis
Electron spectroscopy for chemical analysis confirmed that the surface chemistry for each of the materials was essentially identical (Table III). PTFE has a theoretical 2:1 ratio of fluorine to carbon atoms. The spectra for each of the materials were similar, containing only fluorine and carbon peaks, including a F(1s) peak, a F(2s) peak, two F Auger peaks, and a C(1s) peak. The F:C ratios were consistent with the theoretical 2:1 ratio of fluorine to carbon atoms.
Table III.
Fluorine (F1s) to Carbon (C1s) Ratio of PTFE Materials Analyzed by ESCA
| Material | F/C Ratio ± SD |
|---|---|
| np-PTFE | 2.0 ± 0.08 |
| 1.2-ePTFE | 2.1 ± 0.04 |
| 3.0-ePTFE | 2.0 ± 0.03 |
| 4.4-ePTFE | 2.0 ± 0.07 |
Data represents average ± standard deviation, n = 5 for each material.
Histology of implanted materials
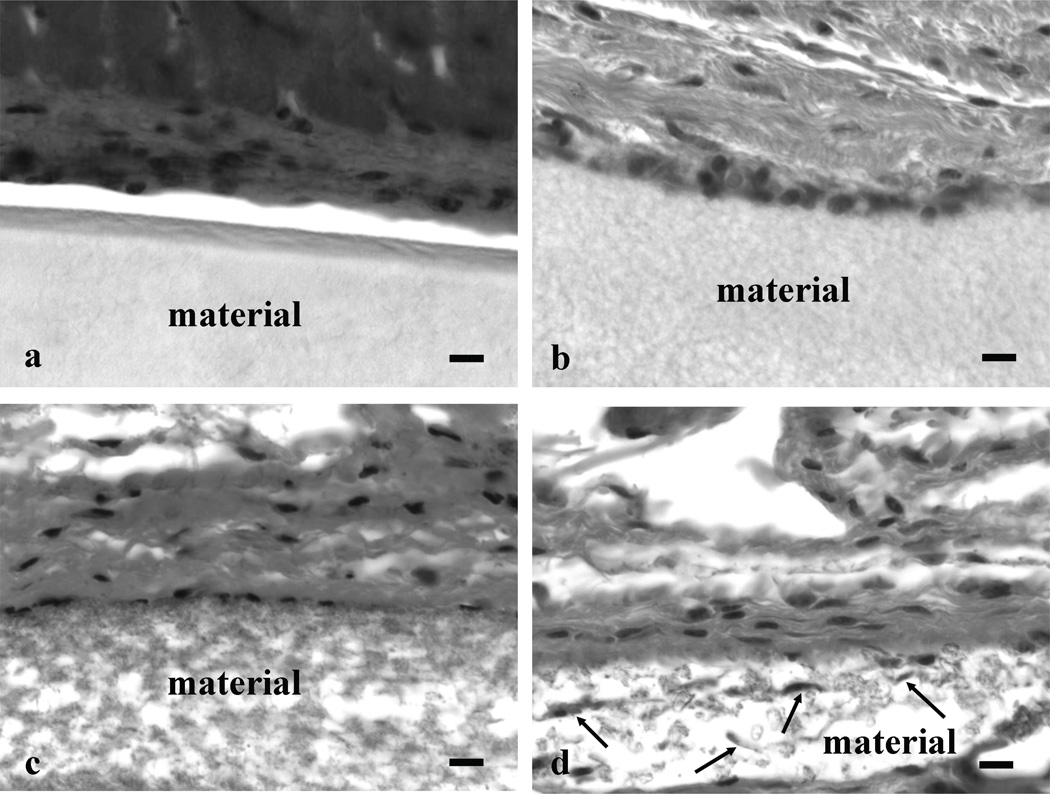
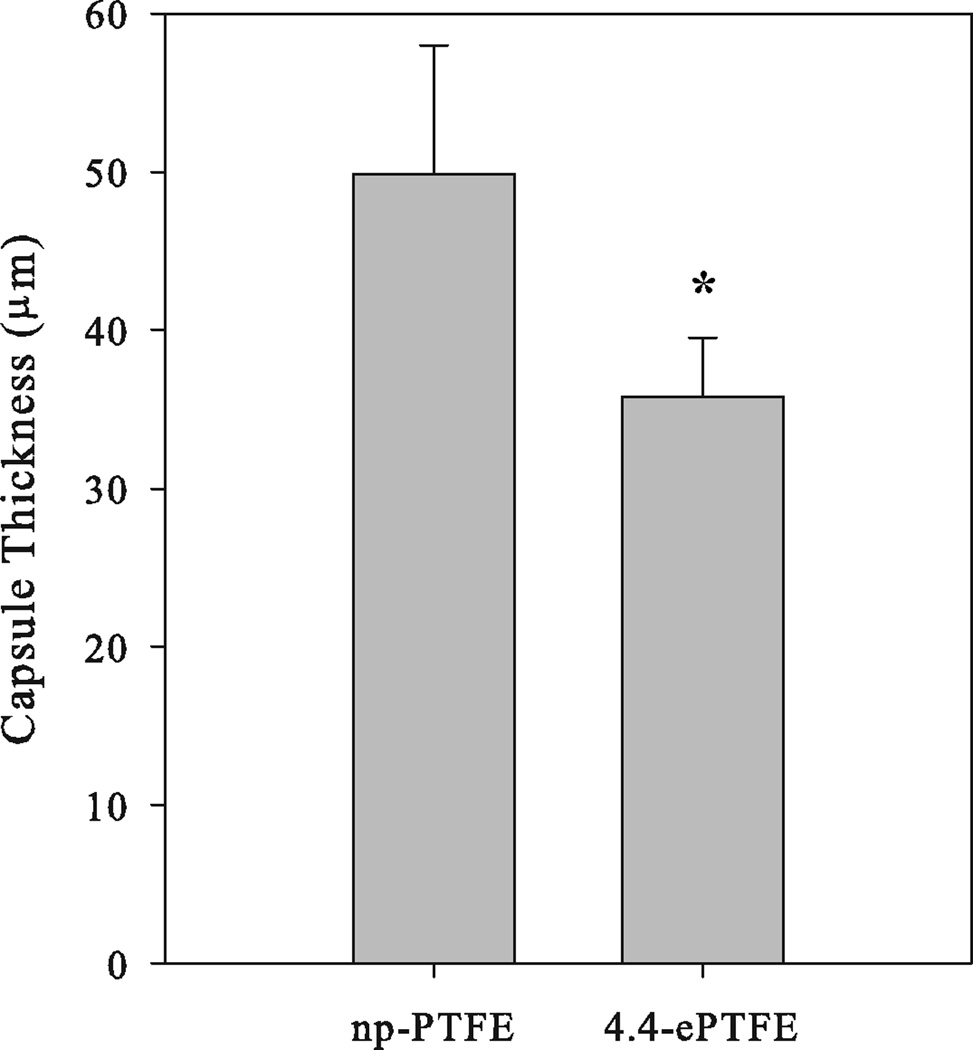
The np-PTFE and ePTFE materials implanted subcutaneously into the backs of mice for four weeks were analyzed histologically to examine interactions with the surrounding dermis (Figure 2). Tissues with np-PTFE implants had gaps between the implant and surrounding dermis, a likely consequence of limited adhesion between the implant and the dermis. In contrast, ePTFE implants remained attached to the dermis after processing. The edges of the 1.2-ePTFE and 3.0-ePTFE materials were lined with cells, and while the pores were too small to allow the cells to invade, the cells did extend processes into the materials (Figure 2b and 2c). The pores in the 4.4-ePTFE were of sufficient size to allow some cellular invasion (Figure 2d) and qualitatively, the capsule was less dense than around the other PTFE materials. Fibrous capsule thickness was measured in slides stained with Masson’s trichrome (Figure 3). The capsule thickness surrounding the 4.4-ePTFE was statistically thinner than the np-PTFE (35.8 ± 3.7 µm and 49.9 ± 8.1 µm, respectively, p<0.01).
Figure 2.
Dermal areas from mice containing PTFE materials implanted subcutaneously for 4 weeks were removed, processed for histology and stained with hematoxylin and eosin. In each panel, the designation material indicates (a) np-PTFE, (b) 1.2-ePTFE, (c) 3.0-ePTFE and (d) 4.4-ePTFE. The 4.4-ePTFE provided pores sufficient in size for some cellular invasion (examples designated with arrows). Scale bar = 10 µm.
Figure 3.
Capsule thickness surrounding PTFE materials implanted subcutaneously for 4 weeks in mice that were removed, processed for histology, and stained with Masson’s trichrome. Average ± sem collected from 5 implants of np-PTFE and 4 implants of 4.4-ePTFE, * denotes p<0.01 in comparison with np-PTFE.
Monocyte/macrophage cytokine secretion in response to ePTFE topography
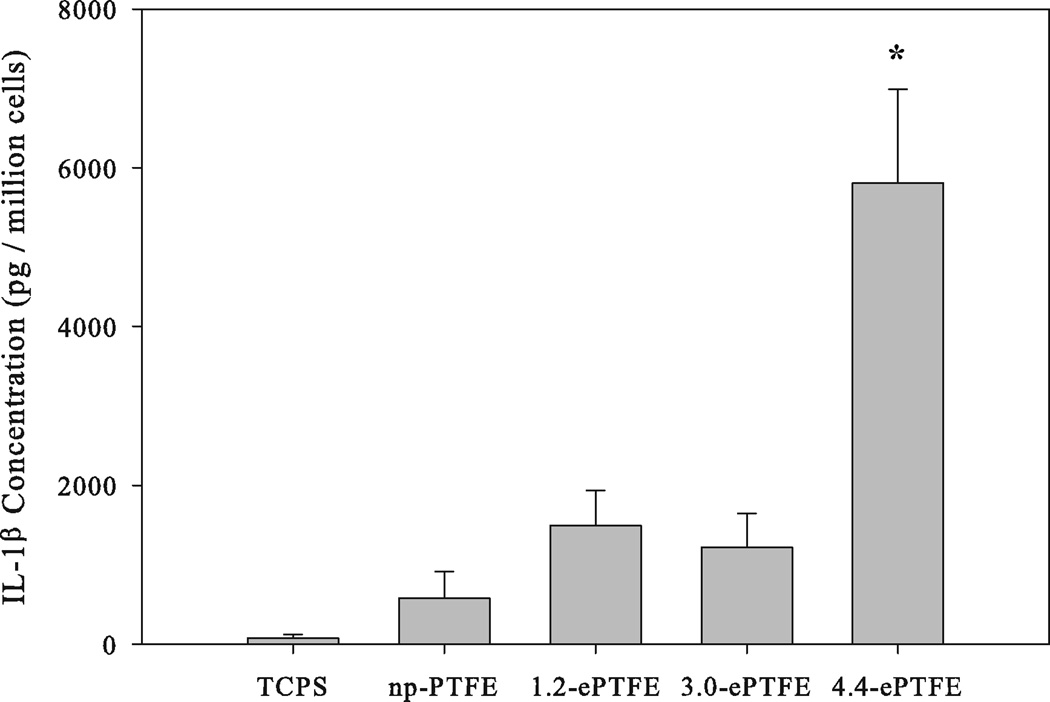
PTFE topography significantly affected MC/MO cytokine production and secretion. Monocytes from four independent donors were seeded onto the series of PTFE materials, and TCPS and the IL-1β levels were ascertained by ELISA 24 hours after cell seeding (Figure 4). The total number of cells per well was not statistically different amongst the various materials (data not shown). MC/MO seeded onto TCPS and np-PTFE samples produced the lowest levels of IL-1β, MC/MO on the 1.2-ePTFE and 3.0-ePTFE samples produced intermediate amounts, and MC/MO on the 4.4-ePTFE produced the highest amount of secreted IL-1β, results demonstrating an increase that was statistically significant over the np-PTFE (p<0.05). IL-1β production without normalization to cell number demonstrated the same trend. The comparison between np-PTFE and the most porous material, 4.4-ePTFE, was repeated with 13 separate donors and demonstrated that the 4.4-ePTFE stimulated a 15-fold increase in IL-1β production over the response to the np-PTFE (np-PTFE stimulated 552 ± 160 pg/106 cells and 4.4-ePTFE stimulated 8241 ± 1130 pg/106 cells, p<0.001).
Figure 4.
MC/MO production of IL-1β in response to TCPS, np-PTFE and a series of porous ePTFE materials expressed as IL-1β concentration for total cell number per well. Average ± sem, n = 4 donors; * denotes p<0.001 in comparison with np-PTFE.
Cytokine gene expression
The transcription of inflammatory genes expressed in MC/MO on np-PTFE or 4.4-ePTFE was examined by microarray in two donors at 4 and 24 hours (Table IV).25 These data paralleled the ELISA-based observations, in that IL-1β was up-regulated in cells cultured on 4.4-ePTFE. Other proinflammatory cytokines were also more highly expressed, including interleukin-1α (IL-1α) and interleukin-6 (IL-6). Significantly higher expression of TNF was seen for one donor, and for the second donor the expression ratio was 1.8, which does not meet the twofold cutoff but showed a similar trend toward higher expression.
Table IV.
Cytokine Array Expression Ratios (4.4-ePTFE/np-PTFE)
| Gene Information | 4 hours | 24 hours | ||||
|---|---|---|---|---|---|---|
| Gene | Accession Number |
Donor #1 |
Donor #2 |
Donor #1 |
Donor #2 |
|
| Interleukin 1-α | IL-1α | AA936768 | 15.3 | 6.3 | No data | 3.8 |
| Interleukin 1–β | IL-1β | W47101 | 2.9 | 2.1 | 36.7 | 4.7 |
| Interleukin 1–β | IL-1β | AA150507 | 2.4 | 2.8 | 7.4 | 2.7 |
| Interleukin 1 Receptor Antagonist | IL1RA | T72877 | 1.5 | 1.3 | 0.7 | 0.5 |
| Interleukin 6 | IL-6 | N98591 | 9.6 | 20.2 | 22.6 | 12.2 |
| Interleukin 10 | IL-10 | - | 1.0 | No data | 0.9 | 3.5 |
| Interleukin 15 | IL-15 | N59270 | 1.2 | 1.8 | 2.4 | 2.9 |
| Colony-Stimulating Factor 1 | CSF1 | T55558 | 2.1 | 1.4 | 2.0 | 0.4 |
| Granulocyte Colony-Stimulating Factor | G-CSF | - | 1.2 | 4.9 | 12.1 | 7.0 |
| Tumor Necrosis Factor | TNF | - | 3.2 | 3.6 | 2.6 | 1.8 |
| Transforming Growth Factor-β1 | TGF-β1 | R36467 | 1.4 | 0.9 | 0.9 | 1.6 |
There were several cytokines that showed differential expression at the 4 hour or 24 hour time points but did not have consistent changes in expression at both time points or in both donors, including interleukin 15 (IL-15), interleukin 10 (IL-10), and interleukin 1 receptor antagonist (IL1RA). There were also several cytokines, the expression of which was similar between the samples at 4 and 24 hours, including transforming growth factor, beta–1 (TGF-β1).
Chemokine gene expression
There were several chemokines that were differentially expressed in MC/MO cultured on the np-PTFE and 4.4-ePTFE (Table V), several of which were MC/MO chemoattractants. Monocyte chemotactic protein 1 (MCP1) and macrophage inflammatory protein 1-β (MIP1-β) were expressed more highly in MC/MO on 4.4-ePTFE at 4 hours and 24 hours, whereas monocyte chemotactic protein 3 (MCP3) was expressed more highly only at 24 hours. Macrophage inflammatory protein 1-α (MIP1-α) was represented by two different sequences on the array. For one sequence, it was expressed more highly in MC/MO on 4.4-ePTFE at 4 and 24 hours. For the second sequence, it was expressed equally between the two samples at 4 and 24 hours. In addition to MC/MO chemoattractants, a neutrophil chemoattractant, GRO1 oncogene (GRO1), was also expressed more highly in the 4.4-ePTFE sample at 4 hours and 24 hours.26
Table V.
Chemokine Array Expression Ratios (4.4-ePTFE/np-PTFE)
| Gene Information | 4 hours | 24 hours | ||||
|---|---|---|---|---|---|---|
| Gene | Systematic Nomenclature |
Accession Number |
Donor #1 |
Donor #2 |
Donor #1 |
Donor #2 |
| MCP1 | CCL2 | AA425102 | 3.5 | 2.1 | 3.2 | 2.2 |
| MIP1-α | CCL3 | AA416584 | 1.7 | 1.0 | 1.0 | 1.0 |
| MIP1-α | CCL3 | AA677522 | 4.1 | 5.1 | 6.0 | 2.8 |
| MIP1-β | CCL4 | H62864 | 4.3 | 3.8 | 4.2 | 2.7 |
| RANTES | CCL5 | AA486072 | 1.1 | 0.9 | 1.1 | 0.9 |
| MCP3 | CCL7 | AA040170 | 1.6 | 1.3 | 2.9 | 2.1 |
| GRO1 | CXCL1 | W46900 | 4.7 | 2.1 | 12.1 | 3.6 |
| GRO1 | CXCL1 | W42723 | 6.6 | 2.1 | 14.4 | 3.6 |
| IL-8 | CXCL8 | AA102526 | 1.1 | 1.6 | 1.6 | 0.9 |
| MIG | CXCL9 | AA131406 | 1.8 | 1.3 | 0.9 | 0.8 |
| IP-10 | CXCL10 | AA878880 | 3.3 | 2.2 | 0.4 | 0.6 |
Abbreviations: Interleukin 8 (IL-8), Monocyte Chemotactic Protein 1 (MCP1), Macrophage Inflammatory Protein 1-α (MIP1-α), Macrophage Inflammatory Protein 1-β (MIP1-β), Regulated Upon Activation, Normally T-expressed, and Presumably Secreted (RANTES), Monocyte Chemotactic Protein (MCP3), GRO1 Oncogene (GRO1), Monokine Induced by Gamma Interferon (MIG), Interferon-Gamma-Inducible Protein 10 (IP-10).
Interestingly interferon-gamma-inducible protein 10 (IP-10), which is a chemoattractant for monocytes and T cells as well as an anti-angiogenic factor, had significantly higher expression in the 4.4-ePTFE sample at 4 hours but a trend was seen toward higher expression in MC/MO on the np-PTFE at 24 hours.27 Although several chemokines demonstrated differential expression by the arrays, several did not show significant changes in expression between the samples at 4 and 24 hours, including IL-8, an inflammatory cytokine that is a chemoattractant for neutrophils, and Regulated Upon Activation, Normally T-expressed, and Presumably Secreted (RANTES), a chemoattractant that acts on several cell types including monocytes.26
Angiogenic factor expression and secretion
Previous studies demonstrated that increasing the porosity of ePTFE correlated with increased levels of post-implantation vascularization in vivo.20,21 Accordingly, we examined the effect of np-PTFE and 4.4-ePTFE on the expression of mRNAs for angiogenic factors known to be released by MC/MO (Table VI).28 These factors included vascular endothelial growth factor (VEGF), TGF-β1, platelet derived growth factor (PDGF), and bFGF. VEGF, TGF- β1, and PDGF subunits A and B were expressed similarly in both samples at 4 hours and 24 hours. However, bFGF was expressed similarly in both samples at 4 hours but at 24 hours was greater in MC/MO cultured on 4.4-ePTFE compared to np-PTFE. Given this differential response, levels of bFGF were examined by ELISA, but no differences were seen in the amount of bFGF secreted by MC/MO cultured for 24 hours on 4.4-ePTFE compared to np-PTFE (data not shown).
Table VI.
Array Expression Ratios for Angiogenic Factors (4.4-ePTFE/np-PTFE)
RT-PCR verification of array results
The mRNA levels of several cytokine and chemokine genes were verified by quantitative RT-PCR on total RNA from Donor 1 (Table VII). Trends from the array data were consistent with the RT-PCR verification for all genes analyzed. While the RT-PCR expression ratios did not reach the same levels observed on the expression array for IL-1β at 4 hours, IL-8 at 24 hours and TNF at 24 hours, the trends were similar. The expression ratios for TNF showed high standard deviations, which could be due to the difficulty of quantifying the low number of transcripts in the original sample.29–31
Table VII.
RT-PCR Expression Ratios (4.4-ePTFE/np-PTFE)
| 4 hours | 24 hours | |||||
|---|---|---|---|---|---|---|
| Gene | Donor #1 RT- PCR Ratio |
Donor #1 Array Ratio |
Donor #2 Array Ratio |
Donor #1 RT-PCR Ratio |
Donor #1 Array Ratio |
Donor #2 Array Ratio |
| TNF | 5.62 +/− 3.25 | 3.2 | 3.6 | 1.30 +/− 0.47 | 2.6 | 1.8 |
| IL-1β | 1.81 +/− 0.43 | 2.9, 2.4 | 2.1, 2.8 | 354.6+/− 37.95 | 36.7, 7.4 | 4.7, 2.7 |
| IL-6 | 4.32 +/− 2.36 | 9.6 | 20.2 | 3.24 +/− 1.42 | 22.6 | 12.2 |
| IL-8 | 0.97+/− 0.20 | 1.1 | 1.6 | 3.22 +/− 0.10 | 1.6 | 0.9 |
| MCP1 | 3.94 +/− 0.78 | 3.5 | 2.1 | 2.07 +/− 0.01 | 3.2 | 2.2 |
| IP-10 | 4.61 +/− 0.31 | 3.3 | 2.2 | 0.45 +/− 0.26 | 0.4 | 0.6 |
RT-PCR expression ratios are the average of triplicate assays +/− the standard deviation.
DISCUSSION
The ability to elicit desirable cell and tissue response to an implanted biomaterial by modification of the surface topography of the material would be a powerful tool in the development of biomaterials and engineered tissues. Although their surface chemistries were identical, the series of biaxially expanded PTFE filters displayed complex topographies that were characterized by a mesh of fibers stretched between nodes of material. Upon implantation, the capsule surrounding the 4.4-ePTFE was qualitatively less organized and less dense than the capsules surrounding the other materials. In addition, the capsule surrounding the 4.4-ePTFE implant was statistically thinner than the capsule surrounding the np-PTFE (p<0.01). These results agree with previous studies using similar materials and highlight that material topography is a crucial variable for the host response to biomaterials.20,21
Results from implantation experiments motivated a mechanistic study to examine MC/MO activation related to PTFE membrane topography in a model in vitro system. MC/MO seeded onto the np-PTFE produced a relatively low level of IL-1β. As the internodal and intranodal distances increased, the production of IL-1β by the MC/MO also increased in a statistically significant manner, data demonstrating an increase in macrophage activation in response to ePTFE topography.
A more global understanding of this MC/MO activation was provided by examination of transcription-level differences between MC/MO cultured on np-PTFE and 4.4-ePTFE at the early time points of 4 and 24 hours that define the initial cell response. The most striking finding was the significantly higher level of pro-inflammatory activation of MC/MO cultured on the 4.4-ePTFE material. In addition to higher levels of inflammatory cytokines such as IL-1β and TNF, there were significantly higher levels of mRNA expression on the 4.4-ePTFE filter for chemokines involved in the recruitment of monocytes and neutrophils. These results indicate that, compared to np-PTFE, the 4.4-ePTFE would have greater recruitment of inflammatory cells into the implant area.
Expanded PTFE with a larger pore size was not available for these studies. However, previous work with the larger pore PTFE materials had shown increased levels of vascularization surrounding the implant.20,21 Of the angiogenic factors examined on the array, only bFGF showed higher expression in MC/MO cultured on the 4.4-ePTFE filter at 24 hours but was shown to have similar amounts of secreted bFGF by MC/MO on the two materials.
In the current study, we have examined the in vivo and in vitro effects of PTFE materials that differ in topography but have consistent surface chemistry. We have shown a topographical effect on early proinflammatory cytokine production and gene transcription in vitro as well as a decrease in fibrous capsule thickness and organization in vivo. It is still unclear to what extent these in vitro, twenty-four hour responses might contribute to the overall foreign body reaction, but it is interesting that the ePTFE surfaces that exhibited a more favorable in vivo response also exhibited a higher initial level of macrophage activation across a broad range of molecular markers. The mechanism behind this reaction is the focus of future work.
Acknowledgments
We gratefully acknowledge financial support from the NSF UWEB ERC (EEC 9529161) and the NIH (GM045873). ESCA was conducted at the NIH NESAC/BIO Center (RR-01296), and SEMs were prepared by Stephanie Lara and Thomas Wight. P.C.S.B was supported by a fellowship from the Whitaker Foundation. A.M.B.C. was supported by fellowships from the Medical Scientist Training Program (GM07266) and the Engineered Biomaterials Training Program (GM065098).
References
- 1.Anderson JM. Biological responses to materials. Annu Rev Mater Res. 2001;31:81–110. [Google Scholar]
- 2.Ziats NP, Miller KM, Anderson JM. In vitro and in vivo interactions of cells with biomaterials. Biomaterials. 1988;9(1):5–13. doi: 10.1016/0142-9612(88)90063-4. [DOI] [PubMed] [Google Scholar]
- 3.Brodbeck WG, Voskerician G, Ziats NP, Nakayama Y, Matsuda T, Anderson JM. In vivo leukocyte cytokine mRNA responses to biomaterials are dependent on surface chemistry. J Biomed Mater Res A. 2003;64(2):320–329. doi: 10.1002/jbm.a.10425. [DOI] [PubMed] [Google Scholar]
- 4.Bauer GJ, Arbabi S, Garcia IA, deHingh I, Rosengart MR, Maier RV. Adherence regulates macrophage signal transduction and primes tumor necrosis factor production. Shock. 2000;14(4):435–440. doi: 10.1097/00024382-200014040-00003. [DOI] [PubMed] [Google Scholar]
- 5.Miller KM, Anderson JM. Human monocyte/macrophage activation and interleukin 1 generation by biomedical polymers. J Biomed Mater Res. 1988;22(8):713–731. doi: 10.1002/jbm.820220805. [DOI] [PubMed] [Google Scholar]
- 6.Yun JK, DeFife K, Colton E, Stack S, Azeez A, Cahalan L, Verhoeven M, Cahalan P, Anderson JM. Human monocyte/macrophage adhesion and cytokine production on surface- modified poly(tetrafluoroethylene/hexafluoropropylene) polymers with and without protein preadsorption. J Biomed Mater Res. 1995;29(2):257–268. doi: 10.1002/jbm.820290217. [DOI] [PubMed] [Google Scholar]
- 7.Dinnes DL, Marcal H, Mahler SM, Santerre JP, Labow RS. Material surfaces affect the protein expression patterns of human macrophages: A proteomics approach. J Biomed Mater Res A. 2007;80(4):895–908. doi: 10.1002/jbm.a.30967. [DOI] [PubMed] [Google Scholar]
- 8.Jones JA, Chang DT, Meyerson H, Colton E, Kwon IK, Matsuda T, Anderson JM. Proteomic analysis and quantification of cytokines and chemokines from biomaterial surface-adherent macrophages and foreign body giant cells. J Biomed Mater Res A. 2007;83(3):585–596. doi: 10.1002/jbm.a.31221. [DOI] [PubMed] [Google Scholar]
- 9.von Recum AF, van Kooten TG. The influence of micro-topography on cellular response and the implications for silicone implants. J Biomater Sci Polym Ed. 1995;7(2):181–198. doi: 10.1163/156856295x00698. [DOI] [PubMed] [Google Scholar]
- 10.Wojciak-Stothard B, Curtis A, Monaghan W, MacDonald K, Wilkinson C. Guidance and activation of murine macrophages by nanometric scale topography. Exp Cell Res. 1996;223(2):426–435. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- 11.Meyle J, Gultig K, Nisch W. Variation in contact guidance by human cells on a microstructured surface. J Biomed Mater Res. 1995;29(1):81–88. doi: 10.1002/jbm.820290112. [DOI] [PubMed] [Google Scholar]
- 12.Campbell CE, von Recum AF. Microtopography and soft tissue response. J Invest Surg. 1989;2(1):51–74. doi: 10.3109/08941938909016503. [DOI] [PubMed] [Google Scholar]
- 13.Sharkawy AA, Klitzman B, Truskey GA, Reichert WM. Engineering the tissue which encapsulates subcutaneous implants. I. Diffusion properties. J Biomed Mater Res. 1997;37(3):401–412. doi: 10.1002/(sici)1097-4636(19971205)37:3<401::aid-jbm11>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 14.Picha GJ, Drake RF. Pillared-surface microstructure and soft-tissue implants: effect of implant site and fixation. J Biomed Mater Res. 1996;30(3):305–312. doi: 10.1002/(SICI)1097-4636(199603)30:3<305::AID-JBM5>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 15.Salthouse TN. Some aspects of macrophage behavior at the implant interface. J Biomed Mater Res. 1984;18(4):395–401. doi: 10.1002/jbm.820180407. [DOI] [PubMed] [Google Scholar]
- 16.Cardona MA, Simmons RL, Kaplan SS. TNF and IL-1 generation by human monocytes in response to biomaterials. J Biomed Mater Res. 1992;26(7):851–859. doi: 10.1002/jbm.820260703. [DOI] [PubMed] [Google Scholar]
- 17.Bonfield TL, Anderson JM. Functional versus quantitative comparison of IL-1 beta from monocytes/macrophages on biomedical polymers. J Biomed Mater Res. 1993;27(9):1195–1199. doi: 10.1002/jbm.820270910. [DOI] [PubMed] [Google Scholar]
- 18.Miller KM, Rose-Caprara V, Anderson JM. Generation of IL-1-like activity in response to biomedical polymer implants: a comparison of in vitro and in vivo models. J Biomed Mater Res. 1989;23(9):1007–1026. doi: 10.1002/jbm.820230904. [DOI] [PubMed] [Google Scholar]
- 19.Bonfield TL, Colton E, Marchant RE, Anderson JM. Cytokine and growth factor production by monocytes/macrophages on protein preadsorbed polymers. J Biomed Mater Res. 1992;26(7):837–850. doi: 10.1002/jbm.820260702. [DOI] [PubMed] [Google Scholar]
- 20.Brauker JH, Carr-Brendel VE, Martinson LA, Crudele J, Johnston WD, Johnson RC. Neovascularization of synthetic membranes directed by membrane microarchitecture. J Biomed Mater Res. 1995;29(12):1517–1524. doi: 10.1002/jbm.820291208. [DOI] [PubMed] [Google Scholar]
- 21.Sharkawy AA, Klitzman B, Truskey GA, Reichert WM. Engineering the tissue which encapsulates subcutaneous implants. II. Plasma-tissue exchange properties. J Biomed Mater Res. 1998;40(4):586–597. doi: 10.1002/(sici)1097-4636(19980615)40:4<586::aid-jbm10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 22.Salzmann DL, Kleinert LB, Berman SS, Williams SK. The effects of porosity on endothelialization of ePTFE implanted in subcutaneous and adipose tissue. J Biomed Mater Res. 1997;34(4):463–476. doi: 10.1002/(sici)1097-4636(19970315)34:4<463::aid-jbm7>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21(19):6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawertz S, Misener S, editors. Bioinformatics methods and protocols: Methods in molecular biology. Totowa, N.J.: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 25.Middleton E, Elllis EF, Yunginger JW, Reed CE, Adkinson NF, Busse WW. Allergy: Principles and practice. St Louis: Mosby-Year Book, Inc.; 1998. [Google Scholar]
- 26.Luster AD. Chemokines--chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338(7):436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 27.Frangogiannis NG. Chemokines in the ischemic myocardium: from inflammation to fibrosis. Inflamm Res. 2004;53(11):585–595. doi: 10.1007/s00011-004-1298-5. [DOI] [PubMed] [Google Scholar]
- 28.Crowther M, Brown NJ, Bishop ET, Lewis CE. Microenvironmental influence on macrophage regulation of angiogenesis in wounds and malignant tumors. J Leukoc Biol. 2001;70(4):478–490. [PubMed] [Google Scholar]
- 29.Overbergh L, Valckx D, Waer M, Mathieu C. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine. 1999;11(4):305–312. doi: 10.1006/cyto.1998.0426. [DOI] [PubMed] [Google Scholar]
- 30.Teo IA, Choi JW, Morlese J, Taylor G, Shaunak S. LightCycler qPCR optimisation for low copy number target DNA. J Immunol Methods. 2002;270(1):119–133. doi: 10.1016/s0022-1759(02)00218-1. [DOI] [PubMed] [Google Scholar]
- 31.Broberg EK, Nygardas M, Salmi AA, Hukkanen V. Low copy number detection of herpes simplex virus type 1 mRNA and mouse Th1 type cytokine mRNAs by Light Cycler quantitative real-time PCR. J Virol Methods. 2003;112(1–2):53–65. doi: 10.1016/s0166-0934(03)00191-5. [DOI] [PubMed] [Google Scholar]