Abstract
During the course of evolution, genomes acquire novel genetic elements as sources of functional and phenotypic diversity, including new genes that originated in recent evolution. In the past few years, substantial progress has been made in understanding the evolution and phenotypic effects of new genes. In particular, an emerging picture is that new genes, despite being present in the genomes of only a subset of species, can rapidly evolve indispensable roles in fundamental biological processes, including development, reproduction, brain function and behaviour. The molecular underpinnings of how new genes can develop these roles are starting to be characterized. These recent discoveries yield fresh insights into our broad understanding of biological diversity at refined resolution.
The tree of life displays vast biological diversity, and genomic variation is a primary source of this phenotypic diversity. Genome evolution is driven by various forces, such as incremental changes to coding and non-coding sequences, as well as the emergence of new genes. The role of new genes as major drivers of organismal evolution is indicated by the great diversity in the numbers and types of genes among species, which mirrors the startling phenotypic diversity across life1,2.
Every gene in an organism was ‘born’ at a certain time in evolution: some are ancient, some are young, and many are in the process of actively being generated or lost. We use a broad definition for new genes that comprises genes which appeared in a species at a certain time during evolution at a locus that did not exist previously. This view encompasses diverse molecular mechanisms of new-gene origination (BOX 1) that have implications for the emergence of novel functionality in the organism. For example, new genes commonly arise through the duplication of existing genes, either at the DNA level or by retrotransposition through an RNA intermediate; in these cases, the new gene may maintain similar functions to the parental gene over a long evolutionary period or may undergo a process of diversification until a completely new function is evolved. New genes with novel functionality can also originate de novo from non-coding DNA or can be derived from various alterations and rearrangements at the existing gene loci. Importantly, new-gene expression and functionality require regulatory elements that may pre-exist at the new gene locus or that may also need to evolve (BOX 1). Further details on origination mechanisms can be found in several review articles (REFS 1–4).
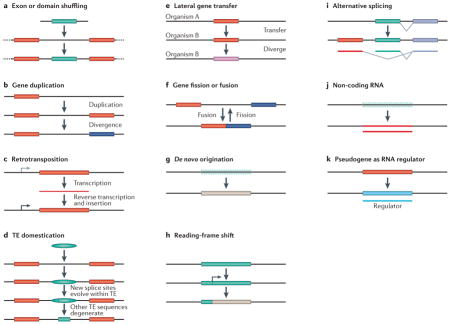
Box 1. Mechanisms of new-gene origination.
Eleven known molecular processes that can generate new gene structures are shown (see the figure), most of which are mentioned in this Review in the cases of new genes that have been analysed for their phenotypic or functional effects. For more detailed discussions of these mechanisms, see REFS 2–4. Here we provide concise explanations of the relevant molecular routes. In exon or domain shuffling, the exons or domains from different genes can be recombined to form many chimeric protein-coding genes (see the figure, part a). Gene duplication is a widely known process that can form new gene structures through DNA-based mechanisms, such as unequal crossover or genome replication in the absence of cell division (see the figure, part b). Duplications can occur on a wide range of scales, from parts of genes to whole genomes. Retrotransposition is an RNA-based duplication in which a transcribed RNA is reverse transcribed and inserted into a new position in the genome (see the figure, part c). This mechanism, although often assumed to produce functionless genetic elements, was found to be important in generating new genes. Transposable element (TE) domestication occurs in several forms (see the figure, part d). In humans and other mammals, hundreds of nuclear genes were found to encode TE-derived peptides and to have evolved new cellular functions. Owing to the multicopy nature of TEs, they can also facilitate unequal-crossover-mediated gene duplication (through recombination involving non-allelic TE copies). Lateral gene transfer was frequently found to play an important part in transferring genes between bacterial species and has recently been found to occur in eukaryotes (see the figure, part e). Gene fission or fusion splits one gene into different genes or fuses neighbouring genes (see the figure, part f). De novo origination of genes has been reported in a number of organisms (see the figure, part g). The mechanism involves generating completely novel proteins by evolving new coding DNA sequences through accumulating mutations in previously non-coding genomic environments. Reading-frame shifts in a protein-coding gene, as seen in human genomes, can generate novel proteins (see the figure, part h). Alternative spicing increases the protein diversity encoded by exons in eukaryotes, and mammals have evolved new exons that result in novel splice variants of proteins (see the figure, part i). Non-coding RNA genes do not encode proteins but can have various important functions (see the figure, part j). Many newly discovered RNAs were found to have originated recently, such as microRNAs in Drosophila spp.123 and the male-functional non-coding RNAs in mice9. Pseudogenes, including some in mammals, were found to be transcribed and to have a role as ‘sponges’ for microRNAs, thus in turn regulating the expression of the functional parental genes (see the figure, part k).
Several mechanisms frequently cooperate in generating a new gene. Furthermore, genes arising from several of these mechanisms — such as retrotransposition and de novo gene origination — must acquire new regulatory systems in order to be expressed and regulated. This can occur by various mutational routes driven by selection. For example, a Drosophila melanogaster retrogene was found to have acquired a regulatory system for testis expression in a mutation selection process124, and new genes that have rapidly evolved insulator sequences and protein-binding sites to set up new regulatory boundaries125 have been identified. Additionally, new vertebrate genes were found to have gradually accumulated U1-binding sites and poly(A) signals to define a productive transcription unit, revealing the evolution of structural elements that control transcriptional directionality after the origination of genes126.
Although various evolutionary and biological insights can be obtained by studying evolutionarily ancient gene families5,6, recently born genes (such as those that appeared within the past 10 million years) are found in one or a few closely related species. These genes provide unique opportunities to study gene formation and the associated phenotypic evolution of organisms because most, if not all, incipient changes during the early stages of origination and evolution are clearly recorded and preserved in their sequences and functions in extant organisms. Since the first young genes, for example, jingwei7, were reported two decades ago, studies of new genes have provided insight into the molecular processes that created the initial structure of genes2,8, the evolution of molecular functions9,10, and global views of the rates, patterns and mechanisms of new-gene origination1,11.
One crucial aspect, and the focus of this Review, is the extent to which new genes contribute to phenotypic evolution. Although such new genes are often assumed to be dispensable12, recent progress in conceptual studies, functional genomics technologies13,14 and accumulation of genome sequence data has provided unprecedented opportunities for the detailed assessment of the phenotypic importance of new genes in various species. TABLE 1 lists some representative cases of new genes with important phenotypic and functional effects that have been documented in Drosophila spp., mammals and plants. Below, we discuss how novel molecular functions derived from new genes can affect disparate phenotypes, including developmental and reproductive processes, brain functions and behaviour. These explorations have converged on a phenomenon in which the genetic systems controlling the related phenotypes have undergone rapid innovation by acquiring new genes, which have often become essential genetic components of the trait. This phenomenon raises interesting and challenging problems that require further understanding from new angles.
Table 1.
Examples of new genes with documented phenotypes and functions in Drosophila spp., mammals and plants
| New genes | Age (million years) | Origination mechanism | Expression | Phenotype | Function | Refs |
|---|---|---|---|---|---|---|
| Drosophila spp. | ||||||
| Sdic family | 0–3 | DNA duplication | Testis | Sperm competition | Cytoskeleton | 69–71 |
| sphinx | 0–3 | Retrotransposition | Neuronal and reproductive tissue | Male courtship | ncRNA | 77,111 |
| jingwei | 0–3 | Retrotransposition | Testis | Recruitment pheromone and hormone | Alcohol metabolism | 7,17 |
| p24-2 | 0–3 | DNA duplication | Multiple stages and tissues | Development, male reproduction | Protein trafficking | 46,47 |
| Xcbp1 | 3–6 | Retrotransposition | Neuronal tissue | Foraging behaviour | Chaperone | 76 |
| Zeus | 3–6 | Retrotransposition | Male reproductive tissues | Male reproduction | Gene regulation | 74,122 |
| nsr | 3–6 | DNA duplication | Imaginal discs, sperm | Sperm maturation | Post-transcriptional regulation | 10 |
| Sflc | 6–11 | DNA duplication | Wide range of tissues | Male and female fecundity | Protease | 127 |
| ms(3)K81 | 6–11 | DNA duplication | Sperm | Zygote development | Paternal chromosome telomere capping | 73 |
| Umbrea | 6–11 | DNA duplication | Multiple stages and tissues | Development | Chromatin binding | 46, 118 |
| Pros28.1A | 6–11 | Retrotransposition | Multiple stages and tissues | Development | Signalling, protein regulation | 46 |
| Desr | 11–25 | Retrotransposition | Neuronal | Foraging behaviour | Transcription regulator | 76 |
| MstProx (also known as Toll3) | 11–25 | DNA duplication | Larva and pupa | Development | Immune response | 46 |
| bicoid | ~150 | Unclear | Early anterior embryo | Embryonic development | Maternal-effect morphogen | 59 |
| Mammals | ||||||
| FGF4 | ~0.01 | Retrotransposition | Distal humerus | Humerus development | FGF signalling | 66 |
| SRGAP2C | 1.0–3.4 | Partial DNA duplication | Brain | Predicted to affect cortex development in a mouse model | Unknown | 109, 110 |
| CDC14C | 7–12 | Retrotransposition | Brain and testis | Unknown | Cell cycle | 21 |
| CYPA | <10 | Retrotransposition | Unknown | Viral infection | HIV-1 resistance | 29 |
| POLDI | 2.5–3.5 | De novo origination | Testis | Knockout reduced testis weight and sperm motility | Unknown | 44 |
| TBC1D3 | <35 | Segmental duplication | Prostate | Insulin modulation | IGF signalling | 128 |
| KRAB–ZNF family | 35–40 | Tandem duplication | Several tissues | Transcriptional suppression | Unknown | 129 |
| INSL4 | 100–160 | Gene duplication | Placental tissue | Fetal development | Regulatory pseudogene | 62 |
| XIST | 100–160 | Pseudogenization | Placental tissue | X-chromosome- inactivation transcript | X-chromosome inactivation | 130 |
| Syncytin (also known as ERVW1) | 100–160 | Recruited viral gene | Placental tissue | Unknown | Mediates placental cytotrophoblast fusion | 131 |
| Primate opsin genes | <60 | Gene duplication | Primates | Trichromatic vision | Trichromatic vision | 132 |
| Olfactory receptor genes | 15–90 | Gene duplication | Mammals, especially mice | Unknown | Odour discrimination | 133 |
| Plants | ||||||
| CYP98A8 | <28 | Retrotransposition | Vascular tissue, pistilis, root tip, etc. | Pollen development | Phenolic synthesis | 19 |
| CYP84A4 | <8 | Gene duplication | Stem and seedling | Unknown | Arabiodopyrone biosynthesis | 20 |
| CYP98A9 | <28 | Retrotransposition | Vascular tissue, | Pollen development | Phenolic synthesis | 19 |
CYP, Cytochrome P450; CYPA, cyclophilin A; Desr, Drosophila Elm2-Sant retrogene; FGF4, fibroblast growth factor 4; IGF, insulin-like growth factor; INSL4, insulin-like 4; KRAB–ZNF, Krüppel-associated box zinc-finger proteins; ms(3)K81, male sterile (3) K81 gene; ncRNA, non-coding RNA; nsr, novel spermatogenesis regulator; Sdic, Sperm-specific dynein intermediate chain; Sflc, Short-lived and few children; SRGAP2C, SLIT–ROBO RHO GTPase-activating protein 2C; TBC1D3, TBC1 domain family member 3; Xcbp1, X-linked calcium binding protein 1 gene; XIST, X-inactive specific transcript gene.
New genes evolve new molecular functions
The origination and evolution of new genes can provide new molecular and cellular functions. Understanding these functions is important for providing insight into how the resultant new gene products assemble into cellular complexes or pathways and ultimately how they affect organismal phenotypes.
New genes evolve novel biochemical functions and pathways
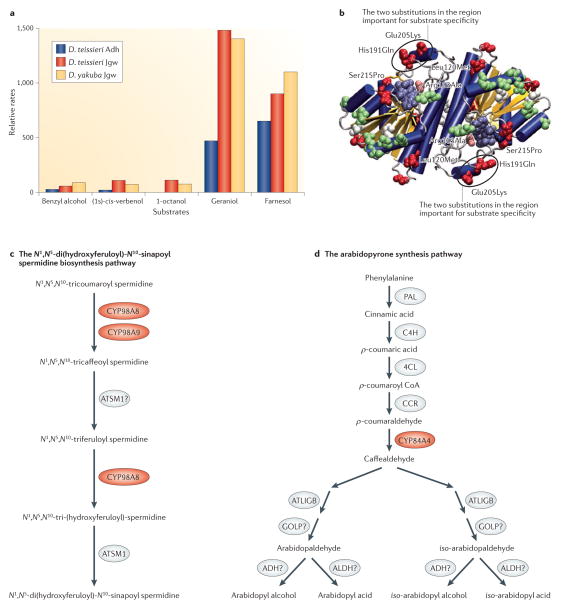
The diversification of ancestral functionality has been a general theme for new genes that originated by gene duplication15. As an example, the chimeric gene jingwei was generated by the retrotransposition of the Alcohol dehydrogenase (Adh) gene in the common ancestor of two African fruitfly species, Drosophila yakuba and Drosophila teissieri, which diverged 3 million years ago (MYA)7,16. Following this retrotransposition, neofunctionalization of jingwei relative to the ancestral Adh gene was driven by adaptive mutations that affected the enzyme active sites to alter substrate specificity in the metabolism of recruitment pheromones and juvenile hormones17,18 (FIG. 1a,b).
Figure 1. Several new genes evolved novel biochemical functions.
a | The protein Jingwei (Jgw) encoded by African Drosophila spp.7 is a dehydrogenase enzyme that has evolved altered substrate specificity compared with the ancestral Alcohol dehydrogenase (Adh). Compared with the parental enzyme, Jgw can use long-chain primary alcohols more efficiently; in particular, Jgw has greater activities than the ancestral Adh towards farnesol (which is involved in juvenile hormone biosynthesis) and geraniol (which is involved in recruitment pheromone biosynthesis)17. b | The locations of the substitutions on the structure of the dimeric Adh. c,d | Three recent duplicates in the cytochrome P450 family in Arabidopsis thaliana led to the assembly of a new pathway of N1,N5-di(hydroxyferuloyl)-N10-sinapoyl spermidine biosynthesis (the new duplicates CYP98A8 and CYP98A9 are shown in red)19 (part c) and a pathway for arabidopyrone biosynthesis (the young duplicated enzyme CYP84A4 is shown in red)20 (part d). D. teissieri, Drosophila teissieri; D. yakuba, Drosophila yakuba. Parts a and b are modified, with permission, from REF. 17 © (2004) US National Academy of Sciences.
As another example, two thorough analyses of biochemical functions of new genes revealed evolutionarily recent origins of de novo biochemical pathways that occur in Arabidopsis thaliana and related species in the Brassicaceae family (FIG. 1c,d). The cytochrome P450-encoding gene CYP98A originated through retrotransposition and was then duplicated at the DNA level into CYP98A9 and CYP98A8. Subsequent adaptive mutations in the catalytic domain of these enzymes led to the origination of a novel pathway for the biosynthesis of N1,N5-di(hydroxyferuloyl)-N10-sinapoyl spermidine, which is a metabolite that is required for pollen development19. Interestingly, another recent duplicate in the cytochrome P450 family, CYP84A4, independently underwent an alteration in its original cytochrome function and became a catalytic enzyme that triggers a new pathway of arabidopyrone biosynthesis in A. thaliana20. Together, these studies revealed examples of biochemical innovation by gene duplication followed by different mechanisms of functional diversification (BOX 1).
New genes evolve new cellular localizations and complexes
The human gene CDC14C (also known as CDC14Bretro) arose 7–12 MYA in the common African ape ancestor of hominoid species, and subsequent adaptive evolution of the cellular localization sequence in the encoded protein has resulted in a change in the localization of this protein from an association with microtubules to a localization in the endoplasmic reticulum21. Additionally, clathrin-coated vesicles, which are essential for endocytosis, evolved by expanding new family members of dynamin proteins; this occurred independently in metazoa and ciliates, depicting a striking and unexpected case of convergent evolution22,23.
New genes can confer stress resistance on organisms
Resistance evolves rapidly under stressful conditions such as extreme temperatures, aberrant pH, and the presence of toxic chemicals and pathogens. In various species, new genes are now known to contribute to drug and pathogen resistance. In microorganisms, antibiotic resistance can occur quickly by the acquisition of resistance genes through lateral gene transfer (also known as horizontal gene transfer) (BOX 1). A good case in point is the ndm1 β-lactamase gene that spread between Enterobacteriaceae spp. and conferred resistance to a broad range of β-lactam antibiotics24. Also, in fruitflies, seven genes that are involved in insecticide resistance (including Cyp6g1 and Cyp6g2) were found to be duplicated in a high proportion of individual flies in a Drosophila melanogaster population, presumably driven by positive selection resulting from insecticide exposure25,26. In plants, adaptive evolution and the gain of new members in the R gene family drove the evolution of pathogen resistance phenotypes, possibly through the gain of new interactions with pathogen virulence genes, or by the evolution of specific recognition of pathogen products27,28. Furthermore, in primates, retrotransposed genes are implicated in pathogen resistance: the owl monkey is highly resistant to HIV-1, and the major resistance factor was found to be a derivative of cyclophilin A (CYPA, also known as PPIA) that had retrotransposed into the TRIM5 locus29,30. These studies suggest that resistance can quickly evolve through the acquisition of new genes, and knowledge of these mechanisms might have various medical and agricultural implications for overcoming resistance to drugs and pesticides or for controlling infectious diseases.
In another striking case, vertebrate species such as Arctic and Antarctic fish convergently evolved new genes encoding antifreeze proteins (AFPs), which are among the first known genes to arise de novo from ancestral non-coding DNA sequences. AFP genes formed through the amplification of repetitive sequences, including microsatellite DNAs. The resultant repeated DNA sequences encode three ring-like peptides that can wrap minuscule ice particles, thus preventing the ice from growing further and enabling survival in frozen environments31–33. These gene originations occurred independently using different precursor DNAs in various species in the past 10–15 million years, when the polar waters became glaciated, thus unveiling how natural selection can result in novelty in a remarkably convergent manner.
The roles of new genes in development
The genetic systems that govern development have been evolving
Studies in evolutionary developmental biology have identified conserved genes and regulatory elements that control early development in diverse taxa ranging from Drosophila spp. to mammals34. Despite this conservation, we are realizing that the genetic systems which control development are rapidly evolving. Since King and Wilson35 suggested that changes to gene regulation have a pivotal role in determining the phenotypic differences between humans and chimpanzees, substantial progress has been made in terms of our mechanistic understanding of gene regulation (for example, the modular structure of cis-regulatory elements) as well as our understanding of its importance in phenotypic evolution36,37. Beyond these regulatory changes, recent explorations of genomics and genetics have continued to add to an ever-growing list of genes that also contribute to the diversification of developmental processes38,39. For example, HOX genes and apoptotic genes (such as apoptotic protease-activating factor 1 (APAF1), BCL2 and caspase genes)40 are ancient metazoan developmental genes that arose ~500–900 MYA. bicoid originated in insects ~100–300 MYA as a duplicate of Hox3, and rapidly evolved as an essential maternal-effect morphogen in the control of gap genes, which are crucial for the initial set-up of the body axis in the D. melanogaster embryo41,42 and in embryos in higher insect taxa43. Therefore, organism body plans have been remodelled as a result of the expansion in the repertoire of developmental genes, and it is interesting to consider the importance and evolution of newly arisen developmental genes.
New genes can rapidly become essential in development
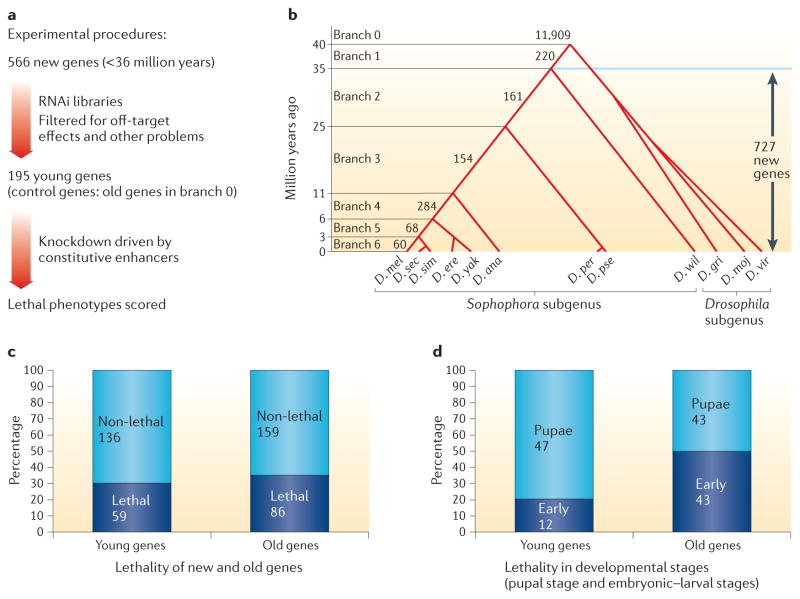
When new genes arise, a reasonable hypothesis is that these new genes will be dispensable or redundant for organismal development or survival12, because the affected genome did not previously encode that gene. Conversely, there have been sporadic reports about the key roles for new genes in developmental pathways or other essential pathways44,45, and a systematic test for the importance of new genes was therefore desirable. The roles of new genes in development had not been extensively studied until recently: a genetic analysis was achieved by generating loss-of-function (LOF) flies — through RNA interference (RNAi)-mediated knockdown or disruption mutagenesis — of each of the ~200 new genes from the set of all new genes that originated within the branches comprising the 36-million-year lineage leading to D. melanogaster46 (FIG. 2). Surprisingly, when silenced, 30% of these young genes showed lethality in various development stages, mostly in the metamorphic pupal stage, indicating that a substantial proportion of new genes have already evolved essential functions. Some new genes that regulate development are very young, existing in only one or a few species. For example, p24-2 exists in a single species, D. melanogaster, and has a vital role in development in this species47, even though the gene is absent in the close sibling species Drosophila simulans, Drosophila mauritiana and Drosophila sechellia46. These observations suggest that these species-specific and lineage-specific genes have rapidly become indispensable components of the genetic control of development.
Figure 2. New genes participate in developmental processes in Drosophila spp.
a,b | The experimental procedures46 (part a) assessed the developmental importance of the new genes that have originated during Drosophila spp. speciation (as mapped in the evolutionary tree by comparing the genomes of different Drosophila spp. (part b))81. The values in the tree indicate the numbers of new genes in the Drosophila melanogaster (D. mel) lineage at each evolutionary branchpoint; 566 genes were chosen for designing knockdown experiments from the total of 727 new genes that have originated since branch 1 in the tree. c,d | A summary of the main experimental results from the 195 new genes tested for the effect on development following gene knockdown. First, knockdown of approximately one-third of either new genes or old genes resulted in lethality, indicating that the proportion of essential new genes is similar to the proportion of essential old genes in branch 0 of the evolutionary tree (part c). Second, knockdown of older genes led to the termination of development at earlier stages than knockdown of new genes, indicating that old genes function earlier in development than new genes, most of which have essential functions in the pupal stages (part d). D. ana, Drosophila ananassae; D. ere, Drosophila erecta; D. gri, Drosophila grimshawi; D. mel, Drosophila melanogaster; D. moj, Drosophila mojavensis; D. per, Drosophila persimilis; D. pse, pseudoobscura; D. sec, Drosophila sechellia; D. sim, Drosophila simulans; D. wil, Drosophila willistoni; D. vir, Drosophila virilise; D. yak, Drosophila yakuba.
An intriguing issue related to this study is whether the age of a gene relates to its developmental importance; that is, do new genes accumulate importance over evolutionary time? The proportion of these young genes that are essential for development is unexpectedly similar to the known proportion of essential genes in genomes48,49 and did not change much during the various stages of evolution towards D. melanogaster (FIG. 3a). These data suggest that, at least within the discussed scale of evolutionary time of Drosophila spp., the age of a gene is not a factor in determining its importance in development, or that gene essentiality evolved much faster in this lineage than conventionally expected. This is despite the expectation that because the majority of essential new developmental genes were generated from duplication events (either DNA based or RNA based), they would be non-essential immediately after their origination. Furthermore, a study in Saccharomyces cerevisiae suggested that newly duplicated genes have evolved differential essentiality even between different laboratory strains within a species50.
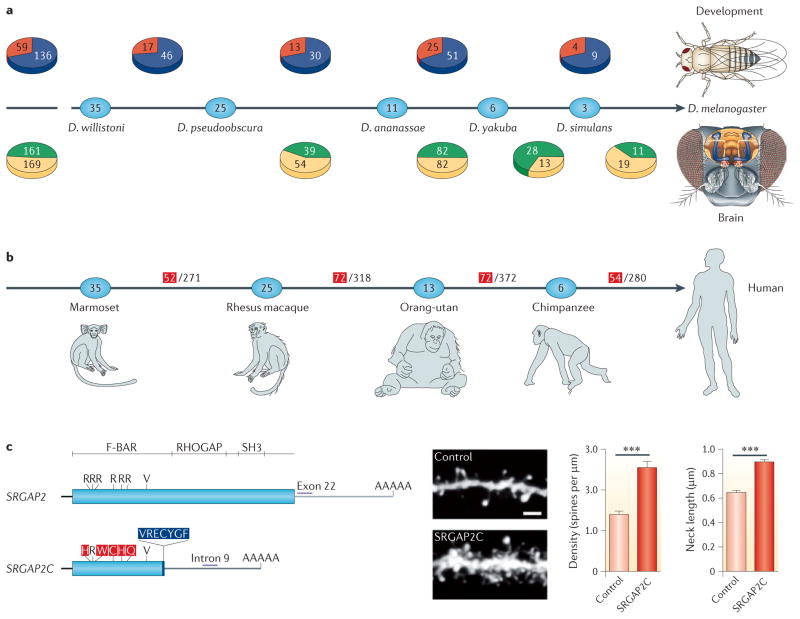
Figure 3. The involvement of new genes in the development of Drosophila spp., and in the brains of Drosophila spp. and humans.
a | The phylogeny shows the distributions of new Drosophila spp. genes involved in development46 (above) and in the brain76 (below) in various evolutionary stages within the past 36 million years81. The numbers in the ovals show the divergence time (in millions of years) between Drosophila melanogaster and various Drosophila spp. Red represents the number of new genes that were found to have essential developmental functions, whereas blue shows the number of new genes that were non-essential in development. Green represents the number of new genes that were expressed in the brain, and yellow shows the number of the non-brain-expressed new genes in reverse transcription PCR screening experiments. The left-most pie charts show the total numbers of new genes across all analysed stages. For the developmental data, the origination events at 3–6 million years ago (MYA) and 0–3 MYA are pooled. These data reveal that older genes are no more likely than newer genes to have evolved essential developmental functions or brain expression, suggesting the rapid evolution of phenotypic effects in new genes. b | The phylogeny shows the numbers of new genes in the human genome that originated at various stages during the divergence of human ancestors from the ancestors of other primates11 through duplication (DNA based and RNA based) and de novo mechanisms (BOX 1). The numbers within the ovals show divergence time (MYA). The numbers in the denominators are the total numbers of new genes that originated in each evolutionary branch, as identified previously67, and the red numerators are the numbers of those new genes that are expressed in the prefrontal cortex (PFC), as detected based on the available microarray expression data. The PFC is the anterior part of the frontal lobe of the neocortex, which is implicated in cognitive functions in the developing human brain. These data suggest that new genes have been frequently acquired into the PFC transcriptome. c | The SLIT–ROBO RHO GTPase-activating protein 2C (SRGAP2C) gene is one of the 54 PFC-expressed human-specific new genes (the end branch of the tree in part b). It was formed by DNA-based duplication and has been subjected to extensive genetic and evolutionary analyses109,110 and functional characterization110. The gene structure shows that SRGAP2C (bottom) is a duplicate of the amino-terminal part of the parental gene, SRGAP2 (top). The transgenic expression of SRGAP2C in cultured mouse cortical neurons induces denser and longer spines, as shown in the dendrite images (scale bar represents 2 μm), and the measured spine density and neck length, shown in the graphs on the right. *** indicates a significance of P<0.001. D. ananassae, Drosophila ananassae; D. pseudoobscura, Drosophila pseudoobscura; D. simulans, Drosophila simulans; D. willistoni, Drosophila willistoni; D. yakuba, Drosophila yakuba. Part c is modified, with permission, from REF. 110 © (2012) Elsevier Science.
New genes in the control of organ formation
New genes also have been found to be essential in the particular process of organogenesis. In one study, LOF experiments were carried out using wing-specific and notum-specific enhancers to drive RNAi-mediated knockdown of new genes in these two organs in D. melanogaster46. Multiple cellular and developmental defects were observed in the LOF flies, such as a signalling defect resembling the Notch phenotype in the wing and the loss of asymmetrical anterior–posterior wing patterning. Consistently, many new genes were strong hits or candidates in genome-wide tissue-specific developmental pathway screens to detect evolutionarily new components in organ development51,52. These data suggest that new genes evolved to control organ development, possibly by integrating into existing pathways.
The importance of new developmental genes across species
The LOF study46 described above indicates that the loss of function of species-specific genes results in the stage-specific termination of developmental processes and tissue-specific morphological defects, and this finding suggests that the genetic control of early development and organ formation is a rapidly evolving system. In different species, although such systems share some conserved components, they also contain the distinct components that are essential for the development of the particular species. For example, Umbrea (also known as HP6) in the D. melanogaster–D. yakuba clade (which diverged just 6 MYA) encodes a chromodomain protein that is essential for embryonic development in these species, but this gene does not exist in outgroup species such as Drosophila ananassae or Drosophila pseudoobscura46. Relatively ancient genes such as bicoid encode the early observed essential maternal morphogens in Drosophila spp. and certain other insects but not in their outgroups42. These studies from Drosophila spp. suggest that the genetic systems governing development have been evolving by recruiting new genes with novel functions. Does this conclusion apply only to Drosophila spp.? Although empirical data from other organisms are lacking, the recently compiled data from mice include many new duplicates that may be involved in development53. A high proportion of gene duplicates (41%) in the mouse are involved in development. In the 240 young gene duplicates identified as having low synonymous substitution rates (Ks <1.5), 22% have essential functions.
New developmental genes and phenotypic diversity
A subsequent question springs to mind: does the rapid evolution of genetic systems driven by new genes lead to the evolution and development of novel morphological characters? It is now clear that new essential genes have integrated into developmental pathways which are responsible for conserved developmental processes controlling organ and tissue development. Related developmental phenotypes, such as embryo size, organ morphology and pigmentation, evolve and diverge as species split54,55. It is therefore intriguing to understand how different genes in different clades regulate species-specific developmental programmes, and how their functional roles stratify in ontogeny and phylogeny. More than a century ago, the similarity and divergence of developing embryos across multiple phyla were observed, although the proposed theory of the recapitulation of phylogeny in ontogeny by Ernst Haeckel was later criticized56. Recent studies in Drosophila spp. and vertebrates (such as zebrafish) have provided extensive evidence that the molecular nature of the phylotypic stage is actually an intermediate developmental phase with strong constraints on the regulation of gene expression and interaction57,58. The combination of phylostratigraphy and stage-specific gene expression data has revealed that the phylotypic stage expresses a transcriptome set representing older genes, whereas earlier and later developmental phases express relatively younger gene sets, in support of the hourglass model57,58. This suggests that both the early and late developmental phases are more divergent than the intermediate phylotypic stage, and that the genetic components of the early and late phases, and their expression, possibly evolve at a faster rate at a global scale. This notion is consistent with the observations that most essential new genes with crucial functions are regulating either early-stage41,42,59 or late-stage developmental processes46.
New genes in reproduction and sexual dimorphism
The effects of new genes have been strongly implicated in the evolution of reproductive functions in metazoans. Furthermore, complementing these detailed case studies, extensive work on new gene populations in various animals has revealed gene traffic that is involved in sexual dimorphism and the evolution of sex chromosomes.
Roles of new genes in reproduction and related processes in mammals
Testicular descent in therian mammals is a unique physiological process induced by the insulin-like 3 (INSL3)–LGR8 (also known as RXFP2) signalling pathway, which ensures that spermatogenesis progresses at a suitable temperature, which is lower than the overall body temperature. The pathway was found to contain a therian-specific duplicate of INSL3 (REF. 60), named relaxin-like 3 (RLN3), which affects testicular descent by modulating the level of luteinizing hormone in the blood61. Additionally, other duplicates in the RLN–INSL gene family have undergone lineage-specific evolution. For example, INSL4 is involved in placental and fetal growth in catarrhine primates62. Other duplication events that have been implicated in mammalian reproduction-related phenotypes include the diversification of fetus-expressed globin genes63, the evolution of milk casein proteins64 and the lineage-specific expansion of SAL1 genes that are involved in pheromone and odorant binding, which is in turn related to the evolution of species-specific mate choice and sexual behaviour65. Furthermore, the artificial selection for dogs with various morphologies in recent human history gave rise to an extremely young functional gene in the fibroblast growth factor family, FGF4, which is responsible for the selected chondrodysplasia morphology66. As an example of a distinct origination mechanism, in Mus musculus, a non-coding RNA gene (BOX 1) that arose 2.5–3.5 MYA has been shown to have important roles in sperm motility and testis size44. Besides these new genes, 5,471 mammal-specific and 1,800–2,900 primate-specific genes were identified in human genomes67, and more than 3,000 rodent-specific genes in mouse genomes67,68. There is a clear pattern of increased testis expression for younger new genes, which is suggestive (albeit not conclusive proof) of functional importance in male reproductive processes. For example, 60–90% of the primate-specific and rodent-specific genes have evolved testis expression.
Roles of new genes in the evolution of the reproductive system in Drosophila spp
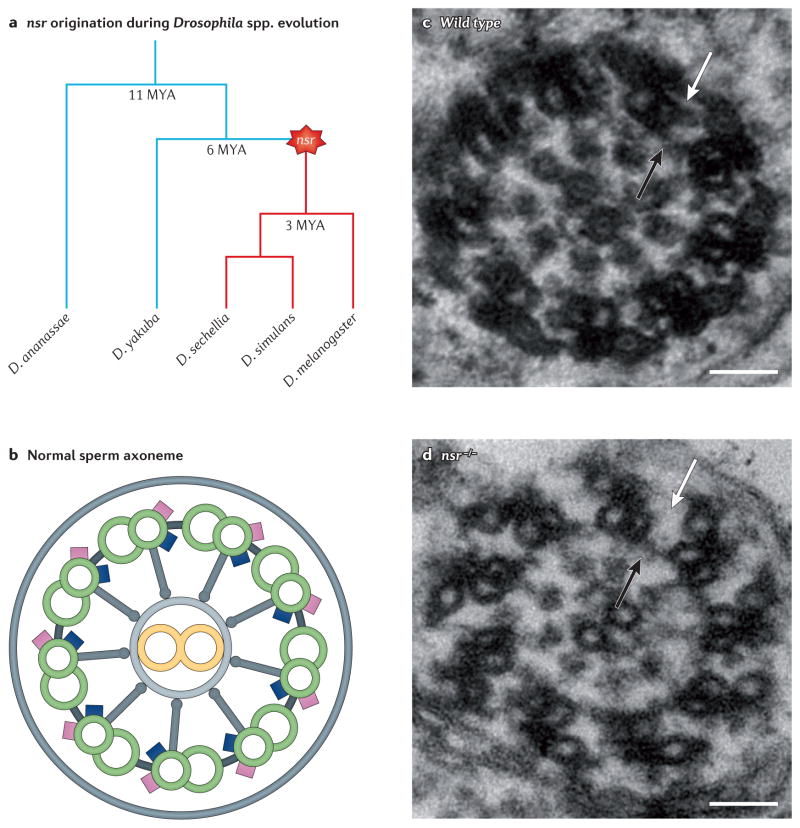
Fruitflies have been a tractable model organism for studying the evolution of sexual reproduction, owing to their species diversity, obvious sexual dimorphism and rapidly evolving reproductive phenotypes. It is now clear that various new genes have key roles in reproductive processes. An early study in D. melanogaster discovered a young species-specific chimeric gene, Sperm-specific dynein intermediate chain 1 (Sdic1), that underwent a strong selective sweep and is expressed specifically in the testis69. When chromosome engineering was used to delete all the copies of the Sdic family genes in D. melanogaster, the resulting sperm competed poorly against sperm from wild-type flies, suggesting that this gene family boosts sperm competition through altered sperm motility70,71. In addition, the gene at the male sterile (3) K81 (ms(3)K81) locus — which originated recently through retrotransposition in species of the D. melanogaster subgroup — was detected as a paternal-effect gene72 that is required for the proper organization of sperm chromatin and for male fertility, suggesting that it evolved clade-specific male germline functions73. Using gene-knockout technology to study the recently expanded kep1 family members, it was found that a young DNA-based duplicate, novel spermatogenesis regulator (nsr), plays an essential part in spermatogenesis, possibly as an RNA-processing factor10 (FIG. 4). Mutational inactivation and RNAi analyses showed that Zeus (also known as Rcd1r), a newly retrotransposed gene that encodes a DNA-binding protein, specifically controls testis gene expression and the reproduction of males but not females74–76. As well as the group of genes that are highly germline specific, many new genes are pleiotropic. RNAi studies of a young serine-type endopeptidase-encoding gene, Short-lived and few children (Sflc; also known as CG4259), have shown that it affects survival and reproduction in both sexes, although the mutant phenotypes are more severe in males than in females127. These studies strongly indicate that new-gene origination is one of the important forces in the evolution of sexual reproduction and dimorphism. Furthermore, similarly to the developmental roles discussed above, new genes can be under positive selection on an evolutionary timescale of a few million years to rapidly evolve key roles in sexual reproduction.
Figure 4. The functions of the young nsr gene in sperm development in Drosophila spp.
a | The novel spermatogenesis regulator (nsr) gene was formed through the DNA-based duplication 3–6 million years ago (MYA) in the common ancestor of the clade including Drosophila melanogaster, Drosophila simulans and Drosophila sechellia. The two outgroups shown are Drosophila ananassae (which separated 11 MYA) and Drosophila yakuba (which separated 6 MYA). b | A schematic cross-section of a normal sperm axoneme with wild-type nsr: the outer (pink) and inner (blue) dynein arms are bounded by nine doublet microtubules (green circles) that surround a central pair of singlet microtubules (yellow circles) through radial spokes (grey). c,d | The functional importance of nsr was shown by P-element-mediated homozygous inactivation of nsr (nsr−/−). As seen from the electron micrographs, inactivation of nsr (part d) results in the loss of the outer dynein arms of the axoneme (white arrows) compared with the wild type (part c), whereas the inner arms remain normal (black arrows); scale bars represent 50 nm. The consequences in nsr−/− sperm are coiled axonemes and deficiencies in individualization. Figure is modified from REF. 10.
Gene traffic and sexually dimorphic gene expression
When examining the chromosomal locations of new genes and their parental duplicates78, a regular pattern, called gene traffic, was observed.
Retrotransposition was the first mechanism (BOX 1) in which a clear gene traffic was observed: new genes were copied from their X-linked parental genes (X) to autosomes (A). Because retrogenes have distinct structures compared with their parental counterparts (such as the absence of introns), it is straightforward to identify the gene pairs involved in retrotransposition-based movements79,80. In Drosophila spp. and mammals, hundreds of retrotransposition events that led to functional new genes have been identified via computational analyses of the gene structures of duplications67,81. Compared with the null hypothesis of random insertion, a significant excess of X→A retrotransposition was first observed in the genome of D. melanogaster79,82 and then in other Drosophila spp.83,84. Remarkably, it was discovered that most of these new retrogenes exhibit testis-specific expression79,83. A similar X→A traffic of male-expressed retrogenes was observed in the human and mouse genomes80. However, in the mammalian genomes, additional traffic in the other direction (A→X) was also observed. Interestingly, most X-linked retrogenes derived from autosomal parents do not show male-biased expression but are expressed fairly equally in both sexes67.
If the retrotransposition-based traffic of male-expressed genes is a result of natural selection acting on the functional consequences of the retrotransposition, rather than just an intrinsic bias towards retrogene insertion in autosomes versus sex chromosomes — as directly tested in humans80,85 and in fruitflies79,86 — similar dynamics in the new genes created by DNA-based duplication would be expected. Indeed, for male-expressed genes, such X→A traffic by DNA-based duplication was detected in Drosophila spp.83 and Teleopsis spp. (stalk-eye flies)87. However, for the small proportion of new genes that were generated during recent evolution (<4 MYA), the expression of X-linked new genes was disproportionately male biased81,67. Interestingly, the expression of X-linked genes becomes increasingly female biased as their age increases. A symmetrical trend was observed in the silkworm, which has a ZW sex determination system in which the female-biased genes tend to move to the autosomes88.
The traffic of male-biased genes provides a mechanistic interpretation for the widely observed distribution of excess male-biased genes in the autosomes and female-biased genes in the X chromosome in Drosophila spp.89,90, the mosquito Anopheles gambiae91–93 and mammals94. The genetic mechanisms underlying gene traffic have been shown to include meiotic sex chromosome inactivation in mammals80,67, Drosophila spp.95–98 and A. gambiae92, as well as sexual antagonism99,67, X-feminization or demasculinization100,99, dosage compensation101,102 and nuclear structure103.
New genes in brain evolution and animal behaviour
The brain is an evolving neuronal complex that controls the behaviour and physiological activities of animals. Recent evidence has shown that animal brain evolution has been accompanied by the recruitment of new genes, as documented in Drosophila spp. and mammals, including humans11,46. Detailed analyses of the expression of many of these new genes have begun to reveal a clear picture of the role of new-gene origination in brain evolution.
New-gene recruitment during the evolution of brain substructure in Drosophila spp
In a screen of new D. melanogaster genes that had originated in the ancestors of this species within the past 25 million years, almost half of these new genes were found to be expressed in the brain76 (FIG. 3a). The proportions of brain-expressed new genes that originated at various stages did not fluctuate significantly from 50%, suggesting that the new brain genes accumulated at a fairly uniform rate throughout this evolutionary period. Similarly, an enhancer trap104 analysis demonstrated that 49% of the new genes are expressed in one or more substructures of the brain, such as the mushroom body, central body complex, antennal lobe, protocerebrum and optic lobe76, suggesting a putative neuronal role for these new genes, although thorough functional analyses will be required to confirm this. Furthermore, neuroanatomy analyses in conjunction with high-throughput RNA sequencing (RNA-seq) revealed that the vast majority of these young brain-expressed genes are expressed in the mushroom body. Interestingly, of the mushroom-body-expressed new genes, almost all are expressed in the recently evolved mushroom body substructure (the α- and β-lobes), whereas only a few are also expressed in other older mushroom body substructures (the α′-, β′- and γ-lobes)76,105,106. Expression profiling of the mushroom body transcriptome by RNA-seq revealed an excess of mushroom-body-enriched young genes under natural selection76, suggesting that this brain substructure evolved by the extensive genomic recruitment of new, mushroom-body-expressed genes during evolution.
An excess of new genes expressed in the developing brain of humans
What are the evolutionary genetic roots for the uniqueness of human brains? Previous studies detected excess human-specific gene duplicates with roles in neuronal development107,108. Examination of new-gene origination in the entire evolutionary period of vertebrates provided evidence that new-gene origination was accelerated in the recent evolution of humans and hominoids11,67 (FIG. 3b). The human genome recruited an unexpectedly large number of new genes in its recent evolution. For example, 1,228 new genes were fixed in the hominoid stage, among which 389 are human specific. Strikingly, transcriptome analyses of the human brain based on available expression data yielded evidence that 198 hominoid-specific and 54 human-specific genes show enriched expression in the prefrontal cortex of the human brain11, which is the most recently evolved structure and is known to be involved in cognitive functions (FIG. 3b). During brain evolution, the transcriptomes of the neocortex have become enriched for the expression of new genes relative to older genes11. Recent studies of the SLIT–ROBO RHO GTPase-activating protein 2 (SRGAP2) genes led a step further towards understanding the genetic mechanisms involved109,110 (FIG. 3c). SRGAP2 has undergone two partial duplications, specifically in the human genome ~1.0–3.4 MYA. One of the human-specific paralogues, SRGAP2C, is expressed in the human brain and antagonizes the ancestral SRGAP2 gene; this antagonism induces neoteny and leads to a higher proportion of the nerve cells growing denser dendritic spines with longer necks (FIG. 3c) to connect with neighbouring neurons better, which may enhance the ‘computing power’ of brains. These studies shed light on the roles of new-gene origination and neuronal function in the human brain.
New brain genes participate in behavioural control
Because brains are in command of behaviours, the action of new genes in the brain can be expected to have consequences for the evolution of behaviours, which had been observed to evolve quickly134. Studies revealed that sphinx — which is a D. melanogaster-specific non-coding RNA gene111 that is expressed in the antennal lobes and the peripheral nervous system — has an impact on male courtship behaviours77,112.
Additionally, a young mushroom-body-expressed protein-coding gene, X-linked calcium binding protein 1 (Xcbp1; also known as CG9906), originated 5 MYA in the ancestor of the D. melanogaster–D. simulans clade, was shown to regulate foraging behaviour and correlates with species-specific behavioural evolution76. Neuroanatomy analysis in D. melanogaster has shown that Xcbp1 is expressed in five glomeruli (VC3m, VC3l, DC4, V, DL5, and DM1) that are innervated by the olfactory receptor neurons, which are probably receptive to food odours113,114. Systemic, pan-neuronal or mushroom-body-specific RNAi-mediated knockdown of Xcbp1 led to significantly reduced foraging behaviour that resembles the behaviour of ancestral Drosophila spp. which do not contain this gene. Enforced synaptic inhibition in Xcbp1-expressing neurons (by driving expression of the temperature-sensitive Shibire-ts1 allele115 in these cells) phenocopied Xcbp1 knockdown. This suggests that Xcbp1 regulates foraging behaviours through neuronal transmission, rather than through unknown pleiotropic functions in non-neuronal tissues. Similar approaches identified another new gene, Drosophila Elm2-Sant retrogene (Desr; also known as CG31875), with mushroom body expression and a foraging-behaviour phenotype; this gene possibly correlates with foraging-behaviour evolution in a relatively older evolutionary period76. These studies suggest that the origin of new genes shaped behavioural evolution through regulating neural circuits.
The rapid emergence of functional importance
One theme of the above discussions is that genetic systems evolved rapidly by recruiting species-specific components that quickly became indispensable (demonstrated by the often lethal effects of knockdowns or knockouts). Thus, it is interesting to consider how the sibling species — the genomes of which do not encode these particular new genes — have evolved to regulate analogous phenotypes, and the contribution that the presence or absence of a specific gene makes to the phenotypic divergence between the species. Another consideration is how a single new gene evolves strong phenotypic effects. In particular, new genes that were created by gene duplication should begin their evolution with trivial phenotypic effects owing to functional redundancy with the parental gene; thus, it is crucial to decipher how they subsequently became essential and what evolutionary process the relevant species have experienced to become ‘addicted’ to the new genes. These issues are only starting to be addressed, and a more complete understanding of the evolutionary change in the functional importance of genes will require thorough comparative analyses of their orthologues in different organisms. Below we outline some preliminary studies in this area.
Evolution of essential functions
Various mechanisms could account for the essentiality of the new gene, such as loss of the parental gene, subfunctionalization (that is, splitting into two essential functions), neofunctionalization or a switch of essentiality from the parental gene to the new gene. In the screen for essential new genes in D. melanogaster mentioned above46, most parental genes persisted with a high level of sequence conservation, whereas the new genes evolved rapidly and independently from their parental counterparts. These findings are consistent with a process of neofunctionalization, in which the new gene copy becomes indispensable. Furthermore, in this data set, 5% of the new genes screened originated de novo (that is, without a parental counterpart), and several of these de novo genes also evolved essential functions, further supporting the notion that essential functions can evolve rapidly46.
On a cellular level, what are the mechanisms that might account for the evolution of essential functions? Next, we propose a hypothetical model based on network biology for how new genes may become central and indispensable components of cellular processes.
Network rewiring as a hypothetical explanatory model
Previous studies have revealed that gene networks evolve and that new genes can be integrated into a network17–20. We further hypothesize that the insertion of a new gene into a gene network can change the topology of the gene network under the force of positive selection. This change in topology can create new links or, more importantly, new pathways by rewiring previously existing gene networks, which in turn could yield substantial phenotypic effects and result in divergence from the species that lack these new genes. A consequence of such topological changes is that the new genes — or other genes that are closely connected to them — can form new hubs in the gene network. According to the centrality–lethality rule116,117, a critical phenotypic effect can be derived from the newly formed hubs, and the subsequent removal of this hub gene is likely to have serious fitness consequences.
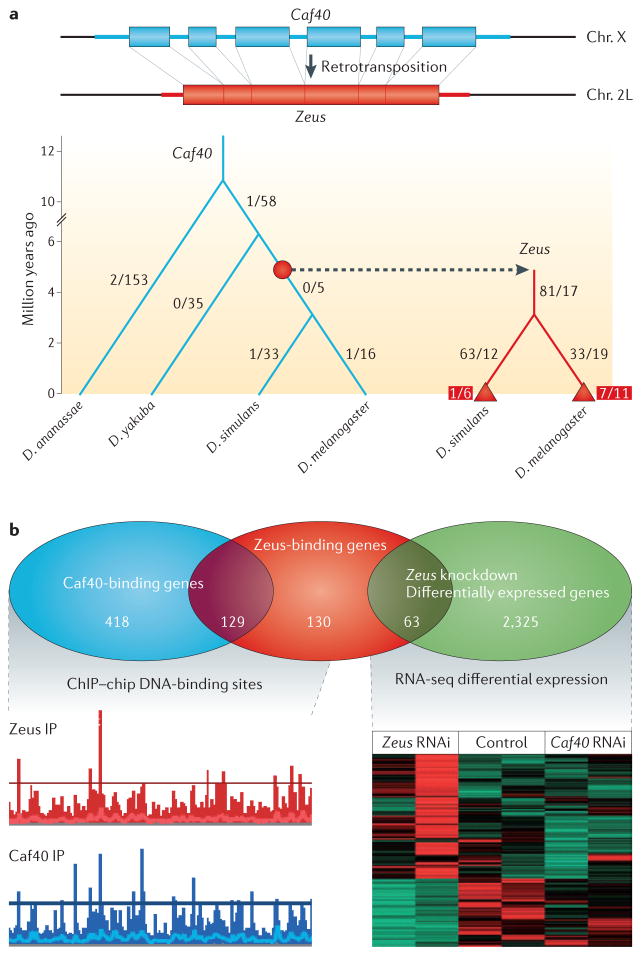
Preliminary testing of the prediction using protein– protein interaction data provided some evidence that new pathways and new hubs can be created by new genes in Drosophila spp.10,46,118,119 and in S. cerevisiae120,121, and new genes were shown to assemble new biochemical pathways for the biosynthesis of new metabolites in A. thaliana 19,20 (FIG. 1c,d). However, further support for the hypothesis is provided by the experimental detection of gene network rewiring caused by the new gene Zeus in D. melanogaster122.
Zeus was created via retrotransposition from a parental gene, Caf40, 4–6 MYA in the ancestor of D. melanogaster, D. simulans, D. mauritiana and D. sechellia122. In the Zeus protein sequence, more than 100 amino acid residues have been substituted under Darwinian positive selection, as detected by nucleotide substitution analyses such as the comparison of the substitution rates at non-synonymous and synonymous sites (the Ka/Ks ratio) and the McDonald–Kreitman test (FIG. 5a). These substitutions led to a novel DNA-binding motif, and chromatin immunoprecipitation followed by microarray (ChIP–chip) analyses revealed that these substitutions have resulted in Zeus binding to 193 genes with male-biased expression, and only one-quarter of the binding sites of the parental protein are bound by Zeus (FIG. 5b). Furthermore, transcriptome profiling following Zeus knockdown revealed that the expression of Zeus is correlated with the differential expression of more than 2,000 genes, with the female-biased genes being mostly downregulated and the male-biased genes upregulated, thus indicating a central role for Zeus in coordinating a male-specific transcriptome (FIG. 5b). The loss of Zeus (and thus the loss of this apparent hub) was shown to have serious physiological consequences such as deformation of testis and sperm, and sterility of adult males. These data suggest that in a short evolutionary time, Zeus has experienced alterations to its DNA-binding specificity and has been under Darwinian positive selection to create thousands of new links with pre-existing genes to alter the global gene expression pattern122. More investigations in the future will be valuable for assessing whether this integration of new genes into pre-existing ancestral gene networks is a widespread route for phenotypic evolution.
Figure 5. The rapid and extensive evolution of gene expression networks by integration of the new gene Zeus in Drosophila spp.
a | Zeus, a new gene derived from the retrotransposition of the parental Caf40 gene, evolved under positive selection, as detected by the McDonald–Kreitman test and by the Ka/Ks ratio (defined as the ratio of the number of substitutions at non-synonymous sites to the number of substitutions at synonymous sites)122. In contrast to the slow evolution of Caf40 (blue), Zeus evolved rapidly in its protein sequences (red). The ratios displayed throughout the tree are Ka/Ks ratios, except those in red, which show the ratio of the number of non-synonymous sites to the number of synonymous polymorphic sites of nucleotides in the alleles of the natural population in Drosophila melanogaster and Drosophila simulans. b | The left panels show a comparison of the DNA-binding sites of Caf40 (blue) and Zeus (Red), as determined by chromatin immunoprecipitation followed by microarray (ChIP–chip). Separate analyses revealed that almost all of these sites have protein-coding genes downstream (not shown). The right panels show the genes for which expression correlates with the expression of Zeus, as determined by high-throughput RNA sequencing (RNA-seq) following RNA interference (RNAi)-mediated knockdown of Zeus, and the raw data is shown in the heatmap. The extensive changes in DNA-binding sites revealed that the integration of new genes reshaped the gene expression networks. D. ananassae, Drosophila ananassae; D. yakuba, Drosophila yakuba; IP, immunoprecipitation. Figure is modified, with permission, from REF. 122 © (2012) Macmillan Publishers Ltd. All rights reserved.
Concluding remarks
Recent studies have identified lineage-specific and species-specific new genes with important phenotypic effects on diverse phenotypes, including development, sexual reproduction, brain functions and behaviour. These findings suggest that the genetic systems controlling these crucial biological processes or structures are rapidly evolving, such that every species has its own repertoire of genetic components that control its biological processes. An area of longstanding interest is to understand how the addition of these new genes into genomes contributes to phenotypes, including their roles in driving phenotypic diversity and evolution. A possible route to address this is to manipulate the new genes (for example, using gene silencing) and to integrate the phenotypic effects that are observed at various biological levels, from molecular to physiological to ecological, thus enabling a deep insight into the mechanisms that drive the origination of new phenotypes. Additionally, although various representative examples of the roles of new genes in physiological processes have been discussed above, it will be interesting to identify the new genes involved in various processes more comprehensively, including a functional assessment of their importance, particularly relative to their ancestral genes.
Acknowledgments
The authors are indebted to their collaborators, A. Dean, K. White, T. Karr, L. Luo and L. Li, for their intellectual and technical support. The authors thank X. Ni, Y. Zhang, M. Vibranovski, M. Spletter and P. Landback for their efforts and contributions to the understanding of new genes and their role in phenotypic evolution. The authors also thank C. Kemkemer, N. VanKuren, S. Newfeld and A. Turkewitz for their discussions. M.L. was supported by the US National Institutes of Health (NIH) and the US National Science Foundation (N S F) grants (1R01100768-01A1, MCB1051826, MCB1026200); S.C. was supported by the NSF dissertation improvement grant (DEB-1110607), and B.H.K. by the NIH Genetics Training grant (T32 GM007197) and a US Department of Education GAANN Fellowship.
Glossary
- Retrotransposition
An RNA-based duplication mechanism that involves the transcription and processing of a gene into an mRNA that is then reverse transcribed and integrated into a new DNA locus in a genome
- Mutation selection
An evolutionary process in which selection acts on a mutation to determine its eventual fate
- Hominoid
A kind of primate species including the great hominoids (humans, chimpanzees, gorillas and orang-utans) and lesser hominoids (gibbons and siamangs)
- Convergent evolution
The evolution of two unrelated lineages such that they share a similar phenotype
- Maternal-effect morphogen
A gene product from a maternal-effect gene; for example, Bicoid in Drosophila spp. is a developmental signal for which the concentration has an impact on embryonic growth
- Clade
A group of closely related species in an evolutionary tree
- Outgroup
A group of reference organisms outside of a group of organisms that are being analysed for their evolutionary relationship
- Ontogeny
The development of an organism
- Phylogeny
A tree of life that records the evolutionary history of a group of species
- Phylotypic stage
A developmental stage in which the embryos of different species resemble each other in morphology
- Phylostratigraphy
A statistical method that traces genes back to their most ancient ancestors to study macroevolutionary patterns
- Hourglass model
A developmental model that uses analogy to the hourglass to describe the interspecies divergence of phenotypes during the entire developmental process. In this model, the initial and final stages are divergent between species, whereas the intermediate phylotypic stage is the most evolutionarily conserved
- Gene traffic
A directional gene duplication process between the sex chromosomes and autosomes. It often confers sex-biased expression on the genes
- Therian mammals
A group of mammalian species consisting of marsupial and placental animals
- Catarrhine primates
A group of primate species that comprises the Old World monkeys, higher apes and hominoids
- Chondrodysplasia
A hereditary skeletal disorder that results in limited development and a deformity in morphology
- Selective sweep
A strong directional selection that happened recently and rapidly elevated the frequencies of advantageous alleles in a population
- Paternal-effect gene
A paternal gene that affects the phenotype of the offspring, irrespective of the offspring genotype
- Pleiotropic
Pertaining to a gene or mutation having multiple phenotypic effects
- Sexual antagonism
The phenomenon whereby a mutation has opposite phenotypic effects on males and females
- Dosage compensation
A process that equalizes the expression levels of the genes on the sex chromosomes, either between males and females or between the sex chromosomes and autosomes
- Enhancer trap
A transgenic construct used to identify enhancer elements for a gene that is expressed in specific tissues. Insertion of the construct near a tissue-specific enhancer results in the tissue-specific expression of the reporter gene
- Hubs
The nodes with many interactions in gene networks
- The centrality–lethality rule
The proposal put forward by Jeong, Mason, Barabasi and Otavi that the genes in the hub positions in gene networks are associated with lethal phenotypes when silenced, suggesting that those genes have essential functions
- Ka/Ks ratio
The ratio of the number of substitutions at non-synonymous sites to the number of substitutions at synonymous sites
- McDonald–Kreitman test
A molecular population genetics test to detect positive selection using a statistical test of the null hypothesis of neutrality
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nature Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 2.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20:1313–1326. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding Y, Zhou Q, Wang W. Origins of new genes and evolution of their novel functions. Annu Rev Ecol Evol Syst. 2012;43:345–363. [Google Scholar]
- 4.Long M, VanKuren NW, Chen S, Vibranovski MD. New gene evolution: little did we know. Annu Rev Genet. doi: 10.1146/annurev-genet-111212-133301. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardison RC. Evolution of hemoglobin and its genes. Cold Spring Harb Perspect Med. 2012;2:a011627. doi: 10.1101/cshperspect.a011627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong J, Fischer WM, Inoue K, Nakahara M, Bauer CE. Molecular evidence for the early evolution of photosynthesis. Science. 2000;289:1724–1730. doi: 10.1126/science.289.5485.1724. [DOI] [PubMed] [Google Scholar]
- 7.Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 8.Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nature Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nature Rev Genet. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, et al. A young Drosophila duplicate gene plays essential roles in spermatogenesis by regulating several Y-linked male fertility genes. PLoS Genet. 2010;6:e1001255. doi: 10.1371/journal.pgen.1001255. This functional analysis of a new gene in D. melanogaster reveals the rapid evolution of an essential role in spermatogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YE, Landback P, Vibranovski MD, Long M. Accelerated recruitment of new brain development genes into the human genome. PLoS Biol. 2011;9:e1001179. doi: 10.1371/journal.pbio.1001179. This reports that an enhanced rate of new-gene origination underlies brain development in humans and other primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krebs JE, Goldstein ES, Kilpatrick ST. Lewin’s Essential Genes. Jones & Bartlett Publishers; 2009. [Google Scholar]
- 13.Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venken KJT, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nature Rev Genet. 2005;6:167–178. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 15.Ohno S. Evolution by Gene Duplication. George Alien & Unwin Ltd; 1970. [Google Scholar]
- 16.Wang W, Zhang J, Alvarez C, Llopart A, Long M. The origin of the Jingwei gene and the complex modular structure of its parental gene, Yellow Emperor, in Drosophila melanogaster. Mol Biol Evol. 2000;17:1294–1301. doi: 10.1093/oxfordjournals.molbev.a026413. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Dean AM, Brunet F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci USA. 2004;101:16246–16250. doi: 10.1073/pnas.0407066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Yang H, Long M, Li L, Dean AM. Evolution of enzymatic activities of testis-specific short-chain dehydrogenase/reductase in Drosophila. J Mol Evol. 2010;71:241–249. doi: 10.1007/s00239-010-9384-5. [DOI] [PubMed] [Google Scholar]
- 19.Matsuno M, et al. Evolution of a novel phenolic pathway for pollen development. Science. 2009;325:1688–1692. doi: 10.1126/science.1174095. [DOI] [PubMed] [Google Scholar]
- 20.Weng JK, Li Y, Mo HP, Chapple C. Assembly of an evolutionarily new pathway for α-pyrone biosynthesis in Arabidopsis. Science. 2012;337:960–964. doi: 10.1126/science.1221614. References 19 and 20 describe the development of a new metabolic pathway as a consequence of new-gene duplication. [DOI] [PubMed] [Google Scholar]
- 21.Rosso L, et al. Birth and rapid subcellular adaptation of a hominoid-specific CDC14 protein. PLoS Biol. 2008;6:e140. doi: 10.1371/journal.pbio.0060140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elde NC, Long M, Turkewitz AP. A role for convergent evolution in the secretory life of cells. Trends Cell Biol. 2007;17:157–164. doi: 10.1016/j.tcb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Kumarasamy KK, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 26.Daborn PJ, et al. A single p450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- 27.Bergelson J, Kreitman M, Stahl EA, Tian DC. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 28.Takken FL, Joosten MH. Plant resistance genes: their structure, function and evolution. Eur J Plant Pathol. 2000;106:699–713. [Google Scholar]
- 29.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 30.Stremlau M, et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, DeVries AL, Cheng CHC. Evolution of antifreeze glycoprotein gene from a trypsinogen gene in Antarctic notothenioid fish. Proc Natl Acad Sci USA. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, DeVries AL, Cheng CH. Convergent evolution of antifreeze glycoproteins in Antarctic notothenioid fish and Arctic cod. Proc Natl Acad Sci USA. 1997;94:3817–3822. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng CH, Chen L. Evolution of an antifreeze glycoprotein. Nature. 1999;401:443–444. doi: 10.1038/46721. [DOI] [PubMed] [Google Scholar]
- 34.Wolpert L, et al. Principles of Development. 2. Oxford Univ. Press; 2002. [Google Scholar]
- 35.King M, Wilson A. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 36.Guss KA, Nelson CE, Hudson A, Kraus ME, Carroll SB. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164–1167. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- 37.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Srivastava M, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 41.Stauber M, Jackle H, Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci USA. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stauber M, Taubert H, Schmidt-Ott U. Function of bicoid and hunchback homologs in the basal cyclorrhaphan fly Megaselia (Phoridae) Proc Natl Acad Sci USA. 2000;97:10844–10849. doi: 10.1073/pnas.190095397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemke S, et al. Maternal activation of gap genes in the hover fly Episyrphus. Development. 2010;137:1709–1719. doi: 10.1242/dev.046649. [DOI] [PubMed] [Google Scholar]
- 44.Heinen TJAJ, Staubach F, Haming D, Tautz D. Emergence of a new gene from an intergenic region. Curr Biol. 2009;19:1527–1531. doi: 10.1016/j.cub.2009.07.049. This reports the remarkable phenotypic effects of a non-coding RNA gene that recently appeared in the house mouse and its close relatives and that, if knocked out, leads to reduced sperm motility and reduced testis weight. [DOI] [PubMed] [Google Scholar]
- 45.Joppich C, Scholz S, Korge G, Schwendemann A. Umbrea, a chromo shadow domain protein in Drosophila melanogaster heterochromatin, interacts with Hip, HP1 and HOAP. Chromosome Res. 2009;17:19–36. doi: 10.1007/s10577-008-9002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. cSience. 2010;330:1682–1685. doi: 10.1126/science.1196380. This article describes a large-scale experiment that investigates the phenotypic effects of 195 new genes in D. melanogaster using RNAi-mediated knockdown, revealing that a high proportion of these recently evolved genes have evolved essential developmental functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saleem S, et al. Drosophila melanogaster p24 trafficking proteins have vital roles in development and reproduction. Mech Dev. 2012;129:177–191. doi: 10.1016/j.mod.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Miklos GL, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 49.Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dowell RD, et al. Genotype to phenotype: a complex problem. Science. 2010;328:469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mummery-Widmer JL, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnorrer F, et al. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- 53.Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ. Canalization of segmentation and its evolution in Drosophila. Proc Natl Acad Sci USA. 2007;104:10926–10931. doi: 10.1073/pnas.0701359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coyne JA, Orr HA. Speciation. Sinauer Associates Sunderland; 2004. [Google Scholar]
- 56.Gould SJ. Ontogeny and Phylogeny. Belknap; 1977. [Google Scholar]
- 57.Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–818. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- 58.Kalinka AT, et al. Gene expression divergence recapitulates the developmental hourglass model. Nature. 2010;468:811–814. doi: 10.1038/nature09634. [DOI] [PubMed] [Google Scholar]
- 59.Lemke S, et al. bicoid occurrence and Bicoid-dependent hunchback regulation in lower cyclorrhaphan flies. Evol Dev. 2008;10:413–420. doi: 10.1111/j.1525-142X.2008.00252.x. [DOI] [PubMed] [Google Scholar]
- 60.Park J-I, et al. Origin of INSL3-mediated testicular descent in therian mammals. Genome Res. 2008;18:974–985. doi: 10.1101/gr.7119108. This evolutionary analysis reveals that an important physiological phenotype in therian mammals — the testicular descent — was derived with a new gene duplicate, INSL3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGowan BM, et al. Relaxin-3 stimulates the hypothalamic-pituitary-gonadal axis. Am J Physiol Endocrinol Metab. 2008;295:278–286. doi: 10.1152/ajpendo.00028.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arroyo JI, Hoffmann FG, Good S, Opazo JC. INSL4 pseudogenes help define the relaxin family repertoire in the common ancestor of placental mammals. J Mol Evol. 2012;75:73–78. doi: 10.1007/s00239-012-9517-0. [DOI] [PubMed] [Google Scholar]
- 63.Johnson RM, et al. Phylogenetic comparisons suggest that distance from the locus control region guides developmental expression of primate β-type globin genes. Proc Natl Acad Sci USA. 2006;103:3186–3191. doi: 10.1073/pnas.0511347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawasaki K, Lafont AG, Sire JY. The evolution of milk casein genes from tooth genes before the origin of mammals. Mol Biol Evol. 2011;28:2053–2061. doi: 10.1093/molbev/msr020. [DOI] [PubMed] [Google Scholar]
- 65.Meslin C, et al. The evolutionary history of the SAL1 gene family in eutherian mammals. BMC Evol Biol. 2011;11:148. doi: 10.1186/1471-2148-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker HG, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. This paper describes a young phenotype in dogs that is derived from a newly retrotransposed gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010;8:e1000494. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 70.Yeh SD, Do T, Abbassi M, Ranz JM. Functional relevance of the newly evolved sperm dynein intermediate chain multigene family in Drosophila melanogaster males. Commun Integr Biol. 2012;5:462–465. doi: 10.4161/cib.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeh S-D, et al. Functional evidence that a recently evolved Drosophila sperm-specific gene boosts sperm competition. Proc Natl Acad Sci USA. 2012;109:2043–2048. doi: 10.1073/pnas.1121327109. This reports an elegant experiment that detects an interesting phenotypic effect of a species-specific locus on sperm competition in D. melanogaster. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yasuda GK, Schubiger G, Wakimoto BT. Genetic characterization of ms(3)K81, a paternal effect gene of Drosophila melanogaster. Genetics. 1995;140:219–229. doi: 10.1093/genetics/140.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. This article describes the first-known essential paternal new gene that originated in the ancestor of the species in the D. melanogaster subgroup. [DOI] [PubMed] [Google Scholar]
- 74.Bai Y, Casola C, Feschotte C, Betran E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quezada-Diaz JE, Muliyil T, Rio J, Betran E. Drcd-1 related: a positively selected spermatogenesis retrogene in Drosophila. Genetica. 2010;138:925–937. doi: 10.1007/s10709-010-9474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen SD, et al. Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep. 2012;1:118–132. doi: 10.1016/j.celrep.2011.12.010. This experimental analysis of a large number of new genes that are expressed in the D. melanogaster brain reveals the significant impact on the evolution of new brain structure and the explicit roles of these genes in shaping foraging behaviours through expression in neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Y, Dai HZ, Chen SD, Zhang LY, Long MY. Highly tissue specific expression of Sphinx supports its male courtship related role in Drosophila melanogaster. PLoS ONE. 2011;6:e18853. doi: 10.1371/journal.pone.0018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long M, Vibranovski MD, Zhang Y. In: Rapidly Evolving Genes and Genetic Systems. Singh R, Xu J, Kulanthinal R, editors. Oxford Univ. Press; 2012. pp. 101–114. [Google Scholar]
- 79.Betran E. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emerson J, Kaessmann H, Betrán E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 81.Zhang YE, Vibranovski MD, Krinsky BH, Long M. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 2010;20:1526–1533. doi: 10.1101/gr.107334.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dai H, Yoshimatsu T, Long M. Retrogene movement within- and between-chromosomes in the evolution of Drosophila genomes. Gene. 2006;385:96–102. doi: 10.1016/j.gene.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 83.Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meisel RP, Han MV, Hahn MW. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol Evol. 2009;1:176–188. doi: 10.1093/gbe/evp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schrider DR, et al. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 2013;9:e1003242. doi: 10.1371/journal.pgen.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schrider DR, Stevens K, Cardeno CM, Langley CH, Hahn MW. Genome-wide analysis of retrogene polymorphisms in Drosophila melanogaster. Genome Res. 2011;21:2087–2095. doi: 10.1101/gr.116434.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker RH, Wilkinson GS. Comparative genomic hybridization (CGH) reveals a neo-X chromosome and biased gene movement in stalk-eyed flies (genus Teleopsis) PLoS Genet. 2010;6:e1001121. doi: 10.1371/journal.pgen.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang J, Long M, Vibranovski MD. Retrogenes moved out of the Z chromosome in the silkworm. J Mol Evol. 2012;74:1–14. doi: 10.1007/s00239-012-9499-y. [DOI] [PubMed] [Google Scholar]
- 89.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 90.Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baker DA, Russell S. Role of testis-specific gene expression in sex-chromosome evolution of Anopheles gambiae. Genetics. 2011;189:1117–1120. doi: 10.1534/genetics.111.133157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magnusson K, et al. Demasculinization of the Anopheles gambiae X chromosome. BMC Evol Biol. 2012;12:69–69. doi: 10.1186/1471-2148-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toups MA, Hahn MW. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics. 2010;186:763–766. doi: 10.1534/genetics.110.118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nature Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 95.Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vibranovski MD, Chalopin DS, Lopes HF, Long M, Karr TL. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics. 2010;186:431–433. doi: 10.1534/genetics.110.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vibranovski MD, et al. Re-analysis of the larval testis data on meiotic sex chromosome inactivation revealed evidence for tissue-specific gene expression related to the Drosophila X chromosome. BMC Biol. 2012;10:49. doi: 10.1186/1741-7007-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. Plos Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu CI, Xu EY. Sexual antagonism and X inactivation--the SAXI hypothesis. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 101.Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- 102.Bachtrog D, Toda NRT, Lockton S. Dosage compensation and demasculinization of X chromosomes in Drosophila. Curr Biol. 2010;20:1476–1481. doi: 10.1016/j.cub.2010.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Diaz-Castillo C, Ranz JM. Nuclear chromosome dynamics in the Drosophila male germ line contribute to the nonrandom genomic distribution of retrogenes. Mol Biol Evol. 2012;29:2105–2108. doi: 10.1093/molbev/mss096. [DOI] [PubMed] [Google Scholar]
- 104.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 105.Strausfeld NJ, Li Y. Organization of olfactory and multimodal afferent neurons supplying the calyx and pedunculus of the cockroach mushroom bodies. J Comp Neurol. 1999;409:603–625. [PubMed] [Google Scholar]
- 106.Strausfeld NJ, Li Y. Representation of the calyces in the medial and vertical lobes of cockroach mushroom bodies. J Comp Neurol. 1999;409:626–646. doi: 10.1002/(sici)1096-9861(19990712)409:4<626::aid-cne8>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 107.Fortna A, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:e207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sudmant PH, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dennis MY, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Charrier C, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. This article details the remarkable neuronal effects of human-specific SRGAP2 duplicates on the mouse brain, implicating a possible function in the human brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang W, et al. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dai H, et al. The evolution of courtship behaviors through the origination of a new gene in Drosophila. Proc Natl Acad Sci USA. 2008;105:7478–7483. doi: 10.1073/pnas.0800693105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 115.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 116.Kim J, Kim I, Han SK, Bowie JU, Kim S. Network rewiring is an important mechanism of gene essentiality change. Sci Rep. 2012;2:900. doi: 10.1038/srep00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 118.Ross BD, et al. Stepwise evolution of essential centromere function in a Drosophila neogene. Science. 2013;340:1211–1214. doi: 10.1126/science.1234393. This reports a clear neofunctionalization process in which a species-specific centromere-targeting gene in D. melanogaster, Umbrea, was generated by a domain-deleted duplicate and a rewiring process under natural selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Konikoff CE, Wisotzkey RG, Stinchfield MJ, Newfeld SJ. Distinct molecular evolutionary mechanisms underlie the functional diversification of the Wnt and TGFβ signaling pathways. J Mol Evol. 2010;70:303–312. doi: 10.1007/s00239-010-9337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li D, et al. A de novo originated gene depresses budding yeast mating pathway and is repressed by the protein encoded by its antisense strand. Cell Res. 2010;20:408–420. doi: 10.1038/cr.2010.31. [DOI] [PubMed] [Google Scholar]
- 121.Capra J, Pollard KS, Singh M. Novel genes exhibit distinct patterns of function acquisition and network integration. Genome Biol. 2010;11:R127. doi: 10.1186/gb-2010-11-12-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen SD, et al. Reshaping of global gene expression networks and sex-biased gene expression by integration of a young gene. EMBO J. 2012;31:2798–2809. doi: 10.1038/emboj.2012.108. This article provides strong experimental evidence that new genes can reshape gene expression networks in short evolutionary time under positive selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu J, et al. The birth and death of microRNA genes in Drosophila. Nature Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 124.Betran E, Long M. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ni X, et al. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol. 2012;10:e1001420. doi: 10.1371/journal.pbio.1001420. This is the first genome-wide experiment to reveal rapid evolution of a regulatory mechanism of insulation with new genes in Drosophila spp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–363. doi: 10.1038/nature12349. This is the first report of a significant linear correlation between the age of genes and two critical signals to define transcription units, U1 small nuclear ribonucleoprotein recognition sites and polyadenylation sites (PAS) in vertebrates. Directional gain of the U1 sites and loss of PAS sites revealed how a new gene acquires its U1–PAS axis for a productive transcription under natural selection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen S, Yang H, Krinsky BH, Zhang A, Long M. Roles of young serine-endopeptidase genes in survival and reproduction revealed rapid evolution of phenotypic effects at adult stages. Fly. 2011;5:345–351. doi: 10.4161/fly.5.4.17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wainszelbaum MJ, et al. TBC1D3, a hominoid-specific gene, delays IRS-1 degradation and promotes insulin signaling by modulating p70 S6 kinase activity. PLoS ONE. 2012;7:e31225. doi: 10.1371/journal.pone.0031225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nowick K, Hamilton AT, Zhang H, Stubbs L. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 2010;27:2606–2617. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duret L, et al. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 131.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 132.Surridge AK, Osorio D, Mondy NI. Evolution and selection of trichromatic vision in primates. Trends Ecol Evol. 2003;18:198–205. [Google Scholar]
- 133.Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE. 2007;2:e708. doi: 10.1371/journal.pone.0000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rollmann SM, et al. Odorant receptor polymorphisms and natural variation behavior in Drosophila melanogaster. Genetics. 2010;186:687–697. doi: 10.1534/genetics.110.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]