Abstract
Gp120 is a critical component of the envelope of HIV-1. Its role in viral entry is well described. In view of its position on the viral envelope, gp120 is a part of the retrovirus that immune cells encounter first and has the potential to influence antiretroviral immune responses. We propose that high levels of gp120 are present in tissues and may contribute to the failure of the immune system to fully control and ultimately clear the virus. Herein, we show for the first time that lymphoid tissues from acutely HIV-1/SIV (SHIV)-KB9-infected macaques contain deposits of gp120 at concentrations that are high enough to induce suppressive effects on T cells, thus negatively regulating the antiviral CTL response and contributing to virus survival and persistence. We also demonstrate that SHIV-KB9 gp120 influences functional T cell responses during SHIV infection in a manner that suppresses degranulation and cytokine secretion by CTLs. Finally, we show that regulatory T cells accumulate in lymphoid tissues during acute infection and that they respond to gp120 by producing TGFβ, a known suppressant of cytotoxic T cell activity. These findings have significant implications for our understanding of the contribution of non-entry-related functions of HIV-1 gp120 to the pathogenesis of HIV/AIDS.
HIV-1-specific CD8+ CTL can be detected in the peripheral blood (PB)3 during the first 4–6 wk following acute retroviral infection and before a neutralizing Ab response is demonstrable. The appearance of these cells in blood coincides with an initial decrease in plasma viremia during acute HIV infection (1, 2) as well as in animal models of simian immunodeficiency virus (SIV) and feline immunodeficiency virus infection (3–5). In this setting, CTL numbers have been shown to negatively correlate with the viral load (6). In the SIV model of infection, CD8+-depleted macaques lose control of SIV replication that is subsequently restored with the reappearance of CD8+ cells (7). However, although CTL responses control viremia, they fail to eradicate the virus.
It is not fully understood why the body fails to eradicate or control the virus, but several phenomena have been described that support the thesis that the retrovirus is capable of evading the host cell-mediated immune response. HIV-1 utilizes various mechanisms to evade the immune response, such as mutation of its envelope protein, maintenance of a latent state, and active infection and killing of immune cells, including CD4+ T cells (8–10). CTLs may also be ineffective because of viral escape and the emergence of resistant strains (1, 11–18).
HIV-specific T cell clones that are present at the time of acute infection have also been shown to rapidly disappear (19). This phenomenon is not thought to be the result of mutations in viral epitopes recognized by these clones (19). It has been proposed that this reflects a deletion process caused by high levels of viral Ag. Clonal exhaustion/deletion is a well-documented phenomenon where an excess of Ag on APCs in the lymphoid organs induces all Ag-specific responsive T cells, resulting in the death of all matured effector T cells within a few days and the deletion of this specific response from the T cell repertoire (20). Both X4 and R5 HIV-1 gp120 itself has been shown in vitro to dysregulate T cell function, including the induction of T cell apoptosis, inhibition of functional T cell responses to recall Ags, and interference with the expression of costimulatory molecules and TCR desensitization (21–25, 26). Additionally, it has been demonstrated that both recombinant and oligomeric forms of gp120 directly influence T cell migration and that high concentrations of the retroviral protein (200 ng/ml) cause active movement of Ag-specific T cells away from the protein via a chemokine (C-X-C motif) receptor 4 (CXCR4)-mediated mechanism, termed fugetaxis or chemorepulsion (22). Consequently, it has been suggested that HIV-specific immune effector cells may fail to migrate to those areas in which HIV proliferation and gp120 expression are high, as in the lymph node (LN) (27). The question therefore arises as to whether high enough concentrations of gp120 exist in vivo to dysregulate T cell migration and suppress their function.
In this study, we used a model of acute chimeric HIV (SHIV) infection to examine the distribution and concentration of gp120 deposits and T cell subpopulations in infected tissues and the effects of the envelope protein on the functional status of T cell populations (28, 29). We chose this time point during the retroviral infection because it occurs before the development of Ab responses that might mask the presence of gp120 by binding it into immune complexes (30). Herein, we demonstrate that during acute infection, a significant amount of the gp120 protein is deposited in the LN where priming and proliferation of lymphocytes occur. Furthermore, we show that T cells from these areas lose their ability to respond to gp120. This defect is Ag-specific, as LN T cells are able to respond to unspecific or SIVmac239 Gag stimulation to a similar extent as do PB T cells. Additionally, we demonstrate that T regulatory cells (Treg cells) accumulate in LNs and produce the immunosuppressive cytokine TGFβ upon stimulation with SHIV gp120. These findings broaden our understanding of the non-entry functions that HIV-1 gp120 may contribute to the pathogenesis of HIV/AIDS.
Materials and Methods
SHIV, animals, and LN and PBMC sampling
The SHIV chimeric virus used in this study was created by using SIVmac239 and the env gene of a cytopathic primary patient isolate of HIV-189.6 that causes an AIDS-like disease in rhesus macaques (28, 29). SHIV-specific CD8+ CTL are known to be present during the early weeks following infection, before a neutralizing Ab response is demonstrable (1, 2, 7). Primary SHIV infection in this study was defined by the presence of SHIV RNA in the plasma of infected macaques and negative or weakly positive gp120 Abs by ELISA.
Eight Indian-origin rhesus macaques (Macaca mulatta) acutely infected with SHIV-KB9 by the i.v. route were involved in this study. PB and LNs from nine healthy, noninfected macaques were used as controls. Blood and LNs from SHIV (KB-9)-infected and uninfected macaques were kindly provided by Dr. Norman Letvin and Dr. Keith Mansfield (see Table I) in accordance with the guidelines of the Harvard Medical School’s Standing Committee on Animals and were collected at the time of sacrifice at day 42 (n = 8). Data from these monkeys have not been published elsewhere. Some of the LNs were snap frozen and stored at −80°C. Mononuclear cells were isolated from PB, and the remaining LNs were enriched by Ficoll gradient centrifugation and frozen at −80°C until used for in vitro assays.
Table I.
Detection of the virological status of the macaquesa
| Macaques | Route of Viral Entry |
Virus | Time of Sacrifice |
Viral Load at Time of Sacrifice |
% of CD4+ T Cells at Time of Sacrifice |
% of CD8+ T Cells at Time of Sacrifice |
|---|---|---|---|---|---|---|
| 179-93 | i.v. | SHIV-KB9 | Day 42 | 55,398 | 8.34 (PB), 11.4 (LN) | 88.8 (PB), 78.2 (LN) |
| 212-92 | i.v. | SHIV-KB9 | Day 42 | 151,841 | 17.1 (PB), 6.77 (LN) | 88.6 (PB), 86.6 (LN) |
| 249-92 | i.v. | SHIV-KB9 | Day 42 | 159,406 | 19 (PB), 12.3 (LN) | 71.1 (PB), 78.6 (LN) |
| 316-90 | i.v. | SHIV-KB9 | Day 42 | 5,258 | 24.9 (PB), 22.3 (LN) | 70.9 (PB), 70.9 (LN) |
| 356-92 | i.v. | SHIV-KB9 | Day 42 | 134 | 46.2 (PB), 55.1 (LN) | 68 (PB), 44.2 (LN) |
| 382-90 | i.v. | SHIV-KB9 | Day 42 | 595,959 | 5.83 (PB), 2.96 (LN) | 84 (PB), 89.3 (LN) |
| 392-91 | i.v. | SHIV-KB9 | Day 42 | 53,934 | 15.2 (PB), 11.7 (LN) | 72.3 (PB), 78.1 (LN) |
| 529-99 | i.v. | SHIV-KB9 | Day 42 | 8,125 | 13.5 (PB), 13.5 (LN) | 80.7 (PB), 79.6 (LN) |
| 248-98 | N/A | Control | N/A | N/A | 57.8 (PB) | 41.6 (PB) |
| 296-93 | N/A | Control | N/A | N/A | 45.4 (PB) | 60.2 (PB) |
| 425-01 | N/A | Control | N/A | N/A | 53.4 (PB) | 54 (PB) |
| 394-91 | N/A | Control | N/A | N/A | 52.6 (PB) | 46.2 (PB) |
| 475-99 | N/A | Control | N/A | N/A | 76.6 (LN) | 21.8 (LN) |
| 295-99 | N/A | Control | N/A | N/A | 56.1 (LN) | 39.1 (LN) |
| 263-01 | N/A | Control | N/A | N/A | 58.0 (LN) | 37.3 (LN) |
| 211-93 | N/A | Control | N/A | N/A | 56.2 (LN) | 37.7 (LN) |
| 165-91 | N/A | Control | N/A | N/A | 70.3 (LN) | 28.4 (LN) |
Eight acutely SHIV-KB-9-infected animals and nine uninfected animals were used in this study. PB and LNs were collected at the time of sacrifice. Plasma SHIV-KB9 RNA levels were measured by an ultrasensitive branched DNA-amplification assay with a detection limit of 125 RNA copies/ml (Bayer Diagnostics). The plasma SHIV-KB9 RNA levels and percentage CD4+ and CD8+ T cells were determined at time of sacrifice.
Immunofluorescent staining of LNs for gp120
Serial 6-μm-thick sections of archived snap-frozen LNs from SHIV-KB9-infected animals were stained as previously described (31, 32). Sections were fixed for 10 min in ice-cold acetone. After washing, nonspecific binding to the Fc receptors was blocked for 15 min in 5% normal goat serum in PBS and purified goat anti-human IgG (H+L) (Invitrogen). After washing in PBS, the primary Ab mAb to HIV-1 V3, 447–52D (obtained through National Institutes of Health AIDS Research and Reference Reagents Program (NARRRP), Division of AIDS, National Institute of Allergy and Infectious Diseases, from Dr. Susan Zolla-Pazner) (33–38), was applied in PBS/1% BSA for 60 min at room temperature. After three washes in PBS, 15 min blocking in 5% goat serum, and another two washes in PBS, a secondary Ab, Alexa Fluor 555 goat anti-human IgG (H+L) (Molecular Probes) in PBS/BSA, was applied for 30 min at room temperature and protected from light. The slides were washed in PBS, mounted, and analyzed with confocal microscopy. A similar procedure was used to stain for CD3+ T cells by using polyclonal rabbit anti-human Ab to CD3 (Dak-Cytomation) and anti-rabbit Ig Alexa 488 (Molecular Probes). Double staining for gp120 and CD3+ T cells was performed, and serial sections were analyzed by confocal microscopy (31, 32). The Ab KK59 directed against SIVmac239 core protein was obtained from the NARRRP (39). Staining for the core protein was done by using the mAb at a 1/50 dilution followed by FITC-conjugated goat anti-mouse secondary Ab (Molecular Probes). Staining of normal, uninfected macaques LNs did not show any significant staining for Gag. The equine infectious anemia virus (EIAV) gp90(A)-86 mAb directed against a conserved peptide in the equine infectious anemia virus surface envelope was used as an unrelated negative control and was also obtained through the NARRRP from Dr. Ronald Montelaro (40–42). Intracellular staining of the cell line CHO-NL-4-3 that constitutively produces gp120 (obtained through the NARRRP from Dr. James Arthos) was used as a positive control.
Confocal microscopy was performed with a Zeiss LSM 5 Pascal laser scanning microscope (Carl Zeiss Microimaging). Individual serial optical slices representing 0.2 μm, and 20–60 optical slices were collected at a 512- by 512-pixel resolution. Colocalization of Ags was indicated by the addition of colors as previously described (31).
Quantifying the amount of gp120 in infected LNs and plasma
To confirm results obtained by immunofluorescent staining and flow cytometry for surface gp120 expression, we quantitated the amount of gp120 in LNs and plasma from four acutely SHIV-infected and four uninfected monkeys by using an ELISA. LNs were fragmented mechanically, and the subsequent mononuclear cell suspension was prepared by Ficoll gradient centrifugation. Cells were counted, resuspended in 1 ml of RPMI 1640, and sonicated on ice. The gp120-producing cell line CHO-NL-4-3 was processed in the same way and used as a positive control, while the parent, a gp120-negative CHO cell line, was used as a negative control. Cell lysates were separated from extracellular debris by centrifugation and the supernatant was tested for presence of gp120 by using an ELISA kit (Immuno-Diagnostics). Plasma samples from infected and uninfected animals were concentrated (10-fold) using Microcon tubes (Millipore). HIV-1 gp120 levels were quantitated as ng/ml of tissue so that levels could be compared between tissue specimens and plasma samples and to make a preliminary determination about the magnitude of the gradient that existed between tissue and plasma. The percentage yield of gp120 from tissues by using this method was determined by spiking normal, uninfected tissues with known amounts of recombinant envelope protein. Cell lyses were performed as previously described, and gp120 was measured after extraction.
Phenotypic and functional characterization of isolated PB and LN lymphocytes
Lymphocytes were isolated from PB and LNs of acutely SHIV-KB-9-infected macaques. Phenotypic characterization was done by multiparameter staining for CD3, CD4, CD8, and CD25 (BD Biosciences) with DIVA software in conjunction with an LSR2 FACS analyzer (BD Biosciences). The source and details of Abs used throughout this study are shown in Table II (under Panel 1). Any significant spillover from one channel to another is indicated by the DIVA software. The negative population for each Ag was established by staining with all of the other Abs except the one of interest and subsequent FlowJo analysis. The same settings were then used for the staining of samples and the quadrant settings established in the negative population were used to analyze the samples.
Table II.
Specificity, manufacturer, clone, and fluorochrome for the panels of mAbs used in this study
| Ab | Manufacturer | Clone | Fluorochrome |
|---|---|---|---|
| Panel 1 (phenotypic characterization) | |||
| Anti-CD3 | BD Biosciences | Sp34-2 | Pacific Blue |
| Anti-CD4 | BD Biosciences | L200 | PerCP-Cy5.5 |
| Anti-CD8 | BD Biosciences | SK1 | Allophycocyanin-Cy7 |
| Anti-CD25 | BD Biosciences | M-A251 | PE-Cy7 |
| Panel 2 (assessment of function) | |||
| Anti-CD4 | BD Biosciences | L200 | FITC |
| Anti-CD3 | BD Biosciences | Sp34-2 | Pacific Blue |
| Anti-CD8 | BD Biosciences | SK1 | Allophycocyanin-Cy7 |
| Anti-IL-10 | Miltenyi Biotec | B-T10 | Allophycocyanin |
| Anti-IFN | BD Biosciences | B27 | Alexa 700 |
| Anti-TNF | BD Biosciences | mAb 11 | PE-Cy7 |
| Anti-CD107a | BD Biosciences | H4A3 | PE-Cy5 |
| Panel 3 (regulatory T cells) | |||
| Anti-CD4 | BD Biosciences | L200 | PE |
| Anti-CD3 | BD Biosciences | Sp34-2 | Pacific Blue |
| Anti-IL-10 | Miltenyi Biotec | B-T10 | Allophycocyanin |
| Anti-TGFβ | BD Biosciences | A75-3 | Biotinylated folled by streptavidin Cascade Yellow |
| Anti-FoxP3 | eBioscience | PHC101 | Alexa 700 |
| Anti-CD25 | BD Biosciences | M-A251 | PE-Cy7 |
| Anti-CD127 | BD Biosciences | hIL-7R-M21 | PE |
The functional capacity of the lymphocytes was assessed following stimulation with the complete set of peptides spanning the entire SHIV 89.6 Env (20-mer) (overlapping by 10 aa) or SIVmac239 Gag (15-mer) (overlapping by 11 aa) from the core of SHIV (NARRRP) and then by performing multiparameter surface and intracellular staining for CD3, CD4, CD8, CD107a (degranulation marker), perforin, IFN-γ, TNF-α, and IL-10 (BD Biosciences). Stimulation and staining were performed as previously published (31, 32, 43–45). SHIV 89.6 is the parent strain of SHIV (KB9), and the sequence of the SHIV 89.6 Env is similar and binds equivalently. The core protein of the SHIV (KB9) virus is equivalent to the SIVmac239 core protein. Nonspecific stimulation with PMA and ionomycin was used as a control and to demonstrate that the cells are functionally capable of degranulating and producing the cytokines.
Isolated and previously frozen PB and LN lymphocytes were thawed and washed. Viability was assessed by trypan blue dye exclusion, and viable cells were counted. Cells were incubated for 1 h at 37°C in 5% CO2 at a concentration of 106/ml in the presence of 1 μl of anti-CD28 (BD Biosciences), 1 μl of anti-CD49d (BD Biosciences), and 1 μg/ml of the complete set of pooled SHIV 89.6 Env peptides. Cells were also stimulated with the complete set of pooled SIVmac239 Gag peptides or 25 ng of PMA (Sigma-Aldrich) and 1 μg of ionomycin (Sigma-Aldrich). PB and LN lymphocytes from any individual monkey were tested in this way at the same time and in parallel with the same amount of peptide. Cells were incubated for an additional 5 h with added GolgiPlug and GolgiStop (BD Pharmingen). Cells were then stained first with surface Abs and then for intracellular IL-10, IFN-γ, or TNF-α. (BD Pharmingen) (Table II, Panel 2) by using a Cytofix/Cytoperm Plus kit (BD Pharmingen) according to the manufacturer’s instructions. At least 5 × 105 cells were acquired per sample. Cells were gated on CD4+CD3+ or CD8+CD3+. The percentage of CD107a, IFN-γ, IL-10, or TNF-α. cells that were induced by a specific stimulant (Env peptides, Gag peptides, or PMA and ionomycin) is equal to the percentage of positive cells in the stimulated sample minus the percentage of positive cells in the unstimulated sample (background). An increased response was therefore determined as the percentage increase in responding cells above the unstimulated background. A positive response to stimulation was defined as being greater than or equal to a 100% increase above background in unstimulated conditions.
Assessment of Treg cell responses to Env was done by stimulating with the complete set of pooled SHIV 89.6 Env peptides, as described above, and multiparameter staining for CD3, CD4, CD25, FoxP3, IL-10, and TGFβ (Table II, Panel 3). Stimulation with anti-CD3 and anti-CD28 was used as a positive control for TGFβ expression. All of the stained cells or at least 1 × 106 cells were acquired. Cells were gated on CD4+ CD3+ and then CD25, and FoxP3 markers.
In vitro exposure of PB lymphocytes to SHIV-KB9 gp120
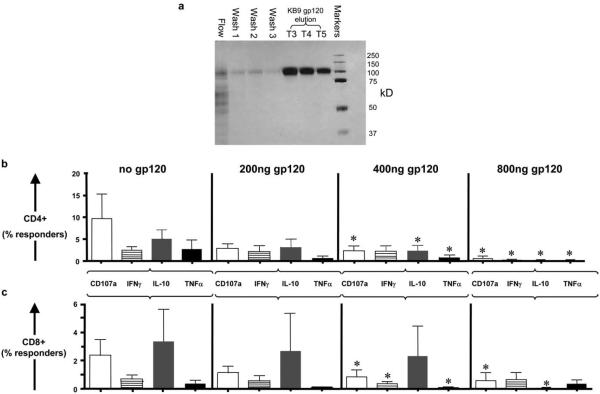
SHIV-KB9 gp120 glycoprotein was cloned, between the KpnI and MfeI sites, into the plasmid pE7-HXBc2(IIIexE7pA-KpnII') (46) in which a stop codon had been introduced at the beginning of the gp41 region, to produce the plasmid pE7-KB9 gp120. The expression of HIV-1 gp120 glycoprotein was achieved in HEK 293T cells cotransfected with the pE7-KB9 gp120 plasmid and a pTat plasmid that express HIV-1 Tat protein. About 14 h after transfection, cells were washed once with PBS and cultured for 72 h in serum-free medium Pro293s-CDM (Cambrex) supplemented with 12 mM glutamine. Sodium butyrate was added to a 3 mM concentration 31 h before harvesting. The medium containing KB9 gp120 glycoprotein was harvested and incubated with 100 μl of IgG1-b12-Sepharose beads for ~16 h at 4°C. After washing the beads three times with washing buffer (Tris 0.1 M (pH 8.0), NaCl 0.5 M), the KB9 gp120 glycoprotein was eluted with Gly 0.1 M (pH 3.0) and neutralized immediately with 1/10 volume of Tris 1 M (pH 8.0). The purity and concentration of KB9 gp120 protein were determined by silver staining of an SDS-PAGE gel and spectrophotometry, respectively (see Fig. 5a).
FIGURE 5.
PB lymphocytes were exposed overnight to SHIV-KB9 glycoprotein gp120. SHIV-KB9 gp120 was generated by expressing it into the plasmid pE7-HXBc2(IIIexE7pA-KpnII'), transfecting the permissible cells, collecting the supernatants, and purifying the KB9 gp120. The purity of gp120 was determined by silver staining of an SDS-PAGE gel (a). (Lanes, left to right, products of flow-through (lane 1), three column washes (lanes 2–4), three lanes of eluted KB9-gp120 protein (lanes 5–7) and a molecular mass marker (lane 8)). Pretreatment of CD3+CD4+ T cells (b) or CD3+CD8+ T cells (c) with recombinant SHIV-KB9 gp120 (gp120) at concentrations of 400 ng/ml and/or 800 ng/ml significantly reduced the percentage of responding cells from the PB of acutely SHIV-KB9-infected monkeys to a complete set of SHIV-KB-9 Env peptides. Graphs show the percentage of CD4+ or CD8+ Env-induced responder cells (% of Env induced = % of the stimulated cells − % of nonstimulated cells). Data sets from PB CD4+ T cells (n = 6 monkeys, b) and CD8+ T cells (n = 6 monkeys, c) are shown. (*, p < 0.05; two-tailed Mann-Whitney U test).
To assess the effect of HIV-1 gp120 on the functionality of T cells previously frozen, PB lymphocytes from acutely SHIV-KB9-infected macaques were thawed and cultured in the absence or presence of 200, 400, and 800 ng/ml of the SHIV-KB9 gp120 glycoprotein. Viability of the cells was assessed by trypan blue exclusion and was >75% after treatment with gp120. Cells were then stimulated with the complete pooled SHIV-89.6 Env peptide mix or SIVmac239 Gag and stained for CD3, CD4, CD8, CD107a, IFN-γ, TNF-α., and IL-10 and analyzed by multiparameter flow cytometry and FlowJo as previously described.
Statistical analysis
All statistical analyses were performed using a Mann-Whitney U test. A value of p < 0.05 was considered significant. p-values were determined with and without single outlier values where present.
Results
LN tissues of acutely SHIV-KB9-infected macaques contain gp120 deposits at levels previously shown to induce T cell dysfunction
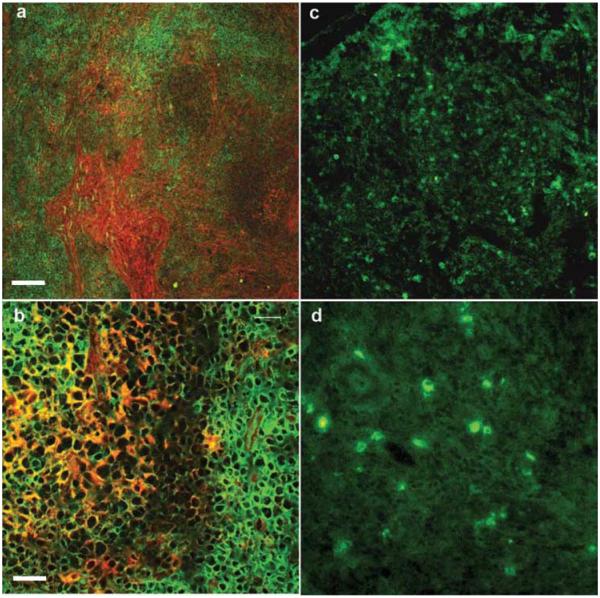
We demonstrated by immunofluorescent staining with V3 447-52D Ab directed to the V3 loop of gp120 a dense and widespread distribution of the envelope protein in LNs derived from acutely SHIV-infected macaques (Fig. 1, a and b). Staining of the same tissues with mAb to the SIVmac239 Gag core protein revealed a different distribution of staining (Fig. 1, c and d), indicating that the gp120 is not only present on infected cells but is also deposited throughout the tissue. In contrast, SIVmac239 Gag was localized on the plasma membrane or within the cytoplasm of scattered cells within T cell areas of the LN (Fig. 1d) as previously reported (47, 48).
FIGURE 1.
Immunofluorescent staining for HIV gp120 and SIV Gag protein was performed on serial sections of LN tissues of macaques in primary SHIV infection. Gp120 expression (red) in the acutely infected LNs did not appear to be related to the CD3+ T cell zones (green) (original magnification ×10 (a) and ×40 (b)). Staining for the SIV Gag protein (green) revealed less SIV Gag-positive cells that were differently distributed (c, original magnification ×20) from gp120. SIV Gag protein was detected at the plasma membrane in most of the positive cells (d, original magnification ×40). The white scale bar represents 100 μm in a and 20 μm in b.
Significant effects of gp120 on T cell function have been previously described (21–26, 49). These effects were only seen at high concentrations of gp120 >20 ng/ml or 200 pM, but the precise relevance of these effects in vivo in HIV-1 infection is not clear. In the first instance we set out to see if a clinically relevant amount of gp120 is present in the LN tissues. We quantitated the amount of gp120 in SHIV-infected LNs and plasma from four acutely SHIV-infected and four uninfected monkeys using an ELISA-based system. Assessment was done on sonicated LN mononuclear cells as described in Materials and Methods. Concentrations of SHIV-KB9 gp120 in LN mononuclear cell lysates varied between 183 and 562 ng/ml (mean of 218 ng/ml) of LN tissue from acutely infected animals, whereas plasma levels varied between 0.33 and 3.73 ng/ml (mean of 1.55 ng/ml). These data indicate that a significant difference exists between the amount of gp120 present in the LN mononuclear cells and plasma. HIV-1 gp120 was detectable at a level of 123 ng/ml from lysed control CHO-NL-4-3 cells engineered to express the envelope protein and was otherwise undetectable in plasma and lysed LN mononuclear cells from uninfected monkeys or the parent CHO cell line (data not shown). Additionally, we measured percentage retrieval of gp120 from normal uninfected tissues spiked with a known amount of the recombinant protein and found that on average 66% (range 50–75%) of gp120 was retrieved by our described method. This measurement does not take into account gp120 bound to extracellular matrix in the LNs, which was detected by immunostaining. Consequently, we think that our measurements of gp120 in infected tissues may be an underestimate of total gp120 in the LN itself. These results demonstrate that gp120 is present during acute SHIV-KB9 infection in the LNs at high levels equivalent to those shown to dysregulate T cell function and migration in vitro.
LN T cells fail to degranulate and secrete less IFN-γ, TNF-α, and IL-10 when stimulated with gp120 peptides compared with PB T cells
The functional fitness of PB and LN T cells from SHIV-infected animals was assessed by determining the secretion of cytokines relevant to T cell function such as IFN-γ and IL-10 and by examining the capacity of T cells to perform cytotoxic function. CTL-mediated lysis of infected targets is initiated by the recognition of a MHC class I exogenous peptide complex on the surface of target cells, and can be achieved by granule-independent triggering of apoptotic processes in the virus-infected cells via TNF-α and CD95 (reviewed in Refs. 50–52) or via a granule-dependent pathway. During the granule-dependent pathway of CTL-mediated lysis, perforin molecules insert themselves into the plasma membrane of target cells, thus enabling granzymes to enter the cell and to activate the precursors of caspases initiating self-destruction of the cell by apoptosis (reviewed in Refs. 50–52). The markers CD107a and CD107b are expressed on the cell surface during degranulation and were used in these studies in conjunction with perforin to assess the capacity of the T cells to degranulate in response to an Ag (53).
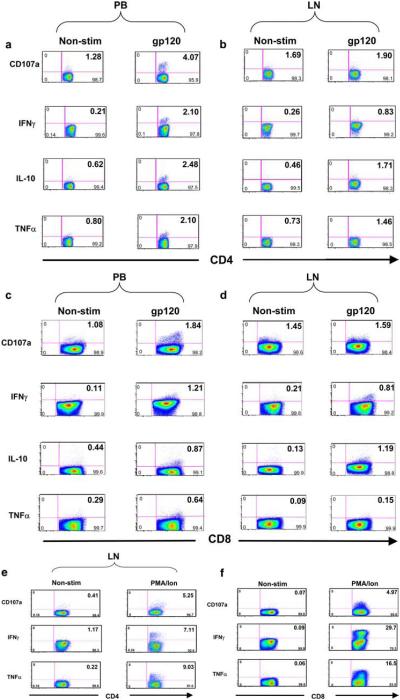
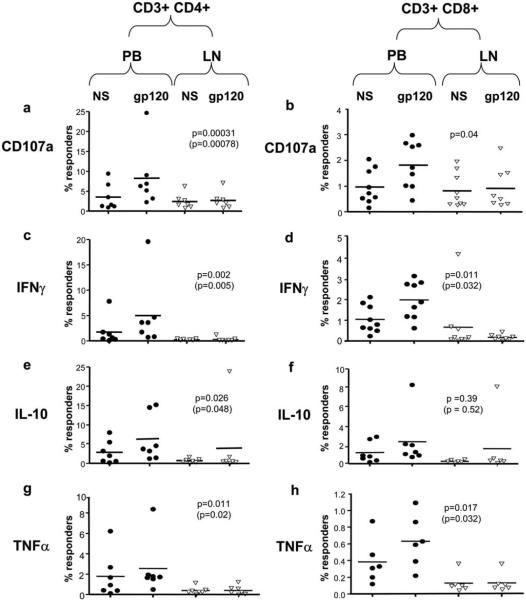
We made the robust finding that while stimulation with the SHIV Env peptide pool induced degranulation as measured by CD107a expression in CD4+ (Figs. 2a and 3a) and CD8+ (Figs. 2c and 3b), PB-derived T cells and CD4+ LN-derived T cells degranulated to a much lesser extent in response to this stimulant (p = 0.00031) (Figs. 2b and 3a). A similar pattern of CD107a expression was seen for CD8+ T cells derived from peripheal blood and LNs (p = 0.04) (Figs. 2, c and d, and 3b). Note that CD107a measurements are prone to fixation artifacts and may be unduly sensitive to excess amounts of peptides. This was internally controlled in this study by comparing responses of PB T cells to LN-derived T cells to identical amounts of peptides and fixation methods. Similarly, while gp120 stimulation induced IFN-γ secretion in both CD4+ and CD8+ PB T cells (Figs. 2, a and c, and 3, c and d), LN CD4+ and CD8+ T cells demonstrated significantly reduced levels of these responses (Figs. 2, b and d, and 3, c and d) (p = 0.002 and p = 0.011, respectively). Secretion of IL-10 was greater in response to gp120 in PB in comparison to LN-derived CD4+ (Figs. 2, a and b, and 3e) (p = 0.026). However, PB- and LN-derived CD8+ T cells demonstrated similar levels of IL-10 responses (Figs. 2, c and d, and 3f) (p = 0.39). This pattern was also evident in TNF-α secretion in response to SHIV gp120 peptide pool. Secretion of TNF-α was greater in PB-derived CD4+ in comparison to LN-derived CD4+ (Figs. 2, a and b, and 3g) (p = 0.011). However, PB- and LN-derived CD8+ T cells demonstrated similar levels of IL-10 responses (Figs. 2, c and d, and 3h) (p = 0.17). In separate experiments nonspecific stimulation of CD3+CD4+ or CD3+CD8+ T cells with PMA and ionomycin showed robust responses as measured by CD107a and intracellular cytokine expression (Fig. 2, e and f).
FIGURE 2.
Representative flow cytometric data from the stimulation of CD4+ (a and b) and CD8+ T cells (c and d) from the SHIV-KB9-infected monkey 212-92. Stimulation with a cocktail of the complete set of SHIV-KB9 Env peptides (gp120) induced an increase in CD107a, IFN-γ, IL-10, and TNF-α. secretion of PB- and LN-derived CD3+CD4+ cells (a) and CD3+CD8+ T cells (c) above background levels of expression in nonstimulated cells (Non-stim). The magnitude of this increase in this specific monkey was reduced in CD3+CD4+ cells from LNs (unstimulated (left), stimulated (right)) as compared with PB-derived CD3+CD4+ T cells for CD107a, IFN-γ, and TNF-α. expression (a and b, respectively). The magnitude of this increase in this monkey was also significantly reduced in CD3+CD8+ cells from LNs as compared with PBMC-derived CD3+CD8+ for CD107a, IFN-γ, and TNF-α. expression (Fig. 2, c and d, respectively). Representative flow cytometric data from the stimulation of LN-derived CD3+CD4+ (e) and CD3+CD8+ (f) T cells from the acutely SHIV-KB9-infected monkey (212-92) in response to PMA/ionomycin (PMA/Ion) stimulation is also shown. CD3+CD4+ or CD3+CD8+ cells show robust responses to PMA/ ionomycin as measured by increases in CD107a, TNF-α., and IFN-γ expression. Five hundred thousand events were acquired for each sample, and quadrant statistics are shown in each dot plot.
FIGURE 3.
Cumulative flow cytometric data from the stimulation of PB-derived and LN-derived CD3+CD4+ (a, c, e, and g) and CD3+CD8+ T cells (b, d, f, and h) from acutely SHIV-KB9-infected monkeys in response to the complete set of SHIV 89.6P Env peptides (gp120). Percentage of Env-specific responder T cells in the context of measurements of CD107a, IFN-γ, IL-10, and TNF-α. are shown for both nonstimulated (NS) and gp120 peptide-stimulated cells. When compared with PB-derived CD3+CD4+ T cells, LN CD4+ T cells (a) showed consistently less degranulation as measured by CD107a expression on their surface (p = 0.00031 or p = 0.00078 without the single outlier value). Secretion of IFN-γ, IL-10, and TNF-α in response to Env peptide stimulation was also significantly lower in the LN CD3+CD4+ (c, e, and g, respectively) T cells as compared with those originating from the PB. These statistically significant differences were maintained when single outlier values where present in the dataset were excluded (bracketed p-value). A similar pattern of reduced Env-induced CD107a and IFN-γ expression in LN-derived CD8+ T cells as compared with PBMC-derived CD8+ T cells was seen (b and d) (CD107a: p = 0.04 and IFN-γ: p = 0.011, respectively). Env-specific IL-10 and TNF-α responses of LN CD3+CD8+ T cells were reduced in comparison to PB-derived CD8+ T cell responses, but this difference did not reach statistical significance. The statistical significance of differences between the responses of PBMC- and LN-derived T cells was measured using a two-tailed Mann-Whitney U test. Each symbol (aaaa, PB-derived T cells; ∇, LN-derived T cells) represents data from an individual monkey.
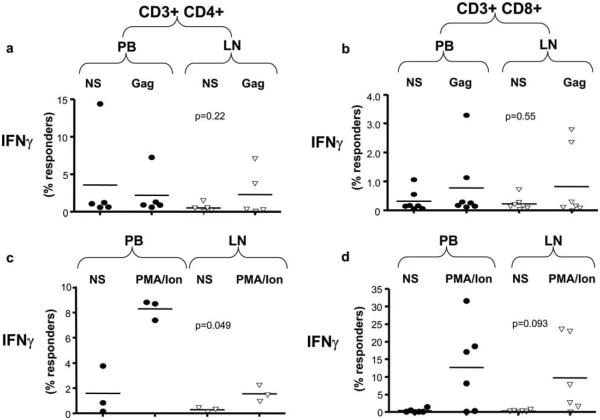
We then set out to examine whether the suppressive effects of gp120 were Ag specific or also occurred in response to other viral Ags including Gag. Stimulation with SIV239 Gag peptide mix induced IFN-γ secretion in both PB and LN CD4+ (Fig. 4a) and CD8+ (Fig. 4b) T cells. Stimulation of lymphocytes from SIVmac251-infected macaques and stimulation with PMA and ionomycin were used as positive controls. Nonspecific stimulation with PMA and ionomycin induced high IFN-γ (Fig. 4, c and d) and TNF-α (data not shown) secretion from both from blood and LN CD8+ T cells. However, when compared with PBMCs, LN CD4+ T cells responded poorly to PMA and ionomycin stimulation, and the percentage of LN CD4+ T cells that were induced to produce both IFN-γ (Fig. 4c) or TNF-α was significantly lower than those in PB (p = 0.049 and p = 0.01) (data not shown).
FIGURE 4.
Cumulative flow cytometric data are shown from the stimulation of PB-derived (PBMC) and LN-derived CD3+CD4+ (a and c) and CD3+CD8+ T cells (b and d) from acutely SHIV-KB9-infected monkeys with the complete set of SIVmac239 Gag peptides peptides (Gag). The percentages of Env specific responder T cells in the context of measurements of IFN-γ are shown for both Gag peptide-stimulated and nonstimulated cells (NS) as well as for PMA/onomycin (PMA/Ion)-stimulated cells (c and d). Stimulation of lymphocytes with SIVGag239-induced IFN-γ responses of similar magnitude in LN and PB CD4+ (a) and CD8+ (b) T cells from acutely SHIV-infected macaques (CD4+ p = 0.22; CD8+ p = 0.55 with outliers). In separate experiments, stimulation of CD4+ or CD8+ T cells with PMA and ionomycin induced IFN-γ responses of similar magnitude in LN and PB CD8+ (d) (p = 0.093; p = 0.2 without the outlying datapoint) but not in CD4+ T cells that were less responsive if they originated from LNs (c) (p = 0.049). Statistical analysis was performed as described in Fig. 3. Each symbol (aaaa, PB-derived T cells; ∇, LN-derived T cells) represents data from an individual monkey.
Exposure of PB-derived lymphocytes to SHIV-KB9 gp120 suppresses CD4+, and CD8+ T cell function in a dose-dependent manner
Having demonstrated that PB lymphocytes from SHIV-KB9-infected macaques responded well to stimulation with the SHIV-KB9 Env peptide pool by degranulating and increasing their secretion of IFN-γ, TNF-α, and IL-10, we set out to examine whether this response could be abolished by the presence of gp120. Isolated PB lymphocytes were incubated overnight with SHIV-KB9 gp120 at concentrations that we had shown above to be present in LNs from acutely SHIV-infected monkeys (200, 400, and 800 ng/ml). The SHIV-KB9 gp120 was generated and purified as previously described (Fig. 5a). Lymphocytes were then stimulated with SHIV-KB9 envelope peptide pool and stained for CD3, CD4, CD8, CD107a, IFN-γ, TNF-α, and IL-10. In these experiments, SHIV-KB9 gp120 at a concentration of 400 ng/ml suppressed CD4+ T cell responses to a mean of 32 ± 21% of those of the nonexposed cells for CD107a, 69 ± 18% (±SEM) for IFN-γ, 45 ± 22% for IL-10, and 35 ± 15% of the responses of nonexposed cells for TNF-α (Fig. 5b). Similarly, preincubation of CD8+ T cells with recombinant KB-9 gp120 led to suppression of Env peptide pool-induced responses to 32 ± 17% for CD107a, 48 ± 20% for IFN-γ, 34 ± 33% for IL-10, and 32 ± 21% for TNF-α (Fig. 5c). In rare instances where suppression of one function of CD4+ or CD8+ cells did not occur, other functions were suppressed. In summary, exposure of the cells to concentrations of 400 and 800 ng/ml of SHIV-KB9 gp120 suppressed all functions in both CD4+ and CD8+ T cells and completely abolished gp120-induced degranulation or cytokine secretion in most of the six samples examined (Table III). This experiment confirmed that, when present at high concentrations, gp120 is able to suppress CTL responses. It is unclear whether other viral proteins are present at high levels in infected tissues and whether they may have a similar effect. We were unable to detect a high amount of the core protein SIVmac239 in acutely infected tissues by immunostaining (see Fig. 1). In control experiments to examine the relative effect of Gag protein, we exposed PB T cells to 0, 2, and 200 ng/ml using the same conditions as for gp120. Exposure to 200 ng/ml of SIVmac Gag protein did not inhibit the cellular responses, but, on the contrary, were slightly increased (data not shown).
Table III.
Quantitation of the suppressive effect of recombinant SHIV-KB9 gp120 on T cell responses to the complete pool of Env peptides by recombinant SHIV-KB9 gp120a
| CD107a | IFN-γ | IL-10 | TNF-α | |
|---|---|---|---|---|
| a) Amount of gp120 (CD4+)b | ||||
| 200 ng/ml | 4/4 | 1/4 | 2/4 | 2/4 |
| 400 ng/ml | 4/4 | 1/4 | 3/4 | 3/4 |
| 800 ng/ml | 4/4 | 4/4 | 4/4 | 4/4 |
| b) Amount of gp120 (CD8+)c | ||||
| 200 ng/ml | 4/4 | 2/4 | 1/4 | 2/4 |
| 400 ng/ml | 4/4 | 3/4 | 2/4 | 4/4 |
| 800 ng/ml | 4/4 | 2/4 | 4/4 | 4/4 |
Reported are the proportions of samples of PB CD4+ (a) and CD8+ (b) T cells from individual monkeys (n = 4) that generated reduced CD107a, IFN-γ, IL-10, and TNF-α responses to Env peptide mix following preexposure to recombinant SHIV-KB9 gp120 at concentrations of 200, 400, and 800 ng/ml as detailed in Fig. 4 (inhibited responder samples/total no. of samples tested). Most T cell samples exposed to concentrations of 400 and 800 ng/ml of recombinant SHIV-KB9 gp120 reported reductions in CD107a and cytokine expression in comparison to T cells, which were not pretreated with recombinant gp120.
No. of inhibited CD4+ T cell responses/total no. of responses measured.
No. of inhibited CD8+ T cell responses/total no. of responses measured.
LNs but not PB of SHIV-KB9 acutely infected macaques contain a significantly higher percentage of regulatory T cells than do those of uninfected macaques
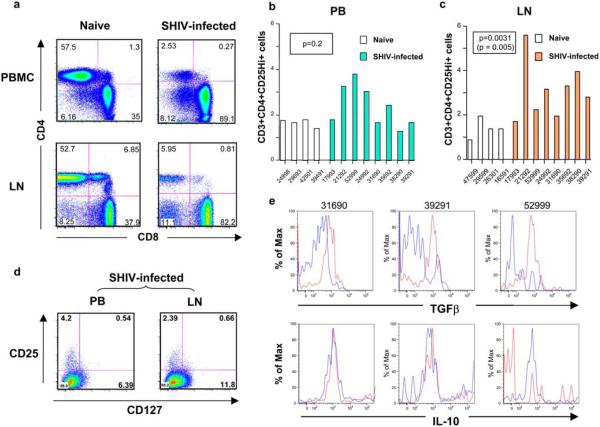
While significant depletion of the CD3+CD4+ cells was seen in both compartments (Fig. 6a), the percentage of regulatory T cells as determined by high-level expression of CD25 on CD3+CD4+ cells was significantly up-regulated in LN lymphocytes (Fig. 6b) but not in PBMCs (Fig. 6c) of SHIV-infected macaques compared with the naive controls. CD4+CD25+ Treg cells normally constitute a small fraction of circulating CD4+ T cells in humans that belong to the memory T cell pool (54–57). They are commonly identified by the expression of the IL-2Ra (CD25) on their surface and by the expression of the transcription factor scurfin that is encoded by the FoxP3 gene. Upon further phenotypic analysis of regulatory T cells from SHIV-infected macaques, we observed that most CD3+CD4+ cells from PB and LNs that expressed high levels of CD25 were CD127low (PB mean of 86.5%; LN mean of 84.0%) (Fig. 6d). We also observed a smaller majority of CD3+CD4+ cells, which were CD25high, that were CD127low in PB (mean of 53.5%) and LN from uninfected monkeys (mean of 72%) (data not shown). These findings are consistent with recent descriptions of the immunophenotype of Treg cells (58, 59). We observed no significant difference in the proportion of CD3+CD4+ CD25+ cells that were CD127low from either blood or LNs. Accumulation of Treg cells has been previously documented and was attributed to the selective promotion of their survival by the HIV virus via a CD4-gp120 interaction (60).
FIGURE 6.
CD4+ Treg cells were quantitated in LNs and PB of acutely SHIV-infected monkeys. Percentages are shown of CD3+ T cells expressing CD4 and CD8 markers in PBMCs (a, upper) and LNs (a, lower) of a representative naive (a, left) and SHIV-infected (a, right) macaque. The differences between PB CD3+CD4+CD25high+ T cells from infected and uninfected animals were not significant (p = 0.2) (b). In contrast, the percentages of CD4+CD25high+ T cells in SHIV-infected LN lymphocytes were significantly higher than in those LN lymphocytes derived from naive macaques (p = 0.0031; p = 0.005 without outlier) (p-values were calculated using a two-tailed Mann-Whitney U test) (c). Flow cytometric analysis of CD3+CD4+ T cells from SHIV-KB9-infected monkeys was also performed to examine the expression of CD25 and CD127 (d). Most (>80%) of the CD3+CD4+CD25high+ cells were CD127low from both PB and LNs. When LN lymphocytes from SHIV-infected monkeys were stimulated with SHIV env peptide mix, the Treg cell population (gated on CD3+CD4+CD25+FoxP3+ cells) was shown to secrete TGFβ but not IL-10 (unstimulated, blue; stimulated, red) (e). Data from three different SHIV-infected monkeys are shown.
Regulatory T cells that accumulate in the LNs of acutely SHIV-infected macaques respond to stimulation with SHIV Env peptide pool by secreting TGFβ
To determine whether the Treg cells that accumulate in the LNs of acutely SHIV-infected macaques respond to gp120 and how gp120 affects their behavior, isolated LN lymphocytes were stimulated with the SHIV Env peptide pool and then stained for CD3, CD4, CD25, FoxP3, IL-10, and TGFβ. Adaptive Treg cells can be induced in the presence of IL-10, and in that case they exert their suppressive activity via the production of IL-10 or by TGFβ (61). Although the first mechanism described above and other potential factors cannot be excluded, in the samples that we tested, the Treg cells population secreted TGFβ but not IL-10 in response to gp120 (Fig. 6e). It is possible that Treg cells contribute to Env-mediated suppression by secreting TGFβ in response to gp120. This cytokine is known to promote Treg cell development and to convert CD4+CD25− T cells into Treg cells in vitro by exerting expression of FoxP3, as reviewed in Romagnani (61). Additionally, TGFβ has been shown to be essential in mediating suppression of CTLs by impairing degranulation (62).
Discussion
Most HIV-infected persons only transiently control viral replication and ultimately progress to AIDS, despite the presence of robust CTL responses (63–65). This could be due in part to the fact that CTL responses documented in vitro do not always correlate with effective effector responses in vivo (12, 66). Additionally, although high HIV-specific CD8 T cell frequencies are maintained until late in disease, many HIV-specific T cells have a restricted ability to function (67, 68). We hypothesized in this study that the presence of high levels of gp120 in the LN contributes to the dysregulation of T cell function and assists the virus in evading the immune response. In this study we describe for the first time that high levels of gp120 are present in the LNs of SHIV-infected macaques. Furthermore, CD8+ and CD4+ T cells originating from those LNs responded poorly to stimulation with the gp120 peptide mix. The same result was not obtained in PBMCs from the same animals that were exposed to significantly lower levels of gp120 in the blood. Our data showed that the CD8+ effector cells were only unresponsive to stimulation with the SHIV Env peptide pool but not to nonspecific stimulation with PMA and ionomycin nor to stimulation with the core protein-derived SIVmac239 Gag peptide mix. The defect seen in CD4+ T cells was demonstrable not only in response to gp120 but also when nonspecific stimulation was used.
While there are extensive published studies on the effects of gp120 on Ab responses, surprisingly little is known about the effects of gp120 on T cells in vivo (30). In vitro studies have shown that gp120 induces movement of T cells toward (chemotaxis) or away from (fugetaxis) the protein in a concentration-dependent and CXCR4 receptor-mediated manner (22). Others have shown that aberrant activation in infected and uninfected CD4+ T lymphocytes by soluble or membrane-bound HIV envelope protein (24, 25) causes apoptosis (21, 23–26, 69). A recent study showed that HIV-1 gp120 activates immature dendritic cells in a manner that abrogates their normal function in host immune responses, and consequently disturbs the homeostatic balance of the immune response of the hosts to the infection (70). These authors suggested that HIV-1 gp120 may support sustained productive infection and transinfection of activated T cells that cluster with gp120-activated dendritic cells (70). In another study, when two L(d)-restricted epitopes derived from HIV-1 envelope gp160 (Env) and from CMV pp89 phosphoprotein were coexpressed, HIV-1IIIB Env, but not HIV-1MN Env variant, impaired recognition by a specific CTL of CMV pp89 epitope (71). It was also recently shown that natural mutations in an immunodominant Th epitope recognized by human CD4 clones specific for the envelope glycoprotein gp120 (from sequences of different HIV strains) escape CD4 T cell recognition. Furthermore, several natural analog peptides derived from gp120 exert an antagonistic function by inhibiting proliferative response of T cells specific to the envelope protein. If similar events occur in vivo, they may represent an additional escape mechanism for HIV (72).
Our findings support previous reports that gp120 suppresses the function of CD4+ T cells (73–75). In our system, CD4+ T cell suppression was observed in acutely SHIV-infected macaques, contrary to previously published reports that exposure to recombinant gp120 inhibits proliferative responses of CD4+ helper T cells to various stimuli (76) in human, but not in chimpanzee, lymphocytes (77). Additonally, we demonstrated both CD4+ and CD8+ T cells had a decreased ability to release cytotoxic granules in response to gp120 as evidenced by CD107a staining. These aberrant responses to the viral envelope might be an important contributing factor to the failure of the cytotoxic lymphocytic response to clear the virus during acute infection. However, caution must be exerted when interpreting the results of this study. These experiments were done in a macaque model that utilizes SHIV-KB9, a chimeric virus that expresses the HIV-1 envelope glycoprotein on a SIVmac239 core (28).
It is unclear at this point to what extent the observed accumulation of Treg cells in the LNs of acutely SHIV-infected animals that we observed contributes to the unresponsiveness of the T cells. Previous studies have shown that removal of Treg cells from PBMCs leads to significant HIV-specific CD4+ and CD8+ T cells secretion of IFN-γ (78) and IL-2 (79). In PB of HIV-infected patients, the proliferative capacity of CD4 T cells to tuberculin, CMV, and p24 significantly increased following depletion of CD4+ CD25+ T cells. Furthermore, addition of increasing numbers of CD4+CD25+ T cells resulted in a dose-dependent inhibition of CD4+CD25− T cell proliferation to tuberculin and p24 (80). Our results demonstrate that the accumulated Treg cells secrete TGFβ in response to stimulation with gp120. Treg cells that are induced by Ag feeding and are TGFβ dependent for their immunosuppressive activity were described for the first time as oral tolerance-mediating Th3 cells (81). These cells secrete TGFβ following Ag triggering, consequently suppressing immune responses and inducing tolerance to orally acquired Ags. TGFβ, on the other hand, can induce FoxP3 gene expression in CD4+CD25− cells mediating their transition toward a Treg cell phenotype with potent immunosuppressive potential (82). Treg cell function beyond cytokine release was not assessed in our study due to limitations of cell numbers. The precise role that the accumulated Treg cells might play in the suppression of anti-HIV responses and the role that gp120 plays in this during acute infection are clearly worthy of further exploration.
The HIV-1 envelope used in this study binds both CCR5 and CXCR4 for viral entry as opposed to mostly CCR5-tropic viruses that are usually detected in early HIV infection. Nevertheless, while most of the viruses isolated from early, asymptomatic HIV infection are CCR5-tropic, emergence of the CXCR4-tropic strains of the virus coincides with CD4 cell depletion and progression to AIDS (83). Additionally, early infection with dual tropic viruses has been associated with CD4 depletion and progression to AIDS (84, 85). The effects of gp120 from purely CCR5-tropic HIV viruses are currently being investigated in our laboratory.
Our data support the thesis that significant amounts of HIV-1 gp120 present in lymphoid organs of acutely infected individuals dysregulate immune cell localization and function and thereby contribute a new mechanism by which the virus evades the immune system. These data also add to a growing body of evidence that supports the view that suggests that Env may have limited utility as a CD8 T cell immunogen (49, 86, 87). Additionally, given the apparent role that high levels of gp120 play in suppressing immune responses in vivo, the data may also contribute to the design of more efficacious vaccines and immunotherapies for HIV/AIDS.
Acknowledgments
We thank Dr. Binxia Wang from the Center for AIDS Research Biostatistics core at Harvard Medical School for her help with the statistical analyses of the data presented herein. We thank Prof. Norman Letvin for his generous donation of tissues from acutely SHIV-infected macaques, and Prof. Joseph Sodroski for his provision of SHIV-KB9 constructs and recombinant gp120 used in this study. We thank Dr. Igor Bagayev for his help with the confocal imaging. We also acknowledge Professors Norman Letvin, R. Paul Johnson, and Ruth Ruprecht for their erudite advice and assistance with this manuscript.
Footnotes
M.C.P. and L.S. were supported by Public Health Service Grants RO1 AI49757 and T32A1007387, respectively.
- PB
- peripheral blood
- LN
- lymph node
- SIV
- simian immunodeficiency virus
- SHIV
- chimeric HIV-1/SIV virus
- Treg
- regulatory T cell
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr. HIV Res. 2003;1:15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- 4.Kaur A, Hale CL, Ramanujan S, Jain RK, Johnson RP. Differential dynamics of CD4+ and CD8+ T-lymphocyte proliferation and activation in acute simian immunodeficiency virus infection. J. Virol. 2000;74:8413–8424. doi: 10.1128/jvi.74.18.8413-8424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 6.Pahwa S, Chitnis V, Mitchell RM, Fernandez S, Chandrasekharan A, Wilson CM, Douglas SD. CD4+ and CD8+ T cell receptor repertoire perturbations with normal levels of T cell receptor excision circles in HIV-infected, therapy-naive adolescents. AIDS Res. Hum. Retroviruses. 2003;19:487–495. doi: 10.1089/088922203766774531. [DOI] [PubMed] [Google Scholar]
- 7.Kuroda MJ, Schmitz JE, Charini WA, Nickerson CE, Lifton MA, Lord CI, Forman MA, Letvin NL. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- 8.Altfeld M, Rosenberg ES. The role of CD4+ T helper cells in the cytotoxic T lymphocyte response to HIV-1. Curr. Opin. Immunol. 2000;12:375–380. doi: 10.1016/s0952-7915(00)00103-5. [DOI] [PubMed] [Google Scholar]
- 9.Brander C, Walker BD. T lymphocyte responses in HIV-1 infection: implications for vaccine development. Curr. Opin. Immunol. 1999;11:451–459. doi: 10.1016/S0952-7915(99)80076-4. [DOI] [PubMed] [Google Scholar]
- 10.Goulder PJ, Walker BD. The great escape: AIDS viruses and immune control. Nat. Med. 1999;5:1233–1235. doi: 10.1038/15184. [DOI] [PubMed] [Google Scholar]
- 11.Couillin I, Culmann-Penciolelli B, Gomard E, Choppin J, Levy JP, Guillet JG, Saragosti S. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 1994;180:1129–1134. doi: 10.1084/jem.180.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 13.Koenig S, Conley AJ, Brewah YA, Jones GM, Leath S, Boots LJ, Davey V, Pantaleo G, Demarest JF, Carter C, et al. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1995;1:330–336. doi: 10.1038/nm0495-330. [DOI] [PubMed] [Google Scholar]
- 14.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 15.Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, Ogunlesi AO, Elvin JG, Rothbard JA, Bangham CR, Rizza CR, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 16.Price DA, Goulder PJ, Klenerman P, Sewell AK, Easterbrook PJ, Troop M, Bangham CR, Phillips RE. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 18.Yang OO, Sarkis PT, Ali A, Harlow JD, Brander C, Kalams SA, Walker BD. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 2003;197:1365–1375. doi: 10.1084/jem.20022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pantaleo G, Soudeyns H, Demarest JF, Vaccarezza M, Graziosi C, Paolucci S, Daucher M, Cohen OJ, Denis F, Biddison WE, et al. Evidence for rapid disappearance of initially expanded HIV-specific CD8+ T cell clones during primary HIV infection. Proc. Natl. Acad. Sci. USA. 1997;94:9848–9853. doi: 10.1073/pnas.94.18.9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zinkernagel RM. Immunology taught by viruses. Science. 1996;271:173–178. doi: 10.1126/science.271.5246.173. [DOI] [PubMed] [Google Scholar]
- 21.Algeciras A, Dockrell DH, Lynch DH, Paya CV. CD4 regulates susceptibility to Fas ligand- and tumor necrosis factor-mediated apoptosis. J. Exp. Med. 1998;187:711–720. doi: 10.1084/jem.187.5.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brainard DM, Tharp WG, Granado E, Miller N, Trocha AK, Ren XH, Conrad B, Terwilliger EF, Wyatt R, Walker BD, Poznansky MC. Migration of antigen-specific T cells away from CXCR4-binding human immunodeficiency virus type 1 gp120. J. Virol. 2004;78:5184–5193. doi: 10.1128/JVI.78.10.5184-5193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbeil J, Richman DD. Productive infection and subsequent interaction of CD4-gp120 at the cellular membrane is required for HIV-induced apoptosis of CD4+ T cells. J. Gen. Virol. 1995;76:681–690. doi: 10.1099/0022-1317-76-3-681. [DOI] [PubMed] [Google Scholar]
- 24.Finkel TH, Tudor-Williams G, Banda NK, Cotton MF, Curiel T, Monks C, Baba TW, Ruprecht RM, Kupfer A. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat. Med. 1995;1:129–134. doi: 10.1038/nm0295-129. [DOI] [PubMed] [Google Scholar]
- 25.Laurent-Crawford AG, Krust B, Riviere Y, Desgranges C, Muller S, Kieny MP, Dauguet C, Hovanessian AG. Membrane expression of HIV envelope glycoproteins triggers apoptosis in CD4 cells. AIDS Res. Hum. Retroviruses. 1993;9:761–773. doi: 10.1089/aid.1993.9.761. [DOI] [PubMed] [Google Scholar]
- 26.Westendorp MO, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debatin KM, Krammer PH. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- 27.Pantaleo G, Soudeyns H, Demarest JF, Vaccarezza M, Graziosi C, Paolucci S, Daucher M, Cohen OJ, Denis F, Biddison WE, et al. Accumulation of human immunodeficiency virus-specific cytotoxic T lymphocytes away from the predominant site of virus replication during primary infection. Eur. J. Immunol. 1997;27:3166–3173. doi: 10.1002/eji.1830271213. [DOI] [PubMed] [Google Scholar]
- 28.Karlsson GB, Halloran M, Li J, Park IW, Gomila R, Reimann KA, Axthelm MK, Iliff SA, Letvin NL, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J. Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klasse PJ, Moore JP. Is there enough gp120 in the body fluids of HIV-1-infected individuals to have biologically significant effects? Virology. 2004;323:1–8. doi: 10.1016/j.virol.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Stevceva L, Alvarez X, Lackner AA, Tryniszewska E, Kelsall B, Nacsa J, Tartaglia J, Strober W, Franchini G. Both mucosal and systemic routes of immunization with the live, attenuated NYVAC/simian immunodeficiency virus SIVgpe recombinant vaccine result in gag-specific CD8+ T-cell responses in mucosal tissues of macaques. J. Virol. 2002;76:11659–11676. doi: 10.1128/JVI.76.22.11659-11676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevceva L, Moniuszko M, Alvarez X, Lackner AA, Franchini G. Functional simian immunodeficiency virus Gag-specific CD8+ intraepithelial lymphocytes in the mucosae of SIVmac251- or simian-human immunodeficiency virus KU2-infected macaques. Virology. 2004;319:190–200. doi: 10.1016/j.virol.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 33.Conley AJ, Gorny MK, Kessler JA, 2nd, Boots LJ, Ossorio-Castro M, Koenig S, Lineberger DW, Emini EA, Williams C, Zolla-Pazner S. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 1994;68:6994–7000. doi: 10.1128/jvi.68.11.6994-7000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- 36.Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- 37.Nyambi PN, Gorny MK, Bastiani L, van der Groen G, Williams C, Zolla-Pazner S. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 1998;72:9384–9391. doi: 10.1128/jvi.72.11.9384-9391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolla-Pazner S, O’Leary J, Burda S, Gorny MK, Kim M, Mascola J, McCutchan F. Serotyping of primary human immunodeficiency virus type 1 isolates from diverse geographic locations by flow cytometry. J. Virol. 1995;69:3807–3815. doi: 10.1128/jvi.69.6.3807-3815.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent KA, Gritz L, Stallard G, Cranage MP, Collignon C, Thiriart C, Corcoran T, Silvera P, Stott EJ. Production and of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. AIDS. 1991;5:829–836. doi: 10.1097/00002030-199107000-00006. [DOI] [PubMed] [Google Scholar]
- 40.Ball JM, Rushlow KE, Issel CJ, Montelaro RC. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J. Virol. 1992;66:732–742. doi: 10.1128/jvi.66.2.732-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeVico A, Montelaro RC, Gallo RC, Sarngadharan MG. Purification and partial characterization of equine infectious anemia virus reverse transcriptase. Virology. 1991;185:387–394. doi: 10.1016/0042-6822(91)90786-b. [DOI] [PubMed] [Google Scholar]
- 42.Hussain KA, Issel CJ, Schnorr KL, Rwambo PM, Montelaro RC. Antigenic analysis of equine infectious anemia virus (EIAV) variants by using monoclonal antibodies: epitopes of glycoprotein gp90 of EIAV stimulate neutralizing antibodies. J. Virol. 1987;61:2956–2961. doi: 10.1128/jvi.61.10.2956-2961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghanekar SA, Nomura LE, Suni MA, Picker LJ, Maecker HT, Maino VC. Gamma interferon expression in CD8+ T cells is a marker for circulating cytotoxic T lymphocytes that recognize an HLA A2-restricted epitope of human cytomegalovirus phosphoprotein pp65. Clin. Diagn. Lab. Immunol. 2001;8:628–631. doi: 10.1128/CDLI.8.3.628-631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hel Z, Nacsa J, Kelsall B, Tsai WP, Letvin N, Parks RW, Tryniszewska E, Picker L, Lewis MG, Edghill-Smith Y, et al. Impairment of Gag-specific CD8+ T-cell function in mucosal and systemic compartments of simian immunodeficiency virus mac251- and simian-human immunodeficiency virus KU2-infected macaques. J. Virol. 2001;75:11483–11495. doi: 10.1128/JVI.75.23.11483-11495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kern F, Surel IP, Brock C, Freistedt B, Radtke H, Scheffold A, Blasczyk R, Reinke P, Schneider-Mergener J, Radbruch A, et al. T-cell epitope mapping by flow cytometry. Nat. Med. 1998;4:975–978. doi: 10.1038/nm0898-975. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 1995;69:4413–4422. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Ingmundson A, Horner SM, Cicchetti G, Allen PG, Pypaert M, Cunningham JM, Mothes W. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 49.Toapanta FR, Craigo JK, Montelaro RC, Ross TM. Reduction of anti-HIV-1 Gag immune responses during co-immunization: immune interference by the HIV-1 envelope. Curr. HIV Res. 2007;5:199–209. doi: 10.2174/157016207780077057. [DOI] [PubMed] [Google Scholar]
- 50.Berke G. The binding and lysis of target cells by cytotoxic lymphocytes: molecular and cellular aspects. Annu. Rev. Immunol. 1994;12:735–773. doi: 10.1146/annurev.iy.12.040194.003511. [DOI] [PubMed] [Google Scholar]
- 51.Henkart PA. Apoptosis: O death, where is thy sting? J. Immunol. 1995;154:4905–4908. [PubMed] [Google Scholar]
- 52.Kagi D, Ledermann B, Burki K, Zinkernagel RM, Hengartner H. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu. Rev. Immunol. 1996;14:207–232. doi: 10.1146/annurev.immunol.14.1.207. [DOI] [PubMed] [Google Scholar]
- 53.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Im- munol. Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 54.Baecher-Allan C, Viglietta V, Hafler DA. Human CD4+CD25+ regulatory T cells. Semin. Immunol. 2004;16:89–98. doi: 10.1016/j.smim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 55.Jonuleit H, Schmitt E. The regulatory T cell family: distinct subsets and their interrelations. J. Immunol. 2003;171:6323–6327. doi: 10.4049/jimmunol.171.12.6323. [DOI] [PubMed] [Google Scholar]
- 56.Mills KH. Regulatory T cells: friend or foe in immunity to infection? Nat. Rev. Immunol. 2004;4:841–855. doi: 10.1038/nri1485. [DOI] [PubMed] [Google Scholar]
- 57.Rouse BT, Suvas S. Regulatory cells and infectious agents: detentes cordiale and contraire. J. Immunol. 2004;173:2211–2215. doi: 10.4049/jimmunol.173.4.2211. [DOI] [PubMed] [Google Scholar]
- 58.Hartigan-O’Connor DJ, Poon C, Sinclair E, McCune JM. Human CD4+ regulatory T cells express lower levels of the IL-7 receptor alpha chain (CD127), allowing consistent identification and sorting of live cells. J. Immunol. Methods. 2007;319:41–52. doi: 10.1016/j.jim.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 59.Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, Gottlieb PA, Kapranov P, Gingeras TR, Fazekas de St Groth B, Clayberger C, Soper DM, Ziegler SF, Bluestone JA. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ Treg cells. J. Exp. Med. 2006;203:1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nilsson J, Boasso A, Velilla PA, Zhang R, Vaccari M, Franchini G, Shearer GM, Andersson J, Chougnet C. HIV-1 driven regulatory T cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. 2006;108:3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Romagnani S. Regulation of the T cell response. Clin. Exp. Allergy. 2006;36:1357–1366. doi: 10.1111/j.1365-2222.2006.02606.x. [DOI] [PubMed] [Google Scholar]
- 62.Mempel TR, Pittet MJ, Khazaie K, Weninger W, Weissleder R, von Boehmer H, von Andrian UH. Regulatory T cells reversibly suppress cytotoxic T cell function independent of effector differentiation. Immunity. 2006;25:129–141. doi: 10.1016/j.immuni.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 63.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gea-Banacloche JC, Migueles SA, Martino L, Shupert WL, McNeil AC, Sabbaghian MS, Ehler L, Prussin C, Stevens R, Lambert L, et al. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 2000;165:1082–1092. doi: 10.4049/jimmunol.165.2.1082. [DOI] [PubMed] [Google Scholar]
- 65.Poli G, Pantaleo G, Fauci AS. Immunopathogenesis of human immunodeficiency virus infection. Clin. Infect. Dis. 1993;17(Suppl. 1):S224–S229. [PubMed] [Google Scholar]
- 66.Brodie SJ, Lewinsohn DA, Patterson BK, Jiyamapa D, Krieger J, Corey L, Greenberg PD, Riddell SR. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat. Med. 1999;5:34–41. doi: 10.1038/4716. [DOI] [PubMed] [Google Scholar]
- 67.Lieberman J, Shankar P, Manjunath N, Andersson J. Dressed to kill? A review of why antiviral CD8 T lymphocytes fail to prevent progressive immunodeficiency in HIV-1 infection. Blood. 2001;98:1667–1677. doi: 10.1182/blood.v98.6.1667. [DOI] [PubMed] [Google Scholar]
- 68.McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980–987. doi: 10.1038/35073658. [DOI] [PubMed] [Google Scholar]
- 69.Holm GH, Gabuzda D. Distinct mechanisms of CD4+ and CD8+ T-cell activation and bystander apoptosis induced by human immunodeficiency virus type 1 virions. J. Virol. 2005;79:6299–6311. doi: 10.1128/JVI.79.10.6299-6311.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams MA, Trout R, Spector SA. HIV-1 gp120 modulates the immunological function and expression of accessory and co-stimulatory molecules of monocyte-derived dendritic cells. J. Hematother. Stem Cell Res. 2002;11:829–847. doi: 10.1089/152581602760404630. [DOI] [PubMed] [Google Scholar]
- 71.Lopez D, Samino Y, Koszinowski UH, Del Val M. HIV envelope protein inhibits MHC class I presentation of a cytomegalovirus protective epitope. J. Immunol. 2001;167:4238–4244. doi: 10.4049/jimmunol.167.8.4238. [DOI] [PubMed] [Google Scholar]
- 72.Fenoglio D, Li Pira G, Lozzi L, Bracci L, Saverino D, Terranova P, Bottone L, Lantero S, Megiovanni A, Merlo A, Manca F. Natural analogue peptides of an HIV-1 GP120 T-helper epitope antagonize response of GP120-specific human CD4 T-cell clones. J. Acquired Immune Defic. Syndr. 2000;23:1–7. doi: 10.1097/00126334-200001010-00001. [DOI] [PubMed] [Google Scholar]
- 73.Diamond DC, Sleckman BP, Gregory T, Lasky LA, Greenstein JL, Burakoff SJ. Inhibition of CD4+ T cell function by the HIV envelope protein, gp120. J. Immunol. 1988;141:3715–3717. [PubMed] [Google Scholar]
- 74.Ruegg CL, Monell CR, Strand M. Inhibition of lymphoproliferation by a synthetic peptide with sequence identity to gp41 of human immunodeficiency virus type 1. J. Virol. 1989;63:3257–3260. doi: 10.1128/jvi.63.8.3257-3260.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinhold KJ, Lyerly HK, Stanley SD, Austin AA, Matthews TJ, Bolognesi DP. HIV-1 GP120-mediated immune suppression and lymphocyte destruction in the absence of viral infection. J. Immunol. 1989;142:3091–3097. [PubMed] [Google Scholar]
- 76.Di Rienzo AM, Furlini G, Olivier R, Ferris S, Heeney J, Montagnier L. Different proliferative response of human and chimpanzee lymphocytes after contact with human immunodeficiency virus type 1 gp120. Eur. J. Immunol. 1994;24:34–40. doi: 10.1002/eji.1830240106. [DOI] [PubMed] [Google Scholar]
- 77.Heeney J, Bogers W, Buijs L, Dubbes R, ten Haaft P, Koornstra W, Niphuis H, Nara P, Teeuwsen V. Immune strategies utilized by lentivirus infected chimpanzees to resist progression to AIDS. Immunol. Lett. 1996;51:45–52. doi: 10.1016/0165-2478(96)02554-0. [DOI] [PubMed] [Google Scholar]
- 78.Aandahl EM, Michaelsson J, Moretto WJ, Hecht FM, Nixon DF. Human CD4+ CD25+ regulatory T cells control T-cell responses to human immunodeficiency virus and cytomegalovirus antigens. J. Virol. 2004;78:2454–2459. doi: 10.1128/JVI.78.5.2454-2459.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kinter AL, Hennessey M, Bell A, Kern S, Lin Y, Daucher M, Planta M, McGlaughlin M, Jackson R, Ziegler SF, Fauci AS. CD25+CD4+ regulatory T cells from the peripheral blood of asymptomatic HIV-infected individuals regulate CD4+ and CD8+ HIV-specific T cell immune responses in vitro and are associated with favorable clinical markers of disease status. J. Exp. Med. 2004;200:331–343. doi: 10.1084/jem.20032069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiss L, Donkova-Petrini V, Caccavelli L, Balbo M, Carbonneil C, Levy Y. Human immunodeficiency virus-driven expansion of CD4+ CD25+ regulatory T cells, which suppress HIV-specific CD4 T-cell responses in HIV-infected patients. Blood. 2004;104:3249–3256. doi: 10.1182/blood-2004-01-0365. [DOI] [PubMed] [Google Scholar]
- 81.Weiner HL. Induction and mechanism of action of transforming growth factor-β-secreting Th3 regulatory cells. Immunol. Rev. 2001;182:207–214. doi: 10.1034/j.1600-065x.2001.1820117.x. [DOI] [PubMed] [Google Scholar]
- 82.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+ CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors: central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 84.Blick G, Kagan RM, Coakley E, Petropoulos C, Maroldo L, Greiger-Zanlungo P, Gretz S, Garton T. The probable source of both the primary multidrug-resistant (MDR) HIV-1 strain found in a patient with rapid progression to AIDS and a second recombinant MDR strain found in a chronically HIV-1-infected patient. J. Infect. Dis. 2007;195:1250–1259. doi: 10.1086/512240. [DOI] [PubMed] [Google Scholar]
- 85.Markowitz M, Mohri H, Mehandru S, Shet A, Berry L, Kalyanaraman R, Kim A, Chung C, Jean-Pierre P, Horowitz A, et al. Infection with multidrug resistant, dual-tropic HIV-1 and rapid progression to AIDS: a case report. Lancet. 2005;365:1031–1038. doi: 10.1016/S0140-6736(05)71139-9. [DOI] [PubMed] [Google Scholar]
- 86.Peut V, Kent SJ. Utility of human immunodeficiency virus type 1 envelope as a T-cell immunogen. J. Virol. 2007;81:13125–13134. doi: 10.1128/JVI.01408-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]