Abstract
Kinesin-12 (also called KIF15) is a microtubule-based motor protein best known for its role in cell division. We previously reported that kinesin-12 is robustly expressed in developing terminally post-mitotic neurons, with levels diminishing as neurons reach maturity. We found that axons of cultured rodent neurons grow faster and longer if kinesin-12 is experimentally depleted, leading us to conclude that kinesin-12 plays a role in modulating axonal growth. Here we used zebrafish to explore whether these results apply to an in vivo system and whether they apply across different kinds of vertebrates. In whole mount in situ hybridization, kinesin-12 mRNA was detectable at 2-cell and 1K-cell stages. At 5.3 and 8 hours post-fertilization (hpf), hybridization signal for kinesin-12 mRNA was observed in the ectoderm. From 14 to 36 hpf, the signal had expanded to the central nervous system. At 60 hpf, the hybridization signal was concentrated in the brain. After 5 days post-fertilization, kinesin-12 expression was reduced. Kinesin-12 knockdown resulted in notably longer fast-growing axons with fewer branches by injection of a splice-blocking morpholino into Tg(huC:egfp) or Tg(hb9:gfp) zebrafish embryos. Kinesin-12 overexpression resulted in shorter axons than controls. These results are consistent with our previous observations on rodents using primary cultures for the experimental manipulations, and suggest a key role of kinesin-12 as a modulator of axonal development.
Keywords: kinesin-12, microtubule, axon, zebrafish, in situ hybridization
Introduction
During development, the microtubule cytoskeleton is essential to the morphogenesis of an organism (Dong, Deng et al. 2011, Pramparo, Libiger et al. 2011). The processes of neurogenesis, neuronal migration and polarization are key to in the embryonic development of the nervous system, and all rely on microtubule function (Chuckowree and Vickers 2003, Kawauchi and Hoshino 2008, Geraldo and Gordon-Weeks 2009, Poulain and Sobel 2010, Pramparo, Libiger et al. 2011, Sakakibara, Ando et al. 2013). The kinesins, which comprise a superfamily of microtubule-based motor proteins, are best known for their roles in transporting vesicles along microtubules, and also in regulating the organization and movements of microtubules themselves in the mitotic spindle apparatus. While vesicle transport is generally considered the main job of microtubule motors in terminally post-mitotic neurons, our work over the past several years implicates kinesins as robust candidates for coordinating and restructuring the microtubule network at the growth cone and axonal shaft during development and also after nerve injury (Liu, Nadar et al. 2010, Lin, Liu et al. 2011, Lin, Liu et al. 2012). In this way, the “mitotic” kinesins serve functions in neurons that are repurposed from their roles in cell division.
Kinesin-12, also called KIF15, KSNL7 or HKLP2, is a microtubule-based plus-end-directed kinesin best known for its role in mitosis. Kinesin-12 has been studied in recent years in terms of its structure and function, with the idea in mind that kinesin-12 might be a useful target for cancer therapy (Drechsler, McHugh et al. 2014, Klejnot, Falnikar et al. 2014). We previously investigated the expression of kinesin-12 in the developing nervous system of rats, and found kinesin-12 is detectable at high levels in both cortex and ganglia at embryonic stages, but progressively diminishes as neurons develop and mature (Liu, Nadar et al. 2010). Using cultured rat neurons, we found that depletion of kinesin-12 affects axonal growth, navigation, and branching, and more recent studies indicate a role for kinesin-12 in dendrite morphology (Lin, Liu et al. 2012) and neuronal migration (Klejnot, Falnikar et al. 2014).
Here we wished to explore two questions, the first being whether the functional results observed in cell culture apply to an intact organism, and the second being whether the role of kinesin-12 in nervous system development is conserved across vertebrate species. To explore these questions, we conducted studies on kinesin-12 in the developing nervous system of zebrafish.
Materials and methods
Zebrafish tissue and embryos
Zebrafish were provided by the Zebrafish Center at Nantong University Jiangsu Key Laboratory of Neuroregeneration. Zebrafish embryos were obtained through natural mating (AB line) and maintained at 28.5°C. Stages of embryonic zebrafish have been previously described (Kimmel, Ballard et al. 1995). Embryos after 24 hours post-fertilization (hpf) were treated with 0.2 mM 1-phenyl-2-thio-urea (PTU, a tyrosinase inhibitor commonly used to block pigmentation and aid visualization of zebrafish development). Zebrafish embryos were collected at various stages, fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) overnight at 4°C or 2 h at room temperature, washed with PBST (PBS plus 0.1% Tween-20), dehydrated in methanol and stored at -20°C until use. Embryos younger than 24 hpf were dechorionated after fixation, prior to storage.
Bioinformatics
The zebrafish kinesin-12 (KIF15) exon information was obtained from Ensemble (http://www.ensembl.org/Danio_rerio/Transcript/Exons?g=ENSDARG00000012073;r=25:36128940-36155903;t=ENSDART00000099866). Predicted protein molecular weight was calculated by the Protein Molecular Weight Calculator (http://www.sciencegateway.org/tools/proteinmw.htm). Conserved domains of the kinesin-12 proteins were localized by the Pfam database (http://pfam.sanger.ac.uk/). Kinesin-12 sequences were aligned by the ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The phylogenetic tree was built by MEGA5 software. Exon-specific primers for real-time PCR were designed by the Primer 3 software online (http://frodo.wi.mit.edu/).
RNA extraction, reverse transcription, and RT-PCR
Tissue was homogenized and frozen in TRIzol Reagent (Invitrogen) and stored at -80°C. Total RNA was extracted following the manufacturer’s instructions. 1 μg of RNA was reverse-transcribed into cDNA by the use of Transcriptor First Strand cDNA Synthesis Kit (Roche) according to the manufacturer’s instructions. Synthesized cDNA was stored at -20°C. The primers for RT-PCR are listed: Left primer, 5′-atcaggcgccaaattttgta-3′; Right primer, 5′-atgtttttagccacgctgga-3′. PCR amplifications were carried out in a total volume of 50μL using specific primers and Advantage2 Polymerase Kit (Clontech).
Riboprobe synthesis and whole-mount in situ hybridization
The 732 bp coding sequence for zebrafish kinesin-12 (GeneBank XM_002666923) was amplified by PCR using the following primers: Left primer, 5′-tgtgctgctggagttaatgc-3′; Right primer, 5′-ttttgtgcgttgcttttctg-3′. Digoxigenin (DIG)-labeled RNA sense and antisense probes were made from the linearized plasmids according to the manufacturer’s protocol using the DIG RNA Labeling Kit (SP6/T7) (Roche). The procedure for in situ hybridization followed our protocol (Huang, Wang et al. 2013) which was modified from a previous study (Thisse and Thisse 2008). The small baskets were not used in our protocol. BM purple AP substrate (Roche) was used instead of the staining solution. We use the BBR (Boehringer blocking reagent, Roche) for blocking.
Morpholino, mRNA synthesis and microinjection
The transgenic zebrafish lines of Tg(huC:egfp) and Tg(hb9:gfp) which was kindly offered by Dr. Jiulin Du (Institute of Neuroscience and State Key Laboratory of Neuroscience) were maintained in the zebrafish center of Jiangsu Key Laboratory of Neuroregeneration. The Morpholinos (MOs) were synthesized by Gene Tools Company. MO antisense oligomers were prepared at a stock concentration of 1 mM according to the manufacturer’s protocol. The sequence of zebrafish kinesin-12 splicing MO in this study was 5′-atg tattaaaaacctcacctggctg -3′ and the standard control MO was 5′-cctcttacctcagttacaatttata -3′. To generate mRNA, zebrafish kinesin-12 was cloned into the PCS2+ vector. Cloning primer sequences were listed below: Left primer: 5′-cgggatccatgaatcttaaaggcaaagcaac-3′, Right primer: 5′-cgg aattctcaagggtgagtgtgttgtagg -3′. Sense-capped mRNA was generated by SP6 mMessagemMachine (Life Technology). MOs or mRNA were injected into the yolk of one-cell stage embryos using borosilicate glass capillaries (world precision Inc. WPI) with PV830 Pneumatic picopump (WPI).
Imaging
At 30hpf or 48hpf, for confocal imaging of neuronal development in Tg(huC:egfp) or Tg(hb9:gfp) zebrafish, embryos were anesthetized with egg water/0.16 mg/mL tricaine/1% 1-phenyl-2-thiourea (Sigma) and embedded in 1% agarose. Images were acquired with an Olympus DP671 camera on an Olympus stereo microscope. Using a 40x objective, confocal stack images of the trunk region were obtained in time intervals of 30 minutes. Confocal imaging was performed with a Leica TCS-SP5 LSM. Analysis was performed using Imaris software.
Statistics
Neurite length between two branches was measured by image J software. The branch number within 100μm of axon length was calculated. All data analysis, statistical comparisons, and graphs were generated using Excel (Microsoft Corp) or GraphPad Prism 5. Data are expressed as mean ± S.E. Statistical analysis were performed using a two-tailed t test (P<0.05). The final figure processing was performed with Adobe Illustrator CS6.
Results
Kinesin-12 is highly conserved across vertebrates
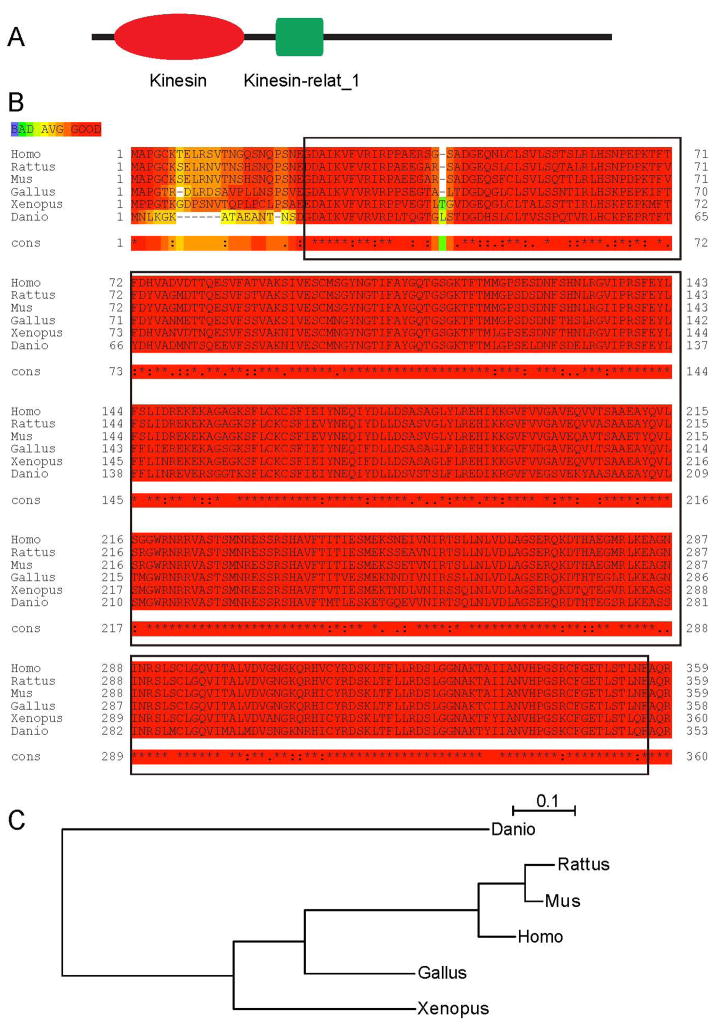
The zebrafish kinesin-12 gene localizes on chromosome 25, according to the zebrafish genome (http://www.ensembl.org, Zv9, Ensembl release 72 - June 2013). Based on the sequence, the gene should encode for a 1378 amino acid protein (NCBI Reference Sequence: XP_002666969.1, GI: 292628456), with a predicted molecular weight of 157 kDa. Kinesin-12 protein contains two conserved domains, kinesin-12 head (motor domain) and kinesin-12-relat_1 (Fig. 1A). Alignment analysis demonstrated an orthologous relationship of zebrafish kinesin-12 to other vertebrate kinesin-12 proteins. The alignment results of zebrafish kinesin-12 with the five matched sequences are displayed in Table 1. The sequences of kinesin-12 functional domains in different species of vertebrates share significant similarities (Fig. 1B). A phylogenetic analysis of several vertebrate kinesin-12 proteins shows that Danio rerio kinesin-12 is most closely related to Xenopus laevis kinesin-12, followed by avian kinesin-12, and has more distant relationships to murine (mice and rats) and other mammalian kinesin-12 (Fig. 1C). The conservation of functional domains of kinesin-12 across vertebrate evolution suggests functional importance of these domains to the protein.
Figure 1.
Functional domains of kinesin-12 are highly conserved during vertebrates. (A)The kinesin-12 sequence is comprised of two conserved domains: kinesin-12_head (motor domain) showed in red, and kinesin-12 stalk showed in green. (B)Alignment of kinesin-12 motor domain amino acid residue sequences of Danio rerio (XP_002666969.1), Xenopus laevis (NP_001081543.1), Gallus gallus (XP_418807.2), Mus musculus (NP_034750.1), Rattus norvegicus (NP_853666.1) and Homo sapiens (NP_064627.1) kinesin-12. The sequences are retrieved from NCBI Protein sequence database. The sequences accessions IDs are labeled in brackets. These protein sequences were aligned using ClustalW2 program. This domain sequence is indicated in red rectangle.(C) Phylogenetic tree of amino acid sequences generated by using the PhyML software (Guindon et al., 2010).
Table 1.
The alignment results of zebrafish kinesin-12 with that of other vertebrate kinesin-12 protein sequences.
| Species | Identities | Positives |
|---|---|---|
| Danio – Xenopus | 258/333 (80 %) | 293/333 (87 %) |
| Danio – Gallus | 264/333 (79 %) | 294/333 (88 %) |
| Danio – Mus | 254/333 (76 %) | 285/333 (85 %) |
| Danio – Rattus | 256/333 (77 %) | 287/333 (86 %) |
| Danio – Homo | 258/333 (77 %) | 291/333 (87 %) |
Kinesin-12 expression in early zebrafish development
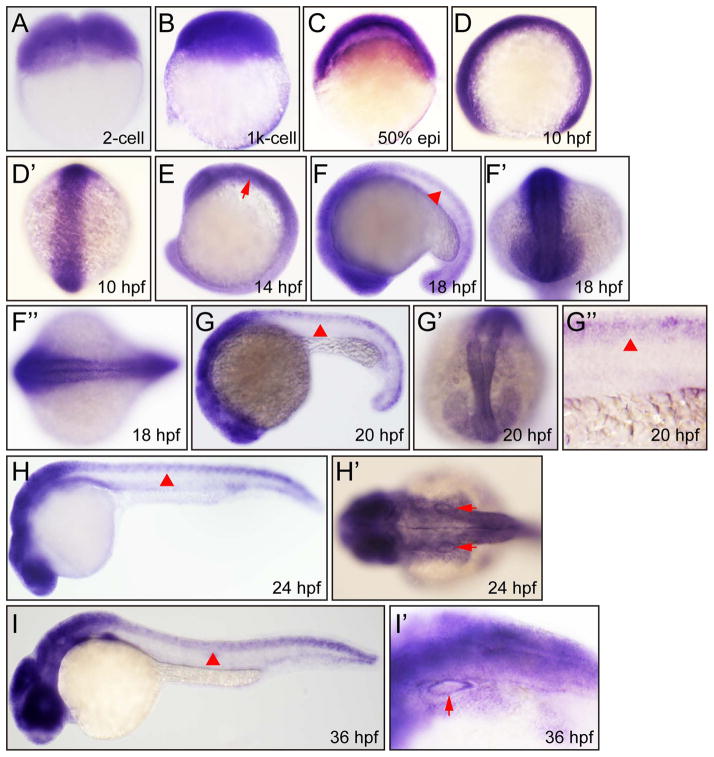
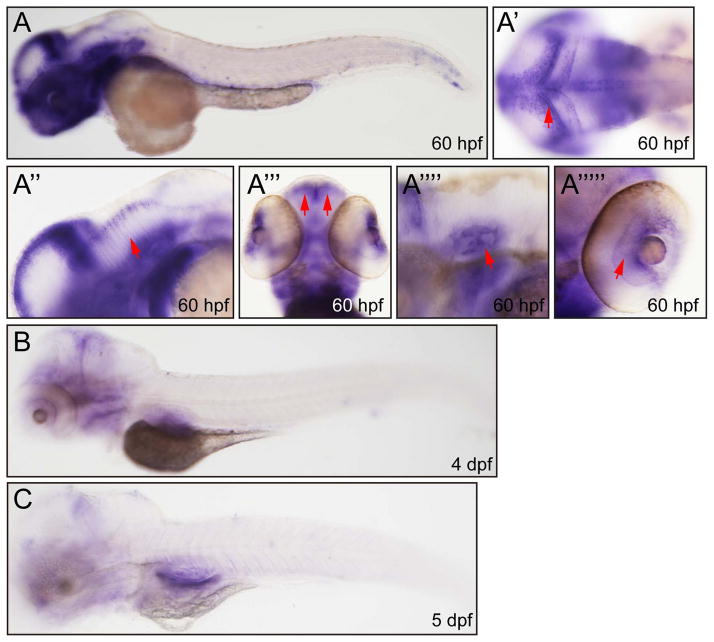
In order to determine spatial distribution of kinesin-12 mRNA, we performed whole mount in situ hybridization (ISH) analyses with kinesin-12-specific antisense probes. The ISH signals were detectable and strong at 2-cell, 1K-cell, 50% epiboly and 10 hpf (Fig. 2A–D′). By 14 hpf to 36hpf, the expression of kinesin-12 had expanded to central nervous system, including telencephalon, cerebellum, diencephalon, hindbrain, and whole spinal cord (Fig. 2E–H, I). At 60 hpf, the expression of kinesin-12 was concentrated in brain, but the ISH signal was not detectable in the spinal cord at this stage (Fig. 3A). In addition, signal was detected in the otic vesicle, retina, fin and peripheral olfactory organ (Fig. 2H′,I′, Fig. 3A″ - A’’’’’). At 4 days post-fertilization (dpf) and 5 dpf, the expression level of kinesin-12 was strongly reduced (Fig. 3B, C). The corresponding sense RNA probe was used at embryos of several stages as negative controls and they produced no detectable signal under the same hybridization and detection conditions (data not shown).
Figure 2.
Whole mount in situ hybridization analysis of zebrafish embryos (2 cell–36 hpf) using antisense Danio rerio kinesin-12 probe. (A) 2 cells, lateral view. (B) 1k cells, lateral view. (C) 50% epiboly, lateral view. (D) 10 hpf, lateral view. (D′) 10 hpf, dorsal view. (E) 14 hpf, lateral view, hind brain (arrow). (F) 18 hpf, lateral view, spinal cord (arrowhead). (F′) 18 hpf, dorsal view. (F″) 18 hpf, dorsal view. (G) 20 hpf, lateral view, spinal cord (arrowhead). (G′) 20 hpf, dorsal view. (G″) 20 hpf, lateral view, spinal cord (arrowhead). (H) 24 hpf, lateral view, spinal cord (arrowhead). (H′) 24 hpf, dorsal view, otic vesicle (arrow). (I) 36 hpf, lateral view, spinal cord (arrowhead). (I′) 36 hpf, lateral view, otic vesicle (arrow).
Figure 3.
Whole mount in situ hybridization analysis of zebrafish embryos (60 hpf–5 dpf) using antisense Danio rerio kinesin-12 probe. (A) 60hpf, lateral view. (A′) 60hpf, dorsal view, brain (arrow). (A″) 60 hpf, lateral view, brain (arrow). (A‴) 60 hpf, dorsal view, olfactory bulb (arrow). (A’’’’) 60 hpf, lateral view, otic vesicle (arrow). (A’’’’’) 60 hpf, dorsal view, retina, (arrow). (B) 4dpf, lateral view. (C) 5dpf, lateral view.
Kinesin-12 impacts neuronal morphology
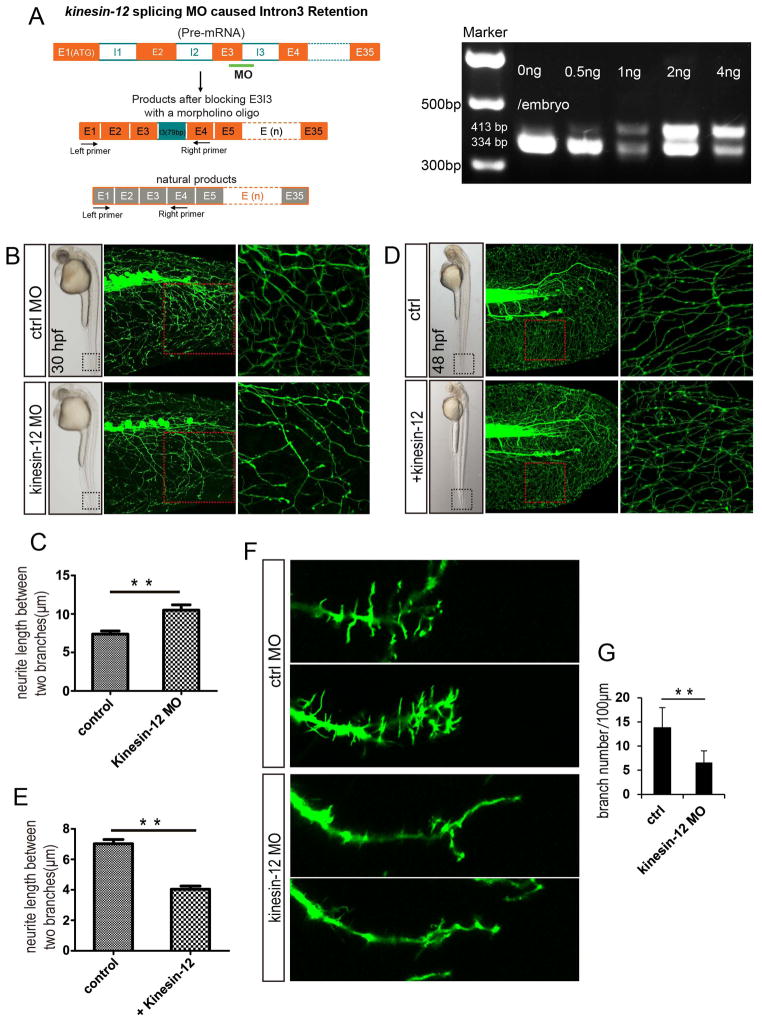
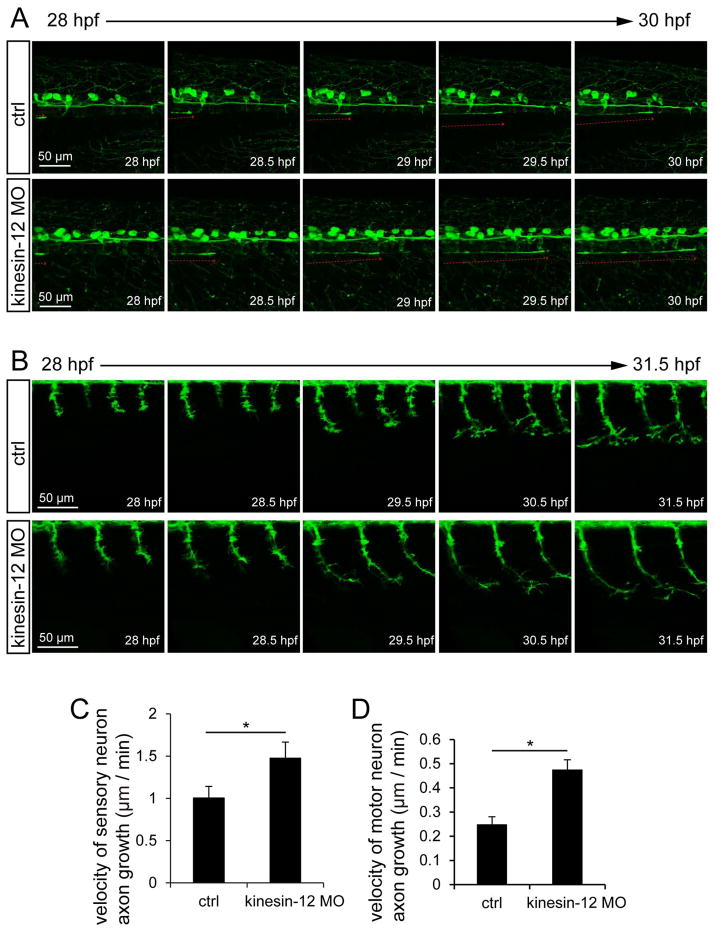
Kinesin-12 was knocked down by injecting a splice-blocking MO into Tg(huC:egfp) or Tg(hb9:gfp) zebrafish embryos. The effects of splice-blocking MO were characterized and quantitated by RT-PCR (Fig. 4A). Kinesin-12 mRNA reading frame shift caused by MO was confirmed by sequencing. For the splice-blocking MO, effects were observed at a dose of 0.5ng, the morphogenesis of the embryo is nearly the same as the control; at higher dosages, we observed dysmorphogenesis of the embryo, and these were therefore not examined in detail. We observed striking changes in neuronal morphology of kinesin-12 morphants, displaying notably longer axons and also less branched (Fig. 4B, C, F, G). Quantification of the morphological changes shown in Figures 4C revealed an almost 42% increase in axonal length, compared to the control. The effect of kinesin-12 overexpression on the neuronal morphology was generally opposite (Fig. 4D, E), resulting in a 43% decrease. We imaged axonal growth in kinesin-12 morphants. We chose a particular neuron, which is likely to be a Rohon-Beard neuron based on previous descriptions (Sato, Takahoko et al. 2006) and with this neuron, we observed a 50% increase in the velocity of axonal growth relative to control (Fig. 5A, C). (Note: While we did not confirm with absolute certainty the identity of this neuron, the comparison with control makes for a valid illustration of the affects of kinesin-12 knockdown). This effect on the motor neuron is similar (Fig. 5B, D). Quantification of the velocity changes shown in Fig. 5D, revealed an almost 88% increase, and the axonal branches were 52% diminished (Fig. 4G), relative to the control, respectively (see figure legend for statistics). Representative movies of the different MO treatments are shown in Movies ctrl MO and kinesin-12 MO; see movie legends for details. Additional details on morphology (such as changes in filopodial length) are described in figure legends.
Figure 4.
Effects of kinesin-12 on the neuronal morphology. (A) Kinesin-12 splicing MO caused intron3 retention. RT-PCR analysis of the effects of splice-blocking MO. Injection of kinesin-12 splice-blocking MO caused intron3 (79 bp) retained in kinesin-12 mRNA, resulting in reading frame shift. (B–C) Injection of kinesin-12 splice-blocking MO into Tg(huC:egfp) zebrafish embryos results in longer filopodia at 30hpf, and the quantification of neurite length revealed significantly longer filopodia compared to those embryos injected control MO. Data are represented as mean ± SE, control MO = 7.4 ± 0.4 μm, n = 56; kinesin-12 MO = 10.5 ± 0.7 μm, n = 38 (** P < 0.01). (D–E) Overexpression of kinesin-12 had an opposite effect compared to the kinesin-12 morphants at 48hpf, and the quantification of filopodia length revealed overexpression of kinesin-12 had an opposite effects compared to the kinesin-12 morphants. Control= 7.0 ± 0.2 μm, n = 33; kinesin-12 overexpression = 4.0 ± 0.2 μm, n = 29 (**P < 0.01). (F–G) Tg (hb9:gfp) marks motor neuron. Injection of kinesin-12 splicing MO into Tg(hb9:gfp) zebrafish embryos. At 30hpf, quantification of motor neuron showed significantly fewer branches in kinesin-12 morphants, control MO = 13.8 ± 1.8/100 μm, kinesin-12 MO = 6.6 ± 1.1/100 μm (**P<0.01). The boxed regions are shown at higher magnification in the right hand panels.
Figure 5.
Effects of Kinesin-12 on axon growth (A, C) Injection of kinesin-12 splice-blocking MO into Tg(huC:egfp) zebrafish embryos, time-lapse in vivo imaging shows higher velocity of axon growth of kinesin-12 morphants from 28 hpf to 30 hpf, control MO = 1.0 ± 0.1 μm/min, kinesin-12 MO = 1.5 ± 0.1 μm/min (n=5, *P < 0.05). The red line represents a neuron which we believe to be Rohon-Beard (but see text for more details). (B, D) Injection of the MO into Tg(hb9:gfp) zebrafish embryos, time-lapse in vivo imaging shows that the effect on velocity change of the motor neuron, control MO = 0.25 ± 0.01 μm/min, kinesin-12 MO = 0.47 ± 0.02 μm/min (n=5, *P < 0.05).
Discussion
Kinesin-12 is thought to contribute to the maintenance of the bipolar microtubule spindle apparatus in dividing cells by regulating/modulating the cross-linking and sliding of microtubules relative to one another (Tanenbaum, Macurek et al. 2009, Vanneste, Takagi et al. 2009, Florian and Mayer 2011). There appears to be overlap in function to some extent with kinesin-5, the motor originally reported to form homotetramers, such that the presence of kinesin-12 can sometimes compensate when kinesin-5 is suppressed (Tanenbaum, Macurek et al. 2009). Recent studies suggest that kinesin-12 can also form tetramers (Drechsler, McHugh et al. 2014). In our work on rodents, we found that kinesin-12 is strongly expressed in the nervous system during development, but that expression diminishes as neurons mature (Liu, Nadar et al. 2010). We found that the axonal phenotype of kinesin-12 depletion is similar to that of kinesin-5 depletion in cultured rodent neurons, but not identical. Depletion of either of these motor proteins yielded longer axons, explicable at least in part by an increase in the transport of microtubules. Depletion of kinesin-12 yielded fewer axonal branches and smaller growth cones, while depletion of kinesin-5 yielded the opposite (Liu et al., 2010), probably because kinesin-12 is able to influence the actin cytoskeleton via a myosin-like domain that is not present in kinesin-5 (Buster, Baird et al. 2003). The neuronal migration phenotype (faster migration) was also similar between kinesin-5 and kinesin-12 depletion (Falnikar, Tole et al. 2011, Klejnot, Falnikar et al. 2014). The dendritic phenotype of kinesin-12 depletion, however, was more similar to the phenotype of kinesin-6 depletion (Lin, Liu et al. 2012).
Do these results apply outside of the culture dish, and across vertebrate species? Our results reported here on zebrafish suggest that the answer to both questions is yes, at least in terms of kinesin-12’s influence on the axon. Kinesin-12 is strongly expressed in developing neurons in zebrafish in a generally similar timetable as in rodent neurons, with the expression diminishing as the neurons mature. Depletion of kinesin-12 results in longer axons with fewer branches, while overexpression produces the inverse phenotype. Additional studies will be required to ascertain whether kinesin-12 plays roles in neuronal migration and dendritic development in vivo, as indicated by the cell culture work. For now, the present studies provide the first in vivo data that confirm the long-standing hypothesis that motor proteins originally believed to be mitosis-specific also have crucial roles to play in terminally post-mitotic neurons.
We envision the microtubules of the neuron as structures bearing various forces imposed upon them by a panoply of molecular motor proteins. These would include cytoplasmic dynein, the mitotic kinesins, as well as actomyosin-based forces. Given the versatility of molecular motors, we would not dismiss the possibility of even the non-mitotic kinesins, which are mainly thought to transport vesicles, potentially participating in the mixture of complementary and opposing forces that push and pull on the microtubules to organize them, transport them and integrate them with other microtubules as well as with the actin cytoskeleton. The fact that notable phenotypes can be achieved by inhibiting just one molecular motor suggests that whatever overlapping motor forces may exist are not sufficient to fully compensate for the loss of that one motor, at least in the case of kinesin-12. As anti-cancer drugs are developed to inhibit mitotic kinesins, caution should be taken to ensure that such drugs to do adversely affect neurons. On a positive note, if there are sufficient levels of kinesin-12 in adult neurons, or if kinesin-12 levels are up-regulated in response to injury, inhibition or depletion of kinesin-12 may prove therapeutically useful as a means for clinically augmenting axonal growth as a treatment for adult nerve injury. We plan on pursuing this.
Supplementary Material
Injection of control MO into Tg(huC:egfp) zebrafish embryos, time-lapse in vivo imaging shows lower velocity of axon growth of control morphants from 28 hpf to 30 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Injection of kinesin-12 splice-blocking MO into Tg(huC:egfp) zebrafish embryos, time-lapse in vivo imaging shows higher velocity of axon growth of kinesin-12 morphants from 28 hpf to 30 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Injection of control MO into Tg(hb9:gfp) zebrafish embryos, time-lapse in vivo imaging shows lower velocity of axon growth of control morphants from 28 hpf to 31.5 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Movie2-MO: Injection of kinesin-12 splice-blocking MO into Tg(hb9:gfp) zebrafish embryos, time-lapse in vivo imaging shows higher velocity of axon growth of kinesin-12 morphants from 28 hpf to 31.5 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Acknowledgments
This study was supported by grants to ML and DL from the National Natural Science Foundation of China (31171007; 31201083), grants from Natural science foundation of Education Department of Jiangsu Province (11KJA180004, 12KJB180010), funding from the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, and a grant to PWB from the National Institutes of Health (R01 NS028785).
References
- Buster DW, Baird DH, Yu WQ, Solowska JM, Chauviere M, Mazurek A, Kress M, Baas PW. Expression of the mitotic kinesin Kif15 in postmitotic neurons: Implications for neuronal migration and development. Journal of Neurocytology. 2003;32(1):79–96. doi: 10.1023/a:1027332432740. [DOI] [PubMed] [Google Scholar]
- Chuckowree JA, Vickers JC. Cytoskeletal and morphological alterations underlying axonal sprouting after localized transection of cortical neuron Axons in vitro. Journal of Neuroscience. 2003;23(9):3715–3725. doi: 10.1523/JNEUROSCI.23-09-03715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Deng W, Jiang D. Distinct cytoskeleton populations and extensive crosstalk control Ciona notochord tubulogenesis. Development. 2011;138(8):1631–1641. doi: 10.1242/dev.057208. [DOI] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. The Kinesin-12 Kif15 is a processive track-switching tetramer. Elife. 2014;3:e01724. doi: 10.7554/eLife.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnikar A, Tole S, Baas PW. Kinesin-5, a mitotic microtubule-associated motor protein, modulates neuronal migration. Mol Biol Cell. 2011;22(9):1561–1574. doi: 10.1091/mbc.E10-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle. 2011;10(20):3533–3544. doi: 10.4161/cc.10.20.17817. [DOI] [PubMed] [Google Scholar]
- Geraldo S, Gordon-Weeks PR. Cytoskeletal dynamics in growth-cone steering. Journal of Cell Science. 2009;122(20):3595–3604. doi: 10.1242/jcs.042309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang XQ, Wang X, Xu M, Liu M, Liu D. Nonmuscle myosin II-B (myh10) expression analysis during zebrafish embryonic development. Gene Expression Patterns. 2013;13(7):265–270. doi: 10.1016/j.gep.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Developmental Neuroscience. 2008;30(1–3):36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klejnot M, Falnikar A, Ulaganathan V, Cross RA, Baas PW, Kozielski F. The crystal structure and biochemical characterization of Kif15: a bifunctional molecular motor involved in bipolar spindle formation and neuronal development. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 1):123–133. doi: 10.1107/S1399004713028721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu M, Mozgova OI, Yu W, Baas PW. Mitotic Motors Coregulate Microtubule Patterns in Axons and Dendrites. Journal of Neuroscience. 2012;32(40):14033–14049. doi: 10.1523/JNEUROSCI.3070-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Liu M, Son YJ, Timothy Himes B, Snow DM, Yu W, Baas PW. Inhibition of Kinesin-5, a microtubule-based motor protein, as a strategy for enhancing regeneration of adult axons. Traffic. 2011;12(3):269–286. doi: 10.1111/j.1600-0854.2010.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, V, Nadar C, Kozielski F, Kozlowska M, Yu W, Baas PW. Kinesin-12, a Mitotic Microtubule-Associated Motor Protein, Impacts Axonal Growth, Navigation, and Branching. Journal of Neuroscience. 2010;30(44):14896–14906. doi: 10.1523/JNEUROSCI.3739-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain FE, Sobel A. The microtubule network and neuronal morphogenesis: Dynamic and coordinated orchestration through multiple players. Molecular and Cellular Neuroscience. 2010;43(1):15–32. doi: 10.1016/j.mcn.2009.07.012. [DOI] [PubMed] [Google Scholar]
- Pramparo T, Libiger O, Jain S, Li H, Youn YH, Hirotsune S, Schork NJ, Wynshaw-Boris A. Global Developmental Gene Expression and Pathway Analysis of Normal Brain Development and Mouse Models of Human Neuronal Migration Defects. Plos Genetics. 2011;7(3) doi: 10.1371/journal.pgen.1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A, Ando R, Sapir T, Tanaka T. Microtubule dynamics in neuronal morphogenesis. Open biology. 2013;3(7):130061–130061. doi: 10.1098/rsob.130061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44(3):136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 Cooperates with Eg5 to Promote Bipolar Spindle Assembly. Current Biology. 2009;19(20):1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature Protocols. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The Role of Hklp2 in the Stabilization and Maintenance of Spindle Bipolarity. Current Biology. 2009;19(20):1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Injection of control MO into Tg(huC:egfp) zebrafish embryos, time-lapse in vivo imaging shows lower velocity of axon growth of control morphants from 28 hpf to 30 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Injection of kinesin-12 splice-blocking MO into Tg(huC:egfp) zebrafish embryos, time-lapse in vivo imaging shows higher velocity of axon growth of kinesin-12 morphants from 28 hpf to 30 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Injection of control MO into Tg(hb9:gfp) zebrafish embryos, time-lapse in vivo imaging shows lower velocity of axon growth of control morphants from 28 hpf to 31.5 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.
Movie2-MO: Injection of kinesin-12 splice-blocking MO into Tg(hb9:gfp) zebrafish embryos, time-lapse in vivo imaging shows higher velocity of axon growth of kinesin-12 morphants from 28 hpf to 31.5 hpf. Images were acquired with a Leica TCS-SP5 LSM confocal microscope and processed by Imaris software.