Abstract
Our understanding of mammalian olfactory coding has been impeded by the paucity of information about the odorant receptors (ORs) that respond to a given odorant ligand in awake, freely behaving animals. Identifying the ORs that respond in vivo to a given odorant ligand from among the ∼1100 ORs in mice is intrinsically challenging but critical for our understanding of olfactory coding at the periphery. Here, we report an in vivo assay that is based on a novel gene-targeted mouse strain, S100a5–tauGFP, in which a fluorescent reporter selectively marks olfactory sensory neurons that have been activated recently in vivo. Because each olfactory sensory neuron expresses a single OR gene, multiple ORs responding to a given odorant ligand can be identified simultaneously by capturing the population of activated olfactory sensory neurons and using expression profiling methods to screen the repertoire of mouse OR genes. We used this in vivo assay to re-identify known eugenol- and muscone-responsive mouse ORs. We identified additional ORs responsive to eugenol or muscone. Heterologous expression assays confirmed nine eugenol-responsive ORs (Olfr73, Olfr178, Olfr432, Olfr610, Olfr958, Olfr960, Olfr961, Olfr913, and Olfr1234) and four muscone-responsive ORs (Olfr74, Olfr235, Olfr816, and Olfr1440). We found that the human ortholog of Olfr235 and Olfr1440 responds to macrocyclic ketone and lactone musk odorants but not to polycyclic musk odorants or a macrocyclic diester musk odorant. This novel assay, called the Kentucky in vivo odorant ligand–receptor assay, should facilitate the in vivo identification of mouse ORs for a given odorant ligand of interest.
Keywords: cell sorting, expression profiling, G-protein coupled receptor, odor detection, olfaction, sensory coding
Introduction
Mammals can detect numerous odorants, and humans may be able to discriminate as many as 1 trillion different odorant mixtures (Bushdid et al., 2014). Remarkably, this enormous capacity for sensory detection and discrimination is not achieved at the expense of specificity. For example, mice have been shown to discriminate every pair of odorant enantiomers tested thus far (Laska and Shepherd, 2007). Underlying this discriminatory power is the expression of a single allele of one odorant receptor (OR) or trace amine-associated receptor (Taar) gene in a mature olfactory sensory neuron (OSN; Mombaerts, 2004; Johnson et al., 2012). In addition, this singularity of expression allows receptor-specific axonal inputs to coalesce into glomeruli of the olfactory bulb (Mombaerts et al., 1996), an anatomical organization that contributes to maximizing the capacity for odor discrimination.
The discriminatory power of olfaction rivals that of the visual and auditory systems, but the patterns of receptor activation by odorant ligands remain elusive. Resolution of this problem has been hampered by the sheer number of ORs (∼1100 intact OR genes in the mouse) and the retention of ORs in the endoplasmic reticulum of heterologous cells as opposed to native mature OSNs (McClintock et al., 1997; Gimelbrant et al., 2001; Lu et al., 2003; Dalton et al., 2013). Nevertheless, odorant ligands for >100 mammalian ORs have been identified. These data indicate that a typical OR is capable of being activated by several structurally related odorants, with some receptors more narrowly and others more broadly tuned (Malnic et al., 1999; Malnic, 2007; Grosmaitre et al., 2009; Kato and Touhara, 2009; Saito et al., 2009; Nara et al., 2011). However, translating these data into an understanding of in vivo OR activation by odorants is difficult. To advance and refine these ideas further will require in vivo assays that go beyond the characterization of individual ORs (Zhao et al., 1998; Malnic et al., 1999; Kajiya et al., 2001; Oka et al., 2006; Shirasu et al., 2014) to assay instead simultaneously the entire repertoire of ORs so that sets of ORs activated by odorants may be identified.
Here, we demonstrate the ability of a novel in vivo assay in mice, supported by heterologous expression data, to identify ORs that respond to a given odorant ligand. With this approach, we identified or re-identified nine eugenol-responsive ORs, including all four previously identified eugenol-responsive ORs, and four muscone-responsive ORs, including the sole OR known previously to respond to muscone. Eugenol is a major component of the oil of several spice plants, especially the clove plant, Szygium aromaticum. Muscone is a large macrocyclic odorant discovered in scent glands of male deer of the family Moschidae and has a long history of use in fragrances (Kraft and Fráter, 2001). The mouse ORs responsive to muscone successfully predicted a human OR that we find to be strongly responsive to macrocyclic ketone and macrocyclic lactone musk odorants.
Materials and Methods
Materials.
Sigma-Aldrich was the source of eugenol [4-allyl-2-methoxyphenol (catalog #35995)], mineral oil (catalog #M5904), Tonalid [Chemical Abstracts Service (CAS) 21145-77-7; catalog #W526401], Astrotone (ethylene brassylate; CAS 105-95-3; catalog #W354309), Exaltone (cyclopentadecanone; CAS 502-72-7; catalog #C111201), and isopropyl myristate (IPM; CAS 110-27-0; catalog #172472). Muscone (racemic mixture, 50% in IPM; CAS 10403-00-6) was purchased from Perfumer's Apprentice. Galaxolide (CAS 1222-05-5) was obtained from International Flavors & Fragrances. Mineral oil was the vehicle for eugenol and IPM for muscone.
S100a5–tauGFP mouse strain.
Using 129/SvJ genomic DNA and the GeneAmp XL PCR kit (Life Technologies), DNA segments flanking the coding exons of S100A5 were cloned after PCR amplification. The upstream arm was amplified using primers 5′-TATATGCGGCCGCTGCCATGATGTGCAATGAATTCTTTGAGGG and 5′-TAGGTGGCGCGCCGATATGTACCCTGGACTAGGAGAGAGGACAATCAC. The downstream arm was amplified using primers 5′-TATATGGCGCGCCAGGACACTGGCAGCTCCTGATCCTG and 5′-CGCGCGTTAATTAAGTGGCTGTCCAACAACGTGGTAAGACCAGG. An upstream 4 kb NotI-Asc1 fragment and a downstream 3.8 kb Asc1–PacI fragment were cloned into pBS–SKII that had been modified to accept these sites. An Asc1 fragment consisting of IRES–tauGFP and a loxP–pgk–neomycin–loxP sequence was subcloned into the Asc1 site. A clone of the correct orientation was identified by sequencing. The targeting vector was linearized with PmeI and electroporated into E14 embryonic stem cells that were cultured and selected using G418 as described previously (Mombaerts et al., 1996). Homologous recombination in embryonic stem cell colonies and in mutant mice was determined by PCR using primers 5′-AGCTTTGGCTCCCATCCACGGTG and 5′-TGTTGGACAGCCAGAGGGTCCCC. The pgk–neomycin fragment was removed by breeding heterozygotes to EIIa–Cre transgenic mice (Lakso et al., 1996), and the Cre recombinase transgene was subsequently removed by intercrossing S100a5–tauGFP mice hemizygous for the transgene. S100a5–tauGFP mice were in a mixed 129 × C57BL/6J background. This strain is publicly available from The Jackson Laboratory as stock number 6709, official strain name B6;129P2-S100a5<tm1Mom>/MomJ. Mice homozygous or heterozygous for the S100a5–tauGFP mutation (hereafter referred to as S100a5–tauGFP mice) have similar numbers of GFP-positive (GFP+) OSNs in the same mosaic pattern across the main olfactory epithelium, 22 ± 8/mm along the olfactory epithelium for homozygotes and 32 ± 5/mm for heterozygotes in 10-μm-thick tissue sections (n = 5; p = 0.0741). Both genotypes were used interchangeably in the experiments described herein. As a precaution for hidden effects of genotype, heterozygous and homozygous mice were distributed equally between vehicle control and odorant treatments. All procedures with mice were done according to protocols approved by the Institutional Animal Care and Use Committees of Rockefeller University and the University of Kentucky.
Odor stimulation in vivo.
Exposing live mice to odor or vehicle was performed with groups of eight S100a5–tauGFP mice of both sexes aged 7–12 weeks. Each mouse was housed individually in chambers under a flow of 1.5 l/min filtered air as described previously (Fischl et al., 2014) for 40 h without food but with ad libitum distilled water. This chamber system minimizes ambient odor sufficiently such that the effect of odors on activity-dependent genes in OSNs, which cannot be detected when introduced in a standard cage environment, can be measured (Fischl et al., 2014). This protocol also minimizes the effects of previous odor exposure by allowing for protein turnover to reduce GFP levels. (Naris occlusion data show that GFP fluorescence is significantly reduced by 24 h and reaches a minimum by 48 h.) After a period of 26 h of filtered air in these chambers, intermittent odor exposure was initiated and performed for a period of 14 h. This 14 h period allows odor-stimulated GFP fluorescence to develop (preliminary experiments revealed that increased GFP expression can be detected as early as 6 h) but is too brief for GFP fluorescence to fade (25% reduction in fluorescence 24 h after naris occlusion). Electronically controlled valves were activated for 10 s to divert the flow of air through the headspace of 30 ml tubes containing 200 μl of odorant (a 1:1 mix of odorant and vehicle) or vehicle alone. This switch was activated every 5 min until the completion of the experiment 14 h later. There were thus 168 odor exposures lasting 10 s each, or 28 min total exposure to the odorant ligand over the course of 14 h. Four mice were exposed to the odorant, whereas the other four were simultaneously exposed to vehicle. When mice from two litters were used, litters were distributed equally between the two treatment groups.
RNA isolation and measurement.
At the completion of odor exposure, olfactory mucosae were dissected and cells dissociated in a procedure involving papain, trypsin, deoxyribonuclease, and low calcium saline as described previously (Yu et al., 2005; Sammeta et al., 2007). Cells from four identically treated mice were pooled, and fluorescence-activated cell sorting (FACS) was performed using an iCyt Synergy cell sorting system (Sony) to collect GFP+ and GFP-negative (GFP−) cell samples in the University of Kentucky Flow Cytometry and Cell Sorting Facility. Total RNA was isolated using the Qiagen RNeasy Micro kit (catalog #74004), and samples were pooled until the GFP+ sample reached at least 50 ng of RNA, which required 8–16 mice per replicate of each treatment. RNA quantity was measured using Affymetrix Mouse GeneChip 1.0 ST arrays in the University of Kentucky Microarray Facility. This microarray contains 1176 probe sets for mRNAs from ORs and Taars, including all of the 1098 intact OR genes and 14 Taar genes. Data were initially processed using Affymetrix GeneChip Command Console software to generate normalized quantities for each OR and Taar transcript cluster. Additional processing to generate GFP+/GFP− ratios was done in Microsoft Excel. No OR or Taar transcript clusters were consistently in the bottom 10% of signals or below 2 SDs of the mean signal for all transcript clusters, so all OR and Taar transcript clusters were used in these analyses. The microarray data are available in the Gene Expression Omnibus under the accession number GSE59336.
Statistical analyses.
The stability of OR GFP+/GFP− ratios makes it possible to screen the entire set of ORs and Taars and identify activated receptors using a relatively small number of replications. We used a Bayesian hierarchical model to obtain normalized measures of odorant effect, accounting for four sources of variation: (1) basal receptor effect, (2) odorant effect, (3) nonspecific effect (change in both odorant and vehicle control), and (4) random measurement error. We find that 86% of the variation in the data are attributable to receptor identity and odorant, and only 14% are attributable to experimental noise. For each odorant effect, the posterior mean divided by the posterior SD provides a measure (Z-statistic) that is approximately normally distributed. Local false discovery rates (FDRs; Efron, 2008, 2010) were used to estimate the probability that each receptor is responsive to the odorant under the conservative assumption that the majority of the receptors are not responsive to the odorant. A 10% FDR was found to be a suitable level of risk for the identification of activated receptors.
Luciferase assay for OR function in Hana3A cells.
Heterologous expression of ORs was performed as described previously (Saito et al., 2004, 2009; Zhuang and Matsunami, 2007; Dey et al., 2011; Li and Matsunami, 2011; Duan et al., 2012). Briefly, this assay is based on OR-driven increases in cAMP to activate CREs that result in expression of firefly luciferase. Plasmids (pCI; Promega) containing cDNAs encoding ORs (C57BL/6 sequences) tagged with the N-terminal 20 aa of rhodopsin were transiently transfected along with RTPS1 (an OR chaperone), Renilla luciferase, CRE–firefly luciferase, and pRL–SV40 into Hana3A cells using Lipofectamine2000 (Life Technologies). Cells were plated into 96-well BioCoat plates (BD Biosciences). One day after transfection, the medium in each well was changed to CD293, a chemically defined medium from Life Technologies (catalog #11913 × 019), for 30 min at 37°C. This medium was then exchanged for 25 μl of CD293 containing either vehicle or the odorant ligand at concentrations ranging from 10−7 to 10−2 m and incubated for 4 h at 37°C and 5% CO2. Each concentration of odorant ligand was tested in triplicate wells. Luciferase activity was then measured using the Dual-Glo Luciferase Assay System (Promega) according to the protocols of the manufacturer. A Wallac Victor 1420 plate reader (PerkinElmer Life and Analytical Sciences) was used to measure luminescence. To control for transfection efficiency, the firefly luciferase luminescence of each well was divided by the Renilla luciferase activity of the well. Normalized luciferase activity was calculated as (LN − Lmin)/Lmax − Lmin), where LN is the corrected firefly luminescence, Lmax is the maximum firefly luminescence on the plate, and Lmin is the minimum firefly luminescence on the plate.
Results
The S100a5–tauGFP mouse strain
To screen for activation of ORs in native mouse OSNs in vivo, we devised an approach that is based on the activity-dependent expression of GFP from the S100a5 locus and the expression of one OR gene per OSN (Fig. 1).
Figure 1.
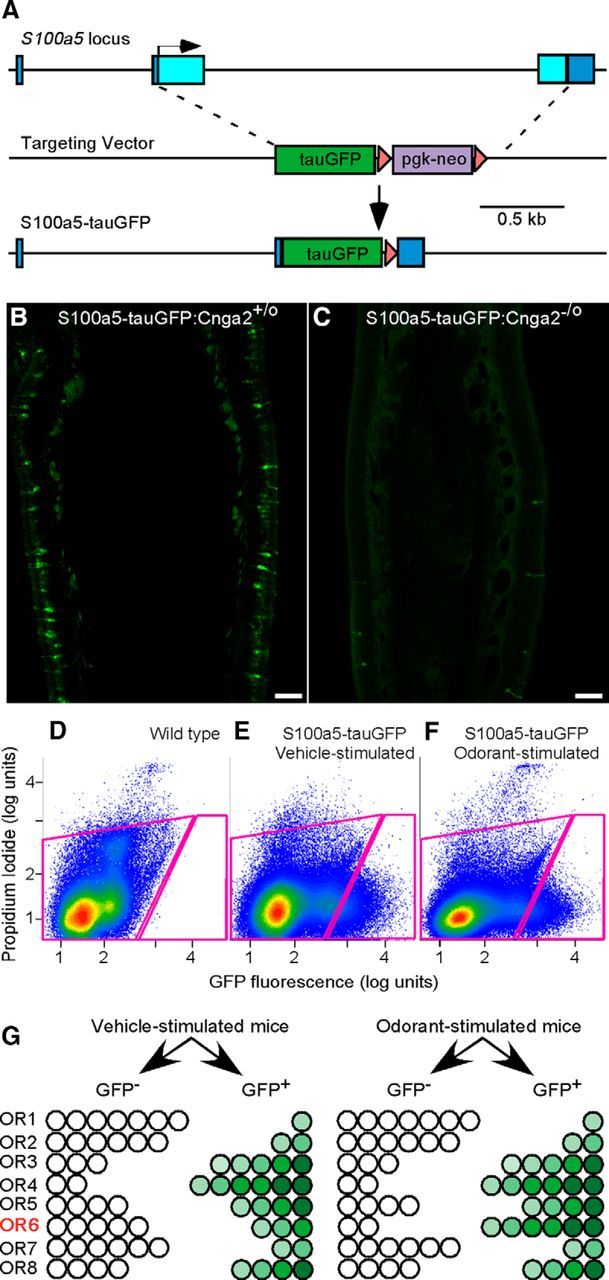
The gene-targeted S100a5–tauGFP mouse strain allows for activity-dependent fluorescent marking of OSNs. A, The design of the targeted mutation of the mouse S100a5 locus, replacing the coding exons with tauGFP. Boxes, Exons; light blue, coding sequence; dark blue, noncoding sequence; triangles, loxP sites; arrow, translation start site. B, Exposure for 4.5 s detects fluorescent OSNs in a 12 μm coronal section at midseptum, showing the mosaic pattern and variable levels of GFP expression in a male mouse, 25 d postnatal, that is wild type for the Cnga2 locus. C, Exposure for 49 s reveals a near absence of fluorescent OSNs in a Cnga2-deficient male mouse, 25 d postnatal. Scale bars, 100 μm. D, FACS of dissociated cells from olfactory mucosae of a wild-type C57BL/6 mouse is used to set sorting gates (circumscribed with magenta lines) for subsequent capture of GFP+ and GFP− cells from S100a5–tauGFP mice. E, F, FACS capture of GFP+ and GFP− cells from a set of four S100a5–tauGFP mice exposed to vehicle (E) and four mice exposed in parallel to muscone (F). G, In the in vivo assay, OSNs that exhibit GFP fluorescence above background are captured in the GFP+ sample. The distribution of each OSN subtype in GFP+ and GFP− FACS samples from S100a5–tauGFP mice exposed to an odorant ligand differs from that of S100a5–tauGFP mice exposed to vehicle only in those few OSN subtypes that express ORs responsive to the odorant ligand, as exemplified by OR6 in this diagram. Odorant-evoked activity in OR6-expressing OSNs results in an increase of the fraction of these OSNs with GFP fluorescence above background, thus causing a redistribution of these OSNs from the GFP− FACS sample into the GFP+ FACS sample. Differing intensities of GFP fluorescence above background exist among OSNs but are not relevant in this assay, which is based on a binary grouping of fluorescence intensities (GFP− vs GFP+).
The S100a5 gene encodes a 93 aa calcium- and zinc-binding protein (Schäfer et al., 2000; Streicher et al., 2010). The abundance of S100a5 mRNA in OSNs is dependent on odor stimulation (Serizawa et al., 2006; Bennett et al., 2010; Fischl et al., 2014). The S100a5 protein is located in both the dendritic cilia and the axons of OSNs (Schäfer et al., 2000; Kuhlmann et al., 2014). The biological function of S100a5 is not known. We generated a novel gene-targeted mouse strain with a tauGFP reporter replacing the two coding exons and intervening intron of the S100a5 gene (Fig. 1A), causing a deletion of the S100a5 gene. Thus far, we have not identified deficits in mice lacking S100a5: homozygous mice are indistinguishable from wild-type littermates. Consistent with the activity dependence of S100a5 expression, GFP expression from the mutant locus is reflected in a mosaic pattern of fluorescent OSNs in the main olfactory epithelium (Fig. 1B). GFP fluorescence is nearly undetectable in a Cnga2-deficient genetic background (Zheng et al., 2000), in which nearly all odorant-evoked electrical activity in OSNs is eliminated (Fig. 1C). Unlike the robust activity-dependent expression of S100a5 in OSNs, neither S100a5 mRNA in C57BL/6 mice nor GFP fluorescence in S100a5–tauGFP mice could be detected in vomeronasal sensory neurons (data not shown).
We used FACS to collect separately GFP-fluorescent (GFP+) OSNs and nonfluorescent cells (GFP−) from olfactory mucosae of S100a5–tauGFP mice that had been maintained in filtered air chambers for 40 h and exposed intermittently to odorant for the last 14 h of this period (Fig. 1D–F). Because the OSNs that respond to the odorant ligand tested represent only a small fraction of the total number of GFP+ OSNs, similar numbers of GFP+ OSNs are captured by FACS from mice exposed to vehicle (Fig. 1E) versus odorant (Fig. 1F). Because of the GFP-dependent capture of more OSNs expressing the few ORs that respond to the odorant ligand, the mRNAs encoding these responsive ORs are the only mRNAs that show an increase in abundance in the GFP+ FACS sample relative to the GFP− FACS sample (Fig. 1G). Odorant-specific effects are confirmed by the absence of effect on GFP+/GFP− ratios in samples from mice exposed to vehicle only. To identify responsive ORs in this assay, we measured their mRNAs via GeneChip microarray.
Eugenol-responsive mouse ORs
We find that, in the minimal odor conditions in which mice are exposed to vehicle only, each OR mRNA has a characteristic GFP+/GFP− ratio (Fig. 2A), which may result from the sum of the distinctive basal, constitutive activity of each OR (Imai et al., 2006; Reisert, 2010; Nakashima et al., 2013) and the effects of odors that may arise from the mouse itself or the chamber system. This characteristic GFP+/GFP− ratio is consistent across experiments: OR identity accounts for 86% of the variation in these data. This consistency is apparent when the mean of GFP+/GFP− ratios taken from four experiments is plotted against the four individual datasets of GFP+/GFP− ratios used to generate these mean values (Fig. 2A). When S100a5–tauGFP mice are exposed to an odorant ligand in the in vivo assay, only the mRNAs encoding the few responsive ORs will shift up from their characteristic positions near the line of equality, as is shown in Figure 2B for exposure to eugenol. The reason for this upward shift is that an odorant ligand acting as an agonist specifically elevates GFP fluorescence only among the OSNs expressing an OR that is activated by this odorant, thereby shifting more OSNs expressing this OR from the GFP− FACS sample into the GFP+ FACS sample. Because most mature OSNs express only one OR, statistically significant increases in the GFP+/GFP− ratio for OR mRNAs thereby allow for the simultaneous identification of the ORs that are activated by the odorant in question.
Figure 2.
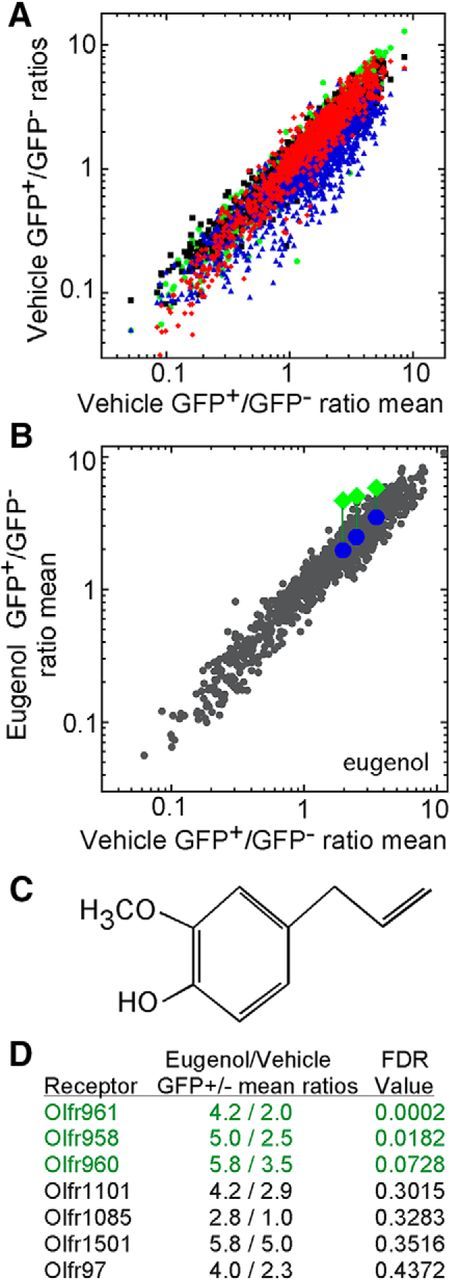
Eugenol exposure has significant effects on three ORs in S100a5–tauGFP mice. A, The distribution of OR mRNAs between GFP+ and GFP− samples is consistent across experiments. GFP+/GFP− ratios from mice exposed to vehicle (mineral oil) from four replications, distinguished by color (red, blue, green, and black), plotted against the mean GFP+/GFP− ratios of these four vehicle treatments. B, Mean OR mRNA GFP+/GFP− ratio values after exposure to eugenol plotted against the mean ratios after exposure to vehicle (n = 4 groups of mice). Three ORs (Olfr961, Olfr958, and Olfr960) show significant elevation (FDR < 0.10) after eugenol exposure. To illustrate the change, their GFP+/GFP− ratio values in this experiment (green diamonds) are compared with the GFP+/GFP− ratio coordinates in other experiments that did not use eugenol (blue circles). C, Chemical structure of eugenol. D, Mean GFP+/GFP− ratios and corresponding FDR probabilities for the seven ORs with FDR probabilities <0.5.
Eugenol (Fig. 2C) is known to activate at least four mouse ORs (Kajiya et al., 2001; Oka et al., 2006). After exposure to eugenol, the GFP+/GFP− ratios of three of the four known eugenol-responsive ORs were significantly elevated: Olfr961, Olfr958, and Olfr960 (Fig. 2B,D). When these three ORs were tested in the Hana3A heterologous expression assay (Saito et al., 2004, 2009; Zhuang and Matsunami, 2007; Dey et al., 2011; Li and Matsunami, 2011; Duan et al., 2012), cells separately expressing these three ORs responded robustly to eugenol (Fig. 3A–C). The fourth eugenol-responsive OR, Olfr73 (mouse olfactory receptor MOR174-9, mOR–EG), gave no increase in its GFP+/GFP− ratio in the in vivo assay, but heterologous cells expressing Olfr73 responded to eugenol (Fig. 3D).
Figure 3.
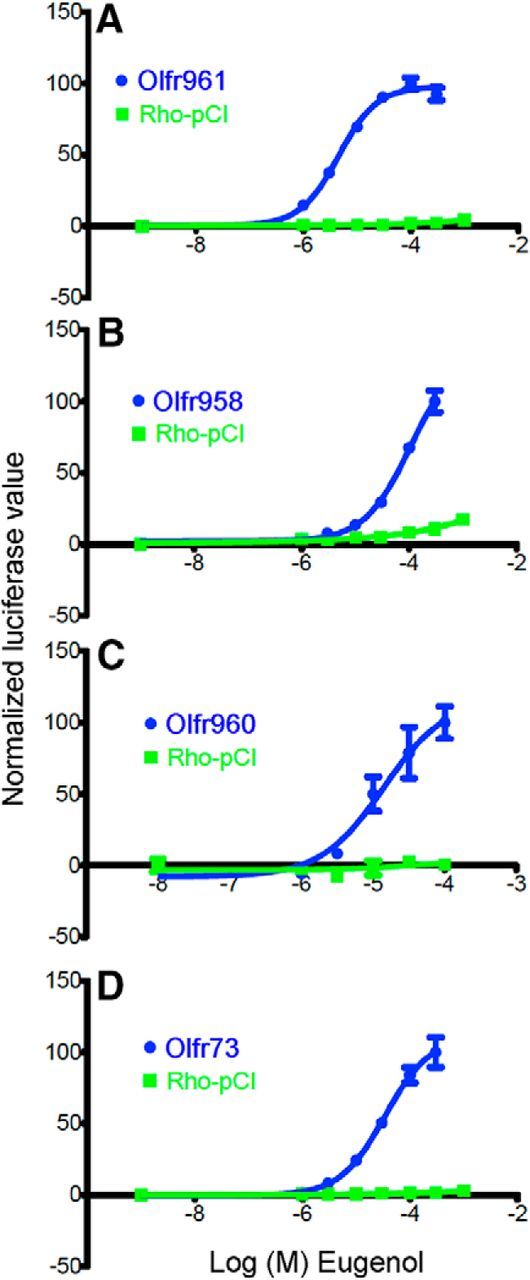
Heterologous expression assay data for ORs responsive to eugenol. A–C, Eugenol concentration–response relationships for Olfr961, Olfr958, and Olfr960, previously identified eugenol-responsive ORs that gave significant differences in the in vivo assay. D, Eugenol concentration–response relationship for Olfr73, the first eugenol-responsive OR to be identified (Kajiya et al., 2001).
Muscone-responsive mouse and human ORs
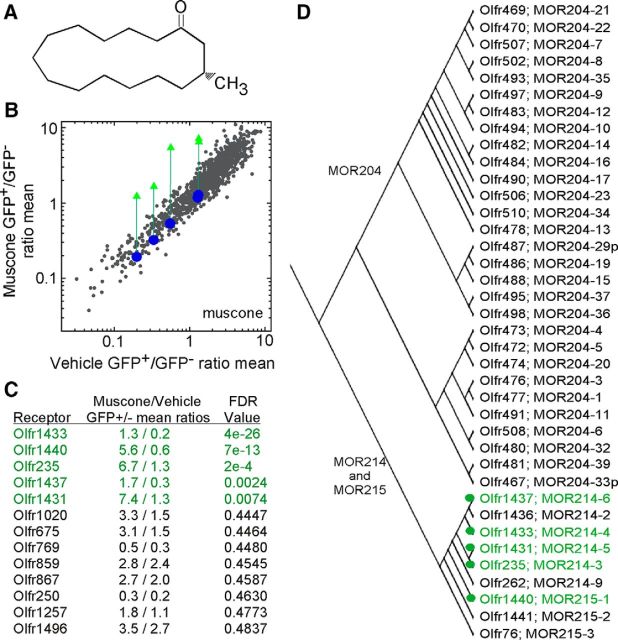
We next exposed S100a5–tauGFP mice to muscone, a macrocyclic ketone musk odorant (Fig. 4A). A single mouse OR, Olfr1440, has been shown recently to respond to muscone (Shirasu et al., 2014). We find that in vivo exposure to muscone has significant effects on five ORs, all from the MOR214 and MOR215 families (Fig. 4B–D). In vitro testing using the heterologous expression assay confirmed the ability of muscone to activate Olfr1440 and Olfr235 (Fig. 5). Limitations in the odorant concentrations that can be applied in this assay prevent these dose–response relationships from reaching saturation. Cells transfected with plasmids encoding Olfr1431, Olfr1437, and Olfr1433 did not respond to muscone (data not shown).
Figure 4.
Muscone exposure has significant effects on five ORs in S100a5–tauGFP mice. A, Chemical structure of muscone. B, Mean OR mRNA GFP+/GFP− ratios after exposure to muscone plotted against the mean ratios after exposure to vehicle (n = 4 groups of mice). Five ORs (Olfr1440, Olfr1433, Olfr235, Olfr1431, and Olfr1437) show significant differences (FDR < 0.10) after muscone exposure (green triangles) compared with vehicle exposure. Blue circles reflect the GFP+/GFP− ratio coordinates of these five ORs in other experiments that did not use muscone. C, Mean GFP+/GFP− ratios and corresponding FDR probabilities for the 13 ORs with FDR probabilities <0.5. D, A portion of the mouse OR phylogenetic tree depicting the relatedness of the members of the MOR214 and MOR215 mouse OR families that gave significant responses to muscone (green).
Figure 5.
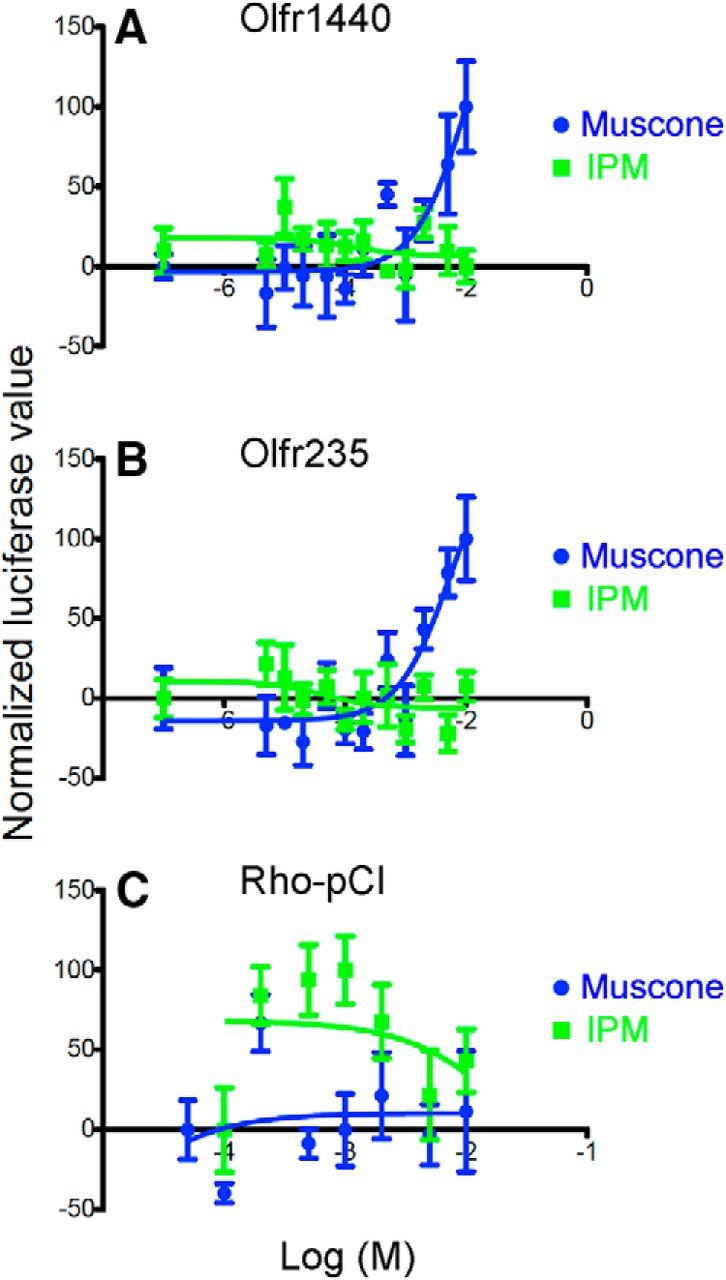
Confirmation of muscone responses from two mouse ORs by heterologous expression. A, Olfr1440. B, Olfr235. C, Rho-pCI, empty plasmid vector control. IPM used as vehicle.
Based on the sequence similarity between the muscone-responsive mouse ORs identified by our in vivo assay, we determined that the most similar human OR is OR5AN1: its percentage identity to the five mouse OR proteins ranges from 68% (Olfr1440) to 81% (Olfr1433). Predicting that OR5AN1 is responsive to musks, we tested both macrocyclic and polycyclic musk odorants against OR5AN1 in vitro. Hana3A cells expressing OR5AN1 responded to three musks: muscone and Exaltone, which are macrocyclic ketone musks, and Exaltolide, a macrocyclic lactone musk (Fig. 6A). OR5AN1 gave robust responses to these macrocyclic musks but did not give responses different from pCI plasmid vector controls when exposed to two polycyclic musks, Galaxolide and Tonalid, or to Astrotone, a macrocyclic diester musk (Fig. 6B–F), confirming and extending previous observations (Shirasu et al., 2014).
Figure 6.
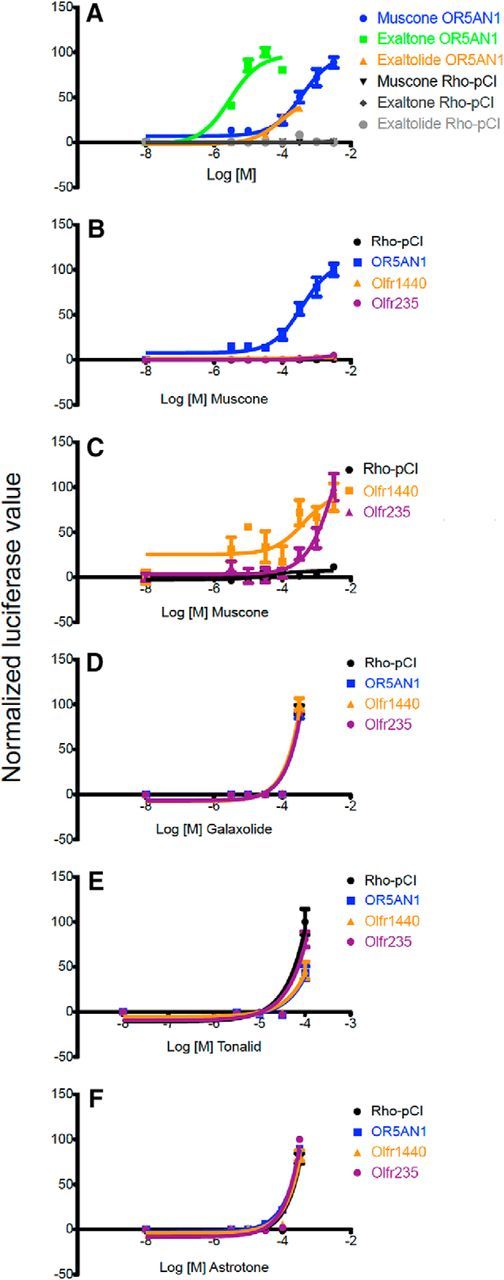
Activation of muscone-responsive mouse and human ORs by certain macrocyclic musk odorants in heterologous expression assays. A, OR5AN1, a human OR, responded to the macrocyclic ketone musks muscone and Exaltone and to the macrocyclic lactone musk Exaltolide. B, The OR5AN1 response to muscone is stronger than responses of mouse ORs. C, Olfr1440 and Olfr235 responses to muscone. D, E, Polycyclic musk odors fail to evoke responses different from the plasmid vector negative control, Rho-pCI, in cells transfected with OR5AN1 or the mouse ORs responsive to muscone. Data for some ORs cannot be seen because of overlap of the curves. F, Astrotone, a macrocyclic diester musk odorant, also fails to evoke responses different from the plasmid vector negative control, Rho-pCI, in cells transfected with OR5AN1 or the mouse ORs responsive to muscone. Data for some ORs cannot be seen because of overlap of the curves.
Weakly eugenol- or muscone-responsive mouse ORs
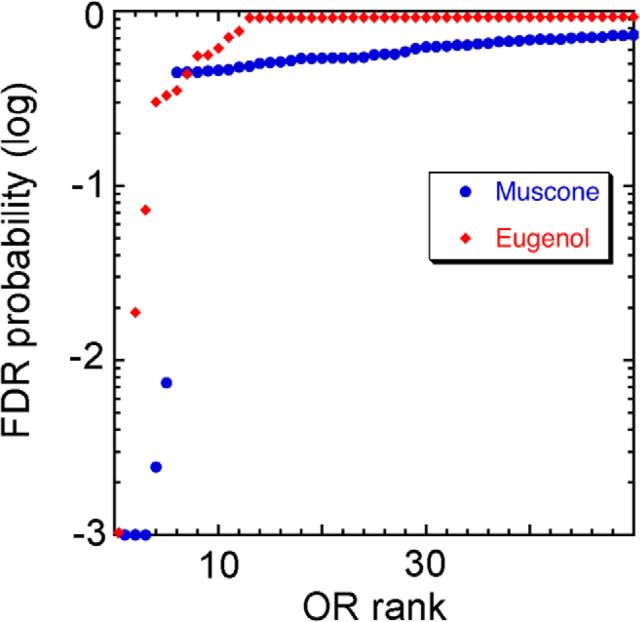
In both the eugenol and muscone experiments, a few dozen ORs had elevated GFP+/GFP− ratios, but only eight exceeded our criterion for statistical significance. The in vivo assay thus produces strong distinctions between responsive ORs and unresponsive ORs, with relatively few ORs showing intermediate responses (Fig. 7). ORs that failed to reach significance in the in vivo assay likely fall into one of three categories: (1) ORs that are insensitive or nonresponsive to the odorant (i.e., true negatives), (2) ORs that are capable of responding to the odorant but responded too weakly in vivo to achieve statistical significance, and (3) ORs that responded in some replications in vivo but too inconsistently to achieve significance. The reasons why ORs fall into the latter two categories are likely attributable to effects related to odorant concentrations. These ORs may require higher concentrations than they can experience in native OSNs during the in vivo assay, and the actual concentrations reached may differ across mice as a result of breathing patterns or other biological variables.
Figure 7.
Distribution of the 50 lowest FDR probabilities from the in vivo eugenol and muscone experiments illustrate the ability of the assay to separate odorant-responsive receptors from nonresponders. Receptors with FDR probabilities <0.001 were truncated at 0.001.
Using heterologous expression, we tested 36 ORs likely to fall into these latter two categories (Table 1). One, Olfr73, is known to respond to eugenol, and we confirmed this (Fig. 3D). Among the other 35 ORs, we identified only seven responsive ORs: five responsive to eugenol (Fig. 8A–E) and two responsive to muscone (Fig. 8F,G). All seven of these were previously orphan receptors. These findings argue that the majority of ORs lacking a significant response in the in vivo assay are not responsive to the odorant ligand and that these nonsignificant in vivo responses are true negative responses, as one would expect from the FDR calculations. In contrast, heterologous expression confirmed the responses of five of the eight significant ORs from the in vivo assay (Figs. 3A–C, 5A,B). These results demonstrate the ability of the in vivo assay to discriminate response from nonresponse.
Table 1.
ORs tested in the Hana3A cell heterologous expression assay
| Eugenol |
Muscone |
||||
|---|---|---|---|---|---|
| Olfr | MOR | FDR | Olfr | MOR | FDR |
| Olfr961* | MOR224-5 | 0.0002 | Olfr1433 | MOR214-4 | 0.0000 |
| Olfr958* | MOR224-9 | 0.0188 | Olfr1440* | MOR215-1 | 0.0000 |
| Olfr960* | MOR188-5 | 0.0728 | Olfr235* | MOR214-3 | 0.0000 |
| Olfr1101 | MOR179-3 | 0.3015 | Olfr1437 | MOR214-6 | 0.0024 |
| Olfr1085 | MOR191-1 | 0.3283 | Olfr1431 | MOR214-5 | 0.0074 |
| Olfr1501 | MOR212-3 | 0.3516 | Olfr1020 | MOR201-2 | 0.4447 |
| Olfr97 | MOR156-2 | 0.4372 | Olfr769 | MOR114-4 | 0.4480 |
| Olfr1160 | MOR173-1 | 0.9179 | Olfr867 | MOR143-2 | 0.4587 |
| Olfr610* | MOR9-2 | 0.9197 | Olfr250 | MOR170-14 | 0.4630 |
| Olfr1052 | MOR172-1 | 0.9245 | Olfr1257 | MOR232-1 | 0.4773 |
| Olfr1234* | MOR231-2 | 0.9832 | Olfr1496 | MOR127-1 | 0.4837 |
| Olfr73* | MOR174-9 | 1.0000 | Olfr74* | MOR174-4 | 0.5204 |
| Olfr151 | MOR171-2 | 1.0000 | Olfr488 | MOR204-15 | 0.5354 |
| Olfr1031 | MOR200-1 | 1.0000 | Olfr816* | MOR113-1 | 0.6397 |
| Olfr178* | MOR184-6 | 1.0000 | Olfr394 | MOR135-8 | 0.8547 |
| Olfr922 | MOR161-3 | 1.0000 | Olfr1213 | MOR233-7 | 1.0000 |
| Olfr1274 | MOR228-4 | 1.0000 | |||
| Olfr821 | MOR109-1 | 1.0000 | |||
| Olfr811 | MOR110-6 | 1.0000 | |||
| Olfr1090 | MOR188-4 | 1.0000 | |||
| Olfr1325 | MOR102-1 | 1.0000 | |||
| Olfr1496 | MOR127-1 | 1.0000 | |||
| Olfr904 | MOR167-3 | 1.0000 | |||
| Olfr591 | MOR24-1 | 1.0000 | |||
| Olfr215 | MOR119-2 | 1.0000 | |||
| Olfr432* | MOR123-2 | 1.0000 | |||
| Olfr1054 | MOR188-2 | 1.0000 | |||
| Olfr1014 | MOR213-5 | 1.0000 | |||
| Olfr360 | MOR159-1 | 1.0000 | |||
| Olfr913* | MOR165-10 | 1.0000 | |||
*Positive responses.
Figure 8.
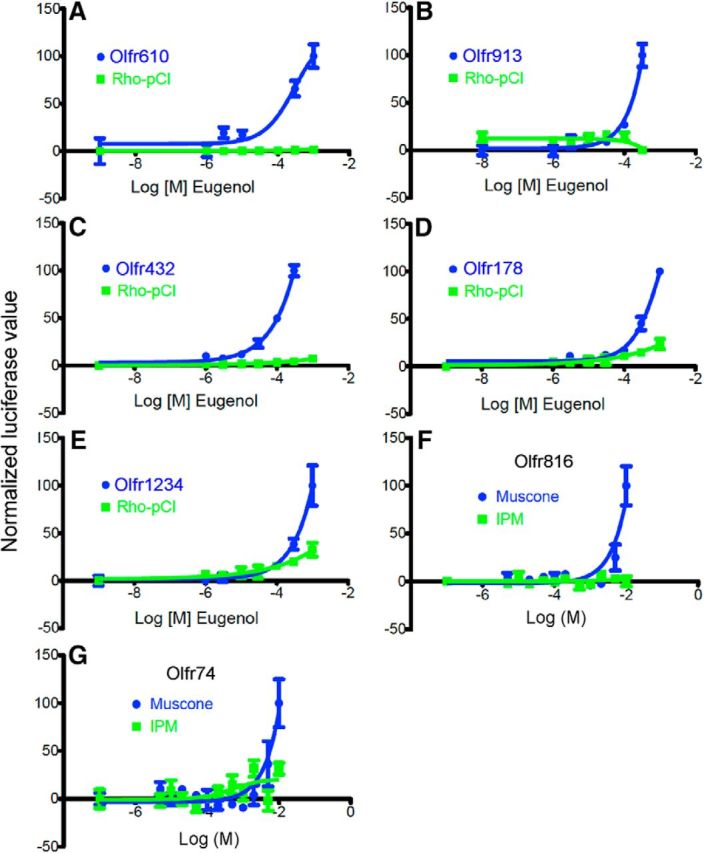
Heterologous expression assay data for additional ORs responsive to eugenol or muscone. A–E, Eugenol concentration–response relationships for five eugenol-responsive ORs that were previously orphan receptors. F, G, Muscone concentration–response relationships for two muscone-responsive ORs that were previously orphan receptors. Rho–pCI, Expression plasmid used as a negative control. IPM used as vehicle.
Discussion
To identify chemosensory receptors that are activated by a given odorant ligand, we developed a novel in vivo assay in live, unanesthetized, freely breathing and freely behaving mice. We demonstrate that this assay re-identified ORs known to be responsive to eugenol and muscone. We identified ORs for muscone and eugenol that respond in vivo and in vitro to these odorants. The ORs that are responsive to eugenol in vivo in S100a5–tauGFP mice encompass three of the four previously identified eugenol-responsive ORs, all of which are related to each other and all of which have been confirmed to respond to eugenol in heterologous expression systems (Kajiya et al., 2001; Katada et al., 2003, 2005; Oka et al., 2006). We do not understand the lack of an in vivo response from the other eugenol-responsive OR, Olfr73, in our assay. We have not yet excluded any of the possible explanations, which range from the presence of an antagonist in the experiments to a polymorphism or differences in expression of the Olfr73 gene in the S100a5–tauGFP strain.
The ORs that are responsive to muscone in vivo encompass five related ORs, and two of these ORs gave muscone responses in heterologous expression assays: Olfr235 and Olfr1440. Our data confirm and extend previous evidence that Olfr1440 is a muscone-responsive receptor and also help explain the activation of a small number of olfactory bulb glomeruli by muscone (Shirasu et al., 2014). In the absence of positive responses in a heterologous expression assay, the other three ORs, Olfr1431, Olfr1433, and Olfr1437, must be considered only as candidate muscone-responsive ORs. The absence of responses does not prove they are unable to respond to muscone, because confounding factors may have prevented them from responding in heterologous cells. Confounding factors, especially involving membrane trafficking, affect a fraction of ORs expressed in heterologous expression systems, even when OR chaperones and sequence modifications are used to enhance delivery of ORs to the plasma membrane (Peterlin et al., 2014). Only if proof of function of these ORs in the heterologous expression assay were obtained, for example by demonstrating responses to a different odorant, would we have evidence sufficient to conclude that these ORs are unresponsive to muscone.
The subsets of ORs we identified as responsive to either muscone or eugenol in vivo each consist of related ORs. Some odorants, including eugenol and muscone, tend to activate clusters of neighboring glomeruli, which are often innervated by OSNs expressing related ORs (Malnic et al., 1999; Oka et al., 2006; Johnson and Leon, 2007; Matsumoto et al., 2010; Nara et al., 2011; Falasconi et al., 2012; Shirasu et al., 2014). However, because these data are based on just two odorants, they are insufficient to conclude that odorants typically activate sets of related ORs. Future experiments will reveal if some odorants instead activate unrelated ORs.
The sequence similarity of the muscone-responsive ORs identified by the in vivo assay predicted an orthologous human OR that is highly sensitive to muscone and some other macrocyclic musk odorants, confirming recent results obtained independently (Shirasu et al., 2014). Musks are a particularly interesting class of odorants because deficits in detecting these odorants are among the most common anosmias and hyposmias in humans (Whissell-Buechy and Amoore, 1973; Gilbert and Kemp, 1996). These sensory deficits are typically not generalized to all musks but rather to subclasses of musks (Triller et al., 2008), consistent with the activation of OR5AN1 by macrocyclic ketone and macrocyclic lactone musks but not polycyclic musks. We speculate that the sets of mouse ORs responsive to other classes of musk odorants, such as polycyclic musk odorants, intersects poorly with those responding to muscone.
Odorant concentration is often positively correlated with the number of OSNs or glomeruli activated (Malnic et al., 1999; Ma and Shepherd, 2000). This correlation predicts that a single moderate concentration of an odorant should show strong activation of a few ORs and weaker activation of other, less sensitive, ORs. Our data are consistent with this prediction. Whether our approach identifies all ORs capable of being activated by any given odorant is difficult to determine, but this completeness can be estimated by comparing the number of responsive ORs to the number of responsive glomeruli per olfactory bulb. We identified four muscone-responsive ORs, which is very similar to the one to three dorsomedial glomeruli and the occasional few ventral glomeruli per olfactory bulb reported to respond to muscone (Shirasu et al., 2014). The nine eugenol-responsive ORs we detected are significantly fewer than the 37–56 eugenol-responsive glomeruli per olfactory bulb reported recently (Shirasu et al., 2014), but the concentration of eugenol was twofold lower in our experiments. The S100a5–tauGFP assay is able to identify many of the ORs capable of responding to an odorant, but it may not identify some ORs, especially weakly responding ORs that are capable of evoking detectable responses in glomeruli because of the amplification effect of axonal convergence, or in heterologous expression assays in which higher concentrations can be applied. When odorants are applied as volatiles in air to live mice, the odorant concentration reaching dendritic cilia of OSNs is not known and is potentially variable because of factors differentially affecting the access of odorants to ORs in the dendritic cilia. In other respects, however, the in vivo assay is resistant to factors that could limit the breadth of the set of responsive ORs detected. The assay gives stable and consistent outcomes in the absence of odor exposure, no OR mRNA is found solely in the GFP+ sample, and even ORs with high GFP+/GFP− ratios after exposure to vehicle are capable of further elevation in their ratio during exposure to an odorant. In fact, ORs with significant responses to the few odorants tested thus far already span control GFP+/GFP− ratios of 0.2–3.9, a range that includes 89% of ORs (T.S.M. and W.B.T., unpublished data). In general, the S100a5–tauGFP assay successfully identifies sets of ORs responsive to an odorant ligand and is resistant to false positives.
In summary, we have developed a novel in vivo assay to identify ORs responding to a given odorant ligand in awake, unanesthetized, freely breathing and freely behaving mice. Mimicking a mouse encountering an ambient odor in its environment, this assay begins with an odorant ligand and returns the identities of ORs that responded. Our assay covers the entire OR repertoire simultaneously in a multiplex manner. Therefore, it has a higher capacity than ex vivo approaches that also rely on the expression of one OR gene per OSN but are based on individual odorant-responsive OSNs (Malnic et al., 1999; Touhara et al., 1999; Kajiya et al., 2001; Nara et al., 2011). Another recently developed in vivo approach differs in that it targets single OSNs and relies on anesthetized mice (Shirasu et al., 2014). Our assay should in principle detect responses from any OR. It is stable and robustly distinguishes a response from the lack of a response. To reduce variability and further increase sensitivity, we are backcrossing the S100a5–tauGFP mice to C57BL/6 mice to place the reporter gene into the inbred genetic background that is the basis for most expression profiling methods. This backcrossing will also facilitate testing of whether other mRNA quantification methods, such as NanoString (Khan et al., 2011) or RNA sequencing, may improve performance. Given the place of origin of our new assay, we have named it the Kentucky in vivo odorant ligand–receptor assay.
Footnotes
This work was supported by National Institutes of Health R01 Awards DC002736 (T.M.) and DC010857 and DC012095 (H.M.) and the University of Kentucky Research Challenge Trust Fund Louis Boyarsky Professorship (T.M.). P.M. acknowledges generous support by Rockefeller University and the Max Planck Society. We thank Dr. G. Frolenkov, M. J. Ni, and Dr. A. M. Fischl for technical assistance, Drs. K. Campbell and Frolenkov for the loan of equipment, and International Flavors & Fragrances (New York, NY) for the gift of Galaxolide. This manuscript is dedicated to the memory of Andreas Walz, without whom this project would not have been possible.
The authors declare no competing financial interests.
References
- Bennett MK, Kulaga HM, Reed RR. Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol Cell Neurosci. 2010;43:353–362. doi: 10.1016/j.mcn.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than 1 trillion olfactory stimuli. Science. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Zhan S, Matsunami H. Assaying surface expression of chemosensory receptors in heterologous cells. J Vis Exp. 2011;48:e2405. doi: 10.3791/2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Block E, Li Z, Connelly T, Zhang J, Huang Z, Su X, Pan Y, Wu L, Chi Q, Thomas S, Zhang S, Ma M, Matsunami H, Chen GQ, Zhuang H. Crucial role of copper in detection of metal-coordinating odorants. Proc Natl Acad Sci U S A. 2012;109:3492–3497. doi: 10.1073/pnas.1111297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Microarrays, empirical Bayes and the two-groups model. Stat Sci. 2008;23:1–22. doi: 10.1214/07-STS236. [DOI] [Google Scholar]
- Efron B. Large-scale inference: empirical Bayes methods for estimation, testing, and prediction. Cambridge, UK: Cambridge UP; 2010. [Google Scholar]
- Falasconi M, Gutierrez-Galvez A, Leon M, Johnson BA, Marco S. Cluster analysis of rat olfactory bulb responses to diverse odorants. Chem Senses. 2012;37:639–653. doi: 10.1093/chemse/bjs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl AM, Heron PM, Stromberg AJ, McClintock TS. Activity-dependent genes in mouse olfactory sensory neurons. Chem Senses. 2014;39:439–449. doi: 10.1093/chemse/bju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert AN, Kemp SE. Odor perception phenotypes: multiple, specific hyperosmias to musks. Chem Senses. 1996;21:411–416. doi: 10.1093/chemse/21.4.411. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Haley SL, McClintock TS. Olfactory receptor trafficking involves conserved regulatory steps. J Biol Chem. 2001;276:7285–7290. doi: 10.1074/jbc.M005433200. [DOI] [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29:14545–14552. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Leon M. Chemotopic odorant coding in a mammalian olfactory system. J Comp Neurol. 2007;503:1–34. doi: 10.1002/cne.21396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Tsai L, Roy DS, Valenzuela DH, Mosley C, Magklara A, Lomvardas S, Liberles SD, Barnea G. Neurons expressing trace amine-associated receptors project to discrete glomeruli and constitute an olfactory subsystem. Proc Natl Acad Sci U S A. 2012;109:13410–13415. doi: 10.1073/pnas.1206724109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K. Odorant response assays for a heterologously expressed olfactory receptor. Biochem Biophys Res Comm. 2003;305:964–969. doi: 10.1016/S0006-291X(03)00863-5. [DOI] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Touhara K. Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell Mol Life Sci. 2009;66:3743–3753. doi: 10.1007/s00018-009-0111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147:907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- Kraft P, Fráter G. Enantioselectivity of the musk odor sensation. Chirality. 2001;13:388–394. doi: 10.1002/chir.1050. [DOI] [PubMed] [Google Scholar]
- Kuhlmann K, Tschapek A, Wiese H, Eisenacher M, Meyer HE, Hatt HH, Oeljeklaus S, Warscheid B. The membrane proteome of sensory cilia to the depth of olfactory receptors. Mol Cell Proteomics. 2014;13:1828–1843. doi: 10.1074/mcp.M113.035378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laska M, Shepherd GM. Olfactory discrimination ability of CD-1 mice for a large array of enantiomers. Neuroscience. 2007;144:295–301. doi: 10.1016/j.neuroscience.2006.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, Matsunami H. Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci Signal. 2011;4:ra1. doi: 10.1126/scisignal.2001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 2003;4:416–433. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- Ma M, Shepherd GM. Functional mosaic organization of mouse olfactory receptor neurons. Proc Natl Acad Sci U S A. 2000;97:12869–12874. doi: 10.1073/pnas.220301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B. Searching for the ligands of odorant receptors. Mol Neurobiol. 2007;35:175–181. doi: 10.1007/s12035-007-0013-2. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Kobayakawa K, Kobayakawa R, Tashiro T, Mori K, Sakano H, Mori K. Spatial arrangement of glomerular molecular-feature clusters in the odorant-receptor class domains of the mouse olfactory bulb. J Neurophysiol. 2010;103:3490–3500. doi: 10.1152/jn.00035.2010. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Landers TM, Gimelbrant AA, Fuller LZ, Jackson BA, Jayawickreme CK, Lerner MR. Functional expression of olfactory-adrenergic receptor chimeras and intracellular retention of heterologously expressed olfactory receptors. Brain Res Mol Brain Res. 1997;48:270–278. doi: 10.1016/S0169-328X(97)00099-5. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004;14:31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/S0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Takeuchi H, Imai T, Saito H, Kiyonari H, Abe T, Chen M, Weinstein LS, Yu CR, Storm DR, Nishizumi H, Sakano H. Agonist-independent GPCR activity regulates anterior-posterior targeting of olfactory sensory neurons. Cell. 2013;154:1314–1325. doi: 10.1016/j.cell.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci. 2011;31:9179–9191. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 2006;52:857–869. doi: 10.1016/j.neuron.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Peterlin Z, Firestein S, Rogers ME. The state of the art of odorant receptor deorphanization: a report from the orphanage. J Gen Physiol. 2014;143:527–542. doi: 10.1085/jgp.201311151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert J. Origin of basal activity in mammalian olfactory receptor neurons. J Gen Physiol. 2010;136:529–540. doi: 10.1085/jgp.201010528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol. 2007;502:1138–1156. doi: 10.1002/cne.21365. [DOI] [PubMed] [Google Scholar]
- Schäfer BW, Fritschy JM, Murmann P, Troxler H, Durussel I, Heizmann CW, Cox JA. Brain S100A5 is a novel calcium-, zinc-, and copper ion-binding protein of the EF-hand superfamily. J Biol Chem. 2000;275:30623–30630. doi: 10.1074/jbc.M002260200. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Shirasu M, Yoshikawa K, Takai Y, Nakashima A, Takeuchi H, Sakano H, Touhara K. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron. 2014;81:165–178. doi: 10.1016/j.neuron.2013.10.021. [DOI] [PubMed] [Google Scholar]
- Streicher WW, Lopez MM, Makhatadze GI. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys Chem. 2010;151:181–186. doi: 10.1016/j.bpc.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci U S A. 1999;96:4040–4045. doi: 10.1073/pnas.96.7.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triller A, Boulden EA, Churchill A, Hatt H, Englund J, Spehr M, Sell CS. Odorant-receptor interactions and odor percept: a chemical perspective. Chem Biodivers. 2008;5:862–886. doi: 10.1002/cbdv.200890101. [DOI] [PubMed] [Google Scholar]
- Whissell-Buechy D, Amoore JE. Odour-blindness to musk: simple recessive inheritance. Nature. 1973;242:271–273. doi: 10.1038/242271a0. [DOI] [PubMed] [Google Scholar]
- Yu TT, McIntyre JC, Bose SC, Hardin D, Owen MC, McClintock TS. Differentially expressed transcripts from phenotypically identified olfactory sensory neurons. J Comp Neurol. 2005;483:251–262. doi: 10.1002/cne.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. Functional expression of a mammalian odorant receptor. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- Zheng C, Feinstein P, Bozza T, Rodriguez I, Mombaerts P. Peripheral olfactory projections are differentially affected in mice deficient in a cyclic nucleotide-gated channel subunit. Neuron. 2000;26:81–91. doi: 10.1016/S0896-6273(00)81140-X. [DOI] [PubMed] [Google Scholar]
- Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282:15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]