Abstract
Background
Polyphosphate (polyP) has bactericidal activity against a gram-negative periodontopathogen Porphyromonas gingivalis, a black-pigmented gram-negative anaerobic rod. However, current knowledge about the mode of action of polyP against P. gingivalis is incomplete. To elucidate the mechanisms of antibacterial action of polyP against P. gingivalis, we performed the full-genome gene expression microarrays, and gene ontology (GO) and protein-protein interaction network analysis of differentially expressed genes (DEGs).
Results
We successfully identified 349 up-regulated genes and 357 down-regulated genes (>1.5-fold, P < 0.05) in P. gingivalis W83 treated with polyP75 (sodium polyphosphate, Nan+2PnO3n+1; n = 75). Real-time PCR confirmed the up- and down-regulation of some selected genes. GO analysis of the DEGs identified distinct biological themes. Using 202 DEGs belonging to the biological themes, we generated the protein-protein interaction network based on a database of known and predicted protein interactions. The network analysis identified biological meaningful clusters related to hemin acquisition, energy metabolism, cell envelope and cell division, ribosomal proteins, and transposon function.
Conclusions
polyP probably exerts its antibacterial effect through inhibition of hemin acquisition by the bacterium, resulting in severe perturbation of energy metabolism, cell envelope biosynthesis and cell division, and elevated transposition. Further studies will be needed to elucidate the exact mechanism by which polyP induces up-regulation of the genes related to ribosomal proteins. Our results will shed new light on the study of the antibacterial mechanism of polyP against other related bacteria belonging to the black-pigmented Bacteroides species.
Keywords: Porphyromonas gingivalis, Polyphosphate, Transcriptome, Microarray, Gene ontology (GO), Protein-protein interaction network analysis
Background
Inorganic polyphosphate (polyP) is a chain of few or many hundreds of phosphate (Pi) residues linked by high-energy phosphoanhydride [1]. polyP has attracted considerable attention as a GRAS (generally recognized as safe) food additive by FDA with antimicrobial properties that can prevent spoilage of food [2],[3]. polyP inhibits the growth of various gram-positive bacteria such as Staphylococcus aureus[4]-[8], Listeria monocytogenes[8],[9], Sarcina lutea[7], Bacillus cereus[10], and mutans streptococci [11],[12], and of fungi such as Aspergillus flavus[5]. The ability of polyP to chelate divalent cations is regarded as relevant to the antibacterial effects, contributing to cell division inhibition and loss of cell-wall integrity [5],[13],[14]. On the other hand, large numbers of gram-negative bacteria including Escherichia coli and Salmonella enterica serovar Typhimurium are capable of growing in higher concentrations, even up to 10% of polyP [5],[7],[15].
Periodontal disease is caused by bacterial infection which is associated with gram-negative oral anaerobes. In our previous study [16], polyP (Nan+2PnO3n+1; n = the number of phosphorus atoms in the chain) with different linear phosphorus (Pi) chain lengths (3 to 75) demonstrated to have antibacterial activity against Porphyromonas gingivalis, a black pigmented, gram-negative periodontopathogen. polyP also showed antibacterial activity against other black-pigmented, gram-negative oral anaerobes such as Prevotella intermedia and Porphyromonas endodontalis[17],[18]. However, the antimicrobial mechanism of polyP against gram-negative bacteria has not yet been fully understood. In the past decade, global genome-wide studies of changes in expression patterns in response to existing and new antimicrobial agents have provided us with a deeper understanding of antimicrobial action [19]. In the present study, we performed the full-genome gene expression microarrays of P. gingivalis, and gene ontology (GO) and protein-protein interaction network analysis of the differentially expressed genes were also performed for elucidating the mechanism of antibacterial action of polyP.
Results and discussion
The complete list of the average gene expression values has been deposited in NCBI’s Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) and is accessible through GEO Series accession number GSE11471. Using filtering criteria of a 1.5 or greater fold-change in expression and significance P-values of <0.05, 706 out of 1,909 genes in P. gingivalis W83 were differentially expressed by polyP75 treatment. The expression of 349 transcripts was increased by polyP treatment while 357 showed decreased expression (Figure 1). To validate the microarray results, quantitative RT-PCR (qRT-PCR) of selected genes was performed. Five of the genes were selected from the up-regulated group and the other five from the down-regulated group in the polyP-treated P. gingivalis cells. We used 16S rRNA as a reference gene for normalization of the qRT-PCR data. There was a high correlation between the expression ratios determined by the microarray and the qRT-PCR (r = 0.926) (Figure 2).
Figure 1.
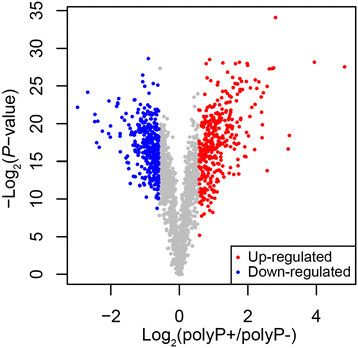
Differential gene expression inP. gingivalisW83 by polyP75 treatment. Differentially expressed genes with 1.5 fold change and P-value < 0.05 were plotted. X-axis presents fold difference between log2 expression of polyP75 treatment and no treatment, and y-axis shows the –log10P -value. Up-regulated genes (over-expressed in polyP75 treatment) were represented as red color and down-regulated genes were colored in blue.
Figure 2.
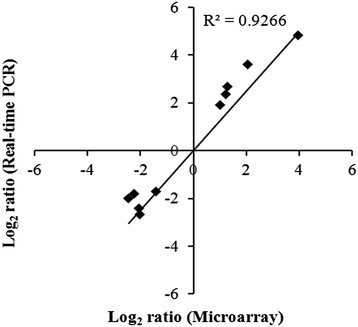
Comparison of transcription measurements by microarray and qRT-PCR. The relative transcription levels for 10 genes are listed in Table 6. The qRT-PCR log2 values were plotted against the microarray data log2 values. The correlation coefficient (r) for comparison of the two datasets is 0. 92.
To broadly characterize the differentially expressed gene (DEG, up- and down-regulated genes) set, GO category enrichment analysis was performed. This analysis identified distinct biological themes associated with each group of the up-regulated and the down-regulated genes. The down-regulated genes were associated with GO terms related to metabolic process (GO:0008152, P = 0.0004), pyridine nucleotide biosynthetic process (GO:0019363, P = 0.0012), regulation of cell shape (GO:0008360, P = 0.002), and polysaccharide biosynthetic process (GO:0000271, P = 0.0015). The up-regulated genes were associated with GO terms related to cellular iron ion homeostasis (GO:0006879, P < 0.0001), ribosome (GO:0005840, P = 0.0032), transposase activity (GO:0004803, P < 0.0001), and DNA binding (GO:0003677, P < 0.0001).
Using 202 DEGs belonging to the above biological themes, we generated the protein-protein interaction network based on a database of known and predicted protein interactions. The network analysis identified 162 DEGs that have direct interaction with one another (Figure 3), and 5 biological meaningful clusters related to 1) iron/hemin acquisition, 2) energy metabolism and electron carriers, 3) cell envelope and cell division, 4) ribosome, and 5) transposon functions.
Figure 3.
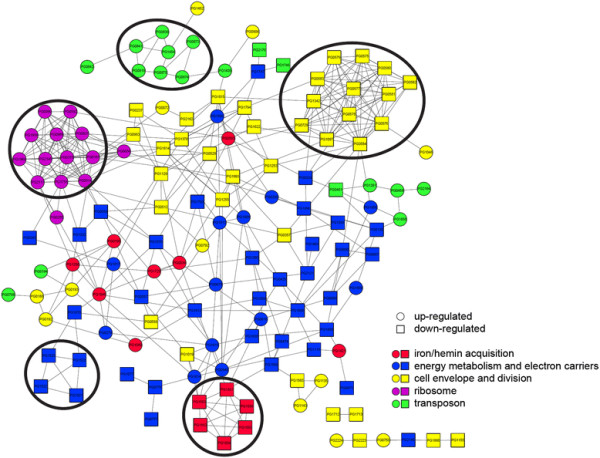
Protein-protein interaction network of differentially expressed functional genes. The network was constructed based on the STRING database. Nodes (symbolized as circles and square) and edges (linking lines) represent DEGs and interactions among DEGs, respectively. Up-regulated genes were represented as a circular shape and down-regulated genes were presented as a square shape. Node color represents the functional annotation of each gene. By applying MCODE clustering algorithm, 5 clusters with the score greater than 3 were obtained.
Hemin acquisition and energy metabolism
In prokaryotic cells, respiration occurs in the cell membrane in which electrons are transferred sequentially through lipoquinones (menaquinones and ubiquinones) and a series of membrane-bound protein carriers such as cytochrome bc1 complex, although the exact organization of enzymes in the respiratory chains varies among different bacteria [20]. P. gingivalis requires hemin as an iron source for its growth [21]. The redox potential of hemin (heme), required as a prosthetic group of cytochrome b, allows it to mediate electron transport with generation of cellular energy [22],[23].
Among 6 genes of hmu locus (PG1551 to PG1556) encoding Hmu YRSTUV, which play a major role in hemin acquisition [24], five genes, but not hmuY, exhibited more than 2-fold decrease in the expression in the presence of polyP75 (Table 1). In addition, genes related to metabolic process including energy metabolism and biosynthesis of lipoquinones, which occupy a central and essential role in electron transport [20], were significantly down-regulated by polyP (Table 2). Genes related to biosynthesis of pyridine nucleotides, known as soluble electron carriers, were also down-regulated (Table 2). These results are compatible with our previous study in which the amount of hemin accumulated on the P. gingivalis surface increased while energy-driven uptake of hemin by the bacterium decreased in the presence of polyP75 [16]. It is conceivable that polyP induce hemin deficiency in P. gingivalis, resulting in disruption of the electron transport occurring in the bacterial membrane. Notably, the up-regulation of oxidative stress response was observed under hemin-limited conditions [25]. Hence, the up-regulation of a series of genes involved in oxidative stress, i.e., 4Fe-4S ferredoxin, rubrerythrin, thioredoxin, Fe-Mn superoxide dismutase, thiol peroxidase, Dps family protein, RprY, ferritin, and HtrA (Table 1), may be due to hemin limitation induced by polyP. However, it is also possible that excessive accumulation of hemin in the vicinity of the bacterial cell surface without formation of μ-oxo bisheme by the bacterium may cause oxidative stress on P. gingivalis[16], as the formation of μ-oxo bisheme protects from hemin-mediated cell damage [23],[26],[27].
Table 1.
Differentially expressed genes related to iron/hemin aquisition and oxidative stress
| Locus no. a | Putative identification a | Cellular role a | Avg fold difference b |
|---|---|---|---|
| PG1551 |
hmuY protein |
Transport and binding proteins: Cations and iron carrying compounds |
−1.19c |
| PG1552 |
TonB-dependent receptor HmuR |
Transport and binding proteins: Cations and iron carrying compounds |
−2.28 |
| PG1553 |
HmuSd |
Hemin acquisitiond |
−2.77 |
| PG1554 |
HmuTd |
Hemin acquisitiond |
−3.44 |
| PG1555 |
HmuUd |
Hemin acquisitiond |
−3.29 |
| PG1556 |
HmuVd |
Hemin acquisitiond |
−2.15 |
| PG1729 |
thiol peroxidase |
Cellular processes : Detoxification |
3.12 |
| PG1421 |
Ferredoxin, 4Fe-4S |
Energy metabolism : Electron transport |
28.54 |
| PG0195 |
Rubrerythrin |
Energy metabolism : Electron transport |
15.49 |
| PG0034 |
Thioredoxin |
Energy metabolism : Electron transport |
2.76 |
| PG1286 |
Ferritin |
Transport and binding proteins: |
2.59 |
| Cations and iron carrying compounds | |||
| PG0090 |
Dps family protein |
Cellular processes: |
2.45 |
| Adaptations to atypical conditions | |||
| PG1545 |
Superoxide dismutase, Fe-Mn |
Cellular processes : Detoxification |
2.34 |
| PG1089 |
DNA-binding response regulator RprY |
Regulatory functions : DNA interactions |
2.00 |
| Signal transduction: Two-component systems | |||
| PG0593 | htrA protein heat induced serine protease | Protein fate: Degradation of proteins, peptides, and glycopeptides | 4.20 |
aLocus number, putative identification, and cellular role are according to the TIGR genome database.
bAverage fold difference indicates the expression of the gene by polyP addition versus no polyP addition.
cThe cut off ratio for the fold difference was < 1.5.
dPutative identification and cellular role are according to Lewis [24].
Table 2.
Differentially expressed genes related to energy metabolism and biosynthesis of electron carriers
| Locus no. a | Putative identification a | Avg fold difference b |
|---|---|---|
| Energy metabolism : Amino acids and amines | ||
| PG1269 |
Delta-1-pyrroline-5-carboxylate dehydrogenase |
−2.02 |
| PG0474 |
Low-specificity L-threonine aldolase |
−1.93 |
| PG1401 |
Beta-eliminating lyase |
−1.74 |
| PG0343 |
Methionine gamma-lyase |
−1.64 |
| PG1559 |
Aminomethyltransferase |
−1.54 |
| PG0324 |
Histidine ammonia-lyase |
−1.53 |
| PG1305 |
Glycine dehydrogenase |
−1.52 |
| PG2121 |
L-asparaginase |
−1.51 |
| PG0025 |
Fumarylacetoacetate hydrolase family protein |
2.11 |
| Energy metabolism : Anaerobic/Fermentation | ||
| PG0687 |
Succinate-semialdehyde dehydrogenase |
−1.76 |
| PG0690 |
4-hydroxybutyrate CoA-transferase |
−1.66 |
| PG0689 |
NAD-dependent 4-hydroxybutyrate dehydrogenase |
−1.58 |
| PG1609 |
Methylmalonyl-CoA decarboxylase, gamma subunit |
−1.87 |
| PG1612 |
Methylmalonyl-CoA decarboxylase, alpha subunit |
−1.71 |
| PG1608 |
Methylmalonyl-CoA decarboxylase, beta subunit |
−1.64 |
| PG0675 |
Indolepyruvate ferredoxin oxidoreductase, alpha subunit |
−1.53 |
| PG1809 |
2-oxoglutarate oxidoreductase, gamma subunit |
2.18 |
| PG1956 |
4-hydroxybutyrate CoA-transferase |
1.74 |
| Energy metabolism : Biosynthesis and degradation of polysaccharides | ||
| PG2145 |
Polysaccharide deacetylase |
−1.94 |
| PG0897 |
Alpha-amylase family protein |
−1.85 |
| PG1793 |
1,4-alpha-glucan branching enzyme |
−1.67 |
| Energy metabolism : Electron transport | ||
| PG0776 |
Electron transfer flavoprotein, alpha subunit |
−2.30 |
| PG0777 |
Electron transfer flavoprotein, beta subunit |
−1.91 |
| PG1638 |
Thioredoxin family protein |
−1.88 |
| PG1332 |
NAD(P) transhydrogenase, beta subunit |
−1.83 |
| PG1119 |
Flavodoxin, putative |
−1.69 |
| PG0429 |
Pyruvate synthase |
−1.64 |
| PG1077 |
Electron transfer flavoprotein, beta subunit |
−1.57 |
| PG1858 |
Flavodoxin |
−2.57 |
| PG2178 |
NADH:ubiquinone oxidoreductase, Na translocating, E subunit |
−1.51 |
| PG0034 |
Thioredoxin |
2.76 |
| PG0195 |
Rubrerythrin |
15.49 |
| PG0548 |
Pyruvate ferredoxin/flavodoxin oxidoreductase family protein |
2.58 |
| PG0616 |
Thioredoxin, putative |
1.52 |
| PG1421 |
Ferredoxin, 4Fe-4S |
28.54 |
| PG1813 |
Ferredoxin, 4Fe-4S |
1.65 |
| Energy metabolism : Glycolysis/gluconeogenesis | ||
| PG0130 |
Phosphoglyceromutase |
−1.68 |
| Energy metabolism : Purines, pyrimidines, nucleosides, and nucleotides | ||
| PG1996 |
Deoxyribose-phosphate aldolase |
−1.73 |
| Energy metabolism : Pentose phosphate pathway | ||
| PG1747 |
Ribose 5-phosphate isomerase B, putative |
−2.45 |
| PG0230 |
Transaldolase |
2.05 |
| PG1595 |
Ribulose-phosphate 3-epimerase |
2.22 |
| Energy metabolism: Sugars | ||
| PG1633 |
Galactokinase |
−1.89 |
| Energy metabolism : TCA cycle | ||
| PG1614 |
Succinate dehydrogenase |
2.25 |
| PG1615 |
Succinate dehydrogenase |
1.60 |
| Energy metabolism : Other | ||
| PG1522 |
Mandelate racemase/muconate lactonizing enzyme family protein |
−2.24 |
| PG0279 |
NADP-dependent malic enzyme |
1.82 |
| PG1017 |
Pyruvate phosphate dikinase |
1.75 |
| PG1513 |
Phosphoribosyltransferase, putative/phosphoglycerate mutase family protein |
3.05 |
| PG1859 |
Glycerate kinase family protein |
1.76 |
| Biosynthesis of pyridine nucleotides | ||
| PG0058 |
Nicotinic acid mononucleotide adenyltransferase |
−1.93 |
| PG1578 |
Quinolinate synthetase |
−1.62 |
| PG0057 |
Nicotinate phosphoribosyltransferase |
−1.61 |
| PG0678 |
Pyrazinamidase/nicotinamidase, putative |
2.00 |
| Biosynthesis of menaquinone and ubiquinone | ||
| PG1870 |
Methlytransferase, UbiE/COQ5 family |
−2.60 |
| PG1467 |
Methlytransferase, UbiE/COQ5 family |
−2.46 |
| PG1523 |
Naphthoate synthase |
−1.89 |
| PG1521 |
O-succinylbenzoic acid--CoA ligase |
−1.78 |
| PG1525 | Isochorismate synthase, putative | −1.50 |
aLocus number, putative identification, and cellular role are according to the TIGR genome database.
bAverage fold difference indicates the expression of the gene by polyP addition versus no polyP addition.
Cell envelope and cell division
Among genes involved in biosynthesis and degradation of surface polysaccharides and lipopolysaccharides, 9 genes were repressed and 5 genes increased by polyP. Among genes related to biosynthesis and degradation of murein sacculus and peptidoglycan, 7 genes were down-regulated (Table 3). For most bacteria, the peptidoglycan cell wall is both necessary and sufficient to determine cell shape [28]. In P. gingivalis W83 genome there is a group of genes called division/cell wall (DCW) cluster, which are involved in cell division and synthesis of peptidoglycan [29]-[31]: PG0575 (penicillin-binding protein 2), PG0576 (murE), PG0577 (mraY), PG0578 (murD), PG0579 (ftsW), PG0580 (murG), PG0581 (murC), PG0582 (ftsQ), PG0583 (ftsA), and PG0584 (ftsZ). Among these, mraY, murD, ftsW, murG, murC, and ftsQ (PG0577- PG0582) were down-regulated by polyP75. It seems that the reduced expression of the genes related to cell envelope biosynthesis in polyP-exposed P. gingivalis may be a result from disruption of the electron transport and reduced production of ATP, since ATP is fundamental for many metabolic processes in bacteria including cell wall biosynthesis and protein synthesis [32]. These transcriptional changes are partially in agreement with the previous report using Bacillus cereus in which polyP inhibited the bacterial cell division [10]. However, unlike B. cereus, formation of elongated aseptate cells and growth phase-dependent bacteriolysis were not observed in P. gingivalis exposed to polyP [16]. It was proposed that polyP, because of its metal ion-chelating nature, may affect the ubiquitous bacterial cell division protein FtsZ, whose GTPase activity is known to be strictly dependent on divalent metal ions. Then, polyP may consequently block the dynamic formation (polymerization) of the Z ring, which would explain the aseptate phenotype of B. cereus[10]. B. cereus exposed to polyP, however, showed normal DNA replication, chromosome segregation, and synthesis of the lateral cell wall [10]. In the present study, P. gingivalis W83 decreased the expression of genes in relation to biosynthesis of cell wall, purine, pyrimidine, nucleoside, and nucleotide, and replication of DNA in the presence of polyP75 (Table 3). These results probably indicate that polyP affects the overall proliferation process including biosynthesis of nucleic acids, DNA replication, biosynthesis of cell wall, and cell division in P. gingivalis.
Table 3.
Differentially expressed genes related to cell envelope and cell division
| Locus no. a | Putative identification a | Avg fold difference b |
|---|---|---|
| Cell envelope : Biosynthesis and degradation of murein sacculus and peptidoglycan | ||
| PG0575 |
Penicillin-binding protein 2 |
−1.41c |
| PG0576 |
UDP-N-acetylmuramoylalanyl-D-glutamyl-2, 6-diaminopimelate ligase |
−1.42c |
| PG0577 |
Phospho-N-acetylmuramoyl-pentapeptide-transferase |
−1.56 |
| PG0578 |
UDP-N-acetylmuramoylalanine--D-glutamateligase |
−1.58 |
| PG0580 |
N-acetylglucosaminyl transferase |
−1.78 |
| PG0581 |
UDP-N-acetylmuramate--L-alanine ligase |
−1.81 |
| PG1342 |
UDP-N-acetylenolpyruvoylglucosamine reductase |
−2.17 |
| PG0729 |
D-alanylalanine synthetase |
−1.80 |
| PG1097 |
Mur ligase domain protein/alanine racemase |
−1.58 |
| Cellular process: Cell division | ||
| PG0579 |
Cell division protein FtsW |
−1.74 |
| PG0582 |
Cell division protein FtsQ |
−1.80 |
| PG0583 |
Cell division protein FtsA |
−1.32 c |
| PG0584 |
Cell division protein FtsZ |
−1.36 c |
| Cell envelope : Biosynthesis and degradation of surface polysaccharides and lipopolysaccharides | ||
| PG1155 |
ADP-heptose--LPS heptosyltransferase, putative |
−1.94 |
| PG1783 |
Glycosyl transferase, group 2 family protein |
−1.87 |
| PG2223 |
Glycosyl transferase, group 2 family protein |
−1.77 |
| PG1815 |
3-deoxy-manno-octulosonate cytidylyltransferase |
−1.73 |
| PG1712 |
Alpha-1,2-mannosidase family protein |
−1.69 |
| PG1345 |
Glycosyl transferase, group 1 family protein |
−1.66 |
| PG2162 |
Lipid A disaccharide synthase |
−1.65 |
| PG1560 |
dTDP-glucose 4,6-dehydratase |
−1.57 |
| PG1880 |
Glycosyl transferase, group 2 family protein |
−1.53 |
| PG0072 |
UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase |
1.83 |
| PG0750 |
Glycosyl transferase, group 2 family protein |
1.51 |
| PG1048 |
N-acetylmuramoyl-L-alanine amidase, family 3 |
2.96 |
| PG1135 |
Bacterial sugar transferase |
5.28 |
| PG1143 |
Sugar dehydrogenase, UD-glucose/GDP-mannose dehydrogenase family |
1.89 |
| Cell envelope : Other | ||
| PG1019 |
Lipoprotein, putative |
−5.47 |
| PG1180 |
Hypothetical protein |
−4.15 |
| PG1713 |
Lipoprotein, putative |
−2.01 |
| PG1767 |
Lipoprotein, putative |
−1.96 |
| PG0490 |
Hypothetical protein |
−1.74 |
| PG1005 |
Lipoprotein, putative |
−1.65 |
| PG1948 |
Lipoprotein, putative |
−1.56 |
| PG0670 |
Lipoprotein, putative |
−1.54 |
| PG2155 |
Lipoprotein, putative |
−1.53 |
| PG1600 |
Hypothetical protein |
−1.52 |
| PG0188 |
Lipoprotein, putative |
1.66 |
| PG0192 |
Cationic outer membrane protein OmpH |
2.68 |
| PG0193 |
Cationic outer membrane protein OmpH |
2.18 |
| PG0717 |
Lipoprotein, putative |
1.95 |
| PG0906 |
Lipoprotein, putative |
1.94 |
| PG1452 |
Lipoprotein, putative |
1.52 |
| PG1828 |
Lipoprotein, putative |
1.87 |
| PG2105 |
Lipoprotein, putative |
1.98 |
| PG2224 |
Hypothetical protein |
2.19 |
| DNA metabolism : DNA replication, recombination, and repair | ||
| PG1814 |
DNA primase |
−2.01 |
| PG1993 |
Excinuclease ABC, C subunit |
−1.77 |
| PG1255 |
Recombination protein RecR |
−1.64 |
| PG1253 |
DNA ligase, NAD-dependent |
−1.62 |
| PG0237 |
Uracil-DNA glycosylase |
−1.58 |
| PG1378 |
A/G-specific adenine glycosylase |
−2.83 |
| PG1622 |
DNA topoisomerase IV subunit A |
−2.02 |
| PG1794 |
DNA polymerase type I |
−1.51 |
| PG2009 |
DNA repair protein RecO, putative |
2.34 |
| Purines, pyrimidines, nucleosides, and nucleotides : 2′-Deoxyribonucleotide metabolism | ||
| PG1129 |
Ribonucleotide reductase |
−2.30 |
| PG0953 |
Deoxyuridine 5′-triphosphate nucleotidohydrolase |
−2.14 |
| Purines, pyrimidines, nucleosides, and nucleotides : Nucleotide and nucleoside interconversions | ||
| PG0512 |
Guanylate kinase |
−1.89 |
| Purines, pyrimidines, nucleosides, and nucleotides : Pyrimidine ribonucleotide biosynthesis | ||
| PG0529 |
Carbamoyl-phosphate synthase small subunit |
−1.70 |
| PG0357 |
Aspartate carbamoyltransferase catalytic subunit |
−1.54 |
| Purines, pyrimidines, nucleosides, and nucleotides : Salvage of nucleosides and nucleotides | ||
| PG0558 |
Purine nucleoside phosphorylase |
−1.51 |
| PG0792 | Hypoxanthine phosphoribosyltransferase | 2.25 |
aLocus number, putative identification, and cellular role are according to the TIGR genome database.
bAverage fold difference indicates the expression of the gene by polyP addition versus no polyP addition.
cThe cut off ratio for the fold difference was < 1.5.
In several transcriptional profiling studies using gram-positive bacteria, a cell wall stress stimulon that includes genes involved in peptidoglycan biosynthesis was induced in the cells challenged with cell wall-active antibiotics [33],[34]. The bacterial cells appeared to respond to the cell wall-active antibiotics by attempting to raise the rate of peptidoglycan biosynthesis in order to compensate for the damaged and partially missing cell wall [35],[36]. Overall, the results indicate that the mode of action of polyP against P. gingivalis may be different from not only that of the cell wall-active antibiotics against gram-positive bacteria, but also that of polyP against gram-positive bacteria.
Ribosomal proteins
In bacteria, production of ribosome requires up to 40% of the cell's energy in rapidly growing bacteria and is therefore tightly regulated on several levels [37]. It seems that bacteria with kinetically impaired ribosomes can to some extent increase the number of ribosomes accumulated under poor growth conditions or under antibiotic challenge in order to compensate for their slower function [38],[39]. It has been reported that antibiotics that target the ribosome or translation factors up-regulate synthesis of ribosomal proteins and accumulate ribosome precursors in Streptococcus pneumoniae[40]. Similarly, in Clostridium difficile, genes encoding many ribosomal proteins were coordinately up-regulated by antibiotics such as amoxicillin, clindamycin, and metronidazole [38]. Therefore, it is conceivable that the up-regulation of the genes encoding ribosomal proteins of polyP- exposed P. gingivalis (Table 4) may reflect a compensatory response for slower or disturbed function of the ribosome.
Table 4.
Differentially expressed genes related to ribosome
| Locus no. a | Putative identification a | Avg fold difference b |
|---|---|---|
| Protein synthesis : Ribosomal proteins | ||
| PG0037 |
50S ribosomal protein L19 |
3.23 |
| PG0167 |
Ribosomal protein L25 |
1.86 |
| PG0314 |
Ribosomal protein L21 |
1.90 |
| PG0315 |
50S ribosomal protein L27 |
1.78 |
| PG0385 |
Ribosomal protein S21 |
3.98 |
| PG0592 |
50S ribosomal protein L31 |
4.01 |
| PG0656 |
50S ribosomal protein L34 |
6.80 |
| PG0989 |
50S ribosomal protein L20 |
3.43 |
| PG0990 |
Ribosomal protein L35 |
1.74 |
| PG1723 |
Ribosomal protein S20 |
2.94 |
| PG1758 |
Ribosomal protein S15 |
6.23 |
| PG1959 |
Ribosomal protein L33 |
2.02 |
| PG1960 |
Ribosomal protein L28 |
2.03 |
| PG2117 |
30S ribosomal protein S16 |
2.93 |
| PG2140 |
Ribosomal protein L32 |
3.40 |
| PG0205 | Peptide chain release factor 3 | 1.50 |
aLocus number, putative identification, and cellular role are according to the TIGR genome database.
bAverage fold difference indicates the expression of the gene by polyP addition versus no polyP addition.
Meanwhile, ribosome biosynthesis of bacteria is governed by transcriptional and translational regulatory mechanisms that provide a balanced and coordinated production of individual ribosomal components [41]. It has been suggested that some free ribosomal proteins act as autogenous feedback inhibitors that cause selective translational inhibition of the synthesis of certain ribosomal proteins whose genes are in the same operon as their own. This inhibition is due to the structural homology between certain ribosomal protein binding regions on 16S rRNA and the mRNA target site for the ribosomal protein [42]-[44]. Although autogenous regulation is known to be a general strategy of balancing ribosomal protein synthesis in bacteria [41], mechanisms for controlling ribosomal protein gene expression in P. gingivalis have not yet been characterized. Further studies will be needed to elucidate the regulatory mechanisms involved in ribosomal protein synthesis in P. gingivalis.
Transposon functions
The majority of the up-regulated genes related to mobile and extrachromosomal element functions were the genes encoding transposases (Table 5). Transposition is generally known to be triggered by cellular stress, i.e., nutritional deficiency, chemicals, and oxidative agents. Little is known about the transposition in P. gingivalis, but up-regulation of transposase-related insertion sequence elements was noticed in P. gingivalis W50 after treatment with H2O2[45]. Thus, it seems quite reasonable to speculate that induction of transposase is associated with oxidative stress-like response which occurred in P. gingivalis W83 due to the presence of polyP.
Table 5.
Differentially expressed genes related to transposon functions
| Locus no. | Putative identification | Avg fold difference |
|---|---|---|
| Mobile and extrachromosomal element functions: Transposon functions | ||
| PG0019 |
ISPg4 transposase |
1.57 |
| PG0050 |
ISPg4, transposase |
1.81 |
| PG0177 |
ISPg4, transposase |
1.87 |
| PG0194 |
ISPg3, transposase |
2.18 |
| PG0225 |
ISPg4, transposase |
1.80 |
| PG0261 |
ISPg3, transposase |
2.20 |
| PG0459 |
ISPg5, transposase |
1.60 |
| PG0487 |
ISPg4, transposase |
1.98 |
| PG0798 |
ISPg3, transposase |
2.11 |
| PG0819 |
Integrase |
1.80 |
| PG0838 |
Integrase |
3.36 |
| PG0841 |
Mobilizable transposon, excision protein, putative |
3.78 |
| PG0842 |
Mobilizable transposon, hypothetical protein, putative |
2.84 |
| PG0872 |
Mobilizable transposon, xis protein |
3.87 |
| PG0873 |
Mobilizable transposon, tnpC protein |
9.34 |
| PG0874 |
Mobilizable transposon, int protein |
2.42 |
| PG0875 |
Mobilizable transposon, tnpA protein |
1.68 |
| PG0970 |
ISPg4, transposase |
1.79 |
| PG1032 |
ISPg3, transposase |
2.23 |
| PG1061 |
ISPg6, transposase |
2.03 |
| PG1261 |
ISPg4, transposase |
2.06 |
| PG1262 |
ISPg3, transposase |
2.11 |
| PG1435 |
Integrase |
2.77 |
| PG1454 |
Integrase |
1.88 |
| PG1658 |
ISPg4, transposase |
1.83 |
| PG1673 |
ISPg4, transposase |
1.77 |
| PG2194 |
ISPg4, transposase |
1.85 |
| PG0461 |
ISPg7, transposase |
−2.77 |
| PG0277 |
ISPg2, transposase |
−1.58 |
| PG0865 |
ISPg2, transposase |
−1.53 |
| PG1746 |
ISPg2, transposase |
−1.63 |
| PG2176 |
ISPg2, transposase |
−1.58 |
| PG1350 | ISPg2, transposase | −1.53 |
Conclusions
We observed that polyP causes numerous events of differential transcription in P. gingivalis. Down-regulated genes were related to iron/hemin acquisition, energy metabolism and electron carriers, and cell envelope and cell division. In contrast, up-regulated genes were related to ribosome and transposon functions. polyP probably exerts its antibacterial effect through inhibition of iron/hemin acquisition by the bacterium, resulting in severe perturbation of energy metabolism, cell envelope biosynthesis and cell division, and elevated transposition. Although the up-regulation of the genes related to ribosomal proteins may possibly reflect autogenous feedback inhibition to regulate the synthesis of certain ribosomal proteins in metabolically disturbed P. gingivalis by polyP, the exact mechanisms underlying this polyP-induced up-regulation of the genes have yet to be elucidated. The current information obtained from the gene ontology and protein-protein interaction network analysis of the differentially expressed genes determined by microarray will shed new light on the study of the antibacterial mechanism of polyP against other related bacteria belonging to the black-pigmented Bacteroides species.
Methods
Chemicals
polyP with a chain length of 75 (polyP75; sodium polyphosphate, glassy, Nan+2PnO3n+1; n = 75) was purchased from Sigma Chemical Co. (St. Louis, MO), dissolved in distilled water to a concentration of 10% (wt/vol), sterilized using a 0.22-μm filter, and stored at −20°C until use.
Bacterial strain and growth conditions
P. gingivalis strain W83 (kindly supplied by Dr. Koji Nakayama, Nagasaki University Graduate School of Biomedical Sciences) was cultured at 37°C anaerobically (85% N2, 10% H2, and 5% CO2) in half-strength brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD) supplemented with 0.5% yeast extract (Difco Laboratories, Detroit, MI), 5 μg/ml of hemin (Sigma), and 1 μg/ml of vitamin K1 (Sigma).
RNA isolation and cDNA synthesis
Use of high concentrations of antibacterial agents for extended periods of time changes the expression of a large set of genes and the effect may be secondary to the action of the drug [46]. Meanwhile, at sub-lethal concentrations, bacteria may sense antibiotics as extracellular chemicals to trigger different cellular responses such as an altered antibiotic resistance/tolerance profile [47]. Hence, we performed the full-genome gene expression microarrays of P. gingivalis W83 exposed to polyP75 at a concentration of 0.03%, which was previously determined to be MIC against the bacterium [16], for a short period of time. P. gingivalis culture grown to early exponential phase (OD600 = 0.3) was divided in half. One aliquot was left untreated, while the other one was treated with 0.03% polyP75. After incubation of both the bacterial cultures for 2 h under anaerobic conditions, the bacterial cells were harvested, and total RNA was extracted from the cells using Trizol Reagent (Invitrogen, Carlsbad, CA). RNA quality was monitored by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and RNA quantity was measured by spectrophotometer. All the samples used in this study exhibited A260/A280 ratio of at least 1.8. cDNA was synthesized with 20 μg of total RNA using SuperScript® II Reverse Transcriptase (Invitrogen).
Microarray analysis
Two individual Cy3-labeled cDNA samples were hybridized into DNA microarrays (Nimblegen Systems, Inc., Madison, WI) containing the whole genome of 1,909 genes of P. gingivalis W83 for 16 h at 42°C. Five replicates of the genome were included per chip. An average of 19 different 60-mer probes which had at least three mismatches compared to other 60-mers represented each gene in the genome. A quality control check (hybridization) was performed for each array, which contained on-chip control oligonucleotides. Data were extracted from the scanned images using an Axon GenePix 4000B microarray scanner and NimbleScan Version 2.3. Quantile normalization was performed across replicate arrays, and RMA (Robust Multichip Average) analysis was performed to generate gene expression values. Genes evidencing statistically significant changes in expression (>1.5-fold difference) were identified via t-tests (P < 0.05).
Assessment of array data quality
To confirm the microarray results using qRT-PCR, 10 genes were selected, and specific primers for the selected genes (Table 6) were designed using Primer3 (http://fokker.wi.mit.edu/primer3/). All quantifications were normalized to the P. gingivalis 16S rRNA gene. The transcriptional ratio from qRT-PCR analysis was logarithm-transformed and then plotted against the average log2 ratio values obtained by microarray analysis [48].
Table 6.
Real-time quantitative RT-PCR confirmation of selected genes
| Locus no. a | Primer sequence (5′-3′) b | Product size (bp) |
|---|---|---|
| 16S rRNA |
F: TGTTACAATGGGAGGGACAAAGGG |
118 |
| R: TTACTAGCGAATCCAGCTTCACGG | ||
| PG0090 |
F: CAGAAGTGAAGGAAGAGCACGAAC |
197 |
| R: GTAGGCAGACAGCATCCAAACG | ||
| PG0195 |
F: TCCACGGCTGAGAACTTGCG |
149 |
| R: TGCTCGGCTTCCACCTTTGC | ||
| PG1545 |
F: CCAAACCCTCAACCACAATC |
142 |
| R: GGTACCGGCTGTGTTGAACT | ||
| PG0593 |
F: CGTGTGGGAGAGTGGGTATTGG |
175 |
| R: CGCCGCTGTTGCCTGAATTG | ||
| PG1089 |
F: CCATCGCGATCGATGATCAGGTAA |
104 |
| R: GGCATAGTTGCGTTCAAGGGTTTC | ||
| PG1019 |
F: TTCGCAGTATCCCATCCAAC |
126 |
| R: TCCGGCTCATAGACTTCCAA | ||
| PG1180 |
F: CAGTCTGCCACAGTTCACCA |
124 |
| R: CCCTACACGGACACTACCGA | ||
| PG1983 |
F: GCTCTGTGGTGTGGGCTATC |
146 |
| R: GGATAACAGGCAAACCCGAT | ||
| PG0885 |
F: CAGATCCAAATCGGGACTGA |
156 |
| R: GTAGAGCAAGCCATGCAAGC | ||
| PG1181 | F: GATGAATTCGGGCGGATAAT |
184 |
| R: CCTTGAAGTGCTCCAACGAC |
aBased on the genome annotation provided by TIGR (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=gpg).
bPrimers were designed using Primer3 program for the study except for the primers of P. gingivalis 16S rRNA and PG1089 [49], which were prepared based on the primer sequences published previously. The 16S rRNA gene was used as the reference gene for normalization. F, forward; R, reverse.
Gene ontology (GO) enrichment analysis
The GO term annotations for P. gingivalis were downloaded from the Gene Ontology website (http://www.geneontology.org/GO.downloads.annotations.shtml, UniProt [multispecies] GO Annotations @ EBI, Apr. 2013). To test the GO category enrichment, we calculated the fraction of gene in the test set (Ftest) associated with each GO category. Then, we generated the random control gene set that has the same number gene of test set. In this process, the random control gene was selected by matching the length of the test gene. The fraction of genes in this randomly selected control set (Fcontrol) associated with the current GO category was calculated. This random sampling process was repeated 10,000 times. Finally, the P-value for the enriched GO category in a test gene set was calculated as the fraction of times that Ftest was lower than or equal to Fcontrol.
Protein-protein interaction network analysis
The protein-protein interaction network data including score were obtained from the STRING 9.1 (http://string-db.org) [50], for P. gingivalis W83. We used Cytoscape software [51] for network drawing, in which nodes and edges represented DEGs and interactions among DEGs, respectively. DEGs with no direct interaction were discarded, and the final dataset consisting of 611 DEGs and 1,641 interactions were used for the network construction. In order to find significant interaction between DEGs, we applied the confidence cutoff as 0.400 (medium confidence).
To understand the biological functions of the DEGs in the network, we annotated 202 DEGs belonging to 8 relevant biological functional clusters and then generated the sub-network using these DEGs in the whole DEGs network constructed above. Cytoscape plug-in MCODE [52] was used to decompose the sub-network and 5 clusters with the score greater than 3 were identified.
Abbreviations
polyP: Inorganic polyphosphate
GO: Gene ontology
DEG(s): Differentially expressed genes (s)
qRT-PCR: Quantitative RT-PCR
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: JHM and JYL. Performed the experiments: JHM. Analyzed the data: JHM, JHL. Wrote the manuscript: JHM, JHL and JYL. All authors read and approve the final manuscript.
Contributor Information
Ji-Hoi Moon, Email: prudence75@khu.ac.kr.
Jae-Hyung Lee, Email: jaehlee@khu.ac.kr.
Jin-Yong Lee, Email: ljinyong@khu.ac.kr.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011–0009233) and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2012R1A5A2051384).
References
- Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: A molecule of many functions. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Marcy JA, Kraft AA, Hotchkiss DK, Molins RA, Olson DG, Walker HW, Merkenich K. Effects of selected commercial phosphate products on the natural bacterial flora of a cooked meat system. J Food Prot. 1998;53:391–393. [Google Scholar]
- Molins RA, Kraft AA, Walker HW, Rust RE, Olson DG, Merkenich K. Effect of inorganic polyphosphates on ground beef characteristics: microbiological effects on frozen beef patties. J Food Sci. 1987;52:46–49. doi: 10.1111/j.1365-2621.1987.tb13969.x. [DOI] [Google Scholar]
- Jen CM, Shelef LA: Factors affecting sensitivity ofStaphylococcus aureus196E to polyphosphates.Appl Environ Microbiol 1986, 52:842–846. [DOI] [PMC free article] [PubMed]
- Knabel SJ, Walker HW, Hartman PA: Inhibition ofAspergillus flavusand selected Gram-positive bacteria by chelation of essential metal cations by polyphosphate.J Food Prot 1991, 54:360–365. [DOI] [PubMed]
- Lee RM, Hartman PA, Olson DG, Williams FD: Bactericidal and bacteriolytic effects of selected food-grade phosphates, usingStaphylococcus aureusas a model system.J Food Prot 1994, 57:276–283. [DOI] [PubMed]
- Post FJ, Krishnamurty GB, Flanagan MD. Influence of sodium hexametaphosphate on selected bacteria. Appl Microbiol. 1963;11:430–435. doi: 10.1128/am.11.5.430-435.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika LL, Kim AH: Effect of sodium polyphosphates on growth ofListeria monocytogenes.J Food Prot 1993, 56:577–580. [DOI] [PubMed]
- Rajkowski KT, Calderone SM, Jones E: Effect of polyphosphate and sodium chloride on the growth ofListeria monocytogenesandStaphylococcus aureusin ultra-high temperature milk.J Dairy Sci 1994, 77:1503–1508. [DOI] [PubMed]
- Maier SK, Scherer S, Loessner MJ: Long-chain polyphosphate causes cell lysis and inhibitsBacillus cereusseptum formation, which is dependent on divalent cations.Appl Environ Microbiol 1999, 65:3942–3949. [DOI] [PMC free article] [PubMed]
- Brown AT, Ruh R Jr: Negative interaction of orthophosphate with glycolytic metabolism byStreptococcus mutansas a possible mechanism for dental caries reduction.Arch Oral Biol 1977, 22:521–524. [DOI] [PubMed]
- Shibata H, Morioka T: Antibacterial action of condensed phosphates on the bacteriumStreptococcus mutansand experimental caries in the hamster.Arch Oral Biol 1982, 27:809–816. [DOI] [PubMed]
- Lee RM, Hartman PA, Olson DG, Williams FD: Metal ions reverse the inhibitory effects of selected food-grade polyphosphates inStaphylococcus aureus.J Food Prot 1994, 57:284–288. [DOI] [PubMed]
- Lee RM, Hartman PA, Stahr HM, Olson DG, Williams FD: Antibacterial mechanism of long-chain polyphosphates inStaphylococcus aureus.J Food Prot 1994, 57:289–294. [DOI] [PubMed]
- Obritsch JA, Ryu D, Lampila LE, Bullerman LB. Antibacterial effects of long-chain polyphosphates on selected spoilage and pathogenic bacteria. J Food Prot. 2008;71:1401–1405. doi: 10.4315/0362-028x-71.7.1401. [DOI] [PubMed] [Google Scholar]
- Moon JH, Park JH, Lee JY: Antibacterial action of polyphosphate onPorphyromonas gingivalis.Antimicrob Agents Chemother 2011, 55:806–812. [DOI] [PMC free article] [PubMed]
- Kong HJ, Choi HY, Min BS, Part SJ, Lee JY, Choi GW: Effect of polyphosphate on the growth of oral bacterium,Prevotella intermedia.Restor Dent Endod 1998, 23:550–560.
- Choi SB, Park SJ, Choi GW, Choi HY: Mechanism in antibacterial activity of polyphosphate againstPorphyromonas endodontalis.J Korean Acad Oper Dent 2000, 25:561–574.
- Song Y, Lunde CS, Benton BM, Wilkinson BJ. Further insights into the mode of action of the lipoglycopeptide telavancin through global gene expression studies. Antimicrob Agents Chemother. 2012;56:3157–3164. doi: 10.1128/AAC.05403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu M, Begari E. Vitamin K2 in electron transport system: are enzymes involved in vitamin K2 biosynthesis promising drug targets? Molecules. 2010;15:1531–1553. doi: 10.3390/molecules15031531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Zheng C: OxyR activation inPorphyromonas gingivalisin response to a hemin-limited environment.Infect Immun 2012, 80:3471–3480. [DOI] [PMC free article] [PubMed]
- Smalley JW, Birss AJ, McKee AS, Marsh PD: Hemin regulation of hemoglobin binding byPorphyromonas gingivalis.Curr Microbiol 1980, 36:102–106. [DOI] [PubMed]
- Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL: Hemoglobinase activity of the lysine gingipain protease (Kgp) ofPorphyromonas gingivalisW83.J Bacteriol 1999, 181:4905–4913. [DOI] [PMC free article] [PubMed]
- Lewis JP, Plata K, Yu F, Rosato A, Anaya C: Transcriptional organization, regulation and role of thePorphyromonas gingivalisW83hmuhaemin-uptake locus.Microbiology 2006, 152:3367–3382. [DOI] [PubMed]
- Dashper SG, Ang CS, Veith PD, Mitchell HL, Lo AW, Seers CA, Walsh KA, Slakeski N, Chen D, Lissel JP, Butler CA, O'Brien-Simpson NM, Barr IG, Reynolds EC: Response ofPorphyromonas gingivalisto heme limitation in continuous culture.J Bacteriol 2009, 191:1044–1055. [DOI] [PMC free article] [PubMed]
- Smalley JW, Birss AJ, Silver J: The periodontal pathogenPorphyromonas gingivalisharnesses the chemistry of the μ-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide.FEMS Microbiol Lett 2000, 183:159–164. [DOI] [PubMed]
- Smalley JW, Birss AJ, Szmigielski B, Potempa J: The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) ofPorphyromonas gingivalispromotes μ-oxo bishaem formation from monomeric iron (III) protoporphyrin IX.Microbiology 2006, 152:1839–1845. [DOI] [PubMed]
- Furchtgott L, Wingreen NS, Huang KC. Mechanisms for maintaining cell shape in rod-shaped Gram-negative bacteria. Mol Microbiol. 2011;81:340–353. doi: 10.1111/j.1365-2958.2011.07616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingorance J, Tamames J, Vicente M. Genomic channeling in bacterial cell division. J Mol Recognit. 2004;17:481–487. doi: 10.1002/jmr.718. [DOI] [PubMed] [Google Scholar]
- Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- Höltje JV, Heidrich C: Enzymology of elongation and constriction of the murein sacculus ofEscherichia coli.Biochimie 2001, 83:103–108. [DOI] [PubMed]
- Samuelsen O, Haukland HH, Kahl BC, von Eiff C, Proctor RA, Ulvatne H, Sandvik K, Vorland LH: Staphylococcus aureussmall colony variants are resistant to the antimicrobial peptide lactoferricin B.J Antimicrob Chemother 2005, 56:1126–1129. [DOI] [PubMed]
- Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ: Transcriptional profiling reveals that daptomycin induces theStaphylococcus aureuscell wall stress stimulon and genes responsive to membrane depolarization.Antimicrob Agents Chemother 2008, 52:980–990. [DOI] [PMC free article] [PubMed]
- Wilkinson BJ, Muthaiyan A, Jayaswal RK: The cell wall stress stimulon ofStaphylococcus aureusand other Gram-positive bacteria.Curr Med Chem Anti-Infect Agents 2005, 4:259–276.
- Pechous R, Ledala N, Wilkinson BJ, Jayaswal RK: Regulation of the expression of cell wall stress stimulon member genemsrA1in methicillin-susceptible or -resistantStaphylococcus aureus.Antimicrob Agents Chemother 2004, 48:3057–3063. [DOI] [PMC free article] [PubMed]
- Bertsche U. In: Bacterial polysaccharides: Current innovations and future trends. Ullrich M, editor. Caister Academic Press, Norfolk, UK; 2009. The polysaccharide peptidoglycan and how it is influenced by (antibiotic) stress; pp. 3–26. [Google Scholar]
- Maguire BA. Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol Mol Biol Rev. 2009;73:22–35. doi: 10.1128/MMBR.00030-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JE, Stabler RA, Wren BW, Fairweather NF: Microarray analysis of the transcriptional responses ofClostridium difficileto environmental and antibiotic stress.J Med Microbiol 2008, 57:757–764. [DOI] [PubMed]
- Mikkola R, Kurland CG. Evidence for demand-regulation of ribosome accumulation in E. coli. Biochimie. 1991;73:1551–1556. doi: 10.1016/0300-9084(91)90190-C. [DOI] [PubMed] [Google Scholar]
- Ng WL, Kazmierczak KM, Robertson GT, Gilmour R, Winkler ME: Transcriptional regulation and signature patterns revealed by microarray analyses ofStreptococcus pneumoniaeR6 challenged with sublethal concentrations of translation inhibitors.J Bacteriol 2003, 185:359–370. [DOI] [PMC free article] [PubMed]
- Leonid VA, Alexandrina AL, Ludmila ST, Nadezda VS, Irina VB: A new regulatory circuit in ribosomal protein operons: S2-mediated control of therpsB-tsfexpression in vivo.RNA 2008, 14:1882–1894. [DOI] [PMC free article] [PubMed]
- Nomura M, Yates JL, Dean D, Post LE: Feedback regulation of ribosomal protein gene expression inEscherichia coli: structural homology of ribosomal RNA and ribosomal protein mRNA.Proc Natl Acad Sci U S A 1980, 77:7084–7088. [DOI] [PMC free article] [PubMed]
- Yates JL, Arfsten AE, Nomura M: In vitro expression ofEscherichia coliribosomal protein genes: autogenous inhibition of translation.Proc Natl Acad Sci U S A 1980, 77:1837–1841. [DOI] [PMC free article] [PubMed]
- Choonee N, Even S, Zig L, Putzer H: Ribosomal protein L20 controls expression of theBacillus subtilis infCoperon via a transcription attenuation mechanism.Nucleic Acids Res 2007, 35:1578–1588. [DOI] [PMC free article] [PubMed]
- Duran-Pinedo AE, Nishikawa K, Duncan MJ: The RprY response regulator ofPorphyromonas gingivalis.Mol Microbiol 2007, 64:1061–1074. [DOI] [PubMed]
- Palzkill T. Antibiotic exposure and bacterial gene expression. Genome Res. 2001;11:1–2. doi: 10.1101/gr.11.1.1. [DOI] [PubMed] [Google Scholar]
- Bernier SP, Surette MG. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol. 2013;4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Zhou D, Pang X, Zhang L, Song Y, Tong Z, Bao J, Dai E, Wang J, Guo Z, Zhai J, Du Z, Wang X, Wang J, Huang P, Yang R: DNA microarray analysis of the heat- and cold-shock stimulons inYersinia pestis.Microbes Infect 2005, 7:335–348. [DOI] [PubMed]
- Hosogi Y, Duncan MJ: Gene expression inPorphyromonas gingivalisafter contact with human epithelial cells.Infect Immun 2005, 73:2327–2335. [DOI] [PMC free article] [PubMed]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


