Abstract
Recombination in the control region (CR) of Mytilus mitochondrial DNA (mtDNA) was originally reported based on the relatively short, sequenced fragments of mitochondrial genomes. Recombination outside the CR has been reported recently with the suggestion that such processes are common in Mytilus. We have fully sequenced a set of 11 different mitochondrial haplotypes representing the high diversity of paternally inherited mitochondrial genomes of Baltic Sea Mytilus mussels, including the haplotype close to the native Mytilus trossulus mitochondrial genome, which was thought to have been entirely eliminated from this population. Phylogenetic and comparative analysis showed that the recombination is limited to the vicinity of the CR in all sequenced genomes. Coding sequence comparison indicated that all paternally inherited genomes showed increased accumulation of nonsynonymous substitutions, including the genomes which switched their transmission route very recently. The acquisition of certain CR sequences through recombination with highly divergent paternally inherited genomes seems to precede and favor the switch, but it is not a prerequisite for this process. Interspecies hybridization in the Baltic Sea during the recent 10,000 years created conditions for both structural and evolutionary mitochondrial instability which resulted in the observed variation and dynamics of mtDNA in Baltic Sea Mytilus mussels. In conclusion, the data shows that the effects of mitochondrial recombination are limited to the CR of few phylogenetic lineages.
Electronic supplementary material
The online version of this article (doi:10.1007/s00438-014-0888-3) contains supplementary material, which is available to authorized users.
Keywords: mtDNA recombination, D-loop, DUI, Evolution
Introduction
Mussels of the genus Mytilus have an unusual system of mitochondrial DNA (mtDNA) inheritance [referred to as doubly uniparental inheritance (DUI)], where the female type (F) is transmitted to all offspring and male type (M) only to the sons (Zouros et al. 1994; Skibinski et al. 1994). This system has also been observed in some other bivalve orders and families, e.g., Unionoida, Veneridae and Donacidae (Liu et al. 1996; Passamonti and Scali 2001; Curole and Kocher 2002; Serb and Lydeard 2003; Theologidis et al. 2008). Divergence between the F and M genomes can be greater than 40 %, but occasionally the M genome can be replaced by the F genome in a process called masculinization (Hoeh et al. 1997). In consequence, the divergence between paternally and maternally inherited genomes can be reduced. The well-documented examples of masculinization come from the Baltic population of Mytilus trossulus mussels. In this population the highly divergent M genome occurs very rarely and both genomes F and M are similar to the F genome of the congeneric M. edulis, and not to the native M. trossulus (Wenne and Skibinski 1995; Burzyński et al. 2003, 2006; Zbawicka et al. 2007). A hybrid zone around the Oresund and Danish belts separate Baltic M. trossulus from North sea M. edulis. Moreover, the Baltic population is composed of individuals of mixed genetic background (Riginos et al. 2002; Bierne et al. 2003; Kijewski et al. 2006, 2011). The hybrid zone apparently allowed complete asymmetric introgression of M. edulis F mtDNA (Rawson and Hilbish 1998; Quesada et al. 1999). The very young age of the Baltic Sea, together with the postglacial timing of M. trossulus invasion from the Pacific (Śmietanka et al. 2013), indicates that this process must have taken place during the last few thousands years. Despite that, all attempts to detect the relict native M. trossulus genomes in Baltic mussels failed, with one possible exception. Quesada et al. (2003) suggested the presence of the native M. trossulus M genome, but only a very short fragment of the genome was sequenced, precluding conclusive identification. Instability of mitochondrial genomes in the Baltic population was also exemplified by heteroplasmy for two and possibly three mitochondrial genomes of low divergence (Quesada et al. 2003; Zbawicka et al. 2003).
Another aspect of Mytilus mitochondrial genome instability is the apparent recombination signature, presented in the control region (CR) of multiple haplotypes (Burzyński et al. 2003, 2006; Rawson 2005; Venetis et al. 2007; Filipowicz et al. 2008). Baltic M. trossulus in particular have a great diversity of structural rearrangements in the CRs (Burzyński et al. 2003), which can be explained by duplication, deletion or intermolecular recombination (Burzyński et al. 2006). Moreover, its paternal lineage is dominated by mosaic haplotypes having M. edulis M-like CR segments, not present in maternally inherited haplotypes. It was hypothesized that the M-like fragment is necessary for a role reversal event (Zouros 2000; Burzyński et al. 2003; Cao et al. 2004). However, the discovery of genomes with mosaic CRs inherited maternally (Śmietanka et al. 2010) as well as the possibility that some masculinized genomes did not have the mosaic CRs (Burzynski et al. 2006) weakened the hypothesis.
Here we present, for the first time, the complete sequences of a representative set consisting of 11 mitochondrial genomes from Baltic Mytilus. Their comparative analysis sheds new light on the timing, mechanisms of emergence and evolution of recombinant mitochondrial genomes in bivalve species exhibiting the unusual system of DUI.
Materials and methods
Samples
A sample of 400 Mytilus sp. mussels collected from the Gulf of Gdańsk, southern Baltic and described previously (Burzyński et al. 2003, 2006) was used. The known taxonomic identity of the specimen, established using nuclear markers by Zbawicka et al. (2007), was typical for the Baltic hybrid population as defined by Kijewski et al. (2006) and Zbawicka et al. (2010). Representative mitochondrial haplotypes were selected for whole mitogenome sequencing, following the methodology described by Zbawicka et al. (2007). Nine haplotypes were derived from sperm, one from eggs and one from female somatic tissues. All haplotypes of M. edulis origin were genotyped at their CR first, using PCR, Southern hybridization and sequencing, as described by Burzyński et al. (2006) (Table 1). Identification of the F haplotype of M. trossulus origin was performed by PCR amplification of the mtDNA fragment spanning the 3’ part of nad2 gene, two tRNA (trnS and trnM) and the 5′ part of cox3 gene with a highly specific primer pair, F1T and U2T (Śmietanka et al. 2013). Initial screening of the whole sample revealed a few positive individuals, and the one with the strongest signal (female 62mc10) was selected. This female had two F haplotypes: one similar to M. edulis and the other similar to M. trossulus. Only the latter was analyzed. To ensure that no contamination influenced this unusual result, the following precautions were taken. All DNA work involving Baltic mussels was done in a separate laboratory, without any contact with mussels having native mtDNA of M. trossulus. Separate sets of equipment and reagents were used. Appropriate negative controls were included in all PCRs. Moreover, the resulting haplotype has unique structural characteristics, not seen in previously isolated genomes. It is therefore highly unlikely that it is a contamination product.
Table 1.
Mitochondrial genomes sequenced during this study
| ID | Sex | Haplogroup | Tissue | Total length | CR length | Accession number |
|---|---|---|---|---|---|---|
| 178mc10 | Female | M. edulis F | Eggs | 16,745 | 1,158 | KM192128 |
| 25mc10 | Male | 1a | Sperm | 16,587 | 1,000 | KM192131 |
| 107mc10 | Male | 1a | Sperm | 16,593 | 1,006 | KM192124 |
| 136mc10 | Male | 1a | Sperm | 16,591 | 1,004 | KM192126 |
| 152mc10 | Male | 1a | Sperm | 16,589 | 1,002 | KM192127 |
| 20mc10 | Male | 11a | Sperm | 17,724 | 2,139 | KM192130 |
| 115mc10 | Male | 11a | Sperm | 17,720 | 2,134 | KM192125 |
| 45mc10 | Male | 1 | Sperm | 16,583 | 996 | KM192132 |
| 46mc10 | Male | 16 | Sperm | >19,291 | >3,704 | KM192134 |
| 195mc10 | Male | M. edulis M | Sperm | 16,632 | 934 | KM192129 |
| 62mc10 | Female | M. trossulus F | Mantle | >17,472 | >1,884 | KM192133 |
PCR and sequencing of mitochondrial DNA
The whole genomes were sequenced in two steps, as described previously (Zbawicka et al. 2007; Śmietanka et al. 2010). First, long-range (LR) PCRs were performed with highly specific primers. Then, overlapping fragments were re-amplified with nested universal primers and sequenced directly. The detailed information on primers used is given in Supplementary Table 1. Additional PCRs with primers covering the CRs were performed, as needed, to fill the gaps. For some haplotypes the structure of the CR was too complex. They contained large arrays of long repeats which were impossible to bridge with current sequencing technology. In such cases, we sequenced the whole coding part and as much of the CR as possible, using a combination of specific LR-PCR with re-amplification and direct sequencing.
For all LR-PCR, Phusion Pfx (Finnzymes Oy) polymerase was used according to the manufacturer’s protocol. For re-amplifications, a 1:800 dilution of the LR-PCR product was used as a template. All re-amplifications were performed as described previously (Zbawicka et al. 2007, 2010; Śmietanka et al. 2010). PCR amplifications were carried out in 15 μl reaction volumes containing 20 ng of template DNA, 0.4 μM of each primer, 200 μM nucleotides, 1.5 mM magnesium chloride, 0.5 unit of high-fidelity DyNAzymeEXT2 DNA polymerase (Finnzymes Oy) and appropriate reaction buffer from Finnzymes. All PCRs were performed in a T-gradient cycler from Biometra (Tampa, FL). PCR products (2 μl of each amplification) were visualized on 1 % agarose gels stained with ethidium bromide. PCR products were purified by alkaline phosphatase and exonuclease I treatment (Werle et al. 1994) and sequenced directly with BigDye™ terminator cycle sequencing method.
Sequence assembly and annotation followed the established protocol (Zbawicka et al. 2007, 2010; Śmietanka et al. 2010). The assembly was facilitated by Phred (Ewing et al. 1998) and performed in Gap4 (Bonfield et al. 1995; Staden 2001). De novo prediction of all protein-coding genes was attempted using a set of algorithms implemented in CRITICA (Badger and Olsen 1999), Glimmer3 (Delcher et al. 1999) and wise2 (Birney et al. 2004). For prediction of RNA genes, Arwen was used (Laslett and Canbäck 2008). All predictions were inspected and critically evaluated after comparison with the closest RefSeq annotations. The assembled and annotated sequences have been deposited in GenBank under accession numbers KM192124-KM192134.
Bioinformatic analysis
For comparative analysis, 30 complete or nearly complete Mytilus mtDNA sequences were used. There were 19 sequences already present in GenBank (Table 2) and 11 newly obtained (Table 1). Individual gene sequences were extracted and aligned in MEGA5 (Tamura et al. 2011), using aminoacid translation as a guide. For most analyses the resulting alignments were concatenated. Genetic distance (K) based on Kimura’s two-parameter model (Kimura 1980) and divergence in synonymous (K s) and nonsynonymous (K a) substitutions, using modified Nei–Gojobori method (Nei and Gojobori 1986) with Jukes–Cantor correction, were calculated in MEGA5, with standard error (SE) computed over 1,000 bootstrap replicates. To identify the sites under selection, nonsynonymous and synonymous changes at individual codons were evaluated using several methods: single likelihood ancestor counting (SLAC), fixed effects likelihood (FEL), random effects likelihood (REL) (Kosakovsky Pond and Frost 2005b), fast, unconstrained Bayesian approximation (FUBAR) (Murrell et al. 2013) and mixed effects model of evolution (MEME) (Murrell et al. 2012), and GA-Branch (Pond and Frost 2005) in HyPhy (Scheffler et al. 2006), as implemented via the datamonkey web server (Kosakovsky Pond and Frost 2005a). Significance levels of 0.05 for SLAC and FEL and Bayes factor criterion of 50 for REL were used.
Table 2.
Published genomes used in comparative analysis
| Accession number | ID | Species | References |
|---|---|---|---|
| AY823625 | M. trossulus | Breton et al. (2006) | |
| AY823623 | M. trossulus | Breton et al. (2006) | |
| AY823624 | M. trossulus | Breton et al. (2006) | |
| FJ890849 | azo20 | M. galloprovincialis | Burzyński and Śmietanka (2009) |
| FJ890850 | ori27 | M. galloprovincialis | Burzyński and Śmietanka (2009) |
| NC_006161 | M. edulis | Boore et al. (2004) | |
| NC_015993 | M. californianus | Cao et al. (2009) | |
| EF434638 | 42ori | M. galloprovincialis | Filipowicz et al. (2008) |
| AY497292 | M. galloprovincialis | Mizi et al. (2005) | |
| AY363687 | M. galloprovincialis | Mizi et al. (2005) | |
| HM462080 | kan12 | M. trossulus | Śmietanka et al. (2010) |
| HM462081 | kan35 | M. trossulus | Śmietanka et al. (2010) |
| DQ399833 | M. galloprovincialis | Venetis et al. (2007) | |
| DQ198231 | 39mc10 | M. trossulus | Zbawicka et al. (2007) |
| DQ198225 | 87mc10 | M. edulis | Zbawicka et al. (2007) |
| GU936625 | 34LE | M. trossulus | Zbawicka et al. (2010) |
| GU936626 | 149LE | M. trossulus | Zbawicka et al. (2010) |
| GU936627 | 117LE | M. trossulus | Zbawicka et al. (2010) |
| JX486124 | M. californianus | GenBank |
To ascertain the recombination and identify recombination breakpoints, recombination detection algorithms: Geneconv (Padidam et al. 1999), MaxChi (Maynard Smith 1992), Chimaera (Posada and Crandall 2001), SiScan (Gibbs et al. 2000), Bootscan (Martin et al. 2005a) and 3SEQ (Boni et al. 2007), as implemented in RDP software (Martin et al. 2005b), were used. Only the recombination events detected with p < 0.05 after Bonferroni correction for multiple tests by at least two methods were considered. This seemed to be the optimal criterion for detecting recombination in this system.
Results
Mitochondrial genome organization
Complete sequences of nine and approximately 70–80 % of two more mtDNA haplotypes of Baltic Mytilus were obtained, including 100 % of coding sequence in all cases. They represent all haplogroups described previously from the Baltic (Burzyński et al. 2006) and a genome very similar (<1 % divergence) to the native M. trossulus F genome, not reported from the Baltic earlier (Table 1). The genomes have the same gene order as the published M. edulis, M. galloprovincialis and M. trossulus F and M genomes (Mizi et al. 2005; Breton et al. 2006; Zbawicka et al. 2007, 2010; Śmietanka et al. 2010). The annotated genes usually have very similar lengths to the related published genomes. Notably, the coding sequences of all haplotypes representing presumably masculinized genomes from 1a, 1 and 16 haplogroups have a length of 15,587 bp, the same as in the typical F genome of M. edulis. However, we noticed an interesting substitution in one of the 11a haplotypes (115mc10). The apparent substitution from T to C occurred in the stop codon of this gene, transforming the termination codon into CAA, encoding glutamine. The next possible stop codon is located 12 bp downstream, extending the nad5 ORF to overlap the neighboring nad6.
There is a certain intra-group length polymorphism associated primarily with the CR. In the case of the four 1a haplotypes, the differences are limited to the CR (1,000–1,006 bp). The length of the single sequenced M haplotype differs by 2 bp from the previously published example of the Baltic M. trossulus M genome (Zbawicka et al. 2007) and this difference is also the result of a longer CR. Similarly, the length of two 11a haplogroup genomes is different (17,720 and 17,724 bp) and the difference is caused by different lengths of their CR (2,134 and 2,139 bp), but in this case there is also 1 bp difference in the length of the lrrna gene. In two cases, we were unable to sequence the CR because of the numerous and long repeats. The CR of one sequenced haplotype 16 (46mc10) was longer than 8 kb, as estimated by PCR products sizing. In the apparently native M. trossulus F-like genome (62mc10), the CR has a complex structure, with the functional trnQ gene translocated into the CR between its M-like and F-like parts, as described previously for other M. trossulus F genomes (Cao et al. 2009; Zbawicka et al. 2010; Śmietanka et al. 2010). However, this CR was also only partially sequenced (1,884 bp), because of the numerous and long repeats. The CR length exceeds 9 kb in this genome; it is 3× longer than in genomes derived from Scotland and Canada (Breton et al. 2006; Śmietanka et al. 2010; Zbawicka et al. 2010).
We propose one minor change in the annotations. In the new interpretation the nad5 gene starts earlier, covering the region previously interpreted as UR5, with a small overlap with the nad4L gene. The nad5 length change resulting from this reinterpretation is 21 bp in F-like and 36 bp in divergent M genomes.
Recombination
The complete mtDNA sequences of six haplotypes: four 1a and one typical F and M have been aligned to elucidate the exact positions of recombination breakpoints. The recombination signal is very strong (Table 3). Surprisingly, the breakpoints detected in particular 1a genomes are not consistent. Two breakpoints are present in each of them, however, not necessarily at the same positions. The results are summarized in Fig. 1, and the alignment details can be seen in Fig. 2. The first breakpoint marking the change from F to M sequence is located in lrrna gene at three possible positions, while the second, marking the change from M to F sequence, is located within the CR, also at three different locations. In effect, the fragment with the high similarity to the M genome has different lengths in the four studied genomes: 1,388 bp (25mc10), 1,317 bp (107mc10), 1,150 bp (152mc10) and 934 bp (136mc10). The remaining parts of the 1a genomes show no signature of recombination and are derived from a typical F genome. Other sequenced genomes were also checked for the presence of recombination signatures. The mosaic or rearranged CR structure of haplotypes 11a, 1 and 16 described by Burzyński et al. (2006) was confirmed (data not shown). No recombination was detected in the coding parts of any of the sequenced genomes, unlike in the recent study of somatic recombination in M. galloprovincalis (Ladoukakis et al. 2011). However, new and unexpected signature of recombination was found in the CR of the native M. trossulus F genome (62mc10) when the sequenced part of the CR was compared with the corresponding part of the Canadian and Scottish genomes (accession numbers AY823625, HM462080, HM462081, GU936625, GU936626) (Table 3; Fig. 3). Apparently, a 706 bp long fragment, very similar in sequence to the native M genome of M. trossulus, replaced the corresponding part of the CR. This fragment has a distance of only 0.0129 from either 149LE or kan35—comparable to the distance between these M genomes and much bigger than the distance from the M. trossulus native F genomes (K = 0.1). This region has been previously shown to be derived from the ancient M genome (Rawson 2005), despite its presence in all native F genomes of M. trossulus. The two recombination breakpoints are located within this M-like part of the CR, making this a case of a secondary recombination: the new fragment of M genome is located within the old fragment of the M genome. The sequenced parts of this genome CR show further signs of sequence instability, such as a relatively big, 105 bp deletion in the 3′ part of the CR (VD2).
Table 3.
Statistical support (average p value) given by recombination detection programs for recombination breakpoints in haplotypes from haplogroup 1a and in the native M. trossulus F genome from the Baltic Sea (62mc10)
| Methods | 1a | 62mc10 |
|---|---|---|
| RDP | 1.649 × 10−86 | 2.538 × 10−21 |
| GENECONV | 9.873 × 10−79 | 1.268 × 10−13 |
| Bootscan | 3.175 × 10−85 | 3.025 × 10−20 |
| MaxChi | 6.591 × 10−36 | 1.778 × 10−19 |
| Chimaera | 5.480 × 10−42 | 4.454 × 10−19 |
| SiScan | 1.234 × 10−47 | 2.137 × 10−18 |
| 3Seq | 1.006 × 10−160 | 4.794 × 10−40 |
Fig. 1.
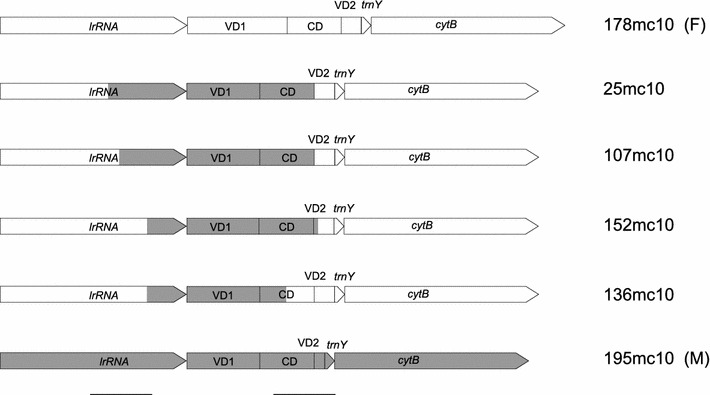
Schematic diagram of the CR structure of the four recombinant haplotypes from the 1a haplogroup (25mc10, 107mc10, 152mc10 and 136mc10). Typical F (178mc10) and M (195mc10) structures are shown for comparison. The shaded boxes represent regional similarities to the M genomes; open boxes represent F-related regions. All haplotypes were aligned at the lrrna. Alignments of the two regions containing recombination breakpoints (indicated by vertical lines below the figure) are presented in Fig. 2
Fig. 2.

Alignment of the region of recombination of the six genomes shown in Fig. 1
Fig. 3.
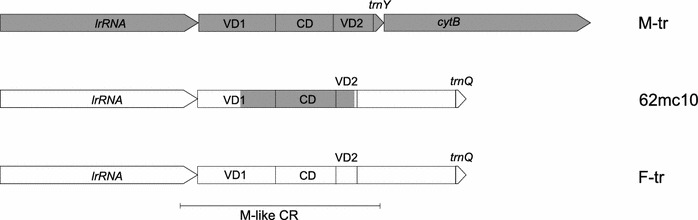
Schematic diagram of the CR of the native M. trossulus haplotype (62mc10). Typical M. trossulus M (GU936625, GU936626, HM462081), and F (AY823625, HM462080) genomes are shown for comparison. The shaded boxes represent regional similarities to the M genome; open boxes represent F-related regions. All haplotypes were aligned at the lrrna
The recombination breakpoints are usually located within fragments of high similarity between putative parental sequences precluding the exact identification of their position and context. However, in the case of the genome belonging to haplogroup 16 (46mc10), an interesting sequence was found at both recombination breakpoints. In this genome, no mosaic M–F structure was found, but an extensive array of repeats of apparently F origin is seen. The repeat array starts at VD1 of the CR and ends within the cob gene. At each of the two recovered breakpoints, the presence of the same structure was discovered (Fig. 4). It is formed by parts of the cob sequence and the short stretch of homology between VD1 and cob. The boxed fragment joining cob and VD1 is not homologous to either parental sequence, but it is perfectly matched to the 10 bp cob fragment upstream from the breakpoint, completing a very large hairpin structure around it with an overall stability of dG = −30 kcal/mol.
Fig. 4.
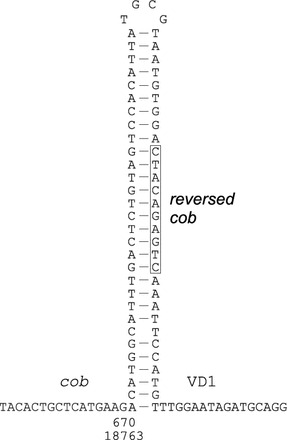
The fragment of the 46mc10 genome (haplogroup 16), with a hairpin structure, present near the two sequenced breakpoints. The boxed fragment is not derived directly from any of the parental sequences (VD1 or cob), but could be synthesized by strand switching during DNA synthesis only
Evolution
To put the sequenced genomes in an evolutionary perspective, a neighbor-joining phylogenetic tree based on the concatenated protein-coding sequences was constructed (Fig. 5). The underlying genetic distances are listed in Supplementary Table 2. The distances separating any haplotype 16 (K = 0.0035, SE = 0.0006) or 1a (K = 0.0040, SE = 0.0006) from the F genomes (39mc10, 178mc10) are similar to the distance between these F genomes (K = 0.0039, SE = 0.0006). Slightly larger distances are observed between haplotypes from the recombinant 11a haplogroup and their sister nonrecombinant haplotype, the M. galloprovincialis F genome from Azov Sea (azo20)(K = 0.0163, SE = 0.0012). The native M. trossulus genome, while very distant from the other F-like genomes from the Baltic, shows only mildly biased affinity toward the published M. trossulus F genome from the West Atlantic (Breton et al. 2006) (K = 0.0046, SE = 0.0007), as compared to the genome from the East Pacific (Śmietanka et al. 2010) (K = 0.0114, SE = 0.0008), with an average distance within this group at 0.0087.
Fig. 5.

Neighbor-joining phylogeny based on concatenated protein-coding alignment (11,958 bp) from the whole mitochondrial genomes of Mytilus mussels. The genomes from the present study are in bold. Genome labels follow the convention outlined in Tables 1 and 2
To assess the degree of selective pressure acting on genomes, the concatenated protein-coding sequences were analyzed in several comparisons. The genomes were sorted into groups consisting of 2–4 sequences and K a/K s were calculated within groups. As expected, the K a values are lower than K s, indicating purifying selection (Table 4). The values observed for the M. edulis M genome groups (average K a/K s = 0.1241) are higher than for F genomes (K a/K s = 0.0765), consistent with relaxed pressure in M groups, first postulated by Stewart et al. (1996). In the Baltic recombinants, this effect is even more pronounced, particularly for haplotypes from the groups 1a (K a/K s = 0.2031) and 1 (K a/K s = 0.2264). Interestingly, the recombinant 42ori genome compared with its closest nonrecombinant genomes revealed the degree of selective constraint similar to that of typical M. edulis or M. galloprovincialis F genomes. Contrary to that, both Baltic representatives of the 11a group did show elevated accumulation of nonsynonymous changes in comparisons involving the 42ori genome. The M. trossulus F genomes have relatively high K a/K s (0.1605).
Table 4.
K a/K s comparisons within and between groups made of groups of 2–4 sequences of mitochondrial proteins
| Comparison | Sequences used in each comparison | Origin of sequences | K a/K s |
|---|---|---|---|
| F genomes | M.edulis, Eur | 39mc10, 178mc10 | 0.0765 |
| M. trossulus, Amer + Eur | AY823625, kan12, 34LE, 62mc10 | 0.1605 | |
| M. galloprovincialis, Eur | azo20, AY497292 | 0.0512 | |
| M genomes | M.edulis, Eur | 87mc10, 195mc10 | 0.0927 |
| M.edulis, Amer | AY823623, AY823624 | 0.1097 | |
| M. galloprovincialis, Eur | ori27, AY363687 | 0.1381 | |
| M. trossulus, Amer + Eur | kan35, 149LE, 117LE | 0.1560 | |
| Recombinant genomes | 1a, Baltic | 25mc10, 107mc10, 136mc10, 152mc10 | 0.2031 |
| 11a, Baltic | 20mc10, 115mc10 | 0.0873 | |
| 1, M. galloprovincialis C | 45mc10, DQ399833 | 0.2264 | |
| 11a and M. galloprovincialis mf2 | 20mc10, 42ori | 0.0996 | |
| 11a and M. galloprovincialis mf2 | 115mc10, 42ori | 0.0799 | |
| 1a, 16, Baltic | 25mc10, 107mc10, 136mc10, 152mc10, 46mc10 | 0.1348 | |
| Recombinant and F genomes | 1a, Baltic vs M.edulis, Eur | 25mc10, 107mc10, 136mc10, 152mc10 vs 39mc10, 178mc10 | 0.1082 |
| 16, Baltic vs M.edulis, Eur | 46mc10 vs 39mc10, 178mc10 | 0.0907 | |
| M. galloprovincialis mf2 vs M. galloprovincialis, Eur | 42ori vs azo20, AY497292 | 0.0466 | |
| 11a, Baltic vs M. galloprovincialis, Eur | 20mc10 vs azo20 | 0.0623 | |
| 11a, Baltic vs M. galloprovincialis, Eur | 115mc10 vs azo20 | 0.0489 |
To further elucidate the observed patterns a codon-by-codon analysis was applied to the concatenated data set. Out of 3,986 codons, 33 showed signature of positive and 3,253 of negative selection by at least one of the methods. An example of the dominance of negatively selected sites in FEL results is shown in Fig. 6. The sites with the strongest signatures of positive selection are marked by arrows and the support is summarized in Table 5. These nonsynonymous substitutions were, however, not distributed consistently on the phylogenetic tree; the GA-Branch analysis revealed accelerated accumulation of these substitutions at terminal branches only, particularly those leading to recombinant haplotypes (data not shown). We searched for group-specific substitutions using individual monophyletic groups or a single polyphyletic “group” identified simply by nature of their being recombinant. There were no substitutions common to any group of the recombinant genomes sequenced.
Fig. 6.

Concatenated protein-coding gene alignment was analyzed by FEL procedure in the context of the phylogenetic tree (Fig. 5).The dN–dS plot capped at −100 is presented. The values above zero indicate a candidate for positively selected sites. The gene position is shown at the top. The support for sites marked by arrows is presented in Table 5
Table 5.
Codons that may be under positive selection
| Codon | Gene | S | NS | SLAC dN–dS | SLAC p value | FEL dN–dS | FEL p value | MEME p value | FUBAR Post. Pr. | FUBAR dN–dS |
|---|---|---|---|---|---|---|---|---|---|---|
| 413 | cob | 0 | 7 | 0.31 | 0.09 | 1.217 | 0.040 | 0.078 | 0.669 | 0.236 |
| 436 | cob | 0 | 5 | 0.203 | 0.229 | 0.862 | 0.051 | 0.055 | 0.589 | 0.085 |
| 1272 | nad4 | 2 | 8 | 0.205 | 0.253 | 0.940 | 0.431 | 0.019 | 0.534 | 0.225 |
| 2085 | nad2 | 1 | 7 | 0.224 | 0.199 | 5.00 × 10−9 | 0.101 | 0.027 | 0.505 | 0.084 |
| 2254 | atp8 | 0.75 | 5.25 | 0.192 | 0.235 | 0.912 | 0.084 | 0.025 | 0.499 | 0.022 |
| 2349 | cox1 | 1 | 6 | 0.269 | 0.093 | 1.198 | 0.322 | 0.514 | 0.485 | 0.134 |
| 3278 | nad5 | 3 | 9 | 0.285 | 0.148 | 2.891 | 0.11 | 0.138 | 0.926 | 2.511 |
| 3669 | nad5 | 1 | 6 | 0.157 | 0.33 | 5.00 × 10−9 | 0.134 | 0.044 | 0.443 | −0.009 |
S observed synonymous changes, NS observed nonsynonymous changes
Discussion
The repertoire of mitochondrial genomes present in the paternal lineage of the Baltic Mytilus population is surprising. They represent all four major F lineages present in the M. edulis species complex (Śmietanka et al. 2009, 2010, 2013). The recent advances in transcriptomics (Chatzoglou et al. 2013) allow more precise annotation of some protein-coding genes within this diverse data set. Our proposal to extend the nad5 gene is consistent with the notion that polycistronic transcript consisting of nad4l, nad5, and nad6 is expressed in Mytilus (Chatzoglou et al. 2013; Sańko and Burzyński 2014). The fragment we propose to include as coding is present within the transcript. The only drawback of this proposal is the generated overlap between nad4 l and nad5. We argue that it is quite possible to have overlapping genes within such a transcript. The extension in nad5 ORF of the 115mc10 genome leads to another example of gene overlap, yet in this case it cannot be avoided by changing annotations.
Native M. trossulus genome in the Baltic Sea
We demonstrate for the first time the presence of the native M. trossulus mtDNA in the Baltic Sea. The haplotype C2 (T-5) reported by Quesada et al. (2003) from Baltic Sea and suggested to be the ancestral European M. trossulus M genomes is in fact most likely one of the recombinant 1a haplotypes, with the first breakpoint at the same positions as in the 107mc10 genome. The overall distance between T-5 and 107mc10 over the whole published sequence is only K = 0.0058 (SE = 0.0041), much lower than its distance from the native M. trossulus genomes (K = 0.0397, SE = 0.0167). Contrary to that, the 62mc10 genome we report here is most certainly closely related to the native M. trossulus F genomes (Fig. 1). The possibility that this genome is an artifact is unlikely for the following reasons. We took utmost care to avoid contamination; therefore, we believe that the obtained sequences are present in the studied animal. The presence of numts in Baltic Sea mussels was negatively verified (Zbawicka et al. 2007), and given the technique used to obtain the sequences it is extremely unlikely that such long parts of mtDNA could be derived from the nuclear genome. The recombinant structure of the CR of this genome is also unlikely to be a product of somatic recombination, because no parental sequences were found in this individual. Interestingly, 62mc10 was a heteroplasmic female. There was no indication of the biased presence of such genomes in males, but the mechanism proposed for this type of heteroplasmy (two divergent genomes in a female) ultimately involves the inheritance of one of the genomes through the paternal lineage in the female ancestor (Obata et al. 2006, 2007; Kyriakou et al. 2010). Therefore we cannot be sure what the transmission route of this genome is, but it certainly is currently very rare in the Baltic Sea. Despite that, it is clearly the part of the same radiation as the other fully sequenced F genomes of M. trossulus (Fig. 1). Since it has been shown that this radiation took place post-glacially (Śmietanka et al. 2013), we can conclude that the age of this genome is in the order of 104 years.
Recombination mechanism
The strong support for the secondary recombination within the 62mc10 genome was unexpected. We have not found the parental native M. trossulus M genome in the Baltic, but the fragment is almost 10× more similar to the contemporary M genomes from other M. trossulus populations than to the respective fragment of the F genomes implying that the native M genome must have also been present. The alternative scenario with the introgression of the genome from some other population in Scandinavia or Scotland can be also considered but will require further screening for similar rearrangements in these populations. Since the same part of the molecule was shown to be involved in a similar recombination event (Rawson 2005; Breton et al. 2006; Cao et al. 2009; Śmietanka et al. 2010; Zbawicka et al. 2010), and no other part of the molecule seems to have any evidence of recombination, it is tempting to speculate that the propensity of the CR to undergo recombination is the driving force of these events. More support for this notion comes from the analysis of recombination breakpoints in the representative haplotype from the 16 haplogroup (46 mc10). This haplogroup was originally considered very young and we speculated that the genomes belonging to this haplogroup could have originated within the individuals we found them in (Burzyński et al. 2006). Given the number of nonsynonymous polymorphisms the sequenced genome accumulated, it seems rather unlikely. However, this genome must still be considered very young, since its distances from its nearest non-rearranged relatives are very small (Fig. 5). We hope that the structure of the breakpoints within this genome are close to their primary, nondegenerated state. The very stable hairpin structure found at the breakpoint has twofold consequences. Such structures by themselves are considered recombinogenic (Chen 2013) and could contribute to the development of long arrays of repeats within the CR of this genome. On the other hand, the sequence at the breakpoint (boxed in Fig. 2) could not possibly be obtained from either of the potential parental fragments. Instead, it is complementary to the fragment of the truncated cob on the other side of the hairpin. The only possibility of obtaining such a fragment is to synthesize it using wrong strand as a matrix for DNA synthesis. Although not conclusive, it suggests the involvement of DNA replication in these rearrangements. Short tandem repeats capable of forming secondary structures are very common in animal mtDNA genomes. These can easily form tandem arrays of several hundred to over a thousand nucleotides (He et al. 2011).
Role reversal and recombinantion
A connection between recombination and masculinization was first hypothesized upon the discovery of CR recombinants (Burzyński et al. 2003). It was suggested that the M-derived fragment of the CR inserted into the otherwise F-like genome caused it to invade the paternal route of inheritance. This hypothesis was formulated based on a limited set of sequence information, covering only the CR and short flanking coding sequences. Mild objections have been raised on the grounds that other parts of these genomes may also have been mosaic (Venetis et al. 2007). With the complete sequences of the haplotypes in question, we can now be sure that recombination in their case is indeed limited to the short part of the CR. It has also been pointed out that the proof for the paternal inheritance of recombinant genomes was indirect only (Cao et al. 2009). The apparently increased rate with which these genomes accumulate nonsynonymous substitutions now provides further argument for their paternal inheritance. The distribution of this feature across the phylogenetic tree (Fig. 5; Table 4) shows that the recombination events predate masculinization. There are two phylogenetic clades hosting the majority of masculinized haplotypes from the Baltic (Fig. 5). The older clade is closely related to the M. galloprovincialis F genome (azo20). It has been dated at approximately 300 KYA (Śmietanka et al. 2010). The two masculinized haplotypes from the Baltic (20mc10 and 115mc10), belonging to the 11a haplogroup (Burzyński et al. 2006) and one recombinant but apparently nonmasculinized haplotype from the Mediterranean Sea (42ori)(Filipowicz et al. 2008) form this well-resolved clade. The two Baltic haplotypes must have independently switched their transmission routes to account for the observed pattern of polymorphism (Table 4). This pattern is consistent with the scenario under which first a recombination event occurs, then the recombinant genome exists in the population for a while (inherited maternally), and later it becomes masculinized and is thereafter inherited through males. Therefore, the masculinization is likely more recent within this clade than the clade age. The distance between the 115mc10 and 42ori haplotypes is 3× lower than the distance from their common nonrecombinant sister haplotype (azo20). It can be concluded that each of the haplotypes, 20mc10 and 115mc10, independently experienced a role reversal (masculinization) event. The alternative interpretation, involving one masculinization event of the ancestral genome to 115mc10 and 20mc10 followed by a feminization event in the formation of 42ori, is equally parsimonious, but would not lead to the observed patterns of nonsynonymous substitutions. The second clade containing haplotypes from 1a haplogroup contains not only recombinant, but also typical F haplotypes. The clade is internally unresolved (Fig. 5; Supplementary Fig. S1–S3) and, given the fact that full mtDNA sequences were used to construct the tree, there is not enough phylogenetic information accumulated within this clade to ever resolve the relationships between haplotypes. The only plausible cause for this is the extremely young age of this group. In our earlier work (Burzyński et al. 2003, 2006), haplotypes from this group were assumed to be a product of a single recombination event, but the first recombination breakpoint was not accounted for. Given the now resolved positions of all the breakpoints (Figs. 1, 2), this assumption is no longer valid. To explain the different extent of the M-like fragment present in the CR of haplotypes from the 1a haplogroup, we must assume a series of homologous recombinations, either independently involving the highly divergent M genome or, what is more likely, sequentially involving the primary recombinants and typical F genomes. In either case these events must be very recent indeed, the divergence between the genomes in question does not exceed a few substitutions (Supplementary Table 2, K < 0.005) and is consistent with the history of this clade confined entirely to the short lifespan of the Baltic Sea (no more than 10,000 years).
Hybridization and masculinization
In the hybrid population of Baltic Mytilus mussels, apparently high level of both CR recombination and masculinization has been taking place in the last few thousands years. A possible explanation could involve the destabilization of mtDNA due to cytonuclear incompatibility. The exclusive presence of M. edulis mtDNA in Baltic Sea Mytilus mussels would not cause functional difficulties if the nuclear background was also dominated by M. edulis component. Several papers suggested this to be the case (Riginos et al. 2002; Bierne et al. 2003; Kijewski et al. 2006, 2011). However, with more nuclear markers it becomes apparent that the Baltic Sea mussels are much closer to M. trossulus than M. edulis (Zbawicka et al. 2014). Therefore, the hybridization could have caused abrupt and rapid increase in the M genome incompatibility, favoring masculinization. Given the presented pattern of polymorphisms, it is tempting the speculate that preexisting recombinant genomes (such as 20mc10 and 115mc10) become masculinized only during the recent history of the Baltic Sea, together with the newly generated recombinants (1a haplotypes). Apparently, the genomes with M-like CRs are preferentially masculinized, but the existence of masculinized genomes with complex CR structures but without M-like sequences (such as 46mc10) suggests that these genomes may spread due to factors other than the recognition of particular sequences within the CR.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was partially funded by project EC ICA1-CT-2002-70022, SPB 127/E-335/SPB/5. PR UE/DIE 63/2005, Polish Ministry of Science and Higher Education to R.W., through a Grant No. NN303 418336 to A.B. and by the statutory topic IV.1 in the IO PAS.
References
- Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- Bierne N, Daguin C, Bonhomme F, David P, Borsa P. Direct selection on allozymes is not required to explain heterogeneity among marker loci across a Mytilus hybrid zone. Mol Ecol. 2003;12:2505–2510. doi: 10.1046/j.1365-294X.2003.01936.x. [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Medina M, Rosenberg LA. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the Scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol Biol Evol. 2004;21:1492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- Breton S, Burger G, Stewart DT, Blier PU. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.) Genetics. 2006;172:1107–1119. doi: 10.1534/genetics.105.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzyński A, Śmietanka B. Is interlineage recombination responsible for low divergence of nad3 in Mytilus galloprovincialis? Mol Biol Evol. 2009;26:1441–1445. doi: 10.1093/molbev/msp085. [DOI] [PubMed] [Google Scholar]
- Burzyński A, Zbawicka M, Skibinski DOF, Wenne R. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol Biol Evol. 2003;20:388–392. doi: 10.1093/molbev/msg058. [DOI] [PubMed] [Google Scholar]
- Burzyński A, Zbawicka M, Skibinski DOF, Wenne R. Doubly uniparental inheritance is associated with high polymorphism for rearranged and recombinant control region haplotypes in Baltic Mytilus trossulus. Genetics. 2006;174:1081–1094. doi: 10.1534/genetics.106.063180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Kenchington E, Zouros E, Rodakis GC. Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.) Genetics. 2004;167:835–850. doi: 10.1534/genetics.103.026187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Ort BS, Mizi A, Pogson G, Kenchington E, et al. The control region of maternally and paternally inherited mitochondrial genomes of three species of the sea mussel genus Mytilus. Genetics. 2009;181:1045–1056. doi: 10.1534/genetics.108.093229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzoglou E, Kyriakou E, Zouros E, Rodakis GC. The mRNAs of maternally and paternally inherited mtDNAs of the mussel Mytilus galloprovincialis: start/end points and polycistronic transcripts. Gene. 2013;520:156–165. doi: 10.1016/j.gene.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Chen XJ. Mechanism of homologous recombination and implications for aging-related deletions in mitochondrial DNA. Microbiol Mol Biol Rev. 2013;77:476–496. doi: 10.1128/MMBR.00007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curole JP, Kocher TD. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol Biol Evol. 2002;19:1323–1328. doi: 10.1093/oxfordjournals.molbev.a004193. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Filipowicz M, Burzyński A, Śmietanka B, Wenne R. Recombination in mitochondrial DNA of European mussels Mytilus. J Mol Evol. 2008;67:377–388. doi: 10.1007/s00239-008-9157-6. [DOI] [PubMed] [Google Scholar]
- Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- He CB, Wang J, Gao XG, Song WT, Li HJ, Li YF, Liu WD, Su H. The complete mitochondrial genome of the hard clam Meretrix meretrix. Mol Biol Rep. 2011;38:3401–3409. doi: 10.1007/s11033-010-0449-8. [DOI] [PubMed] [Google Scholar]
- Hoeh WR, Stewart DT, Saavedra C, Sutherland BW, Zouros E. Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia: Mytilidae) Mol Biol Evol. 1997;14:959–967. doi: 10.1093/oxfordjournals.molbev.a025839. [DOI] [PubMed] [Google Scholar]
- Kijewski TK, Zbawicka M, Väinölä R, Wenne R. Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M. edulis. Mar Biol. 2006;149:1373–1385. doi: 10.1007/s00227-006-0316-2. [DOI] [Google Scholar]
- Kijewski T, Śmietanka B, Zbawicka M, Gosling E, Hummel H, et al. Distribution of Mytilus taxa in European coastal areas as inferred from molecular markers. J Sea Res. 2011;65:224–234. doi: 10.1016/j.seares.2010.10.004. [DOI] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kyriakou E, Zouros E, Rodakis G. The atypical presence of the paternal mitochondrial DNA in somatic tissues of male and female individuals of the blue mussel species Mytilus galloprovincialis. BMC Research Notes. 2010;3:222. doi: 10.1186/1756-0500-3-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladoukakis ED, Theologidis I, Rodakis GC, Zouros E. Homologous recombination between highly diverged mitochondrial sequences: examples from maternally and paternally transmitted genomes. Mol Biol Evol. 2011;28:1847–1859. doi: 10.1093/molbev/msr007. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- Liu HP, Mitton JB, Wu SK. Paternal mitochondrial DNA differentiation far exceeds maternal mitochondrial DNA and allozyme differentiation in the fresh-water mussel, Anodonta grandis grandis. Evol Int J Org Evol. 1996;50:952–957. doi: 10.2307/2410870. [DOI] [PubMed] [Google Scholar]
- Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses. 2005;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Mizi A, Zouros E, Moschonas N, Rodakis GC. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol. 2005;22:952–967. doi: 10.1093/molbev/msi079. [DOI] [PubMed] [Google Scholar]
- Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, et al. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol. 2013;30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Obata M, Kamiya C, Kawamura K, Komaru A. Sperm mitochondrial DNA transmission to both male and female offspring in the blue mussel Mytilus galloprovincialis. Dev Growth Differ. 2006;48:253–261. doi: 10.1111/j.1440-169X.2006.00863.x. [DOI] [PubMed] [Google Scholar]
- Obata M, Sano N, Kawamura K, Komaru A. Inheritance of two M type mitochondrial DNA from sperm and unfertilized eggs to offspring in Mytilus galloprovincialis. Dev Growth Differ. 2007;49:335–344. doi: 10.1111/j.1440-169X.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Passamonti M, Scali V. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia) Curr Genet. 2001;39:117–124. doi: 10.1007/s002940100188. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol Biol Evol. 2005;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada H, Wenne R, Skibinski DOF. Interspecies transfer of female mitochondrial DNA is coupled with role-reversals and departure from neutrality in the mussel Mytilus trossulus. Mol Biol Evol. 1999;16:655–665. doi: 10.1093/oxfordjournals.molbev.a026148. [DOI] [PubMed] [Google Scholar]
- Quesada H, Stuckas H, Skibinski DOF. Heteroplasmy suggests paternal co-transmission of multiple genomes and pervasive reversion of maternally into paternally transmitted genomes of mussel (Mytilus) mitochondrial DNA. J Mol Evol. 2003;57(Suppl 1):S138–S147. doi: 10.1007/s00239-003-0019-y. [DOI] [PubMed] [Google Scholar]
- Rawson PD. Nonhomologous recombination between the large unassigned region of the male and female mitochondrial genomes in the mussel, Mytilus trossulus. J Mol Evol. 2005;61:717–732. doi: 10.1007/s00239-004-0035-6. [DOI] [PubMed] [Google Scholar]
- Rawson PD, Hilbish TJ. Asymmetric introgression of mitochondrial DNA among European population of blue mussels (Mytilus spp.) Evol Int J Org Evol. 1998;52:100–108. doi: 10.2307/2410924. [DOI] [PubMed] [Google Scholar]
- Riginos C, Sukhdeo K, Cunningham CW. Evidence for selection at multiple allozyme loci across a mussel hybrid zone. Mol Biol Evol. 2002;19:347–351. doi: 10.1093/oxfordjournals.molbev.a004088. [DOI] [PubMed] [Google Scholar]
- Sańko T, Burzyński A. Co-expressed mitochondrial genomes: recently masculinized, recombinant mitochondrial genome is co-expressed with the female – transmitted mtDNA genome in a male Mytilus trossulus mussel from the Baltic Sea. BMC Genet. 2014;15:28. doi: 10.1186/1471-2156-15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- Serb JM, Lydeard C. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca) Mol Biol Evol. 2003;20:1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM. Mitochondrial DNA inheritance. Nature. 1994;368:817–818. doi: 10.1038/368817b0. [DOI] [PubMed] [Google Scholar]
- Śmietanka B, Burzyński A, Wenne R. Molecular population genetics of male and female mitochondrial genomes in European mussels Mytilus. Mar Biol. 2009;156:913–925. doi: 10.1007/s00227-009-1137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Śmietanka B, Burzyński A, Wenne R. Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: rapidly evolving atp8. J Mol Evol. 2010;71:385–400. doi: 10.1007/s00239-010-9393-4. [DOI] [PubMed] [Google Scholar]
- Śmietanka B, Zbawicka M, Sańko T, Wenne R, Burzyński A (2013) Molecular population genetics of male and female mitochondrial genomes in subarctic Mytilus trossulus. Mar Biol 160:1709–1721. doi:10.1007/s00227-013-2223-7 [DOI] [PMC free article] [PubMed]
- Staden R, Judge DP, Bonfield JK (2001) Sequence assembly and finishing methods. In: Baxevanis AD, Ouellette BFF (eds) Bioinformatics. A practical guide to the analysis of genes and proteins. Wiley, New York [DOI] [PubMed]
- Stewart DT, Kenchington ER, Singh RK, Zouros E. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics. 1996;143:1349–1357. doi: 10.1093/genetics/143.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evol Int J Org Evol. 2008;62:959–970. doi: 10.1111/j.1558-5646.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- Venetis C, Theologidis I, Zouros E, Rodakis GC. A mitochondrial genome with a reversed transmission route in the Mediterranean mussel Mytilus galloprovincialis. Gene. 2007;406:79–90. doi: 10.1016/j.gene.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Wenne R, Skibinski DOF. Mitochondrial DNA heteroplasmy in European populations of the mussel Mytilus trossulus. Mar Biol. 1995;122:619–624. doi: 10.1007/BF00350683. [DOI] [Google Scholar]
- Werle E, Schneider C, Renner M, Volker M, Fiehn W. Convenient single-step, one tube purification of PCR products for direct sequencing. Nucleic Acids Res. 1994;22:4354–4355. doi: 10.1093/nar/22.20.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbawicka M, Skibinski DOF, Wenne R. Doubly uniparental transmission of mitochondrial DNA length variants in the mussel Mytilus trossulus. Mar Biol. 2003;142:455–460. [Google Scholar]
- Zbawicka M, Burzyński A, Wenne R. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene. 2007;406:191–198. doi: 10.1016/j.gene.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zbawicka M, Burzyński A, Skibinski D, Wenne R. Scottish Mytilus trossulus mussels retain ancestral mitochondrial DNA: complete sequences of male and female mtDNA genomes. Gene. 2010;456:45–53. doi: 10.1016/j.gene.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Zbawicka M, Sańko T, Wenne R. New SNP markers reveal largely concordant clinal variation across the hybrid zone between Mytilus spp. in the Baltic Sea. Aquat Biol. 2014;21:25–36. doi: 10.3354/ab00566. [DOI] [Google Scholar]
- Zouros E. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet Syst. 2000;75:313–318. doi: 10.1266/ggs.75.313. [DOI] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA. 1994;91:7463–7467. doi: 10.1073/pnas.91.16.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


