Abstract
A prophage vB_CibM-P1 was induced by mitomycin C from the epipelagic strain Citromicrobium bathyomarinum JL354, a member of the alpha-IV subcluster of marine aerobic anoxygenic phototrophic bacteria (AAPB). The induced bacteriophage vB_CibM-P1 had Myoviridae-like morphology and polyhedral heads (approximately capsid 60–100 nm) with tail fibers. The vB_CibM-P1 genome is ~38 kb in size, with 66.0% GC content. The genome contains 58 proposed open reading frames that are involved in integration, DNA packaging, morphogenesis and bacterial lysis. VB_CibM-P1 is a temperate phage that can be directly induced in hosts. In response to mitomycin C induction, virus-like particles can increase to 7 × 109 per ml, while host cells decrease an order of magnitude. The vB_CibM-P1 bacteriophage is the first inducible prophage from AAPB.
Temperate phages integrate their nucleic acid into host bacterial genomes in a process termed lysogeny1,2. The genetic material of a bacteriophage in host chromosome, which is referred to as prophage, can be passed to daughter cells during cell division. Prophages may be induced under certain circumstances (such as UV radiation or mitomycin C treatment), which activates a lytic cycle1,3,4,5. Thus, prophages are viewed as “dangerous molecular time bombs”6. DNA could be transferred among different hosts during phage infection through either the lytic cycle or the lysogenic cycle1,2,3,4,5. Previous studies showed that more than 50% of marine isolates harbor prophages, which play a significant role in shaping the phenotypic traits of their hosts1,4,5.
Citromicrobium bathyomarinum strain JL354, a member of aerobic anoxygenic phototrophic bacteria (AAPB), was isolated from surface water in the South China Sea7. The type strain C. bathyomarinum JF-1 was first isolated from the deep-sea hydrothermal vent plume waters8. A complete prophage sequence was found from the preliminary genomic analysis of strain JL354. Two different phototrophic operons and a xanthorhodopsin-like protein co-exist in the same genome7,9,10. Horizontal gene transfer is thought to occur for phototrophic genes of phototrophic microorganisms. The objectives of this study were to 1) induce the prophage from C. bathyomarinum JL354 and 2) compare its genome to other homologous phages.
Results
Discovery of the vB_CibM-P1 phage by induction from C. bathyomarinum JL354
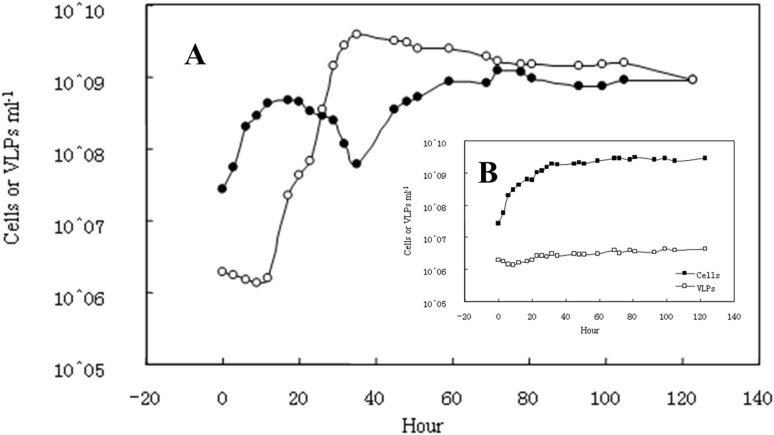
A novel Mu-like phage-related gene cluster was discovered within the genomic sequence of C. bathyomarinum JL3547. To determine whether this phage was active, mitomycin C was used to treat C. bathyomarinum JL354 in the exponential growth phase and flow cytometry was used to measure the induction of virus-like particles (VLPs). Within 30 hours, the quantity of VLPs increased up to three orders of magnitude (Fig. 1), while the number of bacterial cells decreased an order of magnitude. The induced VLPs had Myoviridae-like (i.e., T4-like) morphology and polyhedral heads (approximately capsid 60–100 nm) with tail fibers (Fig. 2).
Figure 1. Viral particle yield following mitomycin C induction of C. bathyomarinum JL354.
Flow cytometry counts of JL354 cells and viral-like particles were performed with (A) a mitomycin C-treated culture and (B) a control culture without mitomycin C.
Figure 2. Virus generated by mitomycin C induction of C. bathyomarinum JL354.
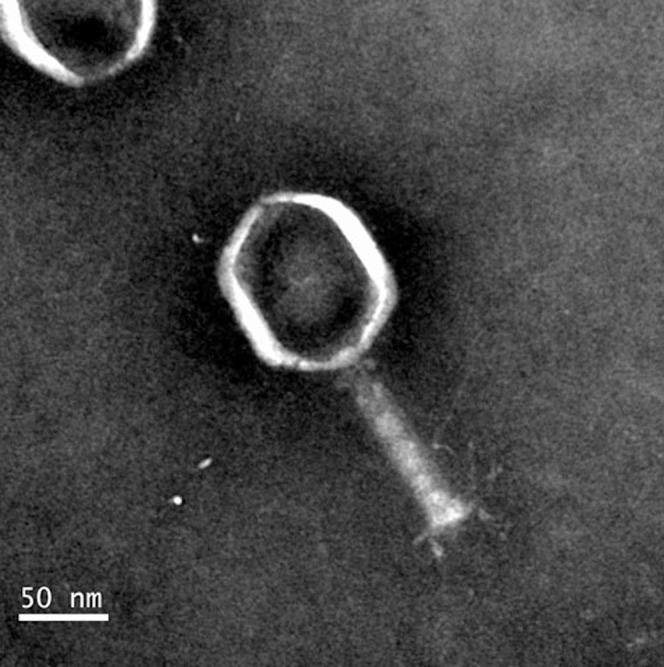
Scale bars, 50 nm.
To confirm whether the induced phage matched the vB_CIBM-P1 prophage observed in the C. bathyomarinum JL354 genome, the induced viral DNA was re-sequenced. Although there was host DNA contamination, the read-rich region mapped to the predicted prophage position (Zheng et al., unpublished data).
Genomic structure of phage vB_CibM-P1
The complete genomic sequence of phage vB_CIBM-P1 is ~38 kb, with a GC content of 66.0%, which is slightly higher than that of its host (65.0%). The vB_CIBM-P1 GC content is much higher than that of bacteriophage Mu (52.0%) from E. coli MH5361 and slightly higher than the prophage found in Hoeflea phototrophica DFL-43 (61.7%). Using PHACTS, a novel tool for predicting whether the lifestyle of a phage is primarily lytic or lysogenic, that- determined vB_CIBM-P1 was temperate11.
In total, the vB_CIBM-P1 genome contains 58 predicted open reading frames (ORFs), representing 97% of the entire genome (Table 1 and Fig. 3). Forty-three ORFs have ATG start codons, whereas 13 ORFs and 2 ORFs start with GTG and TTG codons, respectively. No tRNA genes were detected by tRNAscan-SE12. Thirty-three ORFs yielded significant hits (E value ≤ 1e-5, coverage ≥ 75%) in the GenBank database, and 21 ORFs were assigned as unknown (Table 1).
Table 1. Gene Annotation List for vB_CIBM-P1.
| ORF | Start | Stop | Size(aa) | Start codon | GC content | Predicted function | Significant matches to proteins in GenBank | % aa identity | PSI-BLAST E-value |
|---|---|---|---|---|---|---|---|---|---|
| orf1 | 432 | 1 | 144 | ATG | 0.63 | Transcription regulation | putative phage repressor [Thioalkalivibrio sp. HL-EbGR7] | 42/105 (40%) | 2.00E-12 |
| orf2 | 759 | 980 | 74 | GTG | 0.59 | Transcription regulation | hypothetic protein | ||
| orf3 | 1243 | 1118 | 42 | ATG | 0.57 | hypothetic protein | |||
| orf4 | 1514 | 1209 | 102 | ATG | 0.61 | hypothetic protein | |||
| orf5 | 1509 | 1748 | 80 | ATG | 0.63 | hypothetic protein | |||
| orf6 | 1849 | 2853 | 335 | ATG | 0.65 | ParB protein, nuclease | chromosome partitioning protein parB nuclease [Bartonella tribocorum CIP 105476] | 69/243 (28%) | 4.00E-12 |
| orf7 | 2853 | 3077 | 75 | ATG | 0.60 | hypothetic protein | |||
| orf8 | 3067 | 3762 | 232 | ATG | 0.66 | DNA methyltransferase? | hypothetical phage protein (possible DNA methyltransferase) [Campylobacter phage CPt10] | 52/203 (25%) | 3.00E-10 |
| orf9 | 3759 | 4121 | 121 | ATG | 0.67 | hypothetic protein | |||
| orf10 | 4121 | 4447 | 109 | ATG | 0.69 | hypothetic protein | |||
| orf11 | 4441 | 4668 | 76 | GTG | 0.67 | hypothetic protein | |||
| orf12 | 4671 | 4874 | 68 | ATG | 0.68 | hypothetic protein | |||
| orf13 | 4867 | 5100 | 78 | ATG | 0.62 | hypothetic protein | |||
| orf14 | 5097 | 5291 | 65 | GTG | 0.71 | hypothetic protein | |||
| orf15 | 5288 | 6214 | 309 | ATG | 0.66 | Gp37Gp68 family protein [Gluconacetobacter diazotrophicus PAl 5] | 153/306 (50%) | 8.00E-83 | |
| orf16 | 6207 | 6668 | 154 | ATG | 0.68 | hypothetical protein PPMP29_gp03 [Pseudomonas phage MP29] | 40/107 (37%) | 3.00E-13 | |
| orf17 | 6661 | 8628 | 656 | ATG | 0.68 | Transposase A | Mu DNA binding I gamma subdomain [Xanthobacter autotrophicus Py2] | 262/639 (41%) | 7.00E-134 |
| orf18 | 8699 | 9736 | 346 | ATG | 0.64 | Transposase B | DNA transposition protein gpB [Bordetella petrii DSM 12804] | 108/285 (37%) | 4.00E-44 |
| orf19 | 9733 | 9882 | 50 | GTG | 0.73 | hypothetic protein | |||
| orf20 | 9882 | 10205 | 108 | GTG | 0.65 | Replication initiation protein DnaA | chromosomal replication initiator protein DnaA [Carboxydothermus hydrogenoformans Z-2901] | 31/83 (37%) | 5.00E-07 |
| orf21 | 10253 | 10885 | 211 | ATG | 0.68 | Modulation of host genes | Mu-like prophage protein gp16 [Fulvimarina pelagi HTCC2506] | 62/168 (36%) | 7.00E-24 |
| orf22 | 10882 | 11232 | 117 | ATG | 0.67 | hypothetic protein | |||
| orf23 | 11453 | 11719 | 89 | GTG | 0.69 | conserved hypothetical protein [Rhodopseudomonas palustris TIE-1] | 31/74 (41%) | 3.00E-10 | |
| orf24 | 11709 | 12068 | 120 | ATG | 0.63 | conserved hypothetical protein [Roseomonas cervicalis ATCC 49957] | 35/91 (38%) | 2.00E-11 | |
| orf25 | 12129 | 12704 | 192 | ATG | 0.64 | Lysozyme | phage lysozyme [Rhodobacter capsulatus SB1003] | 51/158 (32%) | 7.00E-31 |
| orf26 | 12785 | 13381 | 199 | ATG | 0.66 | hypothetic protein | |||
| orf27 | 13390 | 13818 | 143 | ATG | 0.68 | hypothetic protein | |||
| orf28 | 13815 | 14249 | 145 | ATG | 0.65 | hypothetic protein | |||
| orf29 | 14246 | 14554 | 103 | ATG | 0.67 | hypothetical protein HPDFL43_14842 [Hoeflea phototrophica DFL-43] | 50/89 (56%) | 5.00E-26 | |
| orf30 | 14554 | 15096 | 181 | ATG | 0.68 | Transmembrane protein | putative ubiquitin-conjugating enzyme E2 transcript variant [Taeniopygia guttata] | 55/178 (30%) | 2.00E-20 |
| orf31 | 15093 | 15671 | 193 | ATG | 0.63 | Terminase small subunit | Mu-like phage gp27 [Fulvimarina pelagi HTCC2506] | 76/187 (40%) | 3.00E-37 |
| orf32 | 15889 | 15683 | 69 | TTG | 0.65 | hypothetic protein | |||
| orf33 | 16006 | 16173 | 56 | TTG | 0.64 | hypothetic protein | |||
| orf34 | 16166 | 17872 | 569 | ATG | 0.67 | Terminase large subunit | Mu-like prophage FluMu protein gp28 [Magnetospirillum magneticum AMB-1] | 312/524 (59%) | 0 |
| orf35 | 17994 | 19565 | 524 | ATG | 0.67 | Portal | Mu-like prophage FluMu protein gp29 [Hoeflea phototrophica DFL-43] | 288/530 (54%) | 0 |
| orf36 | 20037 | 19786 | 84 | ATG | 0.62 | hypothetic protein | |||
| orf37 | 20050 | 21162 | 371 | ATG | 0.68 | Head morphogenesis | bacteriophage Mu GP30-like protein [Roseovarius sp. 217] | 189/374 (50%) | 9.00E-110 |
| orf38 | 21239 | 21730 | 164 | ATG | 0.63 | Virion morphogenesis | virion morphogenesis protein [Roseovarius sp. 217] | 78/171 (45%) | 3.00E-40 |
| orf39 | 21805 | 21939 | 45 | ATG | 0.71 | hypothetic protein | |||
| orf40 | 21936 | 23021 | 362 | GTG | 0.67 | Protease | Mu-like prophage I protein [Hoeflea phototrophica DFL-43] | 162/360 (45%) | 7.00E-80 |
| orf41 | 23023 | 23427 | 135 | ATG | 0.67 | DNA mismatch repair | hypothetical bacteriophage protein [Pseudomonas aeruginosa] | 72/134 (53%) | 9.00E-40 |
| orf42 | 23440 | 24339 | 300 | ATG | 0.62 | Major head subunit | Mu-like prophage major head subunit gpT-like protein [Hoeflea phototrophica DFL-43] | 160/299 (54%) | 4.00E-115 |
| orf43 | 24446 | 25132 | 229 | ATG | 0.70 | hypothetic protein | |||
| orf44 | 25136 | 25564 | 143 | GTG | 0.68 | Mu-like prophage Flumu protein gp36 [Syntrophus aciditrophicus SB] | 57/144 (39%) | 1.00E-16 | |
| orf45 | 25561 | 26019 | 153 | GTG | 0.68 | hypothetical protein RTM1035_05080 [Roseovarius sp. TM1035] | 53/143 (37%) | 2.00E-23 | |
| orf46 | 26016 | 26258 | 81 | ATG | 0.73 | hypothetic protein | |||
| orf47 | 26259 | 27197 | 313 | ATG | 0.67 | hypothetical protein HPDFL43_14907 [Hoeflea phototrophica DFL-43] | 164/309 (53%) | 2.00E-113 | |
| orf48 | 27758 | 27961 | 68 | ATG | 0.72 | phage protein [Silicibacter sp. TrichCH4B] | 30/64 (46%) | 5.00E-12 | |
| orf49 | 27261 | 27590 | 110 | ATG | 0.65 | hypothetical protein Rsph17025_1309 [Rhodobacter sphaeroides ATCC 17025] | 48/109 (44%) | 2.00E-21 | |
| orf50 | 27987 | 30326 | 780 | GTG | 0.64 | Tail tape measure | Tail length tape measure protein [Polymorphum gilvum SL003B-26A1] | 42/118 (36%) | 5.00E-07 |
| orf51 | 30323 | 30991 | 223 | ATG | 0.67 | hypothetical protein Oant_0252 [Ochrobactrum anthropi ATCC 49188] | 57/167 (34%), | 6.00E-07 | |
| orf52 | 30975 | 31583 | 203 | ATG | 0.66 | Tail fiber protein | hypothetical protein Oant_0253 [Ochrobactrum anthropi ATCC 49188] | 75/205 (36%) | 7.00E-23 |
| orf53 | 31598 | 31999 | 134 | ATG | 0.69 | hypothetic protein | |||
| orf54 | 31984 | 34098 | 705 | ATG | 0.67 | Tail fiber protein H | putative tail fiber protein H [Rhizobium phage 16-3] | 187/677 (27%) | 7.00E-47 |
| orf55 | 34111 | 36042 | 644 | GTG | 0.65 | hypothetical protein [uncultured phage MedDCM-OCT-S09-C299] | 124/488 (25%) | 2.00E-05 | |
| orf56 | 36076 | 36867 | 264 | ATG | 0.63 | hypothetic protein | |||
| orf57 | 36876 | 37052 | 59 | GTG | 0.61 | hypothetic protein |
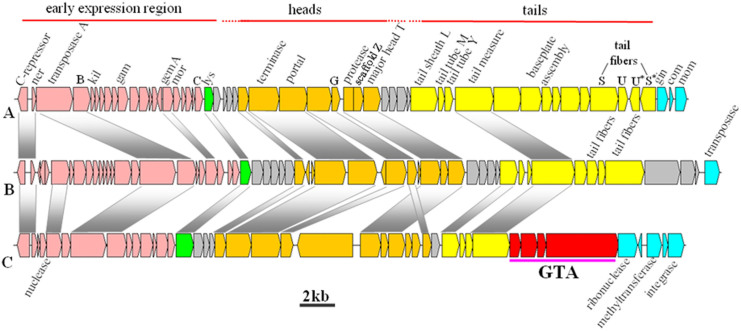
Figure 3. Comparison of bacteriophage Mu (A), vB_CIBM-P1 (B) and prophage in Hoeflea phototrophica DFL-43 (C).
Pink, early expression genes; orange, heads; yellow tails; red, GTA-like region; green, lysozyme genes; light gray, putative proteins.
Early expression region
The early expression region contains regulatory genes with associated lytic genes followed by two structural regions encoding for head and tail assembly (Fig. 3, Table 1)13. The genome starts with a C-repressor and ner (encoded by ORF 1 and ORF 2, respectively), which are involved in the regulation of lysogeny and lytic development. Similar to bacteriophage Mu, if the C-repressor bound, it would repress the lytic cycle and induce the lysogenic cycle instead; whereas if unbound, the lytic cycle would occur13. Transposases A and B, which are also located in this region at position 6661–9736, allow for integration (ORF 17) and transposition (ORF 18), respectively. ORF 17 encodes phage integrase, which is most similar to the integrase in Roseomonas cervicalis ATCC 49957.
Head, tail and tail fiber assembly
The head proteins have high identity to classical Mu or Mu-like bacteriophages, which suggests that heads may appear to be Myoviridae-like. From the structure and composition of the three phages (bacteriophage Mu, vB_CIBM-P1 and the Mu-like prophage from Hoeflea phototrophica DFL-43), the genes encoding head protein assembly are within the most conserved region (Fig. 3). ORF 37 and ORF 42 are the two identifiable head proteins found in the genome (Table 1). ORF 37 is most identical to a Mu-like phage in Roseovarius sp. 217, which is another marine alphaproteobacterial (AAPB) found in the euphotic zone. The final head assembly protein ORF 42 has the highest similarity (54%) with the Mu-like major head subunit of Hoeflea phototrophica DFL-4314.
ORFs 50–54 form the major tail/tail fiber proteins, which are 9 kb in size and represent 24% of the genome. However, only the tail tape measure protein appears to be similar to the Mu-like tail protein, and the rest of the tail proteins have closer identity to Siphoviridae (e.g., Rhizobium phage 16-3)15.
Discussion
The vB_CIBM-P1 genome is highly mosaic in nature, but it is a novel representative of marine Mu-like phages or myoviruses. The genome has 58 predicted ORFs, and 17% of the genome encodes for identifiable Mu-like genes. Forty percent of the genome encoding for predicted proteins has no match in GenBank database, indicating the novelty and lack of knowledge about the genomes of tailed phages. Additional data are needed to confirm this variability in genome structure.
Mu-like phages, in general, have few predicted and conserved tail proteins, which are supported by the hypothesis that tailed phages are genetic mosaics derived by multiple step-wise recombination exchanges that occur within a large gene pool16,17,18,19. Tail proteins are generally more variable due to their role as host-range determinants17,18. Consistent with other Mu-like phages, genes that contribute to tail morphology are quite diverse and may be a red-queen driven process20. If driven by a red-queen process, both phage tail and host entry receptors need to evolve at a highly consistent rate20, which could potentially explain the high diversity and lack of conservation observed in this region.
Phylogenetic analysis of Mu-like phages
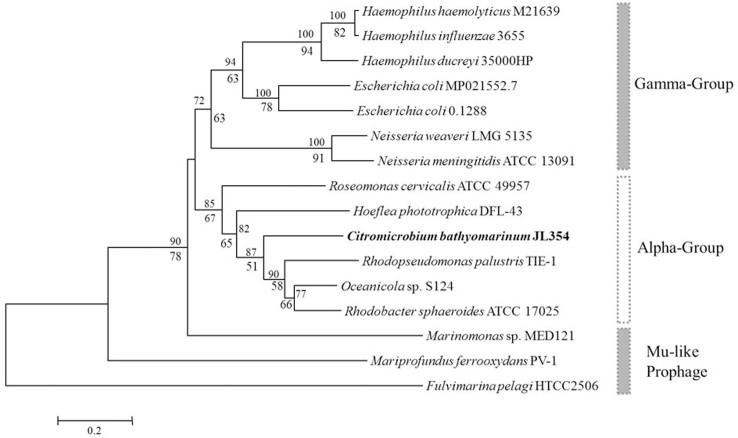
The major head gene was compared to many other Mu-like phages or prophages present in host genomes. The replicative nature of the genome in the phage vB_CIBM-P1 made it difficult to assign all sequences to a meaningful phylogeny due to their high mosaicism and variable genome structure13. Many Mu-like phages or prophages have been found in enteric bacteria, e.g., Gamma group (Fig. 4)13. It appears that AAPB hosts containing Mu-like prophages are mainly found in aquatic environments (freshwater or marine, Alpha group, Fig. 4). The vB_CIBM-P1 host C. bathyomarinum JL354 is a member of the Alpha group, with close identity to Hoeflea phototrophica DFL-43 (Fig. 4).
Figure 4. Neighbor joining phylogenetic tree based on the phage major head gene sequences.
The Mu-like prophage is commonly found in the Neisseria, Escherichia and Haemophilus genera. It has been recently found in marine bacteria genomes. All bacteria had complete Mu-like prophage sequences in their genomes. Haemophilus haemolyticus M21639, NZ_AFQR00000000.1; Haemophilus influenzae 3655, NZ_AAZF00000000.1; Haemophilus ducreyi 35000HP, NC_002940.2; NZ_ AQCL00000000.1; Escherichia coli 0.1288, NZ_AMVJ00000000.1; Neisseria weaveri LMG 5135, NZ_AFWQ00000000.1; Neisseria meningitidis ATCC 13091, NZ_AEEF00000000.1; Roseomonas cervicalis ATCC 49957, NZ_ADVL00000000.1; Hoeflea phototrophica DFL-43, NZ_ABIA00000000.2; Citromicrobium bathyomarinum JL354, NZ_ADAE00000000.1; Rhodopseudomonas palustris TIE-1, NC_011004.1; Oceanicola sp. S124, NZ_AFPM00000000.1; NC_009428.1; Marinomonas sp. MED121, NC_009654.1; Mariprofundus ferrooxydans PV-1, NZ_AATS00000000.1; Fulvimarina pelagi HTCC2506, NZ_AATP00000000.1. Bootstrap percentages (>50) from neighbor joining (above) and maximum likelihood (below) are shown in the tree. The scale bar represents 20% amino acid substitution.
Bacteriophage of vB_CIBM-P1
Mu genome replication occurs by duplicative transposition into host chromosomes by random integration13,19,21,22. At the time of Mu genome packaging, the phage transposase cuts beyond the phage genome and can include approximately 1.8–3.0 Kb of host DNA23,24. In our re-sequencing data of the induced phage vB_CibM-P1, none of the reads contained host DNA fragments at either end of the phage genome. Additional experiments are needed to determine whether our phage has similar transposition properties as the Mu phage. Furthermore, our prophage could be induced by mitomycin C instead of high temperature (42°C), while previous studies showed that the Mu phage is usually induced by high temperature rather than mitomycin C13,24. All of the evidence indicates that the phage vB_CIBM-P1 is a novel myovirus.
Although temperate phages are widely found in marine bacteria2,4,6, this study is the first to report a temperate phage isolated from a marine aerobic anoxygenic phototrophic bacterium. With the increasing number of bacterial genomes in the GenBank database, more marine prophages will be potentially found and characterized, which will improve our understanding of the evolution and function of phage-host interactions. Comparison of phototrophy and closely related non-phototrophy genomes may provide clues to how temperate phages influence the evolution of photosynthetic genes.
Methods
Sequence analysis
The draft genome sequence of C. bathyomarinum JL354 is available under the GenBank accession number NZ_ADAE00000000. The JL354 genome was manually analyzed for the presence of a putative prophage. First, the genome was scanned for phage-related genes. When a phage-related gene cluster (ZP_06862281-ZP_06862290) was encountered, the surrounding genes were also examined. Putative prophage fragments were re-annotated by performing a PSI-BLAST search against the GenBank database for further analysis25. The beginning and end of a specific prophage genome were estimated based on the annotations of surrounding genes. The complete prophage genome was used to predict lifestyle with the Phage Classification Tool Set (PHACTS) (http://www.phantome.org/PHACTS/)11.
A tRNA search was performed in the prophage genome using tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE)12. Homologous prophages from Hoeflea phototrophica DFL-43 (NZ_ABIA00000000) and bacteriophage Mu (NC_000929) were used for comparative genomic analysis with the prophage found in C. bathyomarinum JL354.
Nearly complete major head (>900 bp) gene was used to construct a phylogenetic tree. All sequences collected from the NCBI database were aligned using Clustal X, and phylogenetic trees were constructed using the neighbor-joining and maximum-likelihood algorithms in MEGA software 5.026. The phylogenetic trees were supported by bootstrap for resampling test with 1000 and 100 replicates using neighbor-joining and maximum-likelihood algorithms respectively.
Induction of the C. bathyomarinum JL354 prophage with mitomycin C
C. bathyomarinum JL354 was grown in rich organic (RO, containing 1.0 g yeast extract, 1.0 g Bacto Peptone and 1.0 g sodium acetate per liter artificial seawater with vitamins and trace element.)8 medium at 28°C with a shaking speed of 180 rpm throughout the induction experiment. The induction process and sampling were performed according to the protocol described by Chen et al.3,27. Briefly, 10 ml of JL354 culture in exponential growth phase was transferred to 200 ml of fresh RO medium. After the subculture reached OD600 = 0.25, it was split into two flasks (100 ml in each); one was treated with mitomycin C (final concentration, 0.5 μg ml−1) and the other served as the control. After incubation for 30 min, the control and mitomycin C-treated cells were washed twice, centrifuged at 7,500 × g for 10 min, and resuspended in 100 ml fresh RO broth. Samples (0.5 ml) for viral and bacterial counting were fixed with glutaraldehyde (final concentration: 2.5%) for 15 min in the dark and then stored in liquid nitrogen for flow cytometry analysis28,29.
Viral particles and bacterial count
Virus and bacterial counts were determined by an Altra Epics II flow cytometer (Beckman Coulter), which was equipped with an external quantitative sample injector (Harvard Apparatus PHD 2000). The bacterial and viral particles were identified and counted28. SYBR Green I (Molecular Probes) was used as a nucleic acid stain for bacteria identification in red fluorescence versus green fluorescence plots. Virus samples were analyzed separately from bacteria samples. Briefly, once thawed at 37°C, samples were diluted in 0.02-μm filtered TE (Tris-EDTA, pH = 8) buffer as needed and heated for 10 min in the dark at 80°C after staining with the DNA dye SYBR Green I, and then cooled for 5 min prior to analysis28,29,30. Viruses were discriminated on the basis of their green DNA-dye fluorescence versus 90° angle light scatter28,29,30.
Purification of induced phage
Phage particles in lysates were harvested and purified as described by Chen et al.3,27 with the following modifications. Phage lysates were treated with RNase A (final concentration: 2 μg ml−1) and DNase I (final concentration: 2 μg ml−1) at room temperature for 1 h. One liter of induced phage lysate was centrifuged at 12,000 × g for 15 min in a Hettich Rotina 38R centrifuge. Supernatants were filtered through a 0.45-μm pore size filter (type HA; Millipore) to remove host cells and cellular debris. Phage particles in the filtrate were treated with polyethylene glycol 8000 (final concentration: 100 g l−1) overnight at 4°C and precipitated by centrifugation at 12,000 × g for 90 min. Pellets were re-suspended with 6 ml of SM buffer (10 mM NaCl, 50 mM Tris, 10 mM MgSO4, and 0.1% gelatin) and incubated overnight at 4°C. The phage suspension was mixed with CsCl (final concentration, 0.6 g ml−1) and centrifuged for 24 h at 200,000 × g using an S55A rotor in a Micro-Ultracentrifuge CS-GX series centrifuge. Visible viral bands were extracted and then dialyzed (MWT = 30 KD) twice in SM buffer overnight at 4°C. CsCl-purified phage lysates were stored at 4°C until further analysis. Virus morphology was examined by transmission electron microscopy (JEM 2100 HC).
Extraction of phage DNA
Purified phage were treated with a proteinase K cocktail (100 μg ml−1 proteinase K, 50 mM Tris, 25 mM EDTA, and 1% SDS) at 55°C for 3 h. The phage DNA was extracted using phenol/chloroform/isoamyl alcohol (25:24:1 by volume)3,27. The DNA was then ethanol-precipitated, re-suspended in TE buffer (10 mM Tris, 1 mM EDTA), and re-sequenced with an Agilent 2100 Bioanalyzer (Ion Torrent PGM, Invitrogen, China). One library with average sizes of 350 bps was constructed. After quality control, total 26,064 reads (4,314,206 bps) were used to assemble the viral genome.
Author Contributions
Q.Z., R.Z. and N.J. conceived and designed the experiments; Q.Z., Y.X., R.A.W., Y.W. and T.L. analyzed the data. All of the authors assisted in writing the manuscript, discussed the results and commented on the manuscript.
Acknowledgments
This work was supported by the 973 program (2013CB955700) and the SOA project (GASI-03-01-02-05) to NJ, Fundamental Research Funds for the Central Universities (2013121051) and NSFC project (41306126) to QZ, and the NSFC project (41376132) to RZ.
References
- Ackermann H. W. & DuBow M. S. Viruses of prokaryotes, vol. I. General properties of bacteriophages. Vol. I (CRC Press, Inc., 1987). [Google Scholar]
- Williamson S. J., Houchin L. A., McDaniel L. & Paul J. H. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microb. 68, 4307–4314 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wang K., Stewart J. & Belas R. Induction of multiple prophages from a marine bacterium: a genomic approach. Appl. Environ. Microb. 72, 4995–5001 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. C., Kellogg C. A. & Paul J. H. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microb. 64, 535–542 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitet C., Riemann L. & Hagstrom A. Plasmids and prophages in Baltic Sea bacterioplankton isolates. J. Mar. Biol. Assoc. Uk. 86, 567–575 (2006). [Google Scholar]
- Paul J. H. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J 2, 579–589 (2008). [DOI] [PubMed] [Google Scholar]
- Jiao N. Z., Zhang R. & Zheng Q. Coexistence of two different photosynthetic operons in Citromicrobium bathyomarinum JL354 as revealed by whole-genome sequencing. J. Bacteriol. 192, 1169–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov V. V., Krieger S., Stackebrandt E. & Beatty J. T. Citromicrobium bathyomarinum, a novel aerobic bacterium isolated from deep-sea hydrothermal vent plume waters that contains photosynthetic pigment-protein complexes. J. Bacteriol. 181, 4517–4525 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. K. et al. Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome. Biol. Evol. 5, 187–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. et al. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PloS one 7, e35790 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNair K., Bailey B. A. & Edwards R. A. PHACTS, a computational approach to classifying the lifestyle of phages. Bioinformatics 28, 614–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic. Acids. Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan G. J., Hatfull G. F., Casjens S. & Hendrix R. W. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317, 337–359 (2002). [DOI] [PubMed] [Google Scholar]
- Fiebig A. et al. Genome of the marine alphaproteobacterium Hoeflea phototrophica type strain (DFL-43T). Stand. Genomic. Sci. 7, 440–448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorgai L. et al. The detailed physical map of the temperate phage 16-3 of Rhizobium meliloti 41. Mol. Gen. Genet. 191, 430–433 (1983). [DOI] [PubMed] [Google Scholar]
- Boltner D., MacMahon C., Pembroke J. T., Strike P. & Osborn A. M. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184, 5158–5169 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R. W., Hatfull G. F. & Smith M. C. M. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154, 253–257 (2003). [DOI] [PubMed] [Google Scholar]
- Hendrix R. W., Smith M. C. M., Burns R. N., Ford M. E. & Hatfull G. F. Evolutionary relationships among diverse bacteriophages and prophages: All the world's a phage. P. Natl. Acad. Sci. USA 96, 2192–2197 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste J. L., Paulin J. P. & Expert D. Bacteriophage Mu as a genetic tool to study Erwinia amylovora pathogenicity and hypersensitive reaction on tobacco. J. Bacteriol. 172, 932–941 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez P. & Buckling A. Bacteria-phage antagonistic coevolution in soil. Science 332, 106–109 (2011). [DOI] [PubMed] [Google Scholar]
- Braid M. D., Silhavy J. L., Kitts C. L., Cano R. J. & Howe M. M. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 186, 6560–6574 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne R. et al. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari A. & Taylor A. Influence of insertions on packaging of host sequences covalently linked to bacteriophage Mu DNA. P. Natl. Acad. Sci. USA 72, 4399–4403 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg P. C., Hynes A. P., Digby E., Lang A. S. & Beatty J. T. Characterization of a newly discovered Mu-like bacteriophage, RcapMu, in Rhodobacter capsulatus strain SB1003. Virology 421, 211–221 (2011). [DOI] [PubMed] [Google Scholar]
- Schaffer A. A. et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic. Acids. Res. 29, 2994–3005 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. Searching for a “hidden” prophage in a marine bacterium. Appl. Environ. Microb. 76, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard C. P. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microb. 70, 1506–1513 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D., Partensky F., Jacquet S. & Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microb. 63, 186–193 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D., Brussaard C. P. D., Thyrhaug R., Bratbak G. & Vaulot D. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microb. 65, 45–52 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]