Abstract
In this chapter, we describe a protocol used for stable silencing of chemokine receptor CXCR7 in human cancer cells using shRNA in a lipid transfection setting, previously published by our laboratory. We provide thorough detail and background information about the process of shRNA to clarify the importance of this process. We use CXCR7 shRNA and scrambled sequence shRNA constructs cloned into a pRS plasmid under the control of a U6 promoter for stable expression. Human cancer cells are transfected with shRNA-pRS using Lipofectamine 2000. Cells stably expressing the shRNA are selected from transfected cultures following 2 weeks in medium containing the selection antibiotic puromycin. The emergent cell colonies are evaluated for knockdown of CXCR7 mRNA and protein expression by q-PCR and immunoblotting with rabbit anti-CXCR7 IgG, respectively.
Keywords: shRNA, Lipofectamine 2000, Stable transfection, Selection, CXCR7
1 Introduction
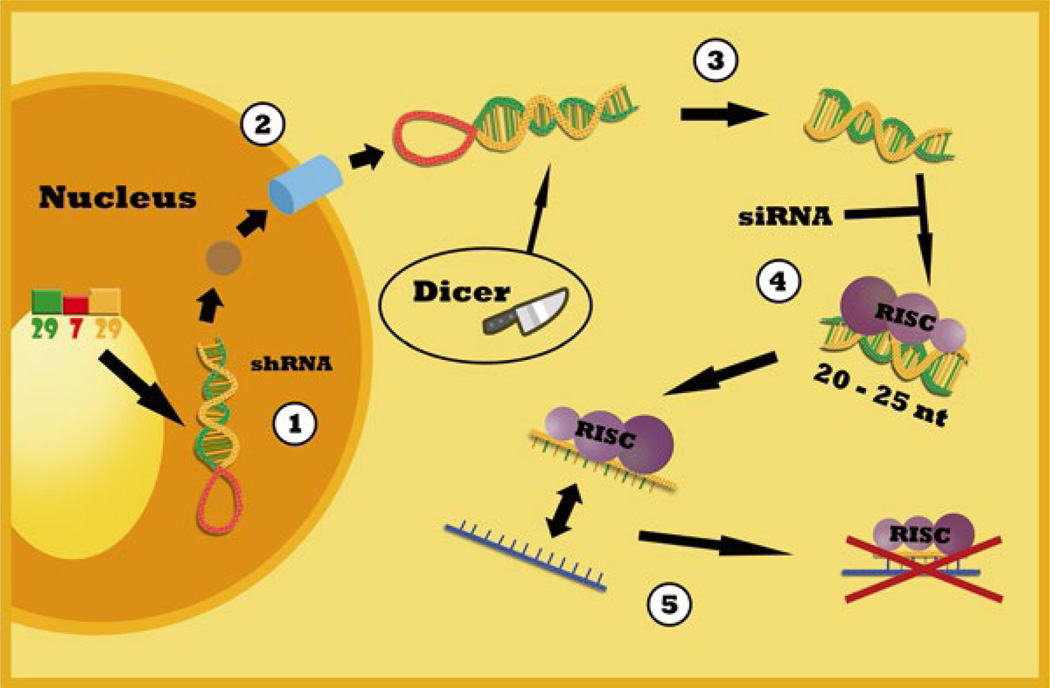
The discovery that double-stranded RNA was orders of magnitude more potent than single-stranded RNA at gene silencing revolutionized modern biology and opened the door to RNAi [1]. This discovery in 1998 earned Fire and Mello the Nobel Prize in Medicine in 2006 [2]. Lee and Ambros described that conserved RNA species undergo processing through a system known as the RNAi machinery, and that the starting product of this process is a stem-loop or short hairpin RNA precursor [3]. This RNA precursor is produced endogenously in the cell as a long double-stranded non-coding RNA transcript known as pri-miRNA. The pri-miRNA forms a hairpin or stem-loop shaped structure as the RNA anneals with itself due to sense and antisense sequences that flank the loop. This double-stranded precursor is then processed by Drosha in the nucleus and exported to the cytoplasm where it is further processed by Dicer to fragment it into pieces of mature microRNA (miRNA) [4, 5]. These short dsRNA sequences are recognized by the RISC complex. The complex combined with the miRNA can recognize and halt targeted mRNA transcripts from being translated. See Fig. 1 for a schematic illustration.
Fig. 1.
Schematic illustration of the use of shRNA for stable suppression of chemokine receptor expression and function in human cancer cell lines. (1) Pri-miRNA endogenously produced in all mammalian nuclei or shRNA is introduced through transfection. (2) Drosha enzyme processes pri-miRNA to pre-miRNA, which is recognized and exported by Exportin V to the cytoplasm. (3) Dicer recognizes pre-miRNA and digests it into short oligos of 20–25 nucleotides of dsRNA called miRNA. (4) RISC complex can bind to this miRNA or to an introduced set of siRNA delivered through transfection. (5) The mi/siRNA-RISC complex then binds to the target mRNA and prevents its translation. Illustrated by Ms. Maite Lopez
Since miRNAs produced by mammalian cells do not have complete complementarity to their targets, it is possible to produce and deliver small interfering RNAs (siRNA) that mimic the function of miRNAs but are designed to have greater specificity to their targets by having complete complementary sequences [6]. One significant drawback to this assay however, is the depletion of siRNA over a few days from delivery. An alternative to this direct delivery method is the development of shRNA and its delivery through a vector-expressing plasmid which contains a selection marker. Expression of the shRNA sequence includes a 29-mer region complementary to the target transcript, followed by a 7-nucleotide loop, followed again, by the antisense sequence of the 29mer region [7]. This produces a dsRNA structure that is similar to the naturally produced pri-miRNAs of the cell and is processed accordingly to its miRNA mimic, the siRNA. This setup allows for the continuous, stable expression of the shRNA for suppression of the target gene [4, 8].
In this protocol we describe an efficient approach to stably silence the chemokine receptor, CXCR7 adapted from the manufacturer’s guide to using the transfection reagents. We use RNA interference (RNAi) implemented with short hairpin RNA (shRNA). Vector expressing shRNA can be used to stably suppress gene expression in cell lines. We used a retroviral silencing plasmid (pRS) that contains the puromycin resistance gene obtained from Origene [7]. Our lab has successfully used these shCXCR7s from Origene to stably down regulate CXCR7 expression in breast and prostate cancer cell lines used for functional assays including in-vivo xenograft tumor assays [9]. Origene provides four different shRNA plasmids for the sequence of interest, fully verified by sequencing, thus guaranteeing that at least one of the four constructs provides over 90 % gene expression inhibition. This allows us to use the vectors for sequence modifications and be able to perform RNA rescue experiments with ease. More information about these and lentiviral shRNA vectors can be found at www.origene.com. Origene now also offers a lentiviral vector for CXCR7 shRNA knockdown, which may provide better transfection efficiency.
2 Materials
2.1 Cell Culture
MCF7 cells (American Type Culture Collection, Manassas, VA).
LNCaP cells (American Type Culture Collection, Manassas, VA).
12-well cluster plates.
RNAase and DNAase-free sterile 1.5 microcentrifuge tubes and barrier-filter tips.
Trypsin-EDTA.
RPMI complete medium: RPMI 1640 with l-glutamine, 10 % fetal bovine serum (FBS), 20 µg/mL of gentamicin sulfate solution.
2.2 Transfection
Transfection medium: RPMI 1640 with l-glutamine; 10 % FBS; no gentamicin.
Complex medium: Opti-MEM I reduced serum medium (Invitrogen, Gibco, 31985–062). If not available, use serum-free medium, such as RPMI.
Plasmid DNA (see Note 1): HuSH 29mer shRNA construct against CMKOR1 (chemokine receptor CXCR7), Origene.
Transfection reagent: Can be lipid based or viral based (see Note 2). Lipofectamine 2000 or Lipofectamine LTX with Plus reagent (Invitrogen, 52887 and 94756).
Rabbit anti-CXCR7 IgG antibody.
E.Z.N.A.® RNA Isolation Kit (101319–242), Omega Bio-Tek.
2.3 Stable Clone Selection
Complete medium with antibiotics as described in Subheading 2.1.
Puromycin stock (1 mg/mL) for kill curve dilutions.
Freezing medium: 70 % RPMI, 20 % FBS, 10 % dimethyl sulfoxide (DMSO; cell culture grade). Add gentamicin sulfate to a final concentration of 20 µg/mL.
3 Methods
3.1 Transfection of Human Cancer Cell Lines LNCaP/MCF-7 with shCXCR7
For transfection of human cancer cell lines (LNCaP/MCF-7) with shCXCR7, perform all transfection experiments in a bio-safety level II cell culture hood. If plating on a smaller plate, vary concentrations proportionately according to well area. Example: In a 12-well plate, plate 1.0 × 105 MCF-7 or LNCaP cells per well in antibiotic-free medium 1 day before transfection. Cells should be healthy, well adhered, and 80–90 % confluent at the time of transfection. Wait a maximum of 24 h after plating to transfect the cells (see Note 3). Refer to Table 1 for an example of calculations.
-
For each transfection sample, prepare the transfection reagent as follows (see Note 4). Also see example in Table 1.
TUBE 1:- Add the required amount of Opti-MEM medium (100 µL final volume minus Lipofectamine volume).
- Add Lipofectamine reagent into Opti-MEM medium.
- Mix/vortex gently.
TUBE 2:- Add the required amount of Opti-MEM medium (100 µL final volume minus DNA volume). Add total volume of DNA required for 0.5 µg (in a 12-well plate well).
- Mix/vortex gently.
- Allow mixtures to incubate at room temperature for 5 min.
After the 5 min incubation, combine the diluted DNA with the diluted Lipofectamine and incubate 25 min at room temperature. You should have 200 µL of DNA/reagent complex. While waiting for the complex to form, remove old media from cells, and add 800 µL of antibiotic-free media per well, so the final volume in the well is 1 mL after the complex is added.
Add the required amount of complex to each well (200 µL per well).
Incubate at 37 °C in a CO2 incubator for 6 h.
Replace with antibiotic-free complete media after 6 h of transfection to minimize cell toxicity.
Incubate for 24–72 h before measuring gene expression by real-time quantitative polymerase chain reaction (RT-qPCR), western blot (WB), or the observed presence of a green fluorescent protein (GFP)-tag signal if you use shGFP as an expression marker. We find that 24 h after transfection, mRNA levels indicate the efficiency of the reaction, and 48 h after transfection is optimal for protein level assessment by western blot analysis (see Note 5). The cells can be evaluated for knockdown of CXCR7 mRNA and protein expression by q-PCR and immunoblotting with rabbit anti-CXCR7 IgG antibody, respectively.
Table 1.
Example: for 3 wells of a 12-well dish, we multiply by a 10 % factor, or 1.1 to account for pipetting errors
| Vector | Concentration (µg/µL) | Tube 1 | Tube 2 | ||
|---|---|---|---|---|---|
| DNA: (0.5 µg/well) × 3 wells × 1.1 (µL) | DNA dilution: Serum-free medium (100 µL per well × 3 wells × 1.1) (100 × 3 × 1.1) − (DNA)= | Lipofectamine 2000: 2.0 µL per well × 3 wells × 1.1 (µL) | Lipo dilution: Serum-free medium (100 µL per well × 3 wells × 1.1) (100 × 3 × 1.1)− (Lipo)= | ||
| Control | 0.588 | (0.5 × 3 × 1.1)/0.588 = 2.81 | 327.19 | (2 × 3 × 1.1) = 6.6 | 323.4 |
| shCXCR7 | 0.441 | (0.5 × 3 × 1.1)/0.441 = 3.74 | 326.26 | (2 × 3 × 1.1) = 6.6 | 323.4 |
3.2 Generating Stable Clones (See Note 6)
Seventy-two hours after transfection, begin antibiotic selection of the cells with puromycin. Pick up cells from the well by trypsinization and resuspend them in enough media to aliquot them into a 96-well plate to generate a kill curve (see Note 7) for the resistance gene; in this case, puromycin.
Plate approximately 1 × 104 cells per well in triplicate wells of the 96-well plate or in duplicate if using a 48-well plate. Use complete media. Freeze the remainder of your transfected cells for future selection needs (see Note 8). You may wait until the cells settle, then change the media to the puromycin containing media, or wait 24 h and change media to selection media.
Make a concentration gradient of puromycin ranging at least 6 concentrations. Make 2.0 µg/mL of antibiotic the middle point of the curve, since Origene recommends this concentration as the selection concentration for its plasmid. For example, use the following concentrations: 0, 0.25, 0.5, 1.0, 2.0, 3.0, 5.0, and 7.5 µg/mL of puromycin.
Change the media every 3 days and replace with fresh complete media with puromycin dilutions. Check for death of non-transfected cells before every media change. Note the lowest and highest concentrations at which the cells survived. The lowest concentration at which the cells survived is the selection concentration (see Note 9).
After 1–2 weeks, you should be able to see colonies forming that are your stable clones. Select at least three separate clones and transfer to a well of a 6-well plate, while continuing selection. Once cultures are 50 % confluent, split them into wells of a 12-well plate for RNA and protein extraction and passage remaining cells into a T25 flask, using the routine Trypsin-EDTA cell detachment procedure.
Assay your stable knockdown efficiency of CXCR7 mRNA and protein expression with RT-qPCR and immunoblotting with rabbit anti-CXCR7 IgG antibody, respectively (see Note 10).
We recommend doing mRNA analysis before protein analysis. If the mRNA results are successful, then proceed to the Western blot analysis.
- mRNA analysis: For mRNA expression analysis of CXCR7, purify RNA and use the isolated RNA as the template. Use CXCR7 mRNA transcript specific primers for the qPCR reaction. You can design the primers using Primer-BLAST from the NCBI Web site and mFOLD (http://mfold.rna.albany.edu/?q=mfold) to ensure no secondary structure interferes with the primer target. The qPCR reaction mix from PerfeCTa SYBR Green FastMix for iQ PPS is as follows:
PerfeCTa SYBR Green FastMix for iQ (2×) 10 µL Forward primer 0.5 µL Reverse primer 0.5 µL Nuclease-free water 4 µL Total volume 15 µL 1 95 °C 30 s 2 95 °C 1 s 3 60 °C 25 s 4 55–95 °C 0.5 °C increments 10 s Repeat steps 2 and 3, 30–45 times Protein analysis: Analyze the expression of specific proteins in treated cells by immunoblotting. We analyze the CXCR7 expression by a 10 % SDS-PAGE, followed by immunoblotting to Immobilon P-(PVDF) membrane. The membrane is blocked with 5 % milk in TBS-T. CXCR7 signal is detected using anti-CXCR7 IgG antibody diluted 1:5,000 in 5 % milk in TBS-T. Signal is detected using ECL. The specific CXCR7 (~42 kDa) band shows up slightly above β-actin and right below the 55 kDa mark. Relative protein band intensities can be quantified using densitometry. Band densities are normalized to those of β-actin.
Acknowledgement
This work was funded by grants from the National Institutes of Health (1R01CA61038-14, 1R01 CA 156778–01, VA MERIT Award No 5312–01 and 5312–02, all to BLL). The authors thank Ms. Maite Lopez for illustration in Fig. 1.
Footnotes
DNA: Using plasmid DNA with a concentration greater than 0.8 µg/µL gives less toxicity. We recommend using 0.5 µg of DNA when working in a 12-well plate. For western blot and RT-qPCR analysis, 12-well plates provide enough material for analysis (2 wells for RNA and 1 well for protein are sufficient) for analysis of transfection efficiency. Use a 1:2–1:4 ratio of DNA (µg) to Lipo (µL) reagent. If using the Lipofectamine LTX reagent, use a 1:1 ratio of DNA (µL) to PLUS (µL) reagent, as in the manufacturer’s instructions.
We recommend lipid transfection as it is fast approach that requires a single transfection round of the target cells. Our transfection efficiency is 50–90 % on transient transfections and 50–80 % on stable transfections. Delivery of shRNA via viral vectors requires two steps. At first, a transfection of virus producing cells with lentiviral plasmids, which results in the production of lentiviral particles is required. Those vectors are then isolated from the supernatant of the virus producing cells and are used for transduction/infection of target cells. Thus, delivery of shRNA via a lentiviral system is more labor intensive compared to lipid mediated transfection. The advantage of viral transfection is its high efficiency, therefore, stable selection using puromycin is rapid (3–5 days versus 1-weeks). However, we have seen similar efficiency in delivery of shRNA via viral vectors compared to delivery of shRNA via lipid transfection as evaluated by RT-qPCR and Western blot analysis. For lipid based transfections-our lab has successfully used Lipofectamine 2000, and Lipofectamine LTX transfection reagents.
Cells should be at a maximum of 80–90 % confluency. This is because the cells should have enough space to divide, in order to be in the growth phase of the cell cycle, allowing for the plasmid DNA to be most efficiently incorporated.
The Lipofectamine LTX manufacturer’s protocol accounts for pipetting variations, so there is no need to multiply by an extra factor. If following the Lipofectamine 2000 protocol multiply all volumes by a factor of 1.1 to account for pipetting error and volume loss. We focus on Lipofectamine 2000 use as Lipofectamine LTX is usually recommended for more delicate and hard to transfect cells, such as slow growing primary cell cultures.
It is a good idea to use GFP as a marker of successful transfection. Counting the number of fluorescent cells versus the total number of cells provides a good measure of the transfection efficiency. A flow-cytometry based selection can also be used for rapid isolation and selection of transfectants.
When generating stable cell lines, it is important to have a positive control and a negative control. For positive control, the cell line transfected with the empty vector backbone is ideal as it will be used to assess biological effect of the specific gene knockdown versus the vector effect. As a negative control, the un-transfected cell lines are used to ensure the puromycin is killing non-transfected cells.
A kill curve is used to determine the minimum optimal antibiotic concentration required to select the cells successfully transfected with the knockdown shRNA from those without it. Selection is seen from 3 days to a week.
To ensure the long-term efficiency of the shRNA in the stable cell lines, it is important to freeze stocks from early passages. It is possible after multiple passages that some cells may lose the shRNA gene silencing effect while maintaining resistance to the selection antibiotic. Stocks from earlier passages can be used to replace the older line, so non-expressing resistant cells do not overtake those expressing the shRNA gene. Such a situation can be caught early by occasionally analyzing the stable line via RT-qPCR or WB and ensuring the expression of the shRNA is consistent at later passages.
We suggest using the concentration of puromycin at twofold the concentration at which no cells survived. For example, if you see no live cells at 0–2 µg/mL puromycin after 7 days, use 4.0 µg/mL of puromycin for selection.
Contributor Information
Nicole Salazar, Sheila and David Fuente Graduate Program in Cancer Biology, University of Miami Miller School of Medicine, Miami, FL, USA; Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL, USA.
Daniel Muñoz, Research Service, Miami VA Medical Center, University of Miami Miller School of Medicine, Miami, FL, USA.
James Hoy, Sheila and David Fuente Graduate Program in Cancer Biology, University of Miami Miller School of Medicine, Miami, FL, USA; Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL, USA.
Bal L. Lokeshwar, Email: blokeshw@med.miami.edu, Sheila and David Fuente Graduate Program in Cancer Biology, University of Miami Miller School of Medicine, Miami, FL, USA; Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, Miami, FL, USA; Research Service, Miami VA Medical Center, University of Miami Miller School of Medicine, Miami, FL, USA; Department of Urology (M-800), University of Miami Miller School of Medicine, 016960, Miami, FL, 33101, USA.
References
- 1.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.“The Nobel Prize in Physiology or Medicine 2006” Nobelprize.org. Nobel Media AB. 2013 http://www.nobelprize.org/nobel_prizes/medicine/laureates/2006/ [Google Scholar]
- 3.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Hutvágner G, McLachlan J, Pasquinelli AE, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke A, Stark RB, Russell SM, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Origene HuSH shRNA plasmid panels application guide [Google Scholar]
- 8.O’Keefe EP. Nucleic acid delivery: lentiviral and retroviral vectors. 2013 Labome.com. [Google Scholar]
- 9.Singh RK, Lokeshwar BL. The IL-8-regulated chemokine receptor CXCR7 stimulates EGFR signaling to promote prostate cancer growth. Cancer Res. 2011;71:3268–3277. doi: 10.1158/0008-5472.CAN-10-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shamaladevi N, Lyn DA, Escudero DO, et al. CXC receptor-1 silencing inhibits androgen-independent prostate cancer. Cancer Res. 2009;69:8265–8274. doi: 10.1158/0008-5472.CAN-09-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh RK, Lokeshwar BL. Depletion of intrinsic expression of Interleukin-8 in prostate cancer cells causes cell cycle arrest, spontaneous apoptosis and increases the efficacy of chemotherapeutic drugs. Mol Cancer. 2009;8:57. doi: 10.1186/1476-4598-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]