SUMMARY
Defects in brain development are believed to contribute towards on-set of neuropsychiatric disorders but identifying specific underlying mechanisms has proven difficult. Here, we took a multi-faceted approach to investigate why 15q11.2 copy number variants are prominent risk factors for schizophrenia and autism. First, we show that human iPSC-derived neural progenitors carrying 15q11.2 microdeletion exhibit deficits in adherens junctions and apical polarity. This results from haploinsufficiency of CYFIP1, a gene within 15q11.2 that encodes a subunit of the WAVE complex, which regulates cytoskeletal dynamics. In developing mouse cortex, deficiency in CYFIP1 and WAVE signaling similarly affects radial glial cells, leading to their ectopic localization outside of the ventricular zone. Finally, targeted human genetic association analyses revealed an epistatic interaction between CYFIP1 and WAVE signalling mediator ACTR2 and risk for schizophrenia. Our findings provide insight into how CYFIP1 regulates neural stem cell function and may contribute to the susceptibility of neuropsychiatric disorders.
INTRODUCTION
Neuropsychiatric disorders, including schizophrenia and autism, are debilitating conditions that are postulated to have a neurodevelopmental aetiology (Geschwind, 2009; Weinberger, 1987). Significant progress has been made to identify the genetic basis of these disorders. In addition to single-nucleotide polymorphisms (SNPs), submicroscopic variations in DNA copy number (CNVs) are also widespread in human genomes and specific CNVs have been identified as significant risk factors for schizophrenia and autism (Malhotra and Sebat, 2012). Because CNVs frequently contain multiple genes and are more difficult to model in mice using traditional gene targeting techniques, we know little about how these CNVs affect neural development. Novel approaches are needed to investigate these genetic risk factors in neural development and identify their signalling mechanisms, which in turn could generate new hypotheses for identification of additional risk factors.
15q11.2 CNVs have emerged as prominent risk factors for various neuropsychiatric disorders, including schizophrenia, autistic spectrum disorder and intellectual disability (Malhotra and Sebat, 2012). 15q11.2 microdeletion (15q11.2del) was identified as one of the most frequent CNVs associated with increased risk for schizophrenia in two large studies (Consortium, 2008; Stefansson et al., 2008), a finding subsequently confirmed in additional cohorts (Kirov et al., 2009; Tam et al., 2010; Zhao et al., 2013). Even in normal subjects, 15q11.2del is associated with cognitive variation and changes in structural measures on MRI scanning (Stefansson et al., 2013). 15q11.2 CNVs encompass four genes, non-imprinted in Prader/Willi Angelman 1 and 2 (NIPA1 and NIPA2), CYFIP1, and TUBGCP5 (Figure 1A). While little is known about functions of these genes in mammalian neural development, CYFIP1 has been shown to interact with Rac1 (Kobayashi et al., 1998), FMRP (Schenck et al., 2001), and eIF4E (Napoli et al., 2008). Biochemical studies have also identified CYFIP1 as a regulator of the WAVE complex, consisting of WAVE1, WAVE2, Nap1 and Abi1, a complex known to regulate Arp2/3-mediated actin polymerization and membrane protrusion formation in non-neuronal cell lines (Kobayashi et al., 1998; Kunda et al., 2003; Steffen et al., 2004). The function of WAVE signaling in mammalian neurogenesis is not well understood.
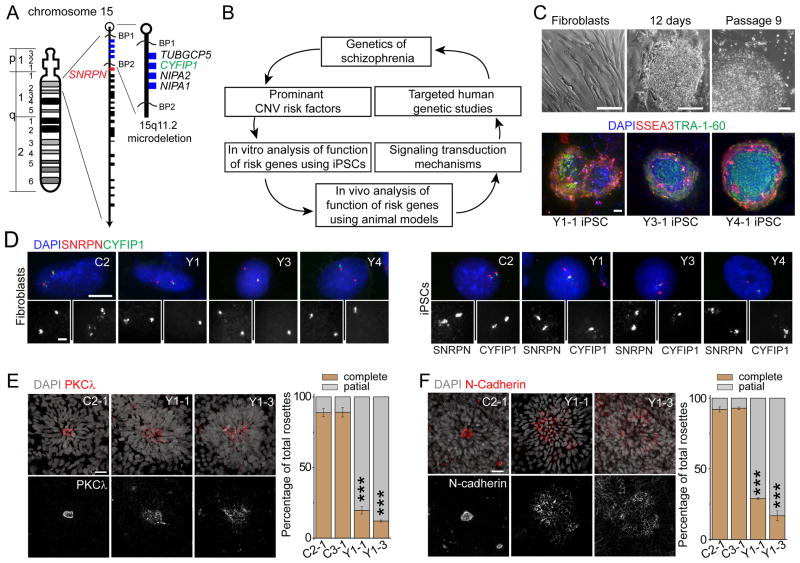
Figure 1. iPSC derivation and aberrant neural rosette formation of hNPCs differentiated from iPSC lines carrying 15q11.2del.
(A) An ideogram of Chromosome 15 with proximal 15q11.2 region expanded. Blue boxes indicate the four deleted genes within 15q11.2del, including CYFIP1.
(B) A schematic summary of the current study design.
(C) Sample light microscopic images and fluorescent images of fibroblasts and iPSC colonies for immunostaining of pluripotency markers. Scale bars, 100 μm.
(D) Sample fluorescence in situ hybridization (FISH) images of donor fibroblasts and derived iPSC lines. Two FISH probes, one for CYFIP1 locus and one for SNRPN locus (outside of 15q11.2; See A for the genomic location), were used. Scale bar, 10 μm.
(E–F) Defects in adherens junctions and apical polarity of hNPCs derived from iPSCs with 15q11.2del. Shown on the left are sample confocal images of immunostaining of atypical PKCλ (E) and N-cadherin (F) for neural rosettes differentiated from iPSCs by the mono-layer method. Scale bars, 20 μm. Shown on the right are quantifications of neural rosettes with complete “apical ring-like structure” (≥ 90% coverage of apical-ring circumference with atypical PKCλ or N-cadherin immunoreactivity) or “partial/scattered” (< 90% coverage). Values represent mean ± SEM (n = 3 cultures; ***p < 0.001; Student’s t-test).
See also Figures S1, S2 and Tables S1, S2.
Patient-derived induced pluripotent stem cells (iPSCs) provide a new means to investigate how risk factors affect nervous system development (Bellin et al., 2013; Christian et al., 2012). Reprogrammed from somatic cells, iPSCs capture identical risk alleles as the donor individual and provide a renewable resource of previously inaccessible, disease-relevant human cell types to facilitate molecular and cellular investigations. In this emerging new field, recent iPSC studies were mostly “proof-of-principle” experiments that confirmed previous findings from animal and post-mortem human studies; its promise as a discovery tool is just beginning to be realized.
While 15q11.2del is linked to schizophrenia, common variants within the deletion region have not shown similar association in case control studies, possibly because of the weak impact of common SNPs on biological functions of individual genes. To mimic the large dose effect of a whole gene deletion, we hypothesized that genetic interactions within the biological network linked to the function of specific genes within 15q11.2del would rise to the level of clinical association and that patient-derived iPSC studies could provide an entry point to identify these networks (Figure 1B). We established iPSC lines from three individuals carrying 15q11.2del and compared them with iPSCs from five individuals without the CNV. Analysis of iPSC-derived neural rosettes with 15q11.2del revealed impairments in adherens junctions and polarity of human neural progenitor cells (hNPCs) due to WAVE complex destabilization. Pinpointing CYFIP1-haploinsufficiency within 15q11.2 as a underlying cause of hNPC defects then guided our investigation of CYFIP1 and its signaling via the WAVE complex in regulating radial glia neural stem cells (RGCs) in the developing mouse cortex in vivo. This, in turn, led to targeted human genetic association analyses, resulting in the identification of an epistatic interaction for risk of schizophrenia. Our integrated analyses from multiple systems provide novel insight into how 15q11.2 CNVs may contribute to defects in neural development and brain disorders.
RESULTS
Defects in adherens junctions and apical polarity of hNPCs derived from human iPSCs carrying 15q11.2del
To determine how 15q11.2del may affect human brain development, we established multiple iPSC lines from skin fibroblasts of 3 individuals carrying 15q11.2del in one chromosome (Y1, Y3 and Y4) and from 3 control individuals (C1, C2 and C3) using non-integrating approaches (Figure 1C and Table S1). We performed detailed quality control analyses of all iPSC lines selected for the current study (Table S1). These iPSCs maintained embryonic stem cell-like morphology, expressed pluripotency-associated markers and exhibited normal euploid karyotypes (Figures 1C, S1 and Table S1, S2). All iPSC lines tested formed teratomas when injected into SCID mice (Table S1). We also included iPSC lines from two neuropsychiatric patients with a DISC1 mutation as another group for comparison (D2 and D3) (Chiang et al., 2011). Using DNA fluorescence in situ hybridization (FISH), we confirmed one copy microdeletion at 15q11.2 locus in Y1, Y3 and Y4, but not in other fibroblasts and iPSC lines we examined (C1, C2, C3, D2, D3; Figure 1A, D and Table S1).
We first differentiated iPSCs into relatively homogenous primitive neural precursor cells (pNPCs) in monolayer using an established protocol (Li et al., 2011). All lines were efficiently differentiated into pNPCs expressing NESTIN and SOX2 (Figure S2A). No consistent differences in the NPC differentiation efficacy or proliferation among different groups of iPSC lines were detected (Figures S2A–B). To partially maintain cell-cell interaction, we next generated cortical neural rosettes using small molecule inhibitors and retinoic acid (Shi et al., 2012). We initially used 4 iPSC lines for pilot phenotypic characterization, including one iPSC line each from two control subjects (C2-1 and C3-1) and two lines from one subject with 15q11.2del (Y1-1 and Y1-3). Neural rosettes from C2-1 and C3-1 iPSCs showed robust expression of atypical PKCλ, an apical polarity marker, as a ring-like structure at the luminal surface of each rosette (Figure 1E), representing typical formation of apical-basal polarity of hNPCs (Shi et al., 2012). Interestingly, the majority of neural rosettes from Y1-1 and Y1-3 iPSCs exhibited scattered expression of atypical PKCλ (Figure 1E). The structure of adherens junctions as revealed by N-cadherin immunostaining was also disrupted in the majority of rosettes from two Y1-iPSC lines (Figure 1F). These results suggest that gene(s) located within 15q11.2del regulate apical polarity and maintaining adherens junctions of hNPCs.
WAVE complex destabilization and polarity defects of hNPCs due to CYFIP1 haploinsufficiency
The actin cytoskeleton acts as a cytoplasmic anchor for cadherin/catenin proteins at adherens junctions and its proper organization is important for maintaining adherens junctions and polarity of neural precursors in the developing mouse cortex (Buchman and Tsai, 2007). Among the four genes within the 15q11.2 region, CYFIP1 is a regulator of the actin-modulating WAVE complex (Kunda et al., 2003, Steffen et al., 2004, Silva et al., 2009). Indeed, co-immunoprecipitation (co-IP) analysis showed that CYFIP1 interacts with WAVE complex components WAVE1, WAVE2 and NAP1 (NCKAP1) in normal hNPCs (Figure 2A). Therefore, we assessed WAVE complex integrity in hNPCs derived from different iPSC lines. Consistent with a haploinsufficiency model, mRNAs of all four genes within 15q11.2 were expressed at ~ 50% levels in all hNPCs carrying 15q11.2del compared to those without the deletion (Figure S2C). CYFIP1 protein was also expressed at ~ 50% levels (Figures 2B, 2D and S2D). Strikingly, the expression of WAVE2 protein, but not its mRNA, in 15q11.2del hNPCs was only ~ 20% of that in control hNPCs (Figures 2B, 2E and S2D). The effect of 15q11.2 microdeletion appeared to be specific, as hNPCs derived from mutant DISC1-iPSC lines showed normal expression of CYFIP1 and WAVE2 proteins (Figures 2D–E and S2D). Together, these biochemical analyses demonstrated a specific defect of WAVE complex stabilization in hNPCs with 15q11.2 microdeletion.
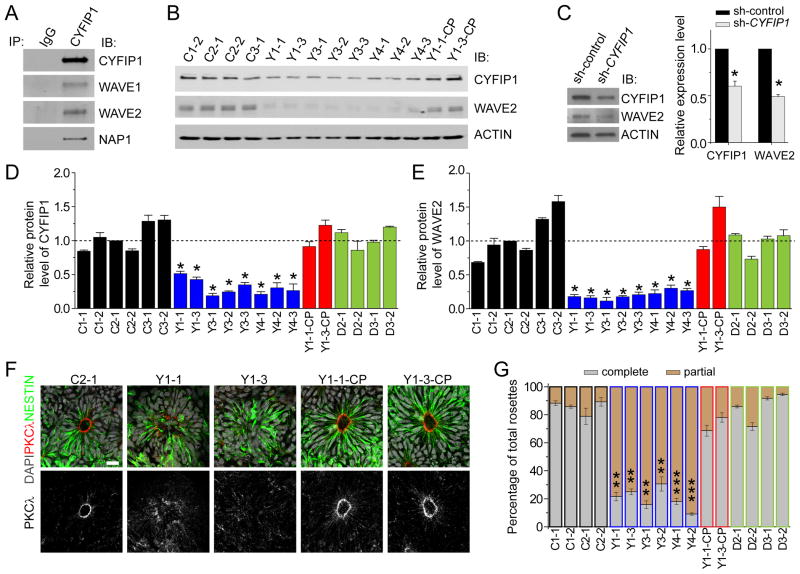
Figure 2. Destabilization of the WAVE complex and polarity defects of hNPCs due to CYFIP1 deficiency.
(A) CYFIP1 is a component of WAVE complex in human pNPCs. Shown are sample Western blot images of co-IP analysis of pNPCs using anti-CYFIP1 antibodies.
(B) Reduced CYFIP1 and WAVE2 protein levels in pNPCs carrying 15q11.2del. Lentiviruses were used to establish two CYFIP1 complementation lines (Y1-1-CP and Y1-3-CP).
(C) Decreased WAVE2 protein levels after CYFIP1 knockdown by lentivirus-mediated shRNA expression in normal pNPCs for 48 hrs.
(D–E) Quantification of protein levels of CYFIP1 and WAVE2 among different pNPCs. All data were normalized to ACTIN levels for loading control and then normalized to CYFIP1 (D) or WAVE2 (E) in C2-1 pNPCs for comparison. Values represent mean ± SEM (n = 3–5 cultures; *p < 0.05; Student’s t-test).
(F–G) Defect in adherens junction and apical polarity of hNPCs carrying 15q11.2del and its rescue by CYFIP1 complementation. Shown on the left are sample confocal images of immunostaining of atypical PKCλ for neural rosettes differentiated from iPSCs using the embryoid body method. Scale bar, 20 μm. Shown on the right are quantifications of neural rosettes similar as in Figure 1E. Values represent mean ± SEM (n = 5 cultures; ***p < 0.001; **p < 0.01; Student’s t-test).
See also Figure S2 and Tables S1, S3.
Is CYFIP1 haploinsufficiency the major cause of observed defects in hNPCs carrying 15q11.2del? First, we performed complementation experiments using lentiviruses to increase CYFIP1 levels in two Y1-iPSC lines. We selected two iPSC lines that gave rise to hNPCs with the total amount of CYFIP1 protein at comparable levels to C3-1 hNPCs (Y1-1-CP and Y1-3-CP; Figures 2B, 2D). Importantly, the WAVE2 protein level in these complemented lines was fully rescued (Figures 2B, 2E), suggesting that CYFIP1 haploinsufficiency is required for WAVE complex destabilization in hNPCs with 15q11.2del. Second, to determine whether decreased CYFIP1 expression is sufficient to cause WAVE complex destabilization in hNPCs, we reduced the endogenous CYFIP1 protein level in control hNPCs to ~ 50% with shRNA (Figure 2C and Table S3). Indeed, expression of shRNA-CYFIP1, but not shRNA-control, led to significantly decreased WAVE2 protein expression (Figure 2C). Finally, we examined whether CYFIP1 haploinsufficiency is the cause of adherens junction and apical polarity impairments observed in neural rosettes from hNPCs with 15q11.2del. We first validated our pilot results using an independent embryoid body protocol (Juopperi et al., 2012), which gave rise to pure PAX6+ neural progenitors (Figure S2E). Scattered expression of atypical PKCλ at the luminal surface was observed for the majority of neural rosettes from multiple iPSC lines with 15q11.2del (Figures 2F–G). Importantly, complementation of CYFIP1 expression to the normal level in two Y1 lines rescued the expression of atypical PKCλ at the luminal surface (Figures 2F–G), whereas reduction of CYFIP1 expression by shRNA in C3-1 hNPCs led to scattered expression of atypical PKCλ (Figure S2F). Consistent with an intact WAVE complex, neural rosettes derived from mutant DISC1-iPSCs exhibited normal distribution of PKCλ at the luminal surface (Figure 2G). Analysis of additional polarity markers, including PAR3 and β-catenin, also showed consistent results across the groups (Figure S2G).
Taken together, this series of biochemical and functional analyses of a collection of 20 iPSC lines established that 15q11.2del, through CYFIP1 deficiency, leads to defects in the maintenance of adherens junctions, apical polarity and WAVE complex stability in hNPCs.
Requirement of CYFIP1 in maintaining adherens junctions and apical polarity of RGCs in developing mouse cortex
Given limitations of in vitro studies of human iPSCs, we next turned to in vivo mouse embryonic cortical development to assess whether the CYFIP1 function we identified in regulating hNPCs is physiologically relevant in vivo and, furthermore, to examine the long-term consequence of CYFIP1 deficiency in cortical development. In the E15.5 dorsal neocortex, CYFIP1 was found to be accumulated at the ventricular surface in the ventricular zone (VZ), with lower expression in migrating neurons in the intermediate zone (IZ; Figure 3A). The VZ of the mid-neurogenic period is mostly occupied by RGCs, which are neural stem cells in the developing cortex (Kriegstein and Alvarez-Buylla, 2009). The apical processes of adjacent RGCs are attached to one another via cadherin-based adherens junctions at the ventricular surface (Loulier et al., 2009; Rasin et al., 2007). Co-immunostaining showed that CYFIP1 was highly expressed at the F-actin-expressing lateral membrane domain, and N-cadherin- and β-catenin-expressing adherens junctions in the apical endfeet of RGCs (Figures 3B–C). With an en face view from the ventricle, CYFIP1 was found as cytosolic puncta inside of the ring-like F-actin structure on the ventricular surface (Figure 3C).
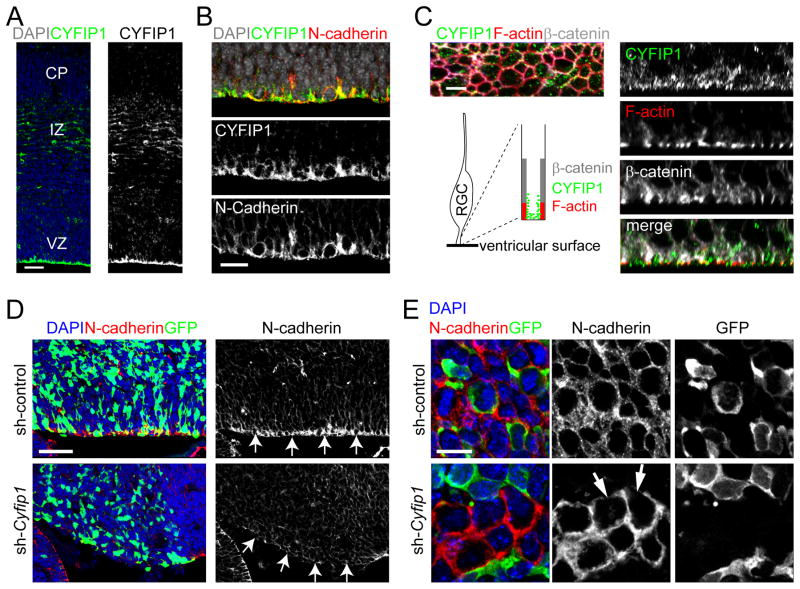
Figure 3. Critical role of CYFIP1 in regulating adherens junctions and apical polarity of RGCs in the developing mouse cortex.
(A–C) Expression of CYFIP1 in the mouse neocortex at E15.5. Shown are sample confocal images of immunostaining of CYFIP1 and N-cadherin. VZ: ventricular zone; IZ: intermediate zone; CP: cortical plate. Also shown is the en face view of CYFIP1 expression at the ventricular surface (C, left panel). The structure of F-actin, which forms the boundary between apical endfeet of RGCs on the ventricular surface, was labelled by Phalloidin-Alexa594 (Red). Cross-section images are shown in right panels. Scale bars, 50 μm (A), 20 μm (B) and 5 μm (C).
(D–E) Expression of CYFIP1 and other apically polarized proteins in the mouse neocortex at E16.5 after in utero electroporation at E13.5 to co-express GFP and sh-control or sh-Cyfip1 (#2). Shown are sample confocal images of immunostaining for GFP and N-cadherin (D) and the en face view at 3 μm from the ventricular surface (E). Scale bars: 50 μm (D) and 10 μm (E).
See also Figure S3 and Tables S2, S3.
To investigate CYFIP1 function in regulating RGCs, we generated effective shRNAs specifically against mouse Cyfip1 (Figure S3A and Table S3). We performed in utero electroporation with vectors co-expressing GFP and shRNA-Cyfip1#2 (sh-Cyfip1), or control shRNA (sh-control), into the E13.5 neocortex, and analyzed 3 days later. GFP+ cells expressing sh-Cyfip1 showed largely absent CYFIP1 immunoreactivity, confirming the shRNA efficiency against endogenous CYFIP1 in vivo (Figure S3B). While GFP+ cells expressing sh-control in the VZ showed robust N-cadherin expression at the ventricular surface, those expressing sh-Cyfip1 did not (Figures 3D–E). En face view near the ventricular surface further showed reduced N-cadherin expression in some GFP− cells at regions in contact with GFP+ cells expressing sh-Cyfip1, suggesting a potential non-cell autonomous effect (Figure 3E). Thus, similar to its function in cultured hNPCs, CYFIP1 maintains adherens junctions and apical polarity of neural stem cells in the developing mouse cortex in vivo.
Ectopic localization of CYFIP1-deficient RGCs outside of the VZ
What is the functional consequence of impairments in adherens junctions and apical polarity of RGCs from CYFIP1 deficiency? We examined RGC cell body distribution by Pax6 immunohistochemistry. Pax6+GFP+ cells expressing sh-control were mostly restricted within the VZ (Figures 4A, left panel, and 4B). In contrast, a significant percentage of Pax6+GFP+ cells expressing sh-Cyfip1 (#2) were ectopically misplaced in the SVZ and IZ, at the expense of VZ localization (Figures 4A, middle panel, and 4B). Aberrant localization of RGCs was also observed with an independent shRNA against mouse Cyfip1 (#1; Figure S4A). Importantly, this defect was rescued by co-expression of shRNA-resistant CYFIP1 cDNA (Figures 4A, right panel, and 4B), confirming the specificity of shRNA experiments.
Figure 4. Ectopic localization of CYFIP1-deficient RGCs outside of the VZ in the developing mouse cortex.
(A–B) Ectopic localization of Pax6+ RGCs in the SVZ and IZ of E16.5 neurocortices after in utero electroporation at E13.5 to co-express GFP and sh-control or sh-Cyfip1, or co-express GFP/sh-Cyfip1 and cDNA for shRNA-Cyfip1 resistant HA-tagged CYFIP1. Shown in (A) are sample confocal images of immunostaining of Pax6, GFP and HA. Co-transfection of GFP/sh-Cyfip1 and shRNA-resistant HA-tagged CYFIP1 was confirmed by co-localization of GFP and HA immunostaining (the right-most panel). Scale bar, 50 μm. Shown in (B) are two different quantifications of the distribution of GFP+Pax6+ cells in the neocortex. Upper panel represents GFP+Pax6+ cells in each of ten equal-size vertical bins expressed as percentages of total GFP+Pax6+ cells (1: the most apical, 10: the most basal). Lower panel represents percentages of GFP+Pax6+ cells in the VZ (VZ) and in other layers (non-VZ). Values represent mean ± SEM (n = 4–5 animals; **p < 0.01; Student’s t-test).
(C) Aberrant localization of proliferating cells expressing sh-Cyfip1 in E16.5 neocortex. Sample confocal images of immunostaining for GFP and an M-phase marker, phospho-HistoneH3 (pH3), are shown. Scale bar, 50 μm. Also shown are quantifications of percentages of GFP+pH3+ cells at the ventricular surface (surface division) and at other locations (non-surface division). Values represent mean ± SEM (n = 5 animals; ***p < 0.001; Student’s t-test)
(D) Proliferation of CYFIP1-deficient RGCs outside of the VZ. E13.5 embryos were electroporated to co-express GFP and sh-Cyfip1 and fixed at 2 hrs after EdU injection at E16.5. Shown are sample confocal images of staining for GFP, Pax6, EdU and DAPI. Note that ectopic GFP+Pax6+ cells in the IZ still incorporated EdU, representing their ability to proliferate far from the ventricular surface. Scale bars, 50 μm (left panel) and 20 μm (insets).
GFP+ mitotic cells labeled with phospho-HistoneH3 were also found to be scattered in the SVZ and IZ (Figure 4C). To determine whether NPC proliferation was affected by CYFIP1 deficiency, proliferating cells were pulsed with EdU and examined 2 hours later. Similar to hNPCs in vitro (Figure S2B), CYFIP1-deficient cells showed EdU incorporation comparable to those expressing sh-control, despite their ectopic localization (Figures 4D and S4B). To examine cell cycle progression, we determined the cell cycle exit index defined as the percentage of EdU+Ki67− cells among all EdU+ cells at 24 hours after EdU administration and did not find any differences (Figure S4C). Together, these results indicate that CYFIP1 is important for proper placement and pattern of mitosis, but not essential for the proliferation and cell cycle progression of RGCs in the developing mouse cortex in vivo.
Ectopic placement of intermediate progenitor cells and cortical neurons generated from CYFIP1-deficient RGCs
We next examined the direct progeny from RGCs, intermediate progenitor cells (IPCs), which express Tbr2 and proliferate transiently in the SVZ to generate neurons (Englund et al., 2005). Tbr2+GFP+ cells expressing sh-Cyfip1 were also scattered in the VZ/SVZ/IZ, while Tbr2+GFP+ cells expressing sh-control mainly resided in the SVZ (Figures 5A–B). On the other hand, the proportion of Pax6+GFP+ cells and Tbr2+GFP+ cells were not altered between those expressing sh-control and sh-Cyfip1 (Figure 5C), suggesting that CYFIP1 is dispensable for the proper differentiation of RGCs into IPCs.
Figure 5. Ectopic placement of intermediate progenitor cells and cortical neurons upon CYFIP1 knockdown in RGCs.
(A–C) Ectopic placement of intermediate progenitor cells (IPCs) upon CYFIP1 knockdown in RGCs. E13.5 embryos were electroporated to co-express GFP and sh-Cyfip1 or sh-control and examined at E16.5. Shown in (A) are sample confocal images of immunostaining for GFP, Pax6, Tbr2 and DAPI. Scale bar, 50 μm. Shown in (B) are quantifications of the distribution of GFP+Tbr2+ cells in the neocortex. The graph represents GFP+Tbr2+ cells in each of ten equal-size vertical bins expressed as the percentage of total GFP+Tbr2+ cells. Values represent mean ± SEM (n = 4–5 animals; **p < 0.01; Student’s t-test). Shown in (C) are quantifications of percentages of GFP+ cells that were Pax6+ or Trb2+. Values represent mean ± SEM (n = 4 animals; p > 0.1; Student’s t-test).
(D–E) Ectopic placement of cortical neurons upon CYFIP1 knockdown in RGCs. Same as in (AC), except that electroporated brains were examined at P5. Shown in (D) are sample confocal images of immunostaining for GFP, Cux1, and CTIP2. Right panels represent two different insets in the left panels. Arrows point to GFP+CTIP2+ or GFP+Cux1+ cells. Scale bars, 100 μm. Shown in (E) are quantifications of percentages of GFP+CTIP2+ cells (top panel) or GFP+Cux1+ cells (bottom panel) that were distributed in upper or deeper layers, respectively. The boundary between upper and lower layers was established at the apical limit of Cux1+ cells. Values represent mean ± SEM (n = 4 animals; *p < 0.05; Student’s t-test).
See also Table S2.
Glutamatergic projection neurons of the adult cortex are generated in a stereotyped temporal order, with deep layer neurons (layer V/VI: CTIP2+) produced first and upper layer neurons (layer II/III/IV: Cux1+) produced later (Gaspard et al., 2008; Leone et al., 2008). Defects in RGCs, which serve as the main radial scaffold for migrating neurons, could potentially lead to failure of neurons to reach their normal position. To examine the long-term consequence of Cyfip1 knockdown on cortical layer formation, we analyzed P5 brains after in utero electroporation of shRNAs at E13.5. CTIP2+ neurons, which normally localize in deeper layers, showed more frequent localization in upper layers after sh-Cyfip1 expression in RGCs (Figures 5D–E). On the contrary, Cux1+ neurons, which normally localize in upper layers, were present in a higher percentage in deep layers after sh-Cyfip1 expression (Figures 5D–E). The ratio of CTIP2+ versus CUX1+ cells among all GFP+ cells was not significantly different between sh-control (0.22 ± 0.03) and sh-Cyfip1 (0.23 ± 0.02; n = 4), suggesting that CYFIP1 is dispensable for neuronal subtype specification. Some mis-localized Cux1+ and CTIP2+ cells appeared to be GFP− (Figure 5D). This could be due to the diluted GFP expression in the P5 brain after multiple rounds of cell division or, alternatively, non-cell autonomous migration defects due to aberrant radial scaffolds of CYFIP1-deficient RGCs. These results demonstrate that CYFIP1 deficiency causes improper placement of IPCs and glutamatergic projection neurons, resulting in cortical layer malformation.
CYFIP1 signaling mechanism in regulating RGCs in the developing embryonic mouse cortex
Similar to findings from hNPCs (Figure 2A), co-IP analysis using E14.5 mouse cortical lysates showed that endogenous CYFIP1 interacted with several WAVE components, including WAVE1, WAVE2, Abi1 and Nap1 (Figure 6A). Knockdown of CYFIP1 in mouse NPCs in vitro also led to marked decrease of WAVE2 and Abi1 proteins (Figure 6B). Immunohistological analysis showed that WAVE2 protein expression at the ventricular surface was drastically decreased after Cyfip1 knockdown in vivo (Figure 6C). These results suggested a conserved role and signaling mechanism of CYFIP1 in regulating WAVE complex stability and adherens junctions in both human and mouse NPCs.
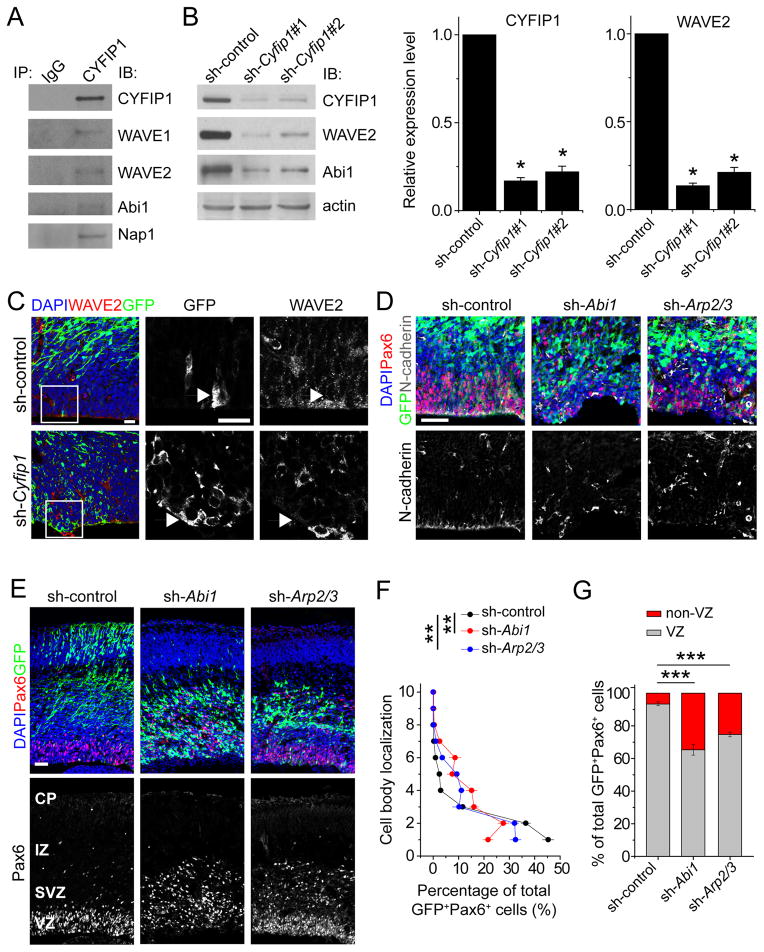
Figure 6. Critical role of CYFIP1 signalling in maintaining adherens junctions and proper placement of RGCs in the developing mouse cortex.
(A) CYFIP1 is a component of WAVE complex in the developing mouse cortex. Shown are sample Western blot images of co-IP analysis using anti-CYFIP1 antibodies and E14.5 forebrain lysates.
(B) Reduced WAVE2 protein levels by CYFIP1 knockdown in mouse NPCs. Mouse NPCs were infected with retroviruses expressing sh-control, or sh-Cyfip1 (#1 or #2) and then subjected to Western blot analyses 3 days later. Shown are sample Western blot images and quantifications of CYFIP1 and WAVE2 protein levels. Values represent mean ± SEM (n = 3; **p < 0.01; Student’s t-test).
(C) Reduced WAVE2 protein levels at the ventricular surface in E16.5 neocortices in vivo. E13.5 embryos were electroporated to co-express GFP and sh-Cyfip1 or sh-control and examined at E16.5. Shown are sample confocal images of immunostaining for GFP, WAVE2 and DAPI. Scale bar, 20 μm.
(D–G) Similar RGC defects among knockdown of CYFIP1, WAVE complex component Abi1, or downstream effectors Arp2/3. E13.5 embryos were electroporated to co-express GFP and sh-Abi1 or sh-Arp2/sh-Arp3 (double knockdown), and examined at E16.5. Shown in (D) are sample confocal images of immunostaining for GFP, Pax6, N-cadherin and DAPI. Shown in (E) are sample confocal images of immunostaining for GFP, Pax6, and DAPI. Scale bars, 50 μm. Also shown are two different quantifications of the distribution of GFP+Pax6+ cells in the neocortex among the different experiments, similar as in Figure 4B. Values represent mean ± SEM (n = 4 animals; **p < 0.01; ***p < 0.001; Student’s t-test).
See also Figure S5 and Tables S2 and S3.
Next, we examined the functional role of CYFIP1-dependent WAVE complex and downstream signaling in regulating RGCs in vivo (Figure S5A). We developed effective shRNAs against mouse Abi1 and downstream mediators Arp2/3 (Figures S5B–C). In utero electroporation analyses showed a lack of N-cadherin expression at the ventricular surface by GFP+ cells expressing sh-Abi1 or sh-Arp2/3 (double knockdown; Figure 6D). GFP+Pax6+ cells expressing sh-Abi1 or sh-Arp2/3 also showed scattered distribution in the VZ/SVZ/IZ (Figures 6E–G). These results suggest that, similar to CYFIP1, WAVE complex-mediated signaling is important for the maintenance of adherens junctions and proper placement of RGCs in the developing cortex.
Epistatic interaction of gene expression-associated variants of the WAVE signalling components for risk of schizophrenia
Our findings of similar roles of WAVE signaling components in regulating RGCs, together with previous findings of association of 15q11.2del with risk for schizophrenia (Malhotra and Sebat, 2012), led to a new hypothesis that common genetic variants within the WAVE signalling pathway might interact to affect risk for schizophrenia even in the absence of association at the individual gene level (Figure 1B). The goal of the clinical genetic association analyses was to model molecular interactions of CYFIP1 and WAVE components identified in iPSC and animal studies. We first examined mRNA expression in the dorso-lateral prefrontal cortex (DLPFC) of post-mortem human brains in order to find specific genetic variants that were associated with expression of genes in the WAVE signalling pathway (i.e. expression quantitative trait loci or eQTLs). We performed a cis-association analysis of SNP variants with gene expression measured by RNA-seq in a group of 64 Caucasian subjects with no history of medical or psychiatric disease (Table S4). Significant associations were found for rs268864 with ACTR2/Arp2 expression (p = 0.02), rs2797930 with ABI1 expression (p = 0.02) and rs7168367 with CYFIP1 expression (p = 0.006). SNP rs4778334, previously associated with risk for schizophrenia in a case-control Han Chinese sample (Zhao et al., 2013), was also associated with CYFIP1 gene expression (p = 0.05). Interestingly, rs4778334 does not show linkage disequilibrium with other genotyped SNPs in the European ancestry populations or in the Chinese, consistent with a potential functional effect of this SNP (Figure S6). We also found that, except for CYFIP1, all genes in this network (ABI1, WASF1/WAVE1, WASF2/WAVE2, NCKAP1/Nap1, ACTR2/Arp2 and ACTR3/Arp3) tend to be co-expressed together in a similar pattern (Table S5).
To search for evidence that eQTLs in the WAVE signalling pathway might be associated with risk for schizophrenia, we performed single genetic association analysis of the selected four expression-associated SNPs (or proxy SNPs) in four independent schizophrenia case-control datasets of European ancestry. No significant single SNP association was found in any of the four cohorts (Table S6). Targeted pair-wise SNP-SNP interaction analyses were carried out among three SNPs - rs268864 (SNP1) at ACTR2, rs4778334 (SNP3) at CYFIP1||NIPA2 and rs7168367 (SNP4) at CYFIP1||NIPA1 as these were only SNPs genotyped in all four cohorts. An interaction was detected between rs268864 at ACTR2 and rs4778334 at CYFIP1||NIPA2 at a marginal significance (p = 0.0553) in the largest American LIBD/CBDB cohort (Figure 7A). The same trend of interaction with these exact alleles was also found in three smaller schizophrenia case-control cohorts of American (GRU; p = 0.147), German (MUN; p = 0.258) and Scottish origin (ABE; p = 0.215). Meta-analysis of the pair-wise interaction in all four cohorts showed significant evidence for interaction (p = 0.00417) between rs268864 at ACTR2 and rs4778334 at CYFIP1||NIPA2; interaction analysis of the combined sample of four cohorts confirmed the interaction in both an additive model (p = 0.0035) and a genotypic model (p = 0.0048; Figure 7A). The results of the meta-analysis are significant after correction for all combinations of two way interactions based on the three SNPs analysed. Moreover, the interactions, which were directionally consistent across all four datasets, were hypothesized based on eQTLs that specifically modelled the directionality of biologic interactions in the model system experiments.
Figure 7. Epistatic interaction of gene expression-associated variants of the WAVE signalling components for risk of schizophrenia.
(A) Interaction analysis of SNPs rs2688674*ACTR2 and rs4778334*CYFIP1-NIPA1 in combined samples of four cohorts in European ancestry population. Analysis was performed adjusting for sex and cohort effect. DF, degree of freedom.
(B) Effect of interaction by genotypes on risk of schizophrenia from a logistic regression model based on genotypic interaction of ACTR2 and CYFIP1 in combined samples of four cohorts. Adj p is p value after adjusting for multiple testing from post-hoc analysis. SNPs were coded as 0, 1 and 2 for number of minor alleles.
See also Figure S6 and Tables S4, S5 and S6.
It is interesting to note that depending on the genotype background of the ACTR2 SNP rs268864, genotypes of rs4778334 at CYFIP1 showed varying effects from negative to positive on risk of schizophrenia (Figure 7B). In the group of rs268864 genotype AA, individuals carrying genotype CC and CT at rs4778334 were less likely associated with risk of schizophrenia in comparison with TT genotype (p = 0.0244), and odds ratio estimates were 0.716 and 0.772 respectively. In contrast, in the group of rs268864 genotype GG, individuals carrying genotype CC and CT at rs4778334 were more likely associated with risk of schizophrenia in comparison with TT genotype (p = 0.0103), and odds ratio estimates were 11.14 and 4.56, respectively. Alternating genotype associations at one locus based on the genotype at another locus are classic epistatic phenomena.
DISCUSSION
Our study identified the functional role and signalling mechanism underlying CYFIP1 regulation of neural stem cellsand provides novel insight into how risk factors for neuropsychiatric disorders regulate neural development. Using human iPSCs as an entry point to investigate a prominent CNV risk factor encompassing multiple genes for schizophrenia and other neuropsychiatric disorders (Figure 1A–B), we uncovered novel cellular phenotypes in derived hNPCs and identified the responsible gene within the CNV. These in vitro findings of developmentally relevant phenotypes in human cells guided our analyses of neural stem cells in the developing mouse cortex in vivo and led to the identification of the underlying signalling mechanism. The mechanistic insight allowed us to generate a new hypothesis and test it with gene expression analyses in human brains and genetic association studies, resulting in the identification of a novel epistatic interaction for risk of schizophrenia. Our study provides an example of how genetic risk factors for complex human disorders can be studied in complementary systems using patient-derived iPSCs as the leading tool for discovery.
15q11.2 CNVs have emerged as a prominent risk factor for several neuropsychiatric disorders (Malhotra and Sebat, 2012). Our results from multiple levels of analyses provide evidence to support a specific gene within this CNV, CYFIP1, as a potential major contributing factor to biological processes implicated in the neurodevelopmental origins of these disorders. 15q11.2del has been identified as one of the three most frequent CNV risk factors for schizophrenia and increases risk 2–4 fold (Consortium, 2008; Stefansson et al., 2008). While none of the SNPs we examined within the CYFIP1 and WAVE signalling pathway showed significant independent risk for schizophrenia in four cohorts of European ancestry, we identified a potential epistatic interaction between CYFIP1 and WAVE signalling mediator ACTR2 /Arp2 for increased risk for schizophrenia with an odds ratio up to 11 (Figure 7B). While these results must be taken as preliminary and in need of further replication as the overall statistics are not particularly strong, our study implicates, for the first time, WAVE signalling in risk for schizophrenia and supports an emergent model that multiple factors within the same signalling pathway interact epistatically to affect the risk for psychiatric disorders. Notably, 15q11.2 CNVs themselves are not specific to schizophrenia (De Wolf et al., 2013). In a large study with over 15,000 patient samples, 15q11.2del was found to be strongly associated with developmental delay in children (Cooper et al., 2011). Studies have also linked 15q11.2del to epilepsy (de Kovel et al., 2010; Jahn et al., 2013; Mullen et al., 2013). Interestingly, CNVs with a duplication of this same region have been associated with autistic spectrum disorder (Nishimura et al., 2007; van der Zwaag et al., 2010; Wegiel et al., 2012). In addition, CYFIP1 is within larger 15q11.2-13.1 CNVs that have also been linked to schizophrenia, autistic spectrum disorder, and bipolar disorder (Malhotra and Sebat, 2012). Therefore, our findings have broad implications for these disorders and identify a new signalling pathway for future targeted investigation.
Our study provides novel insight into how CYFIP1 signaling regulates early mammalian neural development. While several previous studies have investigated roles of CYFIP1 in neurons, its function in neural stem cells was completely unknown. In Drosophila, the fly ortholog of Cyfip1 was shown to regulate neuromuscular junction formation (Schenck et al., 2003; Zhao et al., 2013) and eye morphogenesis (Bogdan et al., 2004; Galy et al., 2011). In mice, CYFIP1 interacts with FMRP and cap protein eIF4E to regulate activity-dependent protein translation in mature neurons (Napoli et al., 2008). Furthermore, Cyfip1 haploinsufficiency in mice produces fragile X-like phenotypes (Bozdagi et al., 2012). By focusing on the earliest stages of cortical development, our study provides evidence for a critical role of CYFIP1 in regulating adherens junctions and apical polarity of both human neural stem cells in the neural rosette model and mouse RGCs in the developing cortex in vivo. Moreover, as a functional consequence of CYFIP1 deficiency, RGCs and their progeny are aberrantly localized in the developing cortex in vivo, resulting in altered stratification of projection neurons and cortical layer malformation. Correct positioning of neurons in the mammalian cortex is a critical determinant of connectivity and neural function, as highlighted by severe neuronal migration disorders in humans (Ross and Walsh, 2001). Deficits in cortical patterning have also been suggested in schizophrenia (Arnold, 1999). A recent study found high incidence of patches of neocortical disorganization in autistic brains (Stoner et al., 2014), reminiscent of what we observed in mouse cortex after in utero exploration to knockdown CYFIP1. Our study therefore provides a new mechanistic model to understand how 15q11.2 CNVs as risk factors may contribute to susceptibility of neuropsychiatric disorders. Our study does not rule out the possibility that other factors within 15q11.2 CNVs affect neural development or that 15q11.2 CNVs also affect functional integrity of mature neurons, as suggested by rodent studies involving eIF4E (Napoli et al., 2008). Future studies of human neurons derived from our iPSC collection will help address the relevance of this pathway in human neuronal function.
Our study also reveals, for the first time, a critical role of the WAVE complex signaling in regulating neural stem cells. Early lethality of knockout mice for the majority of WAVE signaling components, including CYFIP1 (Bozdagi et al., 2012), WAVE2 (Yamazaki et al., 2003; Yan et al., 2003), Abi1 (Dubielecka et al., 2011), and Arp3 (Vauti et al., 2007), supports a requisite role of this pathway for mouse survival, which may have precluded in vivo investigation of its role in neural stem cells in early studies. Given that adherens junctions are rapidly lost in newly committed IPCs and neurons (Itoh et al., 2013; Rousso et al., 2012) and that there is little or reduced CYFIP1 expression in IPCs and immature neurons (Figure 3A), our result suggests that aberrant positioning of cortical projection neurons is caused by CYFIP1-WAVE signaling defects in RGCs. Future studies of cell type specific manipulation of CYFIP1-WAVE signaling in IPCs and/or immature neurons will provide a more definitive answer.
Human iPSC technology provides a new experimental platform to investigate cellular phenotypes and mechanisms in genetically tractable and disease-relevant human cell types. To date, patient-derived iPSCs, especially those related to monogenic disorders, have been successfully used to support models of disease pathology developed from animal studies, to demonstrate conserved cellular function of signalling pathways across species, or to facilitate large-scale screening of compounds to identify novel therapeutics (Bellin et al., 2013). Different from monogenic disorders, psychiatric disorders are often genetically complex and typically present with substantial variations in symptoms and degrees of impairment across individuals. Further, many risk associated genetic mutations are not exclusive to clinical populations, nor a particular disease. A key challenge and opportunity for human iPSC biology is to generate new insight into (patho)physiological phenotypes and mechanisms beyond merely supporting previous findings and concepts. Using human iPSCs as a leading discovery tool, we identified novel and consistent cellular phenotypes of neural stem cells that are specific for 15q11.2del, as they were not present in hNPCs derived from iPSCs with a DISC1 mutation, another risk factor for neuropsychiatric disorders (Thomson et al., 2013). While genetic risk factors for psychiatric disorders do not code for behaviour, we provide an example that they can lead to specific cellular abnormalities of biological processes implicated in the neurodevelopmental origins of these disorders. Using human iPSCs as an entry point enabled identification of novel investigative targets, followed by validation of in vivo physiological relevance and identification of underlying mechanisms using animal models, and finally, a return to human genetic association studies to support the disease-relevance of the identified pathway in humans (Figure 1B).
In summary, by leveraging and integrating information derived from multiple levels of analyses, ranging from cellular processes in human neural stem cells, in vivo animal models, to targeted human genetic association studies, we provide a novel mechanistic understanding of how 15q11.2 microdeletion affects neural developmental processes. Furthermore, our study illustrates the potential of human iPSC-based research to enable a multifaceted approach to tackle the mystery of complex psychiatric disorders.
EXPERIMENTAL PROCEDURES
iPSC Generation, Culture, Characterization and Neural Differentiation
All iPSC lines were derived from human donor dermal skin fibroblasts using integration-free episomal or Sendai virus methods. Fibroblasts with 15q11.2del (Y1, Y3, and Y4) were collected through the NIMH childhood-onset schizophrenia cohort and their family members (Mattai et al., 2011). All procedures were performed in accordance with IRB and ISCRO protocols approved by the Institutional Committees. iPSCs were cultured and characterized as previously described (Chiang et al., 2011; Juopperi et al., 2012).
iPSCs were differentiated into pNPCs according to a published protocol (Li et al., 2011). Neural rosette formation assays were performed using the mono-layer (Shi et al., 2012) and embryoid body methods (Juopperi et al., 2012). Neural rosettes were initially identified based on polarized pattern of DAPI staining and NESTIN immunoreactivity. Only individual non-overlapped neural rosettes that were 50–200 μm in diameter were included for quantification. The number of rosettes showing an intact apical-ring structure (more than 90% of coverage of apical-ring circumference with atypical PKCλ, N-cadherin, PAR3 or β-catenin immunoreactivity) and incomplete/partial apical structure (less than 90% coverage) were quantified.
In utero Electroporation and Quantitative Analysis of Mouse Cortical Development
In utero electroporation was performed as described (Saito, 2006). For quantitative analysis of electroporated neocortices, GFP+ cells localized within the dorso-lateral cortex were examined. A total of 3–6 brain sections were analyzed per animal by taking 3×3 images to cover the electroporated region of each coronal section with a 25× or 40× objective and comparing them with equivalent sections in littermate counterparts. Quantifications were performed using Imaris software (Bitplane). For distribution plots, the distances between GFP+Pax6+ cells or GFP+Tbr2+ cells and the ventricular surface were calculated by using an in-house MATLAB script (The MathWorks, Inc.), and plotted after dividing each distance by total length of the neocortex and subgrouping into ten equal-size vertical bins (1: the most apical, 10: the most basal).
All animal procedures were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee.
mRNA Expression Analysis of Post-mortem Human Brains, SNP Genotyping and Clinical Genetic Association and Interaction Analyses
mRNA expression data were generated from post-mortem DLPFC grey matter from 64 subjects without history or diagnosis of a medical or psychiatric disorder (51 males; mean age: 44 ± 14.9 years), all from European ancestry population and matched on age and various post-mortem tissue characteristics. Detailed methods relating to the Brain Tissue Collection of the Clinical Brain Disorders Branch at NIMH (CBDB/NIMH) and the Lieber Institute for Brain Development (LIBD) have been described elsewhere (Colantuoni et al., 2011).
DNA for genotyping was obtained from the cerebella of samples in the collection using Illumina OMNI 2.5M SNP chips. We used ANCOVAs, with age, sex and RIN (RNA Integrity Number) as covariates, to investigate main effects of SNPs on gene expression.
We carried out clinical genetic association and interaction analyses using logistic regression in four independent sample cohorts of cases with schizophrenia and healthy controls. Final interaction analysis was also assessed in the combined sample of four cohorts while controlling cohort effect in order to gain adequate power to detect interactions. The sample collection, genotyping and quality control have been described elsewhere (Zhang et al., 2011).
Supplementary Material
Highlights.
hiPSC-derived neural rosettes carrying 15q11.2 CNV exhibit polarity defects
CYFIP1 haploinsufficiency causes polarity defects via WAVE complex destabilization
CYFIP1 and WAVE signaling regulate radial glia cells in the developing mouse cortex
CYFIP1 and ACTR2 interact epistatically to affect risk for schizophrenia
Acknowledgments
We would like to thank members of Ming and Song laboratories for discussion, ICE stem cell core and H. Kim for generating some iPSC lines, K. Ahn, T. Andersen, V. Villagomez, L. Liu and Y. Cai for technical support and help. This work was supported by NIH (NS048271, HD069184), NARSAD, and MSCRF to G-l.M., SFARI, NIH (NS047344, MH087874), and IMHRO to H.S., The Lieber Institute for Brain Development to D.R.W., J.E.K and T.M.H., NARSAD and MSCRF to K.M.C., and by fellowships from MSCRF to G.M., N-S.K. and Z.W. and from NIH (F31MH102978) to H.N.N.
Footnotes
Supplementary information includes six tables and six figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold SE. Neurodevelopmental abnormalities in schizophrenia: insights from neuropathology. Dev Psychopathol. 1999;11:439–456. doi: 10.1017/s095457949900214x. [DOI] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2013;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Bogdan S, Grewe O, Strunk M, Mertens A, Klambt C. Sra-1 interacts with Kette and Wasp and is required for neuronal and bristle development in Drosophila. Development. 2004;131:3981–3989. doi: 10.1242/dev.01274. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Dorr N, Pilorge M, Takahashi N, Buxbaum JD. Haploinsufficiency of Cyfip1 produces fragile X-like phenotypes in mice. PLoS One. 2012;7:e42422. doi: 10.1371/journal.pone.0042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. doi: 10.1038/nrn2058. [DOI] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian K, Song H, Ming G. Application of reprogrammed patient cells to investigate the etiology of neurological and psychiatric disorders. Frontiers in Biology. 2012;7:179–188. doi: 10.1007/s11515-012-1216-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, von Spiczak S, Ostertag P, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wolf V, Brison N, Devriendt K, Peeters H. Genetic counseling for susceptibility loci and neurodevelopmental disorders: the del15q11.2 as an example. Am J Med Genet A. 2013;161A:2846–2854. doi: 10.1002/ajmg.a.36209. [DOI] [PubMed] [Google Scholar]
- Dubielecka PM, Ladwein KI, Xiong X, Migeotte I, Chorzalska A, Anderson KV, Sawicki JA, Rottner K, Stradal TE, Kotula L. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc Natl Acad Sci U S A. 2011;108:7022–7027. doi: 10.1073/pnas.1016811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy A, Schenck A, Sahin HB, Qurashi A, Sahel JA, Diebold C, Giangrande A. CYFIP dependent actin remodeling controls specific aspects of Drosophila eye morphogenesis. Dev Biol. 2011;359:37–46. doi: 10.1016/j.ydbio.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Geschwind DH. Advances in autism. Annu Rev Med. 2009;60:367–380. doi: 10.1146/annurev.med.60.053107.121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Moriyama Y, Hasegawa T, Endo TA, Toyoda T, Gotoh Y. Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nat Neurosci. 2013;16:416–425. doi: 10.1038/nn.3336. [DOI] [PubMed] [Google Scholar]
- Jahn JA, von Spiczak S, Muhle H, Obermeier T, Franke A, Mefford HC, Stephani U, Helbig I. Iterative phenotyping of 15q11.2, 15q13.3 and 16p13.11 microdeletion carriers in pediatric epilepsies. Epilepsy Res. 2013 doi: 10.1016/j.eplepsyres.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Grozeva D, Norton N, Ivanov D, Mantripragada KK, Holmans P, Craddock N, Owen MJ, O’Donovan MC. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18:1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulier K, Lathia JD, Marthiens V, Relucio J, Mughal MR, Tang SC, Coksaygan T, Hall PE, Chigurupati S, Patton B, et al. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattai AA, Weisinger B, Greenstein D, Stidd R, Clasen L, Miller R, Tossell JW, Rapoport JL, Gogtay N. Normalization of cortical gray matter deficits in nonpsychotic siblings of patients with childhood-onset schizophrenia. J Am Acad Child Adolesc Psychiatry. 2011;50:697–704. doi: 10.1016/j.jaac.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen SA, Carvill GL, Bellows S, Bayly MA, Berkovic SF, Dibbens LM, Scheffer IE, Mefford HC. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013;81:1507–1514. doi: 10.1212/WNL.0b013e3182a95829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Martin CL, Vazquez-Lopez A, Spence SJ, Alvarez-Retuerto AI, Sigman M, Steindler C, Pellegrini S, Schanen NC, Warren ST, Geschwind DH. Genome-wide expression profiling of lymphoblastoid cell lines distinguishes different forms of autism and reveals shared pathways. Hum Mol Genet. 2007;16:1682–1698. doi: 10.1093/hmg/ddm116. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Ross ME, Walsh CA. Human brain malformations and their lessons for neuronal migration. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Pearson CA, Gaber ZB, Miquelajauregui A, Li S, Portera-Cailliau C, Morrisey EE, Novitch BG. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci U S A. 2001;98:8844–8849. doi: 10.1073/pnas.151231598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. S471. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Meyer-Lindenberg A, Steinberg S, Magnusdottir BB, Morgen K, Arnarsdottir S, Stefansson K. CNVs conferring risk of autism or schizophrenia affect cognition in controls. Nature. 2013 doi: 10.103/nature12818. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TE. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. Embo J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GW, van de Lagemaat LN, Redon R, Strathdee KE, Croning MD, Malloy MP, Muir WJ, Pickard BS, Deary IJ, Blackwood DH, et al. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38:445–451. doi: 10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- Thomson PA, Malavasi EL, Grunewald E, Soares DC, Borkowska M, Millar JK. DISC1 genetics, biology and psychiatric illness. Front Biol (Beijing) 2013;8:1–31. doi: 10.1007/s11515-012-1254-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zwaag B, Staal WG, Hochstenbach R, Poot M, Spierenburg HA, de Jonge MV, Verbeek NE, van ‘t Slot R, van Es MA, Staal FJ, et al. A co-segregating microduplication of chromosome 15q11.2 pinpoints two risk genes for autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:960–966. doi: 10.1002/ajmg.b.31055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauti F, Prochnow BR, Freese E, Ramasamy SK, Ruiz P, Arnold HH. Arp3 is required during preimplantation development of the mouse embryo. FEBS Lett. 2007;581:5691–5697. doi: 10.1016/j.febslet.2007.11.031. [DOI] [PubMed] [Google Scholar]
- Wegiel J, Frackowiak J, Mazur-Kolecka B, Schanen NC, Cook EH, Jr, Sigman M, Brown WT, Kuchna I, Wegiel J, Nowicki K, et al. Abnormal intracellular accumulation and extracellular Abeta deposition in idiopathic and Dup15q11.2-q13 autism spectrum disorders. PLoS One. 2012;7:e35414. doi: 10.1371/journal.pone.0035414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S, Fujiwara T, Yoshida N, Takenawa T. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, Fujiwara Y, Bronson R, Snapper SB, Kirschner MW, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. Embo J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wang D, Wang Q, Rodal AA, Zhang YQ. Drosophila cyfip regulates synaptic development and endocytosis by suppressing filamentous actin assembly. PLoS Genet. 2013;9:e1003450. doi: 10.1371/journal.pgen.1003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Li T, Zhao X, Huang K, Wang T, Li Z, Ji J, Zeng Z, Zhang Z, Li K, et al. Rare CNVs and tag SNPs at 15q11.2 are associated with schizophrenia in the Han Chinese population. Schizophr Bull. 2013;39:712–719. doi: 10.1093/schbul/sbr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.