Abstract
Purpose
The goal of our study was to determine whether genomic copy number abnormalities (deletions and duplications) affecting genes involved in eye development contribute to the etiology of anophthalmia, microphthalmia and coloboma.
Methods
The affected individuals were tested for deletions and duplications in genomic DNA using 2 million probe (HD2) comparative genomic hybridization arrays (aCGH) from Roche-NimbleGen.
Results
Array analysis of 32 patients detected one case with a deletion encompassing the Renal-coloboma syndrome associated gene PAX2. Non-polymorphic copy number changes were also observed at several candidate chromosomal regions, including 6p12.3, 8q23.1q23.2, 13q31.3, 15q11.2q13.1, 16p13.13 and 20q13.13.
Conclusions
This study identified the first patient with the typical phenotype of the Renal-coloboma syndrome caused by a submicroscopic deletion of the coding region of the PAX2 gene. The finding suggests that PAX2 deletion testing should be performed in addition to gene sequencing as a part of molecular evaluation for the Renal-coloboma syndrome. aCGH testing of 32 affected individual showed that genomic deletions and duplications are not a common cause of non-syndromic anophthalmia, microphthalmia and/or coloboma, but undoubtedly contribute to the etiology of these eye anomalies. aCGH testing therefore represents an important and valuable addition to candidate gene sequencing in research and diagnostics of ocular birth defects.
Keywords: array CGH, deletions, duplications, microphthalmia, anophthalmia, coloboma
INTRODUCTION
Three related ocular birth defects–anophthalmia, microphthalmia and coloboma–are an important contributor to childhood visual impairment and blindness, impacting 2 out of 10,000 newborns annually1–3. More than twenty different genetic loci have been implicated in congenital eye malformations, with most associated genes having a role in eye development4–6. However, each known gene is responsible for only a small percentage of cases, and many additional causative genetic factors still await identification5–6. Mutations in the known genes account for merely ~15% of cases of anophthalmia, microphthalmia and coloboma7. For the remaining patients, the lack of a specific molecular diagnosis prevents prediction of long term outcomes, anticipation of systemic complications and estimation of the recurrence risk in their families.
Studies have shown that gene deletions and duplications may comprise up to 15% of mutations underlying monogenic disease 8. High resolution whole genome aCGH testing of patients with genetic diseases can therefore detect copy number abnormalities in genes responsible for their clinical phenotypes. Application of aCGH not only detects abnormalities in known disease-causing genes, but also can identify new candidate genes for specific disorders8. Multiple examples exist of successful application of aCGH in disease gene discovery. These include implication of the TCF2 gene in the etiology of multicystic dysplastic kidneys9, identification of PORCN as the causative gene for focal dermal hypoplasia (FDH; Goltz syndrome)10 and discovery of loci for congenital diaphragmatic hernia 11. aCGH has also contributed to identification of genes associated with congenital eye anomalies. For example, CHARGE syndrome is characterized with a specific set of birth defects which includes coloboma of the iris, retina, choroid and/or optic disc, with or without microphthalmia12. Using aCGH, Vissers et al. implicated the CHD7 gene in the etiology of CHARGE syndrome by detecting a de novo microdeletion of the CHD7 locus at 8q12 in an affected individual 13. The roles of the GDF6 gene at 8q21.2-q22.1, TFAP2A gene at 6p24.3 and TMX3 gene at 18q22.1 in causing ocular developmental anomalies have also been discovered by testing patients who carried deletions of these genes14–16.
Although causative copy number changes have been reported in isolated cases of ocular birth defects, no one has systematically tested large numbers of affected individuals for deletions and duplications in genomic DNA. We hypothesized that ocular birth defects frequently result from copy number abnormalities involving critical genes. To test this hypothesis, we examined a cohort of patients with anophthalmia, microphthalmia and coloboma for submicroscopic deletions and duplications using whole genome high resolution oligo aCGH.
MATERIALS AND METHODS
Patients were enrolled through an ongoing, IRB approved research study “Genetics of Microphthalmia, Anophthalmia and Coloboma” at the University of Minnesota, Department of Pediatrics, Division of Genetics and Metabolism. Written informed consent was obtained from all participants and/or their parents, as appropriate. Thirty-two patients with either isolated anophthalmia, microphthalmia and coloboma (23 cases) or with anophthalmia, microphthalmia and coloboma in association with other congenital anomalies (cases 2, 6, 8, 9, 26, 28, 30, 31, 32) were selected for testing (Table 1). All patients were evaluated by a clinical geneticist or an ophthalmologist and were enrolled in the study if they were lacking genetic diagnosis after clinical examination and standard of care testing. Molecular tests performed in each patient prior to the study (if known) are listed in Table 1. Array CGH analysis was performed using a commercially available HD2 human whole-genome CGH array (Roche NimbleGen Systems Inc., Madison, WI), with total of ~2 million probes at a median interprobe distance of approximately 1169bp. This array can detect small genomic imbalances (deletions and duplications) at the resolution of individual genes (~5–10 kb). Specimen labeling, array hybridization, washing and scanning were performed at NimbleGen service laboratory in Iceland. Data analysis was performed at the UW Cytogenetic Services Laboratory at the University of Wisconsin-Madison, using the NimbleScan and SignalMap softwares from Roche NimbleGen and OneClickCGH software from Infoquant. Regions with copy number changes detected by aCGH were compared against the database of Genomic Variants (http://projects.tcag.ca/variation/), which catalogues known benign copy number polymorphisms (CNPs) in the human genome. Only imbalances that do not correspond to known polymorphisms were evaluated further. Non-polymorphic copy number changes were confirmed by separate aCGH experiments, using a different aCGH platform (EmArray Cyto6000 array (Agilent Technologies, Santa Clara, CA)).
Table 1.
Patients with syndromic and non-syndromic anophthalmia, microphthalmia and coloboma tested by aCGH.
| Sample ID | Ocular Phenotype | Other Phenotypes (if not non-syndromic) | Previous Genetic Testing |
|---|---|---|---|
| 1 | bilateral iris coloboma | No information | |
| 2 | bilateral optic pits | hypothalamic, hypogonadism, mild cognitive disability, hypoplastic mullerian derivatives, absent ovary, left leg spasticity | karyotype |
| 3 | bilateral iris coloboma | No information | |
| 4 | L iris coloboma | No information | |
| 5 | L iris, retinal and optic nerve coloboma | No information | |
| 6 | bilateral colobomatous microphthalmia | developmental delay | No information |
| 7 | R retinal coloboma, L microphthalmia | No information | |
| 8 | L anophthalmia, R microphthalmia | UPJ obstruction and hydronephrosis | No information |
| 9 | bilateral colobomatous microphthalmia | kidney reflux | No information |
| 10 | bilateral iris coloboma | No information | |
| 11 | bilateral iris coloboma | PAX6, SHH, karyotype | |
| 12 | bilateral iris coloboma | Sibling of 4071 | |
| 13 | bilateral colobomatous microphthalmia, PHPV | PAX6, SHH, SIX6, SOX2, karyotype | |
| 14 | bilateral iris coloboma | No information | |
| 15 | bilateral coloboma | No information | |
| 16 | unknown coloboma, grandchildren with aniridia | No information | |
| 17 | unilateral microphthalmia, morning glory | No previous testing | |
| 18 | bilateral iris coloboma | No information | |
| 19 | unilateral severe microphthalmia, inferonasal coloboma and cataract | No information | |
| 20 | unilateral right complete coloboma with microphthalmia | No previous testing | |
| 21 | bilateral iris, retinal coloboma | Agilent 44K array, karyotype | |
| 22 | coloboma unspecified | No information | |
| 23 | bilateral colobomatous microphthalmia | TORCH titres | |
| 24 | unilateral right iris coloboma | No information | |
| 25 | bilateral chorioretinal coloboma | No information | |
| 26 | bilateral colobomatous microphthalmia | renal failure | No information |
| 27 | coloboma unspecificed | No information | |
| 28 | bilateral retinal/iris coloboma | shawl scrotum and glandular hypospadias | No information |
| 29 | coloboma unspecified | No information | |
| 30 | right unilateral microphthalmia | multicystic kidney | No information |
| 31 | morning glory anomaly | bilateral renal failure with renal hypoplasia | No information |
| 32 | optic nerve hypoplasia, | abnormal ears, renal hypoplasia | No previous testing |
RESULTS
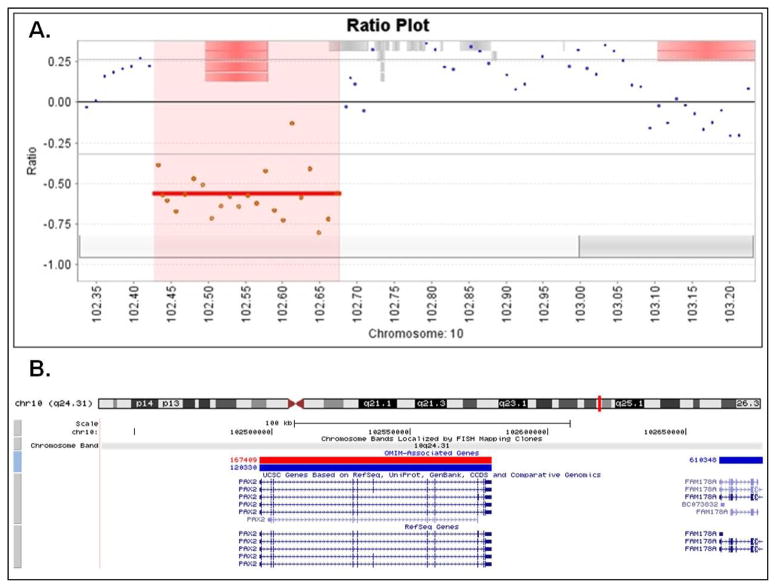
High resolution aCGH analysis of 32 patients with ocular defects revealed a ~240kb deletion on chromosome 10 in a patient with clinical features of Renal-coloboma (Papillorenal) syndrome 17. The deletion included the entire PAX2 coding region and a portion of the FAM178A (C10orf6) gene (Figure 1). This finding provided the molecular confirmation of the patient’s clinical diagnosis and showed that, in addition to point mutations, deletions in the PAX2 gene contribute to the etiology of the Renal-coloboma syndrome.
Figure 1.
A) Array CGH profile of the 10q24.31 deletion affecting the PAX2 gene in the patient with Renal-coloboma (Papillorenal) syndrome; panel A (top) is showing the close view of the deleted segment from the OneClick CGH software. B) A screenshot from the UCSC genome browser showing the deleted region at cytogenetic location 10q24.31, from 102,437,000 to 102,677,000 based on the Human Genome March 2006 (hg18) assembly.
A 5.38Mb duplication was detected on the long arm of chromosome 15 (cytogenetic location 15q11.2q13.1) in a patient with microphthalmia (R), anophthalmia (L), ureteropelvic junction (UPJ) obstruction and hydronephrosis. The following fourteen genes were affected by the duplication: ATP10A, C15orf2, GABRA5, GABRB3, GABRG3, GOLGA8E, HERC2, MAGEL2, MKRN3, NDN, OCA2, SNRPN, SNURF and UBE3A. Deletions of the same region, when inherited maternally, result in the phenotype of Prader-Willi Syndrome, while paternal deletions of this region lead to the Angelman Syndrome phenotype. Duplications of the 15q11.2 region, particularly when inherited on the maternal chromosome, are associated with hypotonia, autistic behavior, developmental delay, mental retardation, seizures and mild dysmorphic features18. However, eye anomalies have not previously been reported in patients with the 15q11.2q13.1 duplications.
In addition to the deletion of the PAX2 gene at 10q24.31 and duplication of the Prader-Willi/Angelman Syndrome critical region at 15q11.2q13.1, non-polymorphic copy number changes were detected at several candidate regions including 6p12.3, 8q23.1q23.2, 13q31.3, 16p13.13 and 20q13.13. Based on the potential function of the genes in the regions, deletions at 13q31.3 and 8q23.1q23.2 were selected for further follow-up.
The 13q31.3 deletion was detected in a patient with bilateral iris coloboma. The deletion was approximately 240kb in size and included only one gene, glypican 5 (GPC5). The GPC5 gene was a plausible positional and functional candidate gene for causing congenital eye defects in our patient; however the 13q31.3 deletion was not present in the patient’s cousin who was also affected with bilateral iris coloboma (parental samples were not available).
The 8q23.1q23.2 deletion was observed in a patient with unilateral severe microphthalmia. The deletion was 1.5Mb in size and affected the following four known genes: PKHD1L1, KCNV1, EBAG9 and GOLSYN. Parental samples were tested by aCGH for the presence of the deletion detected in the proband and the unaffected father was found to carry the same deletion on chromosome 8.
A summary of all detected non-polymorphic copy number changes, including their genomic location, gene content, presence in additional family members and likely clinical significance is provided in Table 2.
Table 2.
Summary of copy number abnormalities detected by aCGH testing.
| Sample | Copy Number Change | Chromosome | Start | Stop | Size in bp | Genes/Proteins | Associated phenotype | Presence in other family members | Significance for Ocular Defects |
|---|---|---|---|---|---|---|---|---|---|
| 7 | deletion | 20 | 47,586,000 | 47,610,000 | 24,000 | PTGIS-prostaglandin I2 (prostacyclin) synthase | unknown | unknown | unknown |
| 8 | duplication | 15 | 20,850,000 | 26,298,000 | 5,448,000 | GOLGA8E-golgi autoantigen, golgin subfamily a, 8E, C15orf2-hypothetical protein LOC23742, SNRPN-small nuclear ribonucleoprotein polypeptide N, UBE3A- ubiquitin protein ligase E3A, AP10A- ATPase, class V, type 10A, GABRG3- gamma-aminobutyric acid (GABA) A receptor, gamma, MKRN3-makorin ring finger protein 3, MAGEL2-MAGE-like protein 2, NDN-necdin, GABRB3-gamma- aminobutyric acid (GABA) A receptor, beta, GABRA5-gamma-aminobutyric acid (GABA) A receptor, alpha, OCA2- oculocutaneous albinism II | Maternal deletions: Prader-Willi syndrome; Paternal deletions: Angelman syndrome; Maternal duplications: autism, developmenttal delay, behavioral issues | unknown | unknown (likely unrelated) |
| 10 | deletion | 13 | 90,870,000 | 91,098,000 | 228,000 | GPC5-glypican 5 | unknown | not present in affected cousin | unknown (likely unrelated) |
| 15 | deletion | 16 | 12,018,000 | 12,030,000 | 12,000 | RUNDC2A-RUN domain containing 2A | unknown | not present in affected daughter | unknown (likely unrelated) |
| 19 | deletion | 8 | 110,454,000 | 110,898,000 | 444,000 | PKHD1L1-fibrocystin L, KCNV1- potassium channel, subfamily V, member 1, EBAG9-estrogen receptor binding site associated, GOLSYN-Golgi-localized syntaphilin-related protein | unknown | present in unaffected father | unknown (likely unrelated) |
| 31 | deletion | 6 | 45,258,000 | 45,378,000 | 120,000 | SUPT3H-suppressor of Ty 3 homolog isoform 2, hsa-miR-586 | unknown | unknown | unknown |
| 32 | deletion | 10 | 102,438,000 | 102,678,000 | 240,000 | PAX2-paired box protein 2 isoform c | Renal- coloboma Syndrome | unknown | causative |
DISCUSSION
aCGH testing of 32 individuals with ocular birth defects detected one deletion responsible for the eye phenotype in the tested individual, one disease associated duplication that was unlikely the cause of the patient’s eye anomalies, two deletions affecting strong candidate genes for eye anomalies and three changes of completely unknown clinical significance.
The causative deletion was detected in a patient with clinical features of Renal-coloboma (Papillorenal) syndrome and it affected the known gene for this disorder, PAX2. The deletion also included a portion of the FAM178A(C10orf6) gene, which codes for a hypothetical protein of unknown function. The partial deletion of the FAM178A gene most likely did not significantly contribute to the patient’s phenotype. The patient was a 9-year-old boy with typical features of the Renal-coloboma syndrome, including optic nerve hypoplasia, secondary strabismus, mild deafness, dysplastic ear helices and renal hypoplasia. PAX2 gene sequencing had been performed previously and no point mutations had been found. To our knowledge, Renal-coloboma syndrome due to a deletion of the PAX2 gene has been reported in only one other patient who had a large interstitial 10q deletion encompassing the PAX2 locus detected by high resolution chromosome analysis19. We report the first submicroscopic deletion affecting the coding region of the PAX2 gene. No other genes likely to contribute to the patient’s phenotype were affected by the rearrangement. Identification of this deletion stresses the importance of incorporating deletion/duplication testing together with the PAX2 gene sequencing into molecular diagnostics of the Renal-coloboma syndrome.
Our study detected a duplication of the Prader-Willi Syndrome critical region on chromosome 15 in a patient with multiple eye and kidney anomalies. Duplications of the 15q11.2q13.1 region have been well described in the literature; when inherited on the maternal chromosome they are associated with hypotonia, autistic behavior, developmental delay, mental retardation, seizures and mild dysmorphic features18. Eye anomalies have not been reported in patients with 15q11.2q13.1 duplications, although a locus for the autosomal dominant colobomatous microphthalmia has been mapped to an overlapping but more distal region on chromosome 1520. Therefore, this duplication may not be the cause of the eye anomalies in our patient, but its detection helps to explain his other clinical findings, like developmental delay and behavioral issues.
Among the detected non-polymorphic copy number changes, two appeared to involve likely candidate genes for eye anomalies: the 13q31.3 deletion and the 8q23.1q23.2 deletion. Deletion of the 13q31.3 region was initially considered as clinically significant based both on the chromosomal position and the function of the affected gene. Coloboma, microphthalmia and anophthalmia have previously been reported in association with deletions at q31–q33 region of the long arm of chromosome 1321. The deletion detected in our patient included the glypican 5 (GPC5) gene, which belongs to a family of glycosylphosphatidylinositol (GPI)-anchored, membrane-bound heparan sulfate (HS) proteoglycans. Glypicans play a role in modulating the activity of HS-binding growth factors22. Their involvement in developmental morphogenesis and growth regulation has been shown by Drosophila mutants, as well as human genetic disorders like Simpson-Golabi-Behmel syndrome23 and autosomal-recessive omodysplasia24. Dally, drosophila ortholog of the gene deleted in our patient GPC5, is known to affect cell division patterning in developing eye22. The GPC5 gene was therefore considered a plausible candidate gene for causing the congenital eye defects in our patient, but testing additional family members did not detect the 13q31.3 deletion in the patient’s cousin with the identical ocular defect. Since it does not segregate with the eye anomalies, the deletion is unlikely the cause of the ocular defects in this family. However, this deletion has not been reported as a benign variant (http://projects.tcag.ca/variation/) and has been detected by another laboratory in two unrelated individuals with developmental delay and cognitive impairment (personal communication). Therefore, the clinical significance of the 13q31.1 deletion requires further investigation.
The 8q23.1q23.2 deletion was observed in a patient with unilateral severe microphthalmia and small inferonasal coloboma. He also had a dense posteriorly subluxated crystalline lens and Persistent Hyperplastic Primary Vitreous (PHPV). His head MRI and development have been normal. The deletion affected four known genes, including GOLSYN, which is known to play an important role in neuronal development. Since the unaffected father was found to carry the same deletion on chromosome 8, it was concluded that the deletion was unlikely the cause of the eye anomalies diagnosed in the proband. However, it is possible that the detected deletion has incomplete penetrance and that some of the carriers do not express abnormal phenotype. Alternatively, cases have been reported where benign copy number variants contribute to abnormal phenotypes by unmasking mutations in non-deleted alleles25,26.
The 13q31.1 and 8q23.1q23.2 deletion cases illustrate difficulties in interpreting clinical significance of copy number abnormalities detected by high resolution aCGH testing. For example, the causative role cannot be assumed solely based on the function of the affected genes. Although plausible candidate genes mapped within both deleted regions, the role of the 13q31.1 and 8q23.1q23.2 deletion in causing eye anomalies in the probands was not supported by testing additional family members. These cases therefore also demonstrate the value of having clinical information and DNA samples available from patients’ parents and other affected and unaffected members of their families.
Our study showed that aCGH could detect deletions and duplications associated with ocular birth defects. However, copy number abnormalities did not appear to be a common cause of isolated anophthalmia, microphthalmia and coloboma. Although gene deletions and duplications significantly contribute to pathogenesis of genetic disorders, the vast majority of disease causing mutations are nucleotide changes in genomic DNA. We propose that a combination of aCGH analysis with high throughput sequencing methods that allow detection of base changes (point mutations) in a large number of candidate genes for eye malformations will be the most successful strategy for identification of new genetic causes of anophthalmia, microphthalmia and coloboma.
Acknowledgments
This project was supported in part by the University of Wisconsin Institute for Clinical and Translational Research through an NIH Clinical and Translational Science Award, grant number 1UL1RR025011, and by funding from the University of Minnesota, Department of Pediatrics.
We gratefully acknowledge the patients, families, and clinicians who have submitted clinical samples for the microphthalmia, anophthalmia and coloboma study.
References
- 1.Kallen B, Robert E, Harris J. The descriptive epidemiology of anophthalmia and microphthalmia. Int J Epidemiol. 1996;25:1009–16. doi: 10.1093/ije/25.5.1009. [DOI] [PubMed] [Google Scholar]
- 2.Morrison D, FitzPatrick D, Hanson I, et al. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet. 2002;39:16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoll C, Alembik Y, Dott B, Roth MP. Congenital eye malformations in 212,479 consecutive births. Ann Genet. 1997;40:122–128. [PubMed] [Google Scholar]
- 4.Gregory-Evans CY, Williams MJ, Halford S, Gregory-Evans K. Ocular coloboma: a reassessment in the age of molecular neuroscience. J Med Genet. 2004;41:881–891. doi: 10.1136/jmg.2004.025494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzpatrick DR, van Heyningen V. Developmental eye disorders. Curr Opin Genet Dev. 2005;15:348–353. doi: 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Chang L, Blain D, Bertuzzi S, Brooks BP. Uveal coloboma: clinical and basic science update. Curr Opin Ophthalmol. 2006;17:447–470. doi: 10.1097/01.icu.0000243020.82380.f6. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez-Rodriguez J, Pelcastre EL, Tovilla-Canales JL, Garcia-Ortiz JE, Amato-Almanza M, Villanueva-Mendoza C, Espinosa-Mattar Z, Zenteno JC. Mutational screening of CHX10, GDF6, OTX2, RAX and SOX2 genes in 50 unrelated microphthalmia–anophthalmia–coloboma (MAC) spectrum cases. Br J Ophthalmol. 2010;94:1100–1104. doi: 10.1136/bjo.2009.173500. [DOI] [PubMed] [Google Scholar]
- 8.Vissers LE, Veltman JA, van Kessel AG, Brunner HG. Identification of disease genes by whole genome CGH arrays. Hum Mol Genet. 2005;14:R215–223. doi: 10.1093/hmg/ddi268. [DOI] [PubMed] [Google Scholar]
- 9.Mefford HC, Clauin S, Sharp AJ, et al. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–1069. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Reid Sutton V, Omar Peraza-Llanes J, et al. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nat Genet. 2007;39:836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- 11.Zayed H, Chao R, Moshrefi A, et al. A maternally inherited chromosome 18q22. 1 deletion in a male with late-presenting diaphragmatic hernia and microphthalmia-evaluation of DSEL as a candidate gene for the diaphragmatic defect. Am J Med Genet. 2010;152A:916–923. doi: 10.1002/ajmg.a.33341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanlaville D, Verloes A. CHARGE syndrome: an update. Eur J Hum Genet. 2007;15:389–399. doi: 10.1038/sj.ejhg.5201778. [DOI] [PubMed] [Google Scholar]
- 13.Vissers L, Evan Ravenswaaij CM, Admiraal R, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 14.Asai-Coakwell M, French CR, Berry KM, et al. GDF6, a novel locus for a spectrum of ocular developmental anomalies. Am J Hum Genet. 2007;80:306–315. doi: 10.1086/511280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milunsky JM, Maher TA, Zhao G, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao R, Nevin L, Agarwal P, et al. A Male with unilateral microphthalmia reveals a role for TMX3 in eye development. PloS ONE. 2010;5:e10565. doi: 10.1371/journal.pone.0010565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eccles MR, Schimmenti LA. Renal-coloboma syndrome: a multi-system developmental disorder caused by PAX2 mutations. Clin Genet. 1999;56:1–9. doi: 10.1034/j.1399-0004.1999.560101.x. [DOI] [PubMed] [Google Scholar]
- 18.Browne CE, Dennis NR, Maher E, et al. Inherited interstitial duplications of proximal 15q: genotype-phenotype correlations. Am J Hum Genet. 1997;61:1342–1352. doi: 10.1086/301624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benetti E, Artifoni L, Salviati L, et al. Renal hypoplasia without optic coloboma associated with PAX2 gene deletion. Nephrol Dial Transplant. 2007;22:2076–2078. doi: 10.1093/ndt/gfm187. [DOI] [PubMed] [Google Scholar]
- 20.Michon L, Morle L, Bozon M, et al. Physical and transcript map of the autosomal dominant colobomatous microphthalmia locus on chromosome 15q12–q15 and refinement to a 4. 4Mb region. Eur J Hum Genet. 2004;12:574–578. doi: 10.1038/sj.ejhg.5201197. [DOI] [PubMed] [Google Scholar]
- 21.Lansink PJ, Moll AC, Imhof SM, et al. Variable expression of ophthalmological findings in the 13q deletion syndrome. Arch Ophthalmol. 2005;123:127–128. doi: 10.1001/archopht.123.1.127-c. [DOI] [PubMed] [Google Scholar]
- 22.Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108:497–501. doi: 10.1172/JCI13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeBaun MR, Ess J, Saunders S. Simpson Golabi Behmel syndrome: progress toward understanding the molecular basis for overgrowth, malformation, and cancer predisposition. Mol Genet Metab. 2001;72:279–286. doi: 10.1006/mgme.2001.3150. [DOI] [PubMed] [Google Scholar]
- 24.Campos-Xavier AB, Martinet D, Bateman J, et al. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am J Hum Genet. 2009;84:760–770. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flipsen-ten Berg K, van Hasselt PM, Eleveld MJ, et al. Unmasking of a hemizygous WFS1 gene mutation by a chromosome 4p deletion of 8. 3 Mb in a patient with Wolf-Hirschhorn syndrome. Eur J Hum Genet. 2007;15:1132–1138. doi: 10.1038/sj.ejhg.5201899. [DOI] [PubMed] [Google Scholar]
- 26.Coman DJ, Gardner RJM. Deletions Revealing Recessive Genes: Deletions that reveal recessive genes. Eur J Hum Genet. 2007;15:1103–1104. doi: 10.1038/sj.ejhg.5201919. [DOI] [PubMed] [Google Scholar]