Abstract
Background
The aim of this study was to analyze the CYP2C19 genetic polymorphism among Han and Uyghur patients with cardiovascular and cerebrovascular diseases in the Kashi area of Xinjiang.
Material/Methods
We enrolled 1020 patients with cardiovascular and cerebrovascular diseases, including 220 Han subjects and 800 Uyghur subjects. We used the gene chip method to detect polymorphisms in CYP2C19. The allele frequencies of CYP2C19 and the metabolic phenotype frequencies were then compared between the 2 ethnic groups.
Results
The frequency of CYP2C19 *1 was 0.6454 in Han subjects and 0.7869 in Uyghur subjects, and the difference was statistically significant (P<0.05). The frequency of CYP2C19 *2 was 0.3273 in Han subjects and 0.1837 in Uyghur subjects (P<0.05). The frequency of the homozygous extensive metabolizer phenotype was 42.72% and 62.13% in Han and Uyghur subjects, respectively (P<0.01). The frequency of the heterozygous extensive metabolizer phenotype was 43.64% and 33.13% in Han and Uyghur subjects, respectively (P<0.01). The frequency of poor metabolizers in Han and Uyghur subjects was 13.64% and 4.76%, respectively (P<0.01).
Conclusions
Among patients with cardiovascular and cerebrovascular diseases located in the Kashgar Prefecture of Xinjiang, there is a differential distribution of CYP2C19 genotypes between the Han and Uyghur populations. Uyghur patients showed higher frequencies of extensive metabolizer genotypes than Han patients, while Han patients showed higher frequencies of poor metabolizer genotypes than Uyghur patients.
MeSH Keywords: Cytochrome-c Oxidase Deficiency; Genes, vif; Genotyping Techniques
Background
Cytochrome P450 (CYP450) is a superfamily of isoenzymes involved in energy conversion in biological organisms. CYP2C19 is an important member of the CYP450 superfamily. It has been shown that the metabolism of a number of drugs, including platelet aggregation antagonists, proton pump inhibitors, anti-diabetes drugs, and anti-cancer drugs, all depend on this enzyme [1]. The CYP2C19 gene exhibits genetic polymorphism and differences among races, as evidenced by variations in drug metabolism. Clopidogrel is an anti-platelet drug commonly used in cardiovascular and cerebrovascular diseases that inhibits platelet aggregation by blocking the platelet ADP receptor pathway [2]. However, a recent study [3] has revealed clopidogrel resistance in some patients. Another study suggested that clopidogrel resistance is associated with CYP2C19*2 and CYP2C19*3 genes and that there is a positive correlation between CYP2C19*2/CYP2C19*3 carriers and clopidogrel resistance [4]. It has also been observed [5,6] that the distribution of CYP2C19 genotypes and allele frequencies are different among healthy Han and Uyghur populations. However, there have been few studies on the CYP2C19 genotype and allele frequencies within populations with cardiovascular and cerebrovascular diseases. The objective of this study was to explore the distribution of CYP2C19 genotypes among Han and Uyghur patients with cardiovascular and cerebrovascular diseases in the Kashi area of Xinjiang and to identify any differences.
Material and Methods
Patient information
We selected 1020 patients hospitalized with cardiovascular and cerebrovascular diseases between May 2011 and July 2013. Patients with severe heart failure or liver or kidney diseases were excluded. All selected patients had undergone clopidogrel therapy and had no familial relations with one another. Two hundred and twenty patients were of Han ethnicity and 33–80 years of age. The remaining 800 patients were of Uyghur ethnicity and were 23–88 years of age. This study was approved by the ethics committee of the Kashgar 1st People’s Hospital and informed consent was obtained from all subjects.
Reagents and devices
We used a blood genomic DNA extraction kit (Shanghai Baio Co.); a genetic testing kit, hybrid staining kit (Shanghai Baio Co.); a Linegene9640 PCR thermocycler (Hangzhou Bioer Co.); a BaioR-Hyb automated hybridization instrument and BaioRBE-2.0 gene chip reading instrument (Shanghai Baio Co.); and a Smart spec3000 UV spectrophotometer (BIORAD Co., USA).
Detection of CYP2C19 genetic polymorphism
Blood genomic DNA purification
Firstly, 2 ml of peripheral venous blood was taken and anti-coagulated using EDTA. Genomic DNA was purified based on manufacturer’s instructions. Then, the UV spectrophotometer was used to measure the concentration and purity of DNA. The target concentration of DNA was 10–60 ng/μl, and the target OD260/OD280 ratio was 1.5–2.0.
PCR amplification
Detection of the CYP2C19 polymorphism required the amplification of 2 sites (636G/A and 681G/A). The PCR amplification proceeded as follows: reaction system: 5 μL genomic DNA; 19 μl CYP2C19 amplification solution 1 or 2; 1 μl reaction solution A; and total volume 25 μl. The amplification solution contained biotinylated primer pairs. Positive and negative controls were amplified simultaneously with the samples. Amplification conditions were 50ºC for 5 min, 94ºC for 5 min, and 35 cycles of 94ºC for 25 s, 48ºC for 40 s, and 72ºC for 30 s.
Hybrid staining
Required materials were: 1200 μl CYP2C19 pre-hybridization solution; 200 μl hybridization reaction solution (180 μl hybridization buffer + 10 μl CYP2C19 amplification product 1 + 10 μl CYP2C19 amplification product 2); 800 μl washing solution A; 1600 μl washing solution B; 400 μl washing solution C; 200 μl antibody solution (with alkaline phosphatase-labeled avidin); developing solution (BCIP\NBT). We placed the pre-hybridization solution in the hybridization container for 5 min at 42ºC and aspirated the remaining solution from the container. We added hybridization reaction solution into the container and quickly inserted the gene chip. After incubation for 30 min at 42ºC, we aspirated the solution from the container, and added washing solution A for 6 min at 42ºC. Then we aspirated the solution from the container and repeated the process once more. Antibody solution was added and incubated at room temperature for 20 min, then the solution from the container was aspirated. We added washing solution B and incubated it at room temperature for 5 min, then aspirated the solution from the container and repeated the process once more. We added washing solution C and incubated it at room temperature for 3 min, then aspirated the solution from the container and repeated the process once again. The developing solution was added and incubated at 42ºC for 30 min. After aspirating any remaining solution from the container, the gene chip was rinsed with distilled water and heat dries.
Gene chip scanning and genotype analysis
We placed the gene chip into the gene chip reading instrument and analyzed it using BaioRBE-2.0 software, which was used for image scanning, data analysis, and data output. Each experiment was accompanied by a positive control (CYP2C19*1/*3) and a negative control (blank). A schematic representation of the CYP2C19 gene chip detection and different genotypic distributions on the chips are shown in Figures 1 and 2.
Figure 1.

Schematic of CYP2C19 gene chip detection.
Figure 2.
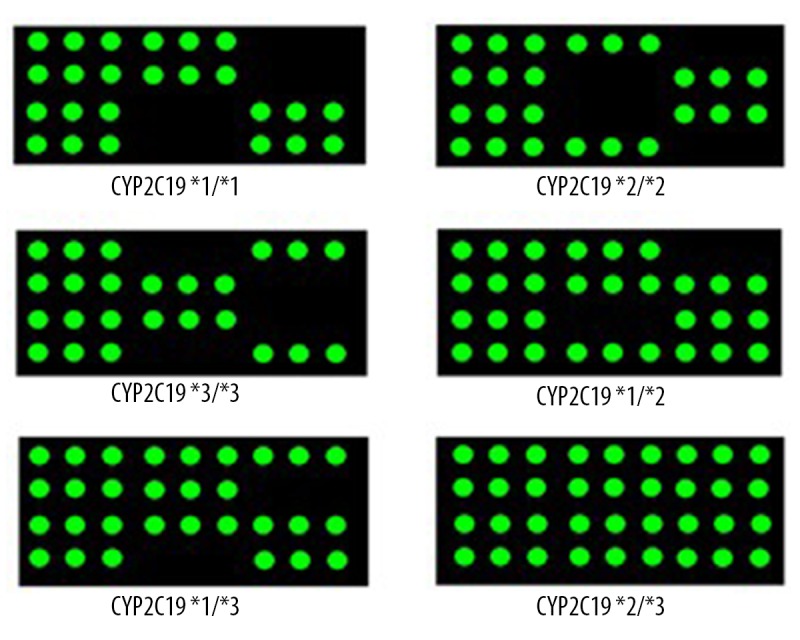
Schematic representation of results obtained from different genotypes.
Statistical analysis
SPSS17.0 was used for statistical analysis. Genotype frequencies and allele frequencies were calculated with the gene frequency counting method. Frequencies for each allele were calculated using the Hardy-Weinberg equilibrium and the data obtained were tested for their fit to the H-W equilibrium (χ2 test) to validate data reliability. The comparison of genotypic frequencies between the 2 ethic groups was performed using the χ2 test, with P≤0.05 as the statistical significance cut-off.
Results
CYP2C19 genotype frequencies in Han and Uyghur subjects
The CYP2C19 genotype frequency in Han and Uyghur populations satisfied the Hardy-Weinberg equilibrium (P>0.05), which signified it was a reliable group representative, as shown in Table 1.
Table 1.
CYP2C19 genotype frequencies for Han and Uyghur patients with cardiovascular and cerebrovascular diseases.
| Gene | Genotype | Han (n=220) | Uyghur (n=800) | ||||
|---|---|---|---|---|---|---|---|
| n | Observed frequency | Expected frequency* | n | Observed frequency | Expected frequency* | ||
| CYP2C19 | *1/*1 | 94 | 0.4272 | 0.4165 | 497 | 0.6213# | 0.6192 |
| *1/*2 | 89 | 0.4046 | 0.4225 | 226 | 0.2825# | 0.2892 | |
| *2/*2 | 25 | 0.1137 | 0.1072 | 31 | 0.0388# | 0.0338 | |
| *1/*3 | 7 | 0.0318 | 0.0351 | 39 | 0.0488 | 0.0463 | |
| *2/*3 | 5 | 0.0227 | 0.0178 | 6 | 0.0075 | 0.0108 | |
| *3/*3 | 0 | 0.0000 | 0.0007 | 1 | 0.0013 | 0.0009 | |
Expected probability calculated using the Hardy-Weinberg equilibrium (P>0.05);
compared with the Han population (P<0.05).
CYP2C19 allele frequencies for the Han and Uyghur subjects
The frequencies of the CYP2C19 allele*1 were 0.6454 and 0.7869 in the Han and Uyghur populations, respectively, and were statistically different (P<0.05). The frequencies of the CYP2C19 allele*2 were 0.3273 and 0.1837 in the Han and Uyghur populations, respectively (P<0.05), and the frequencies of the CYP2C19 allele*3 were 0.0273 and 0.0294 in the Han and Uyghur populations, respectively, but the difference was not significant. The results are listed in Table 2.
Table 2.
CYP2C19 allele frequencies for Han and Uyghur patients with cardiovascular and cerebrovascular diseases.
| Gene | Allele | Allele frequency | |
|---|---|---|---|
| Han | Uyghur | ||
| CYP2C19 | *1 | 0.6454 | 0.7869* |
| *2 | 0.3273 | 0.1837* | |
| *3 | 0.0273 | 0.0294 | |
Compared with the Han population (P<0.05).
CYP2C19 metabolic phenotype distribution for the Han and Uyghur subjects
Based on genetic polymorphism, the CYP2C19 metabolic phenotypes were classified into 3 groups. The homozygous extensive metabolizers included the genotype *1/*1. The heterozygous extensive metabolizers included the genotypes *1/*2 and *1/*3. The poor metabolizers included the genotypes *2/*2, *2/*3, and *3/*3. The frequencies of homozygous extensive metabolizers within the Han and Uyghur population were 42.72% and 62.13%, respectively (P<0.01). The heterozygous extensive metabolizers were present within the Han and Uyghur population at frequencies of 43.64% and 33.13%, respectively (P<0.01). Similarly, the frequencies of poor metabolizers within the Han and Uyghur populations were 13.64% and 4.76%, respectively (P<0.01). The results are shown in Table 3.
Table 3.
CYP2C19 genotype mutated site (%).
| Genotype | Gene mutated sites | Han | Uyghur | |
|---|---|---|---|---|
| Homozygous extensive metabolizers | CYP2C19*1/*1 | None | 42.72 | 62.13* |
| Heterozygous extensive metabolizers | CYP2C19*1/*2 | 681G>A | 43.64 | 33.13* |
| CYP2C19*1/*3 | 636G>A | |||
| Poor metabolizers | CYP2C19*2/*2 | 681G>A | 13.64 | 4.76* |
| CYP2C19*3/*3 | 636G>A | |||
| CYP2C19*2/*3 | 681G>A, 636G>A | |||
Compared with the Han population (P<0.01).
Discussion
The CYP2C19 enzyme is mainly found in the liver microsome and is encoded by a gene located on chromosome 10q24. The protein consists of 490 amino acids. The gene has been sequenced and found to contain 9 exons and 8 introns. CYP2C19 genetic polymorphism is a major cause of individual variability in reactions to drugs and shows considerably differential distribution between races or ethnic groups. A recent publication [7] reported that the CYP2C19 genetic polymorphism involved 25 sites in the human genome, among which CYP2C19*1 (wild-type), CYP2C19*2, and CYP2C19*3 alleles are common in the Asian population. Additionally, it has been found that CYP2C19*2 and CYP2C19*3 allele mutations are major causes of the poor metabolizer phenotype among the Chinese population [8]. It has been demonstrated [9,10] that the products of CYP2C19*2 and CYP2C19*3 mutants are both non-catalyzing enzymes, which result in a reduced ability to metabolize drugs and lead to severe adverse effects that undermine clinical treatment. By assessing the CYP2C19 genotype, not only can the efficacy of clopidogrel on patients be evaluated, but the risk of cardiovascular and cerebrovascular disease may be predicted as well. In clinical applications of clopidogrel, the drug dose can be adjusted based on the patients’ genotype or metabolic phenotype. For instance, poor metabolizers should be given higher doses or anti-coagulation substitutes [11,12]. Furthermore, relevant Chinese and international handbooks have suggested that CYP2C19 mutation analysis should be performed on patients before clopidogrel therapy to determine the selection of anti-platelet drugs [13,14].
This study analyzed the CYP2C19 genetic polymorphism among 1020 patients with cardiovascular and cerebrovascular diseases undergoing clopidogrel therapy (220 Han patients and 800 Uyghur patients). In total, 6 different genotypes were observed and compared between the 2 ethnic groups. Our results show that the Han subjects have lower wild-type homozygote frequency, lower allele *1 frequency, and higher allele *2 frequency than the Uyghur subjects. The difference in genotypic distribution between the 2 groups was statistically significant, which is consistent with the previous results reported by Tao et al. [15]. Our results show racial difference in CYP2C19 genetic polymorphism and also indicate a lower frequency of homozygous extensive metabolizers and a higher frequency of poor metabolizer frequency among Han subjects, which is similar to results reported in related studies [6,16]. The frequency of poor metabolizers among Han subjects is within the distribution range of Asians [16], while that of the Uyghur subjects (4.67%) is similar to the distribution range of Caucasians [17]. This may be explained by the genetic influence of Caucasian immigrants along the Silk Route on the present day Uyghur population. Therefore, we propose that CYP2C19-metabolized clopidogrel is more appropriate for Uyghur patients, who show a lower incidence of clopidogrel resistance. In contrast, Han patients are more sensitive to CYP2C19 genetic polymorphisms than Uyghur patients. Because we were limited by the scope of the experiment and research conditions, this study may be biased in terms of subject selection. Further studies should be performed to consolidate information on CYP2C19 genetic polymorphism in healthy populations from these 2 ethic groups.
Conclusions
This study examined the CYP2C19 genetic polymorphism among Han and Uyghur patients with cardiovascular and cerebrovascular diseases and observed differential distributions of genotypes and metabolic phenotypes between the 2 ethnic groups. This finding may provide a theoretical basis and technical support for determining drug compatibility, dose optimization, and in predicting adverse effects with the goal of reducing individual variability, enhancing drug efficacy, and ultimately promoting the development of individualized drug therapy among ethnic minority populations.
Footnotes
Declaration of conflict of interest
None.
Source of support: This work was supported by grant No. 201291168 from the Science and Technology Supporting Project of Xinjiang Uygur Autonomous Region
References
- 1.Wei DQ, Wang JF, Chen C, et al. Molecular modeling of two CYP2C19 SNPs and its implications for personalized drug design. Protein Pept Lett. 2008;15(1):27–32. doi: 10.2174/092986608783330305. [DOI] [PubMed] [Google Scholar]
- 2.Sabatini MS, Cannon CP, Gibson CM, et al. Addition of clopidogrel to aspirin and fibrinolytic therapy for myocardial infarction with ST-segment elevation. N Engl J Med. 2005;352(12):1179–89. doi: 10.1056/NEJMoa050522. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Yang Z. Research status of clopidogrel resistance on CYP2C19 gene polymorphism. Journal of Clinical Cardiology. 2012;28(3):163–65. [Google Scholar]
- 4.Li-ping Y, Jing X, Yao L, et al. Correlation between the Genetic Polymorphism of CYP2C19*2, *3 and the Clinical Efficacy of Clopidogrel: A Systematic Review. Chinese Journal of Evidence-Based Medicine. 2012;12(9):1063–70. [Google Scholar]
- 5.Zhou J, Hong LV, Kang X-x. Genetic polymorphism analysis of cytochrome P450 2C19 among gender, age and body mass index in Chinese Han population. Chinese Journal of Clinical Pharmacology and Therapeutics. 2007;12(2):208–13. [Google Scholar]
- 6.Niu C-yN, Guan L, Luo J-y. Genetic polymorphism analysis of cytochrome CYP2C19 in Uigur population. Journal of Xinjiang Medical University. 2005;28(3):247–49. [Google Scholar]
- 7.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–23. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He N, Yan FX, Huang SL, et al. CYP2C19 genotype and S-mephenytoin 4′-hydroxylation phenotype in a Chinese Dai population. Eur J Clin Pharmacol. 2002;58(1):15–18. doi: 10.1007/s00228-002-0425-x. [DOI] [PubMed] [Google Scholar]
- 9.Shirai N, Furuta T, Moriyama Y, et al. Effects of CYP2C19 genotypic differences in the metabolism of omeprazole and rabeprazole on intragastric pH. Aliment Pharmacol Ther. 2001;15(12):1929–37. doi: 10.1046/j.1365-2036.2001.01108.x. [DOI] [PubMed] [Google Scholar]
- 10.Shimatani T, Inoue M, Kuroiwa T, et al. Effect of omeprazole 10 mg on intragastric pH in three different CYP2C19 genotypes, compared with omeprazole 20 mg and lafutidine 20 mg, a new H2-receptor antagonist. Aliment Pharmacol Ther. 2003;18(11–12):1149–57. doi: 10.1046/j.1365-2036.2003.01804.x. [DOI] [PubMed] [Google Scholar]
- 11.Jinnai T, Horiuchi H, Makiyama T, et al. Impact of CYP2C19 polymorphisms on the antiplatelet effect of clopidogrel in an actual clinical setting in Japan. Circ J. 2009;73(8):1498–503. doi: 10.1253/circj.cj-09-0019. [DOI] [PubMed] [Google Scholar]
- 12.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6(8):1439–41. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]
- 13.Wenger NK. 2011 ACCF/AHA focused update of the guidelines for the management of patients with Unstable Angina/Non-ST-Elevation Myocardial Infarction (updating the 2007 Guideline): highlights for the clinician. Clin Cardiol. 2012;35(1):3–8. doi: 10.1002/clc.20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardiology branch of Chinese Medical Association. Syndrome diagnosis and treatment guidelines for non ST segment elevation acute coronary artery. Chinese Journal of Cardiology. 2012;40(5):353–67. [Google Scholar]
- 15.Guo T, Zuo J, Xia D, et al. Geneic polymorphism analysis of cytocgrome CYP3A4, CYP2C9, CYP2C19, CYP2D6 in Chinese Han and Mongolian population. Chin Pharm J. 2012;47(24):2017–22. [Google Scholar]
- 16.Yan C, Zhan J, Chen S. Study on cytochrome p-450 2C19 genetic polymorphism in Zhejiang Han and She nationalities. Chinese Pharmaceutical Journal. 2005;39(11):866–68. [Google Scholar]
- 17.Hoskins JM, Shenfield GM, Gross AS. Relationship between proguanil metabolic ratio and CYP2C19 genotype in a Caucasian population. Br J Clin Pharmacol. 1998;46(5):499–504. doi: 10.1046/j.1365-2125.1998.00807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


