Abstract
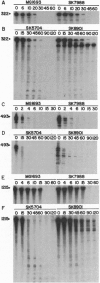
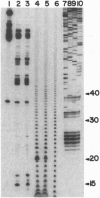
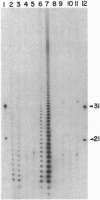
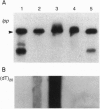
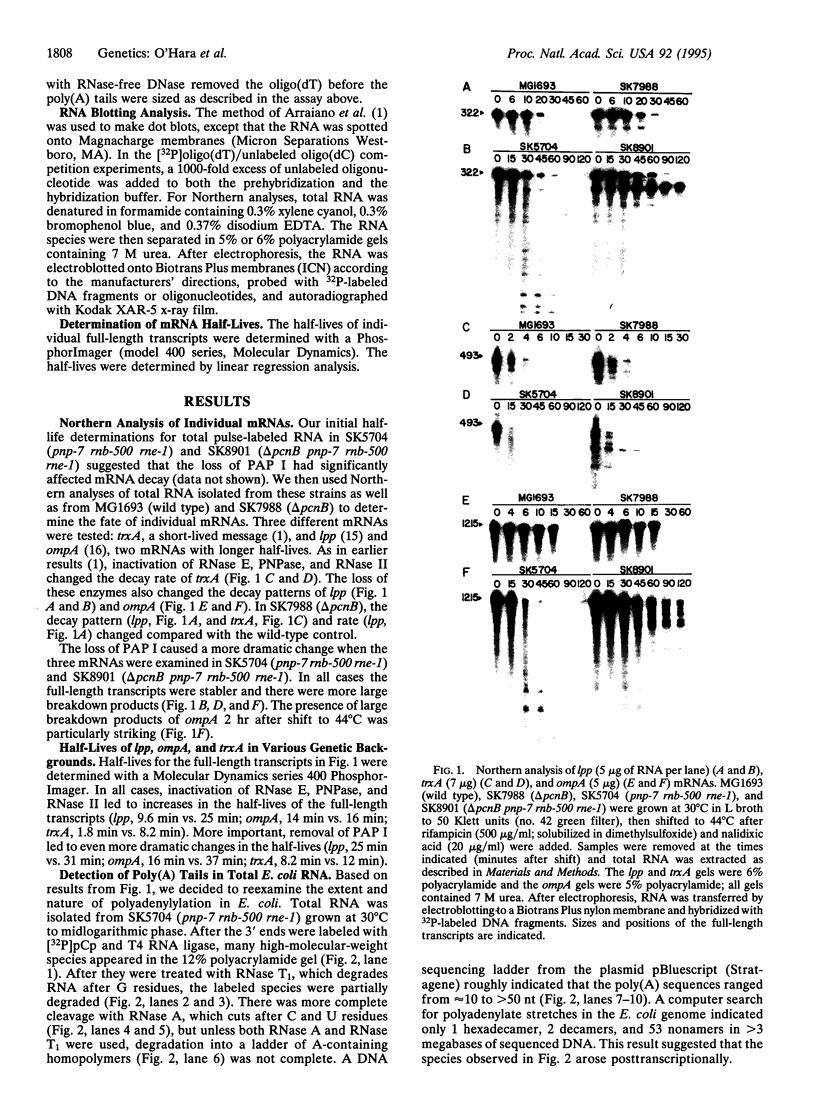
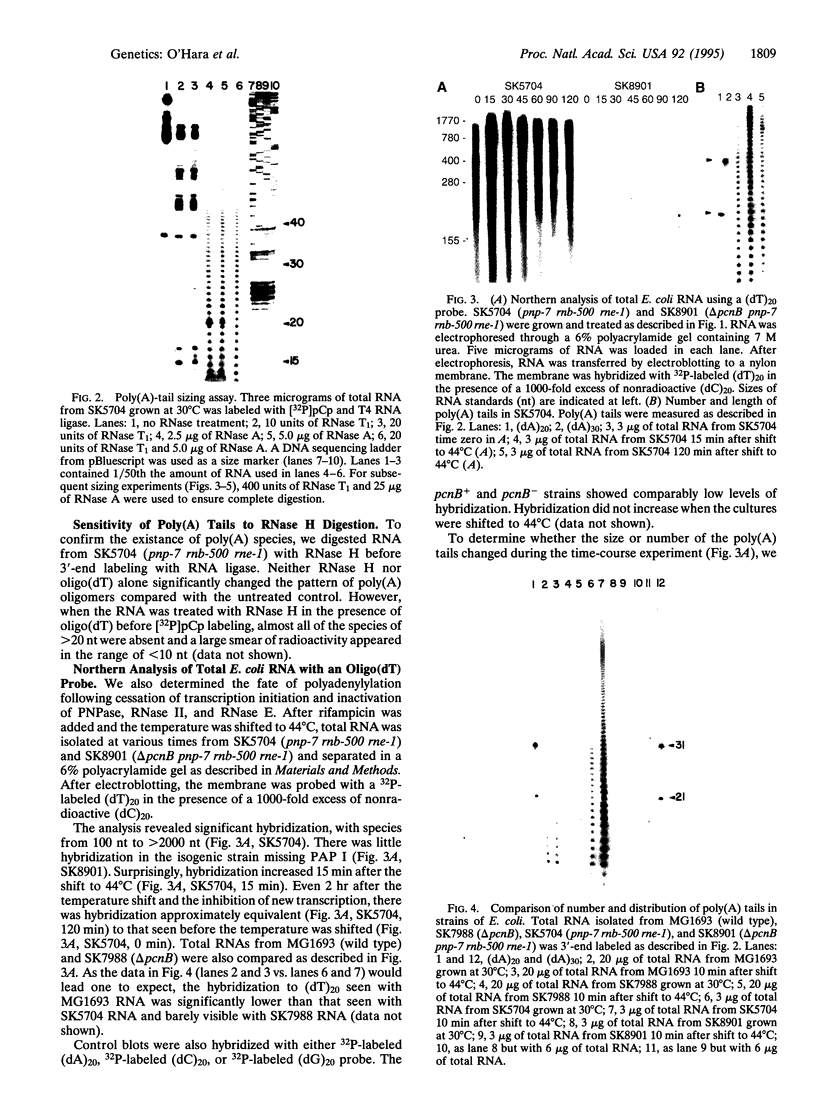
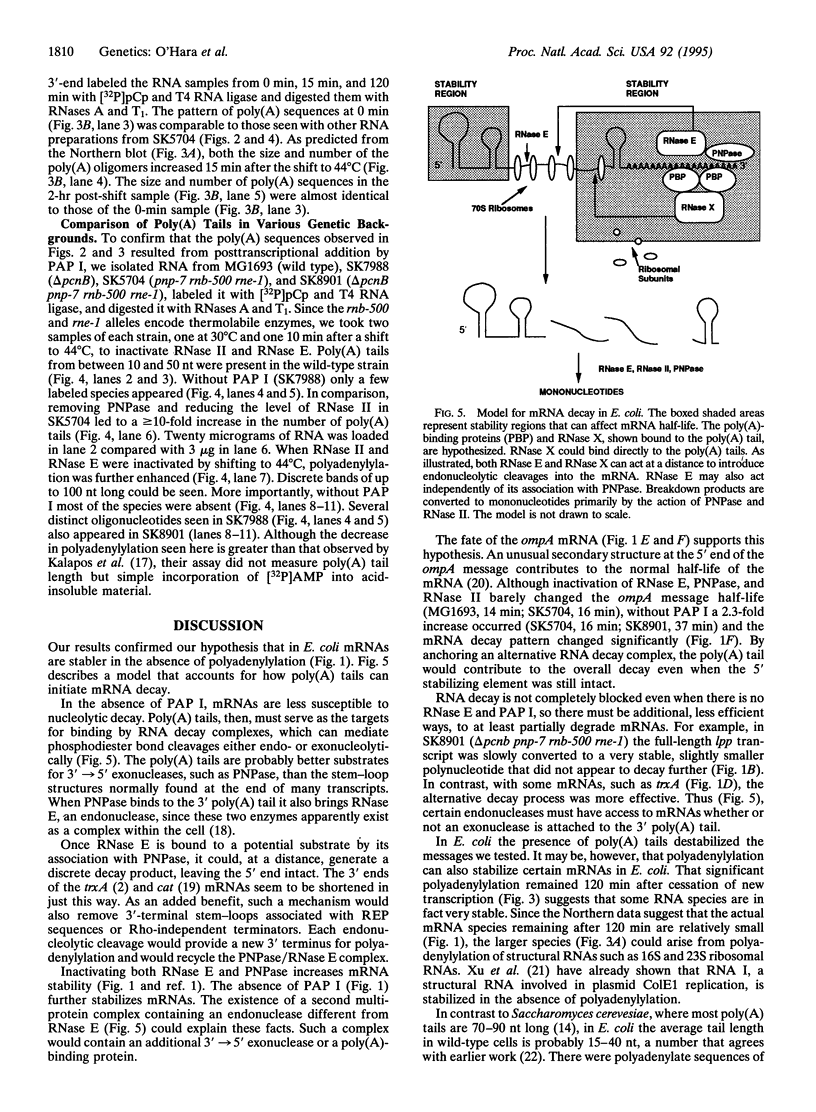
As part of our genetic analysis of mRNA decay in Escherichia coli K-12, we examined the effect of the pcnB gene [encoding poly(A) polymerase I] on message stability. Eliminating poly(A) polymerase I (delta pcnB) dramatically stabilized the lpp, ompA, and trxA transcripts. The half-lives of individual mRNAs were increased in both a delta pcnB single mutant and a delta pcnB pnp-7 rnb-500 rne-1 multiple mutant. We also found mRNA decay intermediates in delta pcnB mutants that were not detected in control strains. By end-labeling total E. coli RNA with [32P]pCp and T4 RNA ligase and then digesting the RNA with RNase A and T1, we showed that many RNAs in a wild-type strain contained poly(A) tails ranging from 10 nt to > 50 nt long. When polynucleotide phosphorylase, RNase II, and RNase E were absent, the length (> 100 nt) and number (10- to 20-fold) of the poly(A) tails increased. After transcription initiation was stopped with rifampicin, polyadenylylation apparently continued. Deleting the structural gene for poly(A) polymerase I (pcnB) reduced the amount of 3'-terminal poly(A) sequences by > 90%. We propose a model for the role of polyadenylylation in mRNA decay.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AUGUST J. T., ORTIZ P. J., HURWITZ J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962 Dec;237:3786–3793. [PubMed] [Google Scholar]
- Ahlquist P., Kaesberg P. Determination of the length distribution of poly(A) at the 3' terminus of the virion RNAs of EMC virus, poliovirus, rhinovirus, RAV-61 and CPMV and of mouse globin mRNA. Nucleic Acids Res. 1979 Nov 10;7(5):1195–1204. doi: 10.1093/nar/7.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C., Yancey S. D., Kushner S. R. Identification of endonucleolytic cleavage sites involved in decay of Escherichia coli trxA mRNA. J Bacteriol. 1993 Feb;175(4):1043–1052. doi: 10.1128/jb.175.4.1043-1052.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aström J., Aström A., Virtanen A. Properties of a HeLa cell 3' exonuclease specific for degrading poly(A) tails of mammalian mRNA. J Biol Chem. 1992 Sep 5;267(25):18154–18159. [PubMed] [Google Scholar]
- Babitzke P., Granger L., Olszewski J., Kushner S. R. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993 Jan;175(1):229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. J., Sarkar N. Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7546–7550. doi: 10.1073/pnas.89.16.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994 Mar 11;76(5):889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Decker C. J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993 Aug;7(8):1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna Y., Langley D., Sarkar N. Detection of high levels of polyadenylate-containing RNA in bacteria by the use of a single-step RNA isolation procedure. Nucleic Acids Res. 1981 Jul 24;9(14):3545–3554. doi: 10.1093/nar/9.14.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishna Y., Sarkar N. The synthesis of DNA complementary to polyadenylate-containing RNA from Bacillus subtilis. J Biol Chem. 1982 Mar 25;257(6):2747–2750. [PubMed] [Google Scholar]
- Hirashima A., Childs G., Inouye M. Differential inhibitory effects of antibiotics on the biosynthesis of envelope proteins of Escherichia coli. J Mol Biol. 1973 Sep 15;79(2):373–389. doi: 10.1016/0022-2836(73)90012-0. [DOI] [PubMed] [Google Scholar]
- Kalapos M. P., Cao G. J., Kushner S. R., Sarkar N. Identification of a second poly(A) polymerase in Escherichia coli. Biochem Biophys Res Commun. 1994 Jan 28;198(2):459–465. doi: 10.1006/bbrc.1994.1067. [DOI] [PubMed] [Google Scholar]
- Karnik P., Gopalakrishna Y., Sarkar N. Construction of a cDNA library from polyadenylated RNA of Bacillus subtilis and the determination of some 3'-terminal sequences. Gene. 1986;49(1):161–165. doi: 10.1016/0378-1119(86)90397-5. [DOI] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991 Aug;173(16):4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell J. E., Rudner D. Z., Sachs A. B. 3'-UTR-dependent deadenylation by the yeast poly(A) nuclease. Genes Dev. 1992 Nov;6(11):2088–2099. doi: 10.1101/gad.6.11.2088. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Schottel J. L. Characterization of cat messenger RNA decay suggests that turnover occurs by endonucleolytic cleavage in a 3' to 5' direction. Mol Microbiol. 1992 May;6(9):1095–1104. doi: 10.1111/j.1365-2958.1992.tb01547.x. [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev. 1994 Apr 1;8(7):855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Nakazato H., Venkatesan S., Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975 Jul 10;256(5513):144–146. doi: 10.1038/256144a0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989 Sep 8;58(5):857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Deardorff J. A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992 Sep 18;70(6):961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sarkar N., Langley D., Paulus H. Isolation and characterization of polyadenylate-containing RNA from Bacillus brevis. Biochemistry. 1978 Aug 22;17(17):3468–3474. doi: 10.1021/bi00610a007. [DOI] [PubMed] [Google Scholar]
- Shyu A. B., Belasco J. G., Greenberg M. E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991 Feb;5(2):221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- Srinivasan P. R., Ramanarayanan M., Rabbani E. Presence of polyriboadenylate sequences in pulse-labeled RNA of Escherichia coli. Proc Natl Acad Sci U S A. 1975 Aug;72(8):2910–2914. doi: 10.1073/pnas.72.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs J. D., Hall B. D. Level of tryptophan messenger RNA in Escherichia coli. J Mol Biol. 1968 Oct 28;37(2):289–302. doi: 10.1016/0022-2836(68)90268-4. [DOI] [PubMed] [Google Scholar]
- Taljanidisz J., Karnik P., Sarkar N. Messenger ribonucleic acid for the lipoprotein of the Escherichia coli outer membrane is polyadenylated. J Mol Biol. 1987 Feb 5;193(3):507–515. doi: 10.1016/0022-2836(87)90263-4. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Mount D. W. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J Bacteriol. 1969 Nov;100(2):923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Lin-Chao S., Cohen S. N. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6756–6760. doi: 10.1073/pnas.90.14.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]