Abstract
Vegetation history in tropical Africa is still to date hardly known and the drivers of population differentiation and speciation processes are little documented. It has often been postulated that population fragmentations following climate changes have played a key role in shaping the geographic distribution patterns of genetic diversity and in driving speciation. Here we analyzed phylogeographic patterns (chloroplast-DNA sequences) within and between eight (sister) species of widespread rainforest herbs and lianas from four genera of Marantaceae (Halopegia, Haumania, Marantochloa, Megaphrynium), searching for concordant patterns across species and concordance with the Pleistocene refuge hypothesis. Using 1146 plastid DNA sequences sampled across African tropical lowland rainforest, particularly in the Lower Guinean (LG) phytogeographic region, we analyzed intra- and interspecific patterns of genetic diversity, endemism and distinctiveness. Intraspecific patterns of haplotype diversity were concordant among most species as well as with the species-level diversity pattern of Marantaceae. Highest values were found in the hilly areas of Cameroon and Gabon. However, the spatial distribution of endemic haplotypes, an indicator for refuge areas in general, was not congruent across species. Each proposed refuge exhibited high values of endemism for one or a few species indicating their potential role as area of retraction for the respective species only. Thus, evolutionary histories seem to be diverse across species. In fact, areas of high diversity might have been both refuge and/or crossing zone of recolonization routes i.e., secondary contact zone. We hypothesize that retraction of species into one or the other refuge happened by chance depending on the species' distribution range at the time of climate deterioration. The idiosyncratic patterns found in Marantaceae species are similar to those found among tropical tree species, especially in southern LG.
Keywords: endemism, distinctiveness, genetic diversity, trnC-petN1r, refugia, Lower Guinea
Introduction
Vegetation history in tropical Africa is still to date hardly known and the drivers of speciation and population differentiation processes are little documented. Hypotheses on the diversification of the Afrotropical flora include allopatric differentiation/speciation driven by population fragmentation following Pleistocene climate changes (Robbrecht, 1994; Sosef, 1994; Maley, 1996) and parapatric differentiation/speciation across ecological gradients (e.g., temperature and precipitation gradients; Fjeldsa and Lovett, 1997; Vande Weghe, 2004; Heuertz et al., 2013). Phylogeographic studies within and between closely related species might shed new light on this matter as indicated by similar studies in temperate regions (Taberlet et al., 1998; Schönswetter et al., 2005).
Palynological studies (Maley and Brenac, 1998; Dupont et al., 2000; Bonnefille, 2007; Ngomanda et al., 2009; Dupont, 2011) and palaeo-environmental reconstructions (Anhuf et al., 2006) suggest a repeated fragmentation of the tropical forest in Africa due to (glacial-interglacial) climate oscillations for the last million years. For example, during the African Humid Holocene period (c. 6000–9000 years BP) a single forest block extended from West to Central Africa beyond the current forest cover limit, while the forest was presumably highly fragmented and reduced in size during the last glacial maximum (c. 19000–26000 years BP). This might have led to population fragmentation followed by the independent evolution of the isolated populations through mutation and drift and ultimately the establishment of species. Alternatively, and/or simultaneously, isolated populations might have adapted to different climatic conditions ultimately forming ecologically different species. Indeed, within Lower Guinea (LG, i.e., the western part of the Central African rainforest block, identified as a phytochorion by White, 1979, Figure 1), climatic heterogeneity is characterized by a marked W-E precipitation gradient from the Coast to the inland and a North-South seasonal inversion at a latitude c. 2°N (Leroux, 1983; Vande Weghe, 2004).
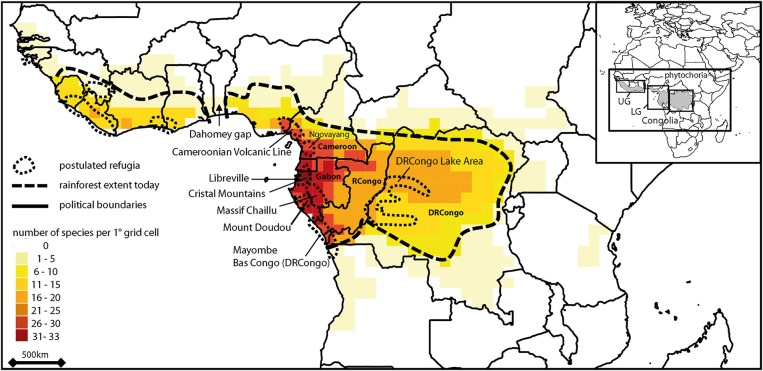
Figure 1.
Pattern of species diversity of Marantaceae in tropical Africa based on Schnell (1957), Dhetchuvi (1996), Jongkind (2008) and Ley and Claßen-Bockhoff (2012). Postulated refugia after (Maley, 1996); DRCongo, Democratic Republic of Congo; RCongo, Republic of the Congo. Inset shows phytochoria after (White, 1979): UG, Upper Guinea; LG, Lower Guinea; C, Congolia.
Recently, comparative phylogeographic studies of central African trees revealed a partial congruence of phylogeographic patterns with postulated refugia (Hardy et al., 2013; Heuertz et al., 2013; Dauby et al., 2014a). However, despite the occurrence of some common phylogeographic features, each species displayed an original pattern, especially in Gabon, suggesting idiosyncratic evolutionary histories. It has been hypothesized that this is due to less severe changes in forest cover reduction in this area during climate oscillations (Dupont et al., 2000; see also Holstein and Renner, 2011).
Here we investigate whether similar genetic patterns as so far detected in tropical African trees might also be found in perennial herbs and lianas from the forest understorey. We might expect that, compared to trees, phylogeographic patterns of herbs and lianas mirror younger historical events due to presumably shorter life cycles (Putz, 1990; Gerwing, 2004; Brandes et al., 2011). Furthermore, general patterns might be more structured in herbs than in trees (see e.g., GST in nuclear markers in Nybom, 2004) due to a more patchy community structure and potentially smaller dispersal distances of pollinators and dispersers in the tropical understorey (for trees: <14 km, Ward et al., 2005; 100 m–100 km, Carbone et al., 1999; for understorey shrub: 10–20 m, Zeng et al., 2012).
More specifically, we perform in Lower Guinea a comparative phylogeographic study of eight perennial herbs and lianas of the family Marantaceae. We search for (1) congruent patterns across species that might have been driven by a common vegetation history, and (2) congruence of these patterns with postulated rainforest refugia that might support the importance of these areas for species survival and population differentiation.
Materials and methods
Species studied and sampling
The Marantaceae (30 genera) are a pantropical family of perennial herbs and lianas of the understorey and gaps of lowland rainforest (0–1500 m) with highest species diversity found in America (~450 spp.) followed by Asia (~50 spp.) and Africa (~40 spp.) (Dhetchuvi, 1996; Andersson, 1998; Kennedy, 2000; Suksathan et al., 2009; Ley and Claßen-Bockhoff, 2011). Each genus of the family is endemic to one continent except Halopegia and Thalia (Andersson, 1998). Phylogenetic investigations suggest a split of this family from its sister family Cannaceae some 95 ± 5 Ma ago. The family then started to diversify ca. 63 ± 5 Ma ago (Kress and Specht, 2005) in the late cretaceous with the establishment of the first tropical everwet habitats in the current tropics (Willis and McElwain, 2002). The Marantaceae are thus probably not a Gondwanan group, i.e., Marantaceae are not distributed pantropically due to vicariant events ca. 110 Ma ago (Kearey and Vine, 1996). Biogeographic analyses suggest instead the occurrence of several independent dispersal events between continents followed each time by intra-continental speciation resulting in several independent species clades per continent (Prince and Kress, 2006). In continental Africa the current distribution of the Marantaceae family ranges from Senegal in the West to Tanzania in the East following today's limits of the tropical rainforest. Highest species numbers are found in Gabon and Cameroon (Figure 1). Distribution ranges of individual species vary from widespread (equaling the distribution of the whole Marantaceae family in Africa) to restricted, either to the West or East of the Dahomey gap and/or to Cameroon and/or Gabon (Dhetchuvi, 1996).
The Marantaceae species differ from their sister family Cannaceae by a pulvinus and an explosive pollination mechanism (Claßen-Bockhoff, 1991; Kennedy, 2000). It is a highly diverse family with regard to species number and adaptations to different pollinators and dispersal agents (Kennedy, 2000; Clausager and Borchsenius, 2003; Locatelli et al., 2004; Ley, 2008; Ley and Claßen-Bockhoff, 2009). The species of Marantaceae show typical characteristics of plants from the tropical understory such as self-compatibility (Halopegia azurea even autogamous, Ley and Claßen-Bockhoff, 2013), clonality via rhizomes (Marantochloa congensis additionally via vivipary (bulbils), Kennedy, 2000) and animal pollination and dispersal (Ley, 2008; Ley and Claßen-Bockhoff, 2009).
For the current study eight species from four different genera with different growth forms, distribution ranges and pollinators were chosen (Table 1). Sampling of leaf material for genetic analyses was envisioned to cover the whole distribution area of each species. However, for all species sampling was better in Cameroon and Gabon and only fragmentary in West Africa (i.e., Upper Guinean phytochorion) and the Congo Basin (i.e., Congolian phytochorion). We thus here used the entire dataset including West Africa and the Congo Basin for the description of the phylogeographic pattern of each species and then limited the dataset to Lower Guinea, when comparing the phylogeographic pattern qualitatively and quantitatively among species.
Table 1.
Ecological information on the eight study species from the family Marantaceae.
| Genera | Species | Abbr. | N of species studied (total per genus°+#) | Distribution range+ | Growth form+ | Pollinator* | Dispersal agent1° |
|---|---|---|---|---|---|---|---|
| Halopegia | azurea | HaloAzu | 1(1) | GC | herb | Bee (Halictidae) | no data |
| Haumania | danckelmaniana liebrechtsiana | HauDanck HauLieb | 2(3) | C, LG | liana | Bee (Xylocopa) | gravity |
| Marantochloa | congensis incertifolia monophylla | MarCong MarIncert MarMono | 3(16) | GC, LG, C, LG | herb | Bee (Amegilla) | water, monkeys |
| Megaphrynium | macrostachyum trichogynum | MegaMacro MegaTrich | 2(4) | GC, LG | herb | Bee + Bird | monkeys |
DNA extraction and amplification
For the eight species we used sequences from the chloroplast (cp) inter-genic spacer trnC-petN1r using the primers trnC 5′-CCAGTTCAAATCTGGGTGTC-3′ (modified from Demesure et al., 1995) and petN1r 5′-CCCAAGCAAGACTTACTATATCC-3′ (Lee and Wen, 2004). For Marantochloa congensis an additional marker (psbA-trnH) was amplified to increase resolution, using the primers psbA 5′-GTTATGCATGAACGTAATGCTC-3′ and trnH2 5′-CGCGCATGGTGGATTCACAATCC-3′ (Sang et al., 1997; Tate and Simpson, 2003). The genetic data for the genera Haumania and Marantochloa was updated from Ley and Hardy (2010, 2014). For the third species of the genus Haumania (H. leonardiana) only sequences from six individuals from the Democratic Republic of Congo (DRCongo) could so far be obtained and were added to the haplotype network to show intragenus relationships but were not analyzed any further due to the scarcity of available sequences. The phylogeographic patterns of three species from the genera Halopegia and Megaphrynium were characterized here for the first time. The production of sequences for these species followed the protocol of DNA extraction, amplification and sequencing described in Ley and Hardy (2010).
Geographic distribution of chloroplast haplotypes and phylogenetic networks
For each species chloroplast haplotypes were analyzed in DnaSP Version 5.10 (Librado and Rozas, 2009) and their geographic distribution mapped. DNA haplotypes were submitted to Genbank (for accession numbers see Supplementary Table 1). To obtain the minimum number of mutations between haplotypes, a network was established with the software Network 4.5.1.0 (www.fluxus-engineering.com; Bandelt et al., 1999) using a maximum parsimony method based on a median joining algorithm (MJ). Networks were established per species and for entire genera to identify possible plastid captures between closely related species (Ley and Hardy, 2010, 2014). Nucleotide diversity, which represents the average number of nucleotide differences per site between two sequences, was calculated in Arlequin (Excoffier et al., 2009).
Grid-based standardized measures of genetic diversity, endemism and distinctiveness
For the comparison of geographic patterns of genetic diversity between species at different scales in Lower Guinea we subdivided the region into three different grid systems with cell sizes of 0.75°-, 1.5°- and 3°-sides (Supplementary Figure 1 for 0.75° and 1.5°; 3° not shown). Given that a minimum of three samples was necessary per species and grid cell to compute diversity indices (see below), smaller cells allowed higher spatial resolution but at the cost of lower precision and higher loss of data in areas were sampling was less dense (for numbers of individuals per grid cell 0.75° and 1.5° see Supplementary Tables 2, 3).
Within cell diversity, endemism and distinctiveness
We computed several statistics quantifying genetic diversity for each species within each cell: Nielsen's estimator of the effective number of haplotypes NAe (Nielsen et al., 2003), the gene diversity corrected for sample size He (Nei, 1978) and the mean phylogenetic distance between individuals v (gene diversity with ordered alleles, Pons and Petit, 1996). The different statistics were computed with SPAGeDi Version 1.4 (Hardy and Vekemans, 2002). The degree of endemism of each haplotype was assessed by the maximal distance between individuals carrying that haplotype. We quantified the degree of haplotypic endemism, End, per cell and species as the proportion of individuals carrying haplotypes with a maximal geographic extension of 200 km. Finally, for each species, the level of phylogenetic distinctiveness of each cell with respect to the other ones was computed following Dauby et al. (2014a). To this end, for each pair of cells (i and j), the mean phylogenetic distance between individuals drawn from i and j (vij) was computed, as well as the spatial distance between the centroids of individuals belonging to i and j (dij). S'ij, the residuals of the regression of vij on ln(dij), or the centered vij values themselves if there was no significant positive correlation between vij and ln(dij) according to a Mantel test, were then averaged over all pairs involving one particular cell, S'i, providing a measure of the phylogenetic distinctiveness of that cell above or below the average across all cells (Petit et al., 2003; Dauby et al., 2014a).
Differentiation statistics
Global differentiation statistics GST and NST among cells (for cells with at least three individuals) were computed for each species. GST accounts for differences in haplotype frequencies while NST additionally accounts for the phylogenetic distances between haplotypes. To test if there was a phylogeographic signal, characterized by NST > GST, permutation tests were performed in SPAGeDi Version 1.4 (Hardy and Vekemans, 2002).
Congruence of phylogeographic patterns among species
Congruence of phylogeographic patterns for each pair of species was evaluated (i) by comparing within cell diversity and endemism metrics using Pearson correlation tests, and (ii) by comparing matrices of pairwise standardized distinctiveness among grid cells (S'ij) using Mantel tests (see Dauby et al., 2014a). To obtain a multispecies test of overall geographic congruence of local diversity, endemism or distinctiveness, these metrics were first centered (i.e., minus their mean value) and reduced (i.e., divided by their standard deviation) within species, and then differences among grid cells were tested using a One-Way ANOVA where grid cells were used as factor (due to missing data, species could not be added as another factor). To represent diversity patterns on a map, centered and reduced diversity and endemism metrics were shown per cell and species, or were averaged over species to represent multi-species trends.
Results
Genetic polymorphism
The numbers of individuals sequenced per species ranged from 75 to 166, totaling 1046 individuals (991 in Lower Guinea, Table 2) sequenced for trnC-petN1r and 110 for psbA-trnH (Lower Guinea only, Table 2).
Table 2.
Sample sizes and genetic diversity at the trnC-petN1r and psbA-trnH2 region in the eight study species from the Marantaceae.
| Species | N (whole tropical Africa/Lower Guinea) | SNP (incl.indels) | Nucleotide diversity × 103 | N of haplotypes | |
|---|---|---|---|---|---|
| whole tropical Africa (total/private) | Lower Guinea (total/private) | ||||
| trnC-petN1r | |||||
| HaloAzu | 121/115 | 11 | 0.846 ± 0.690 | 11 | 8 |
| HauDanck | 121/121 | 16 | 1.575 ± 1.060 | 14/10 | 14/10 |
| HauLieb | 110/110 | 20 | 3.268 ± 1.894 | 10/6 | 10/6 |
| MarCong | 194/169 | 61 | 3.619 ± 2.149 | 15/11 | 15/9 |
| MarIncert | 81/81 | 22 | 6.901 ± 3.755 | 13/6 | 13/6 |
| MarMono | 118/109 | 60 | 7.289 ± 3.925 | 19/16 | 19/15 |
| MegaMacro | 162/162 | 22 | 4.897 ± 2.852 | 16/15 | 16/15 |
| MegaTrich | 131/131 | 13 | 1.022 ± 0.900 | 7/6 | 7/6 |
| Total | 1038/998 | ||||
| psbA-trnH2 | 110/110 | 9 | 5.754 ± 3.152 | 7 | 7 |
| MarCong | |||||
For abbreviations of species names see Table 1. N, number of individuals; SNP, number of single nucleotide polymorphisms. Haplotypes (total/private): “total” includes haplotypes shared between sister species from the same genus. “Private” considers only haplotypes found in the respective species.
The trnC-petN1r region had an average length of about 800 bp (including indels). The number of SNPs (counting indels as single mutations) per species varied between 11 (Halopegia azurea) and 27 (Marantochloa monophylla) and the number of haplotypes per species varied between seven (Megaphrynium trichogynum) and 19 (Marantochloa monophylla, see Table 2). Overall nucleotide diversity ranged from 0.000846 ± 0.000690 in Halopegia azurea to 0.007289 ± 0.003925 in Marantochloa monophylla (Table 2). Average genetic diversity per grid cell measured as He was highest in Haumania danckelmaniana (Table 3). Genetic diversity measured as v taking genetic distance between haplotypes into account was highest in Marantochloa monophylla and endemism per grid cell (End) was highest in M. incertifolia. Measures of genetic diversity were independent of grid cell size (Supplementary Table 4).
Table 3.
Within cell diversity pattern at the trnC-petN1r region in eight Marantaceae species in Lower Guinea for grid cell size 0.75° (average across grid cells, [range]).
| Species | Total sample size | N cells with >2 individuals | Mean [range] per cell of | ||||
|---|---|---|---|---|---|---|---|
| N individuals | NAe | He | v | End | |||
| HalAzu | 91 | 18 | 5.06 [3,11] | 1.41 [1,3.13] | 0.19 [0,0.71] | 0.26 [0,0.86] | 0.09 [0,1] |
| HauDanck | 115 | 19 | 6.05 [3,15] | 1.92 [1,10] | 0.37 [0,1] | 0.63 [0,2.2] | 0.16 [0,0.8] |
| HauLieb | 98 | 15 | 6.53 [3,16] | 1.36 [1,3.04] | 0.21 [0,0.67] | 0.81 [0,5] | 0.11 [0,0.54] |
| MarCong | 141 | 21 | 6.71 [3,15] | 1.61 [1,5.05] | 0.30 [0,1] | 0.84 [0,7.33] | 0.10 [0,1] |
| MarIncert | 73 | 9 | 8.11 [3,20] | 1.61 [1,3.57] | 0.24 [0,0.8] | 1.04 [0,3.69] | 0.55 [0,1] |
| MarMono | 95 | 18 | 5.28 [3,12] | 1.76 [1,4.61] | 0.27 [0,1] | 1.29 [0,8] | 0.12 [0,0.78] |
| MegaMacro | 140 | 22 | 6.36 [3,22] | 1.72 [1,4.61] | 0.34 [0,1] | 0.99 [0,4] | 0.17 [0,1] |
| MegaTrich | 113 | 21 | 5.38 [3,11] | 1.50 [1,2.27] | 0.29 [0,0.67] | 0.38 [0,2.16] | 0.02 [0,0.2] |
N, number; NAe, Effective N of alleles (Nielsen et al., 2003); He, gene diversity corrected for sample size (Nei, 1978); v, mean phylogenetic distance between individuals (Pons and Petit, 1996); End, mean proportion of individuals carrying endemic alleles (haplotype range < 200 km). For abbreviations of species names see Table 1.
For M. congensis psbA-trnH sequences reached a length of about 900 bp (including indels and reverse mutations). Reverse mutations were excluded in the following analyses leaving nine mutations, seven haplotypes (Table 2) and a network without loops with a maximum of two mutations between adjacent haplotypes (Supplementary Figure 2).
Haplotype networks based on trnC-petN1r required 11–27(32) mutations (without torso) within species and 11–38(56) mutations within genera. Haplotypes did never show a high divergence neither within nor between congeneric species (Figures 2, 3). Within species haplotypes differed from the closest other haplotype generally by one mutation. Only few exceptions presented a distance of up to three mutations between closest haplotypes within species. All species networks included loops except for Halopegia azurea, Haumania liebrechtsiana and Marantochloa incertifolia. Marantochloa monophylla was the only species that showed two intraspecific divergent lineages. Between species, maximum distances between nearest haplotypes ranged between two to three mutations. In all networks we found a few individuals that belonged to one morphological species but exhibited the same haplotypes as individuals from the other morphological species.
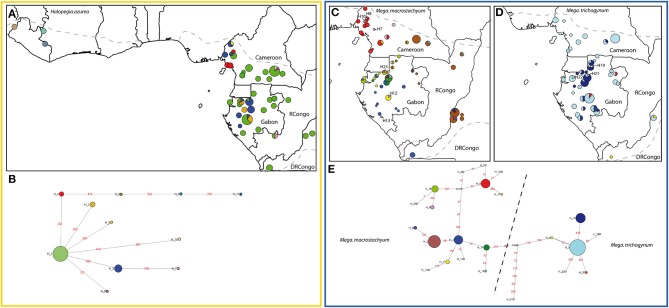
Figure 2.
Geographic distribution of chloroplast haplotypes and generic haplotype networks based on trnC-petN1r for the following Marantaceae species. Halopegia azurea (A,B), Megaphrynium macrostachyum (C,E), Mega. trichogynum, (D,E). Gray hatched line: species distribution range. Sizes of circles are proportional to sample sizes at each locality in the geographic maps and proportional to haplotype frequency in the haplotype network. Frequent haplotypes are color-coded, rare haplotypes are number-coded. Stippled lines throughout the networks delineate groups of haplotypes according to the species in which they are usually found, but haplotypes can also be shared among species. Red numbers along branches are IDs of mutations, mv1 to mv2 indicate median vectors (Bandelt et al., 1999).
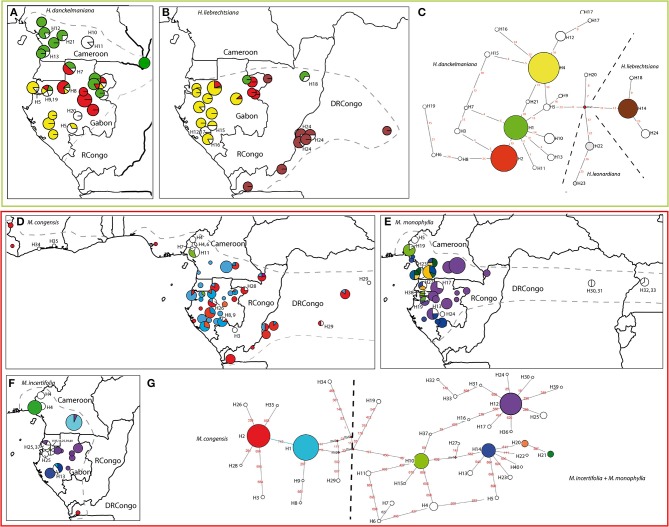
Figure 3.
Geographic distribution of chloroplast haplotypes and generic haplotype networks based on trnC-petN1r updated from Ley and Hardy (2010, 2014). Haumania danckelmaniana (A, C), H. liebrechtsiana (B, C), Marantochloa congensis (D, G), M. monophylla (E, G), M. incertifolia (F, G). Gray hatched line: species distribution range. Sizes of circles are proportional to sample sizes at each locality in the geographic maps and proportional to haplotype frequency in the haplotype network. Frequent haplotypes are color-coded, rare haplotypes are number-coded. Stippled lines throughout the networks delineate groups of haplotypes according to the species in which they are usually found, but haplotypes can also be shared among species. Red numbers along branches are IDs of mutations, mv indicate median vectors (Bandelt et al., 1999).
Phylogeographic patterns within each genus and species
Halopegia azurea was the species with the lowest haplotype diversity (11 haplotypes) resulting in a simple network without loops (Figure 2B). The only frequent haplotype was distributed over the whole Lower Guinean-Congolian range of the species (Figure 2A). Localities with additional one to several rare geographically restricted haplotypes divergent by one mutation from the single widespread haplotype were found around the Cameroonian Volcanic Line and the Chaillu Massif in Gabon. In West Africa three divergent haplotypes were found. They were most closely related (different by three mutations) to the rare haplotype of the Cameroonian Volcanic Line.
The two species from the genus Megaphrynium presented very different phylogeographic patterns. In Mega. trichogynum (Figure 2D) there was one widespread haplotype covering the whole distribution area of the species and another frequent haplotype restricted to Gabon. The diversity center in this species was found in the North of Gabon where the frequent haplotypes overlapped in their distribution and three rare haplotypes also occurred. Mega. macrostachyum presented four haplotypes (H1, 2, 9, 16) exclusive to different, large geographic areas (Southwest Cameroon, Southwest Gabon to DRCongo (Bas Congo), North to Northwest Gabon, East Gabon/East Cameroon/Congos, Figure 2C). Each widespread haplotype was co-occurring with closely related and geographically restricted haplotypes. This resulted in five areas of increased haplotype diversity: the Cameroonian volcanic line, western DRCongo, northern Gabon, coastal northwestern Gabon (near Libreville) and the Cristal Mountains area in Gabon. Only five Megaphrynium individuals out of 282 carried a haplotype typical of the other species.
The spatial genetic structure of species from the genera Haumania and Marantochloa were already discussed in previous publications (Ley and Hardy, 2010, 2014) but updated here (Figure 3). Haumania danckelmaniana (Figure 3A) exhibited three haplotypes each covering a different large geographic area (Cameroon + northern Gabon, eastern Gabon, western Gabon). Additionally, there were several geographically very restricted haplotypes in localities found almost all over the species' distribution range. H. liebrechtsiana (Figure 3B) carried the same haplotypes as H. danckelmaniana in Gabon where both species occur in sympatry. In the Congo basin H. liebrechtsiana carried specific haplotypes: one widespread haplotype occurring from the Atlantic coast in DRCongo to the Center of the Congo Basin and several rare haplotypes being concentrated at the middle course of the Congo river.
In the genus Marantochloa there were two distinct patterns when comparing species. M. congensis (Figure 3D), the most widespread species, had two widespread haplotypes found across its entire distribution range and a few rare (locally restricted) haplotypes concentrated along the coast of Ivory Coast, in the Cameroonian Volcanic Line, in East DRCongo and in a corridor from the southern Chaillu Massif in Gabon to eastern Cameroon. M. monophylla (Figure 3E) in contrast exhibited a strong geographic pattern of two genetically distinct haplogroups, one distributed along the Atlantic coast, the other one east of that toward the Congo Basin. Major diversity centers were found in mountain ranges: Cameroonian Volcanic Line and Ngovayang (Cameroon); Cristal Mountains and the southern Chaillu Massif (Gabon); and in the Albertine Rift Valley (Uganda) (for localities compare Figure 1). Whereas, M. congensis and M. monophylla did hardly share any haplotypes (only one at Mount Cameroon), M. incertifolia (Figure 3F) shared half of its haplotypes with either one or the other sister species, M. congensis and M. monophylla. In western Cameroon and the Cristal Mountains there was one haplotype (H10) shared between all three Marantochloa species occurring there.
The fixation indices (GST: 0.15–0.77 and NST: 0.14–0.83, Table 4) were higher in Haumania liebrechtsiana, Marantochloa incertifolia and Megaphrynium macrostachyum and rather low in Megaphrynium trichogynum. Fixation indices varied somewhat according to grid cell size but the ranking of species was generally fairly consistent. There was always a marked difference in GST and NST between congeners: Mega. macrostachyum > Mega. trichogynum; H. liebrechtsiana > H. danckelmaniana; M. incertifolia > M. monophylla > M. congensis. In most species a marginally significant to very significant phylogeographic signal (NST > GST) could be detected at least at one scale (0.75°, 1.5°, and/or 3°) (Table 4). The signal was most clear in M. congensis and M. monophylla. No phylogeographic signal could be detected in Halopegia azurea, Haumania liebrechtsiana and Megaphrynium trichogynum.
Table 4.
Global GST and NST for the eight study species in Lower Guinea.
| Species | Total Number of grid cells | 0.75° | 1.5° | 3° | |||||
|---|---|---|---|---|---|---|---|---|---|
| 0.75° all(>3indiv) | 1.5° all(>3indiv) | 3° all(>3indiv) | GST | NST | GST | NST | GST | NST | |
| HalAzu | 39 (18) | 24 (16) | 12 (10) | 0.66 | 0.60 | 0.53 | 0.45 | 0.47 | 0.34 |
| HauDanck | 25 (19) | 15 (12) | 7 (7) | 0.54 | 0.60 | 0.54 | 0.65* | 0.58 | 0.60 |
| HauLieb | 24 (15) | 12 (9) | 9 (6) | 0.67 | 0.70 | 0.58 | 0.65 | 0.59 | 0.67 |
| MarCong | 42 (21) | 24 (18) | 11 (10) | 0.52 | 0.64* | 0.57 | 0.83** | 0.42 | 0.71* |
| MarIncert | 16 (9) | 9 (8) | 9 (7) | 0.75 | 0.82(*) | 0.72 | 0.77(*) | 0.77 | 0.83 |
| MarMono | 32 (18) | 19 (12) | 11 (7) | 0.58 | 0.70(*) | 0.54 | 0.71** | 0.42 | 0.47 |
| MegaMacro | 39 (22) | 23 (17) | 11 (11) | 0.60 | 0.69* | 0.58 | 0.68* | 0.62 | 0.68 |
| MegaTrich | 33 (21) | 15 (12) | 9 (8) | 0.33 | 0.26 | 0.26 | 0.24 | 0.15 | 0.14 |
Test of significance for NST > GST:
, marginally significant (p < 0.1);
, significant (p < 0.05);
, highly significant (p < 0.01). All the GST values were significantly higher than 0 (p < 0.001). For abbreviations of species names see Table 1.
Congruence of genetic diversity pattern among species
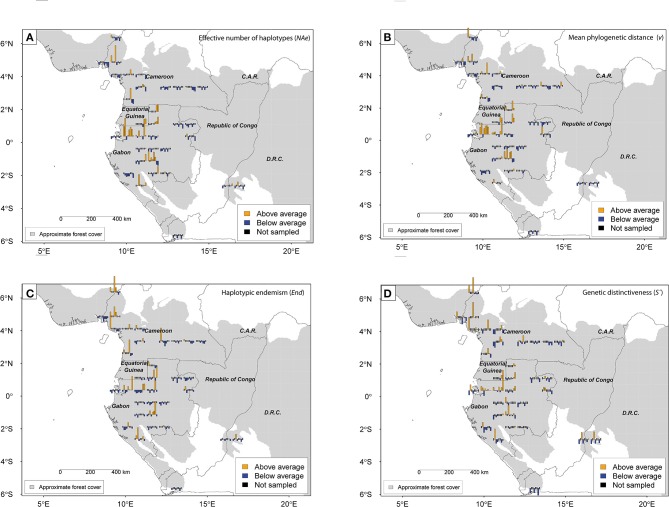
As general geographic patterns of diversity were independent of grid cell size, only results based on 0.75° grid cells are reported here. Standardized effective numbers of alleles, gene diversity and phylogenetic diversity per grid cell and species revealed a significant geographic effect according to the ANOVA analyses (NAe: F = 1.79, P = 0.01; He: F = 2.59, P < 0.001; v: F = 12.64, P < 0.001). By contrast, the ANOVA test was non-significant for the mean frequency of endemic haplotypes (End; F = 0.99, P = 0.49) and the genetic distinctiveness per grid cell (S'i; F = 0.86, P = 0.69; for 1.5°: F = 1.47, P = 0.12). Averaged standardized effective numbers of haplotypes per cell across species showed that diversity is highest in Cristal Mountains (Gabon) followed by northern and southern Gabon and Cameroonian volcanic line (Supplementary Figure 3, 4). By contrast south-western and eastern Gabon and coastal western and eastern Cameroon displayed below-average diversity values. South-western Cameroon and the DRCongo displayed close to average values. The multi-species pattern for phylogenetic diversity (v) was similar (Supplementary Figures 3, 4).
Comparing diversity patterns pairwise between species, the Pearson correlation tests revealed congruence between Halopegia azurea, Haumania danckelmaniana, M. incertifolia, M. congensis and M. monophylla (Table 5, Supplementary Table 5) with two main common centers of diversity in Gabon: the western Cristal Mountains area close to Libreville and the northern Chaillu Massif (Figure 4, Supplementary Tables 6, 7). Diversity centers of M. congensis are beside the Cameroonian volcanic line in Cameroon, the Cristal Mountains area, the northern Gabon and only observed in this species: the southern Chaillu Massif of Gabon and the northern part of the RCongo (see Figure 4, Supplementary Tables 6, 7). Furthermore, Mega. macrostachyum and Mega. trichogynum were inter-correlated for NAe. These two species showed many centers of genetic diversity well distributed across Cameroon and Gabon. They shared the center of diversity in the southwest of Gabon with H. liebrechtsiana and M. congensis. Concerning pattern of endemism there is congruence between Marantochloa monophylla and Haumania liebrechtsiana and between M. monophylla and M. incertifolia (the latter only at 0.75°) as well as between Halopegia azurea and M. congensis and between Halopegia azurea and H. danckelmaniana (but the latter only detectable at 1.5° grid because not enough shared cells at 0.75°). Interestingly, there was no congruence in the patterns of haplotypic endemism across species (Supplementary Table 8).
Table 5.
Pearson correlation of the effective number of haplotypes (NAe, lower diagonal) and within-cell phylogenetic diversity (v, upper diagonal) between species for grid cell size of 0.75°.
| HaloAzu | HauDanck | HauLieb | MarCong | MarIncert | MarMono | MegaMacro | MegaTrich | |
|---|---|---|---|---|---|---|---|---|
| HaloAzu | 0.93** | 0.63 | 0.42 | 0.18 | 0.69* | 0.04 | 0.05 | |
| HauDanck | 0.95** | −0.07 | 0.85** | 0.97** | 0.62(*) | 0.30 | 0.04 | |
| HauLieb | 0.13 | −0.30 | −0.12 | 0.44 | 0.45 | −0.33 | −0.04 | |
| MarCong | 0.51(*) | 0.47 | 0.43 | 0.34 | 0.76* | 0.30 | −0.09 | |
| MarIncert | 0.79* | 0.95* | 0.17 | −0.05 | 0.71 | 0.52 | 0.03 | |
| MarMono | 0.76* | 0.55 | −0.15 | 0.59(*) | 0.81(*) | 0.01 | −0.33 | |
| MegaMacro | 0.20 | 0.25 | 0.06 | 0.01 | 0.17 | −0.03 | 0.39 | |
| MegaTrich | −0.12 | 0.05 | 0.14 | −0.10 | 0.32 | −0.25 | 0.53(*) |
, marginally significant (p < 0.1);
, significant (p < 0.05);
, highly significant (p < 0.01). For abbreviations of species names see Table 1.
Figure 4.
Geographic distribution of standardized (i.e., centered and reduced) genetic diversity and endemism for eight Marantaceae species in Lower Guinea for grid cell size 0.75°. Effective number of haplotypes (NAe) (A); mean phylogenetic distance between individuals (v) (B); haplotypic endemism (haplotype range <200 km, End) (C); Genetic distinctiveness of each grid cell (S'i) (D). Distinctiveness above or below average is based on standardized pairwise genetic distance (S'kij computed for each species) among populations where genetic distance is estimated as the number of mutational steps between two individuals drawn from two populations (vij). Species along barplots from left to right are: Halopegia azurea, Haumania danckelmaniana, H. liebrechtsiana, Marantochloa congensis, M. incertifolia, M. monophylla, Megaphrynium macrostachyum and Mega. trichogynum.
Congruence of genetic distinctiveness patterns among species
The genetic distinctiveness per grid cell for each species is presented in Figure 4D. Above average levels of population distinctiveness for three or more species are reached in the Cameroonian volcanic line and in north-western Gabon (Libreville/coastal Gabon and western Cristal Mountains). In contrast, South and East Gabon, East Cameroon and DRCongo displayed always low levels of distinctiveness for most species.
There were only very few species pairs that showed statistically significant congruent patterns of genetic distinctiveness among grid cells (S'ij); there were three at grid cell size 0.75° (Table 6, Megaphrynium macrostachyum with Marantochloa congensis and Marantochloa monophylla, and Marantochloa monophylla with Marantochloa congensis) and three at grid cell size 1.5° (Supplementary Table 9).
Table 6.
Results of Mantel test comparing pairwise standardized distinctiveness among grid cells (S'ij) between species pairs for grid cell size of 0.75°.
| HaloAzu | HauDanck | HauLieb | MarCong | MarIncert | MarMono | MegaMacro | MegaTrich | |
|---|---|---|---|---|---|---|---|---|
| HaloAzu | 7 | 7 | 12 | 7 | 9 | 14 | 9 | |
| HauDanck | 0.15 | 6 | 10 | 5 | 9 | 9 | 12 | |
| HauLieb | −0.23 | 0 | 9 | 4 | 5 | 7 | 10 | |
| MarCong | 0.4 | −0.22 | −0.39 | 7 | 10 | 12 | 9 | |
| MarIncert | −0.31 | −0.02 | −0.05 | −0.23 | 5 | 6 | 6 | |
| MarMono | 0.08 | 0.14 | 0.02 | 0.35* | −0.44 | 12 | 11 | |
| MegaMacro | −0.03 | −0.21 | −0.11 | 0.6** | 0.26 | 0.4* | 14 | |
| MegaTrich | 0.17 | 0.09 | −0.07 | −0.1 | 0.21 | 0.1 | 0.1 |
Upper diagonal: number of grid cells shared between species; lower diagonal: correlation coefficient. Significant values (p < 0.05) and highly significant values (p < 0.01) are written in bold and indicated by
and
, respectively. For abbreviations of species names see Table 1.
Discussion
In this study, phylogeographic patterns of the plastid genome of eight herb and liana species from the family Marantaceae were compared in Lower Guinea. We expected that profound vegetation changes might have left their imprints in the distribution pattern of genetic diversity of species, and that similar species responses would lead to congruent phylogeographic patterns. In our study, however, we did not find overall congruence in the pattern of genetic diversity, endemism and distinctiveness across all study species but rather multiple patterns characteristic for one or a few species. Thus, there was not a uniform congruence of genetic pattern with the putative rainforest refugia proposed by Maley (1996). Our results indicate either idiosyncratic histories of the chosen taxa, or that once congruent genetic patterns resulting from similar species responses to particular climatic changes are already overlain by younger historical events (Alexandre et al., 1998; Maley and Brenac, 1998; Maley, 2002) leaving new individual imprints in the genetic patterns of species. Here, compared to tree species, phylogeographic patterns in herbs might reflect younger evolutionary events due to their shorter life cycles (for life cycles in perennial herbs/lianas see Putz, 1990; Gerwing, 2004; Brandes et al., 2011).
Genetic diversity and differentiation between sister species in lower guinea
In the eight Marantaceae species studied, the level of genetic diversity at the plastid gene sequenced (7–19 haplotypes per species, see also nucleotide diversity) was similar to that found for the same plastid marker in tree species from Lower Guinea (6–24 haplotypes per species, (Dauby et al., 2014a); for nucleotide diversity see Heuertz et al., 2013). The high molecular diversity found in Marantochloa monophylla was congruent with its high morphological diversity – an exceptional morphological and genetic diversity (NAe, v) was found in Ngovayang Mountain in Cameroon. By contrast, genetic diversity was especially low in Halopegia azurea, a selfing species (Ley and Claßen-Bockhoff, 2013). Although selfing should not per se affect the diversity of maternally inherited genomes, it might enhance selective sweeps by generating a global linkage between nuclear and cytoplasmic genomes (Glemin et al., 2006), a possible explanation for the low diversity observed in the plastid genome.
The divergence of haplotypes within and between Marantaceae sister species was rather low (1–2 mutations) indicating a low degree of interspecific molecular divergence, potentially due to relatively recent speciation events. Species seem not to have yet established strong species boundaries which was probably the reason for the observation of recurrent hybridization events in sympatric regions in almost all sister species pairs considered here (see also Ley and Hardy, 2014).
Spatial genetic structure and phylogeographic signal within species
A spatial genetic structure was found in all species (see significant GST values) indicating intra-specific population differentiation. In addition, a significant phylogeographic pattern (NST > GST) could be detected in five of the eight Marantaceae species. This implies that, for these species at least, some of their populations have evolved in isolation for long enough to generate related haplotypes that tend to co-occur locally. Such phylogeographic pattern is expected if species survived in multiple isolated refugia. Only Halopegia azurea, Haumania liebrechtsiana and Megaphrynium trichogynum do not show such a signal. In all three species the low number of haplotypes (<10) prevents sufficient testing power.
Genetic differentiation between areas (GST) in the eight Marantaceae species were comparable to values found in maternally inherited markers in many other angiosperm taxa, including tropical African trees (see Duminil et al., 2007; Dauby et al., 2014a). This is in contrast to our expectation of more sub-structuring in (perennial) herbs/lianas than in trees and may indicate rather similar dispersal and population structure in both growth form groups.
Within the Marantaceae, GST values seem to correlate superficially with dispersal ability (see also Petit et al., 2003): the GST was lowest in Megaphrynium trichogynum whose red fleshy fruits are ape/monkey dispersed (Williamson et al., 1990; Tutin and Fernandez, 1993; White and Abernethy, 1997), in Halopegia azurea (dispersal mode still unknown) and in M. congensis (see interpretation below), while it was highest in species of Haumania liebrechtsiana with large, probably gravity dispersed fruits, and in Marantochloa incertifolia with rather isolated occurrences and an extremely low production of flowers and fruits (5–15 flowers per inflorescence flowering sparsely over a month with a fruit set of 3–6%, see Ley, 2008; Ley and Claßen-Bockhoff, 2013). Fruits are here dispersed by small birds and/or water (Tutin, 1998; Ley, 2008). A rather low GST value in the Marantaceae is presented by Mega. trichogynum (GST = 0.33). However, there is no indication that its fruits are better dispersed than the ones of its sister species Mega. macrostachyum (fruit morphology compare in Dhetchuvi, 1996) which presents a much higher GST value. There are other contrasting GST values between sister species pairs (Table 4). However, we see a possible explanation in terms of dispersal ability difference only for the lower GST value found in M. congensis compared to its sister species. The three investigated Marantochloa species produce rather small amounts of fruits (see Ley and Claßen-Bockhoff, 2013). M. congensis is the only species which additionally frequently propagates by vegetative means, producing large quantities of bulbils which might be dispersed by water and animals, potentially contributing to an efficient gene flow between populations and a rapid clonal expansion of the species distribution range (Kennedy, 2000; Ley, 2008). M. congensis is the species with the largest current distribution range of the three investigated Marantochloa species, occurring from West Africa to eastern DRCongo (Dhetchuvi, 1996). The observation of a shared, possibly ancestral haplotype in the Cameroonian Volcanic Line and Cristal Mountains might suggest that the three species originated there. We favor ancestral haplotype over chloroplast capture, as we are dealing here with a haplotype in the center of the haplotype network between the three species. Under this assumption, the much larger distribution range of M. congensis might be explained by better dispersal capacities.
Concordance of observed genetic patterns across species and with postulated pleistocene refugia in lower guinea
We demonstrated congruent geographic patterns of diversity across species: local genetic diversity is congruently high for six out of eight study species in the Cristal Mountains area and low for seven out of eight study species in eastern Cameroon, eastern and southern coastal Gabon, and Bas Congo (Mayumbe) in DRCongo. By contrast, values of above-average frequency of endemic haplotypes can almost be found in every grid cell when taking all species together. However, few species pairs display correlated patterns of endemic haplotype frequencies. Similarly, there is no general correlation in distinctiveness indices among Marantaceae species, while such a correlation was reported among five of eight tree species for Lower Guinea (Dauby et al., 2014a).
To interpret these patterns, it is worth noting that endemic haplotypes are potentially the best indicators for refuge areas in general (stable populations are expected to accumulate endemic haplotypes that have not the opportunity to emigrate), and distinctiveness indices might be best indicators of refuge areas that are not source of adjacent areas. High diversity is expected in refuge areas with historically large population sizes (but not under small but stable population size), but also in areas recolonized from multiple differentiated populations (secondary contact zones), a situation where phylogenetic diversity (v) should peak (Petit et al., 2003). As we found interspecific congruence in genetic diversity but not in endemism and distinctiveness, the data do not support a hypothesis whereby the different Marantaceae species would have primarily survived in the same set of refugia during periods of climate deterioration. Nevertheless, the correlation in diversity indices might indicate that there are some shared secondary contact and/or refuge areas. In fact, some high diversity areas, like the Cristal Mountains, might have been both a refuge for some species and a secondary contact zone for other species, or even the two for some species (if we imagine that a refuge area becomes “invaded” by an expanding population from another origin).
Overall, we found marked differences in patterns of haplotype distribution across species: (i) Some species are characterized by mostly parapatric distributions of their frequent haplotypes (H. danckelmaniana, Mega. macrostachyum and M. monophylla; plus H. liebrechtsiana though in this case the pattern may be due to plastid capture). Here a common pattern becomes apparent distinguishing Cameroon from south-western Gabon and eastern Gabon. Frequent haplotypes often overlap in their distribution range in the Mount Cristal area. Each of these individual distribution ranges overlaps with a different postulated refugium allowing two different scenarios: either an expansion of each of the frequent haplotypes from a central refugium in the Cristal Mountains area into different directions to Cameroon, south-western Gabon and eastern Gabon, or the other way round with an expansion from three different refugia with an overlap today in the Cristal Mountains area. A similar pattern of restricted haplotype distribution ranges was also found in some tree species and has here been attributed to the retraction of these species to different refugia. In trees it might additionally have been coupled with an adaptation to different climatic conditions evoked by the East-West rainfall gradient and the North-South seasonal gradient in Lower Guinea (Duminil et al., 2013; Heuertz et al., 2013). (ii) A second haplotype distribution pattern is characterized by a wide distribution of one or a few frequent haplotypes over the entire distribution range of a species (Halopegia azurea, M. congensis, Mega. trichogynum). These species might have a single locality of origin from where an expansion took place across the current distribution range (though a second refugium but without visible expansion would explain the endemic haplotypes found in the Cameroonian Volcanic Line for M. congensis). So far the detected diversity and endemism pattern in these three species suggest a refugium within Lower Guinea based on high diversity and endemism e.g., Cristal Mountains with or without Chaillu Massif and/or Cameroonian Volcanic Line and/or eastern Cameroon depending on species. The only study species so far showing evidence of a refugium outside Lower Guinea in DRCongo is H. liebrechtsiana (see also Ley and Hardy, 2010, 2014). As already found in trees (see Hardy et al., 2013) major rivers seem not to play an important role as barriers to gene flow in Marantaceae in contrast to evidence found in animals (e.g., Gonder and Disotell, 2006; Anthony et al., 2007; Nicolas et al., 2011).
There is repeatedly genetic evidence in species for the Cristal Mountains being a refuge area (see Koffi et al., 2011; Dauby et al., 2014b). The fact that in the Marantaceae the high diversity of the Cristal Mountains area is not associated with high endemism for at least half of the species suggests that the high diversity is best explained by a recolonization from several sources, rather than by a refuge effect, at least for these species. Note however that an area might be a refuge and at the same time have been “invaded” by other sources.
The Cameroonian volcanic line is a locality well-known for its high species diversity and endemism level, which has been interpreted as a signature of a past forest refuge (see Sosef, 1994; Maley, 1996). In the Marantaceae, only species with large distribution ranges from Lower Guinea to West Africa present high genetic diversity and/or distinctiveness and/or endemism values here. This is in accordance with patterns found in widespread trees (see also Lowe et al., 2010) and might be due to a refuge effect, i.e., accumulation of mutations in stable populations, and/or to a topographical effect, i.e., differentiation between geographically close populations isolated by mountainous barriers (see also Dauby et al., 2014a).
Assuming that the limited concordance between phylogeographic patterns of Marantaceae species in Lower Guinea reflects their idiosyncratic histories of past population fragmentation, one may question the relative importance of chance (species survived by chance in one or several refugia following forest fragmentation) and ecological adaptations (e.g., species survived only in refugia reflecting the optimum of their climatic tolerance). As all species are currently co-occurring in all potential refugia without a marked adaptation to different habitats and/or climate regimes (see Dhetchuvi, 1996) we favor the hypothesis that demographic stochasticity affecting population survival as well as rare long distance dispersal driving recolonization routes played a major role in the resulting phylogeographic patterns. For tree species, congruence of genetic distinctiveness patterns was observed in northern Lower Guinea but not in southern Lower Guinea (Dauby et al., 2014a). This pattern was tentatively explained by a less drastic forest cover reduction in southern Lower Guinea where multiple micro-refugia (e.g., gallery forests) would have remained (see Kingdon, 1980; Dupont et al., 2000; Leal, 2001). The high ecological drift associated with these micro-refugia would imply that each one would have hosted a limited number of typical rainforest species, which might have led to the observed idiosyncratic demographic histories of species. This hypothesis might also hold for our Marantaceae species.
A current limitation for the interpretation of our data is the difficulty to date population divergence or admixture and provide a confirmation that such events are concomitant with Pleistocene climate changes and not earlier or later events of climate change. Plastid markers are not ideal for this purpose due to their relatively low mutation rate. Additional studies based on nuclear sequencing should bring new insights.
Beyond lower guinea—the role of upper guinea and congolia
Assessing the importance of the areas adjacent to the East and the West of Lower Guinea (Congolia and Upper Guinea, respectively) for speciation and population differentiation is still difficult due to a lack of sufficient data. Patterns so far documented indicate that these areas have widespread haplotypes but also endemic haplotypes. In Upper Guinea several refugia were postulated by Maley (1996, see Figure 1) and the dry Dahomey gap in Benin might play an important role in isolating Upper and Lower Guinea (see Hardy et al., 2013), although the two forest blocks were probably connected during the Humid Holocene period (c. 6–9 kr BP). This can explain why several species are still restricted to Western Africa today (White, 1979; for Marantaceae see Schnell, 1957; Jongkind, 2008) and endemic haplotypes are found there (e.g., Halopegia azurea, see also Duminil et al., 2013) advocating the uniqueness of this area. Similarly, the Congo Basin and the adjacent eastern mountain range are interesting areas. Preliminary data suggest overall genetic diversity to be low in this area for most species, defining this region rather as an area of expansion. Some authors have suggested that Marantaceae species could have been spread to east Cameroon/RCongo due to human activities (Maley, 2001; Brncic et al., 2009). However, despite a rather fragmentary sampling, endemic haplotypes have also been detected in the Congo Basin and in the Albertine Rift Valley (see M. monophylla). This suggests a rather long existence of those species in that area.
Author contributions
Alexandra C. Ley has been conducting research on Marantaceae since her PhD starting in 2004. She did most of the field collections and genetic laboratory manipulations, analyzed the chloroplast haplotype distribution and diversity pattern and took a lead in the editing of the article. Gilles Dauby developed the analyses of divergence of haplotype distribution across species in a previous project and applied his knowledge here on the Marantaceae dataset. Julia Köhler and Catherina Wypior conducted a “Forschungsgruppenpraktikum” on the acquisition and analyses of the genetic data of one species of Megaphrynium each. Martin Röser and Olivier J. Hardy are both group leaders. The latter conducted the analyses of spatial genetic pattern and endemism and significantly contributed to the discussion of results. All co-authors gave their final approval of the version to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
For support in collecting specimens in Gabon, we thank the National Herbarium in Libreville (LBV), World Wildlife Fund (WWF), T. Stevart and M. Leal both from Missouri Botanical Garden (MBG), the Institut de Recherche en Ecologie Tropical (IRET), O. Hymas from Wildlife Conservation Society (WCS), the Conservators employed by the National Parks Council (CNPN) and IPHAMETRA and CENAREST for granting mission orders and research permits. For the collections in Cameroon, we are grateful for support by J.M. Onana, O. Sene from the Herbarium Cameroon in Yaounde (Y), by the staff of Limbe Botanical Garden as well as by A. Enang, M. Cheek, M. Etuge, P. Mambo and S. Moses. For the collections in DRCongo, we are thankful for the support of A. Serckx and WWF Congo, as well as for additional collections made in RCongo, DRCongo-Salonga and West Africa by J.-F. Gillet, G. Hohmann and C. Jongkind, respectively. B. Hildebrandt kindly supported our genetic work in the laboratory. The collection missions to Gabon and Cameroon were financed by the German Exchange Service (DAAD). Following this, the first author was granted a 2-year postdoctoral fellowship by the Germany Research Foundation (DFG). The laboratory work was financed by the Belgian Fund for Scientific Research (F.R.S-FNRS, grants FRFC 2.4.577.10 and MIS 4.519.10), through the project “C3A” financed by the French ANR (Agence Nationale de la Recherche) under the ANR-BIODIV program as well as the University Halle-Wittenberg.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2014.00403/abstract
References
- Alexandre A., Meunier J. D., Lezine A. M., Vincens A., Schwartz D. (1998). Phytoliths: indicators of grassland dxnamics during the late holocene in intertropical Africa. Palaeogeogr. Palaeocl. 136, 213–229 10.1016/S0031-0182(97)00089-8 [DOI] [Google Scholar]
- Andersson L. (1998). Marantaceae, in The families and Genera of Vascular Plants. IV: Flowering plants. Monocotyledons. Alismatanae and Commelinanae (except Gramineae), ed Kubitzki K. (Berlin: Springer Verlag; ), 278–474. [Google Scholar]
- Anhuf D., Ledru M.-P., Behling H., Da Cruz F. W., Jr., Cordeiro R. C., Van der Hammen T., et al. (2006). Paleo-environmental change in Amazonian and African rain forest during the LGM. Palaeogeogr. Palaeocl. 39, 510–527 10.1016/j.palaeo.2006.01.017 [DOI] [Google Scholar]
- Anthony N. M., Johnson-Bawe M., Jeffery K., Clifford S. L., Abernethy K. A., Tutin C. E., et al. (2007). The role of pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc. Natl. Acad. Sci. U.S.A. 104, 20432–20436. 10.1073/pnas.0704816105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.-J., Forster P, Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- Bonnefille R. (2007). Rain forest responses to past climate changes in tropical Africa, in Tropical Rain Forest Responses to Climate Change, eds Bush M. B., Flenley J. R., Gosling W. D. (Chichester: Praxis Publishing; ), 117–170. [Google Scholar]
- Brandes A. F. N., Lisi C. S., Barros C. F. (2011). Dendrochronology of lianas of the leguminosae family from the Atlantic forest, Brazil. Trees 25, 133–144 10.1007/s00468-010-0529-3 [DOI] [Google Scholar]
- Brncic T. M., Willis K. J., Harris D. J., Telfer M. W., Bailey R. M. (2009). Fire and climate change impacts on lowland forest composition in northern Congo during the last 2580 years from palaeoecological analyses of a seasonally flooded swamp. Holocene 19, 79–89 10.1177/0959683608098954 [DOI] [Google Scholar]
- Carbone I., Anderson J. B., Kohn L. M. (1999). Patterns of descent in clonal lineages and their multilocus fingerprints are resolved with combined gene genealogies. Evolution 53, 11–21 10.2307/2640916 [DOI] [PubMed] [Google Scholar]
- Claßen-Bockhoff R. (1991). Untersuchungen zur Konstruktion des Bestäubungsapparates von Thalia geniculata (Marantaceen). Bot. Acta 104, 183–193 10.1111/j.1438-8677.1991.tb00215.x [DOI] [Google Scholar]
- Clausager K., Borchsenius F. (2003). The Marantaceae of Sabah, northern Borneo. Kew Bull. 58, 647–678 10.2307/4111147 [DOI] [Google Scholar]
- Dauby G., Duminil J., Heuertz M., Koffi G., Stévart T., Hardy O. J. (2014a). Congruent phylogeographic patterns of eight tree species in Atlantic Central Africa provide insights into the past dynamics of forest cover. Mol. Ecol. 23, 2299–2312. 10.1111/mec.12724 [DOI] [PubMed] [Google Scholar]
- Dauby G., Hardy O. J., Leal M., Breteler F., Stévart T. (2014b). Drivers of tree diversity in tropical rain forests: new insights from a comparison between littoral and hilly landscapes of Central Africa. J. Biogeogr. 41, 574–586 10.1111/jbi.12233 [DOI] [Google Scholar]
- Demesure B., Sodzi N., Petit R. J. (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol. Ecol. 4, 129–131. 10.1111/j.1365-294X.1995.tb00201.x [DOI] [PubMed] [Google Scholar]
- Dhetchuvi J. B. (1996). Taxonomie et phytogéographie des Marantaceae et des Zingiberaceae de l'Afrique Centrale (Gabon, Congo, Zaire, Rwanda et Brundi). Ph.D. thesis, Université Libre de Bruxelles, Belgique. [Google Scholar]
- Duminil J., Brown R. P., Ewédjè E. B. K., Mardulyn P., Doucet J.-L., Hardy O. J. (2013). Large-scale pattern of genetic differentiation within African rainforest trees: Insights on the roles of ecological gradients and past climate changes on the evolution of Erythrophleum spp (Fabaceae). BMC Evol. Biol. 13: 195. 10.1186/1471-2148-13-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duminil J., Fineschi S., Hampe A., Jordano P., Salvini D., Vendramin G. G., et al. (2007). Can population genetic structure be predicted from life-history traits? Am. Nat. 169, 662–672. 10.1086/513490 [DOI] [PubMed] [Google Scholar]
- Dupont L. (2011). Orbital scale vegetation change in Africa. Quat. Sci. Rev. 30, 3589–3602 10.1016/j.quascirev.2011.09.019 [DOI] [Google Scholar]
- Dupont L. M., Jahns S., Marret F., Shi N. (2000). Vegetation change in equatorial West Africa: time-slices for the last 150 ka. Palaeogeogr. Palaeocl. 155, 95–122 10.1016/S0031-0182(99)00095-4 [DOI] [Google Scholar]
- Excoffier L., Laval G., Schneider S. (2009). Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fjeldsa J., Lovett J. C. (1997). Geographical patterns of old and young species in African forest biota: the significance of specific montane areas as evolutionary centres. Biodivers. Conserv. 6, 325–346 10.1023/A:1018356506390 [DOI] [Google Scholar]
- Gerwing J. J. (2004). Life history diversity among six species of canopy lians in an old-growth forest of the eastern Brazilian Amazon. For. Ecol. Manag. 190, 57–72 10.1016/j.foreco.2003.10.006 [DOI] [Google Scholar]
- Glemin S., Bazin E., Charlesworth D. (2006). Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc. R. Soc. B 273, 3011–3019. 10.1098/rspb.2006.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonder M. K., Disotell T. R. (2006). Contrasting phylogeographic histories of chimpanzees in Nigeria and Cameroon: a multi-locus genetic analysis, in Fleagle Primate Biogeography, Progress and Prospects, eds Lehman S. M., Fleagle J. G. (US: Springer; ), 135–168 10.1007/0-387-31710-4_5 [DOI] [Google Scholar]
- Hardy O. J., Born C., Budde K., Dainou K., Dauby G., Duminil J., et al. (2013). Comparative phylogeography of African rainforest trees: a review of genetic signatures of vegetation history in the Guineo-Congolian domain. C. R. Geosci. 345, 284–296 10.1016/j.crte.2013.05.001 [DOI] [Google Scholar]
- Hardy O. J., Vekemans X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2, 618–620 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- Heuertz M., Duminil J., Dauby G., Savolainen V., Hardy O. J. (2013). Comparative phylogeography in rainforest trees from Lower Guinea, Africa. PLoS One 9:e84307. 10.1371/journal.pone.0084307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein N., Renner S. (2011). A dated phylogeny and collection records reveal repeated biome shifts in the African genus Coccinia (Cucurbitaceae). BMC Evol. Biol. 11, 28. 10.1186/1471-2148-11-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongkind C. C. H. (2008). Two new species of Hypselodelphys (Marantaceae) from West Africa. Adansonia, ser. 30, 57–62. [Google Scholar]
- Kearey P., Vine F. J. (1996). Global Tectonics, 2nd Edn. Oxford: Blackwell Science. [Google Scholar]
- Kennedy H. (2000). Diversification in pollination mechanisms in the marantaceae, in Monocots: Systematics and Evolution, eds Wilson K. L., Morrison D. A. (Melbourne: Morrison CSIRO; ), 335–343. [Google Scholar]
- Kingdon J. S. (1980). The role of visual signals and face patterns in African forest monkeys (guenons) of the genus cercopithecus. Trans. Zool. Soc. 35, 425–475 10.1111/j.1096-3642.1980.tb00062.x [DOI] [Google Scholar]
- Koffi K. G., Hardy O. J., Doumenge C., Cruaud C., Heuertz M. (2011). Diversity gradients and phylogeographic patterns in a widespread African tree typical of mature rainforests, Santiria trimera (Burseraceae). Am. J. Bot. 98, 254–264. 10.3732/ajb.1000220 [DOI] [PubMed] [Google Scholar]
- Kress J. W., Specht C. (2005). Between cancer and Capricorn: phylogeny, evolutionand evology of the primarily tropical Zingiberales, in Plant Diversity and Complexity Patterns. Local, Regional and Global Dimensions, eds Friis I., Balslev H. (Proceedings of an international symposium, Royal Danish Academy of Sciences and Letters in Copenhagen, Denmark: ), 459–478. [Google Scholar]
- Leal M. E. (2001). Microrefugia, small scale ice age forest remnants. Syst. Geogr. Pl. 71, 1073–1077 10.2307/3668739 [DOI] [Google Scholar]
- Lee C., Wen J. (2004). Phylogeny of Panax using chloroplast trnC–trnD intergenic region and the utility of trnC–trnD in interspecific studies of plants. Mol. Phylo. Evol. 31, 894–903. 10.1016/j.ympev.2003.10.009 [DOI] [PubMed] [Google Scholar]
- Leroux M. (1983). Le Climat de I' Afrique Tropicale. Vol. 1, 2 Paris: Champion Slatkine. [Google Scholar]
- Ley A. C. (2008). Evolution in African Marantaceae—evidence from Floral Morphology, Ecology and Phylogeny. Ph.D. thesis. Institut für Spezielle Botanik, University of Mainz, Germany. [Google Scholar]
- Ley A. C., Claßen-Bockhoff R. (2009). Pollination syndromes in African Marantaceae. Ann. Bot. 104, 41–56. 10.1093/aob/mcp106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley A. C., Claßen-Bockhoff R. (2011). Evolution in African Marantaceae - evidence from phylogenetic, ecological and morphological studies. Syst. Bot. 36, 277–290 10.1600/036364411X569480 [DOI] [Google Scholar]
- Ley A. C., Claßen-Bockhoff R. (2012). Five new species of marantaceae from Gabon. Adansonia 34, 37–52 10.5252/a2012n1a4 [DOI] [Google Scholar]
- Ley A. C., Claßen-Bockhoff R. (2013). Breeding system and fruit set in African Marantaceae. Flora 208, 532–537 10.1016/j.flora.2013.07.011 [DOI] [Google Scholar]
- Ley A. C., Hardy O. J. (2010). Species delimitation in the Central African herbs Haumania (Marantaceae) using georeferenced nuclear and chloroplastic DNA sequences. Mol. Phyogenet. Evol. 57, 859–867. 10.1016/j.ympev.2010.08.027 [DOI] [PubMed] [Google Scholar]
- Ley A. C., Hardy O. J. (2014). Contrasting patterns of gene flow between sister plant species in the understorey of African moist forests - the case of sympatric and parapatric Marantaceae species. Mol. Phyogenet. Evol. 77, 264–274. 10.1016/j.ympev.2014.04.026 [DOI] [PubMed] [Google Scholar]
- Librado P., Rozas J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Locatelli E., Machado I. C., Medeiros P. (2004). Saranthe klotzschiana (Koer.) Eichl. (Marantaceae) e seu mecanismo explosivo de polinizacao. Rev. Bras. Bot. 27, 757–765 10.1590/S0100-84042004000400014 [DOI] [Google Scholar]
- Lowe A. J., Harris D., Dormontt E., Dawson I. K. (2010). Testing putative African tropical refugia using chloroplast and nuclear DNA phylogeography. Trop. Plant Biol. 3, 50–58 10.1007/s12042-010-9045-2 [DOI] [Google Scholar]
- Maley J. (1996). The African rain forest: main characteristics of changes in vegetation and climate from the Upper Cretaceous to the Quaternary. P. Roy. Soc. Edinb. B. 104B, 31–73. [Google Scholar]
- Maley J. (2001). The impact of arid phases on the African rainforest through geological history, in African Rain Forest Ecology and Conservation, eds Weber W., White L., Vedder A., Naughton-Treves L. (New Haven, CT: Yale University Press; ), 68–87. [Google Scholar]
- Maley J. (2002). A catastrophic destruction of African Forests about 2,500 Years ago still exerts a major influence on present vegetation formations. IDS Bull. 33, 14–30 10.1111/j.1759-5436.2002.tb00003.x [DOI] [Google Scholar]
- Maley J., Brenac P. (1998). Vegetation dynamics, palaeoenvironments and climatic changes in the forests of western Cameroon during the last 28,000 years B.P. Rev. Palaeobot. Palynol. 99, 157–187 10.1016/S0034-6667(97)00047-X [DOI] [Google Scholar]
- Nei M. (1978). Estimation of average heterozygosity and genetic distance for small number of individuals. Genetics 89, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngomanda A., Neumann K., Schweizer A., Maley J. (2009). Seasonality change and the third millennium BP rain forest crisis in southern Cameroon (Central Africa). Quat. Res. 71, 307–318 10.1016/j.yqres.2008.12.002 [DOI] [Google Scholar]
- Nicolas V., Missoup A. D., Denys C., Kerbis Peterhans J., Katuala P., Couloux A., et al. (2011). The roles of rivers and pleistocene refugia in shaping genetic diversity in praomys misonnei in tropical Africa. J. Biogeogr. 38, 191–207 10.1111/j.1365-2699.2010.02399.x [DOI] [Google Scholar]
- Nielsen R., Tarpy D. R., Kern Reeve H. (2003). Estimating effective paternity number in social insects and the effective number of alleles in a population. Mol. Ecol. 12, 3157–3164. 10.1046/j.1365-294X.2003.01994.x [DOI] [PubMed] [Google Scholar]
- Nybom H. (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 13, 1143–1155. 10.1111/j.1365-294X.2004.02141.x [DOI] [PubMed] [Google Scholar]
- Petit R. J., Aguinagalde I., de Beaulieu J.-L., Bittkau C., Brewer S., Cheddadi R., et al. (2003). Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300, 1563–1565. 10.1126/science.1083264 [DOI] [PubMed] [Google Scholar]
- Pons O., Petit R. J. (1996). Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics 144, 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince L. M., Kress J. (2006). Phylogenetic relationships and classification in marantaceae: insights from plastid DNA sequence data. Taxon 55, 281–296 10.2307/25065578 [DOI] [Google Scholar]
- Putz F. E. (1990). Liana stem diameter growth and mortality rates on Barro Colorado Island, Panama. Biotropica 22, 103–105 10.2307/2388725 [DOI] [Google Scholar]
- Robbrecht E. (1994). Geography of African Rubiaceae with reference to glacial rain forest refugia, in The Biodiversity of African Plants: Proceedings XIVth AETFAT Congress, eds van der Maesen L. J. G., van der Burgt X. M., van Medenbach de Rooy J. M. (Wageningen: Kluwer Academic Publishers; ), 564–581. [Google Scholar]
- Sang T., Crawford D. J., Stuessy T. F. (1997). Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 84, 1120–1136. 10.2307/2446155 [DOI] [PubMed] [Google Scholar]
- Schnell R. (1957). Clef empirique des principales Marantacees ouestafricaines (Guinée, Côte dÍvoire et territoires limitrophes). Bull. Inst. Fond. Afr. Noire. 4, 1124–1134. [Google Scholar]
- Schönswetter P., Stehlik I., Holderegger R., Tribsch A. (2005). Molecular evidence for glacial refugia of mountain plants in the European Alps. Mol. Ecol. 14, 3547–3555. 10.1111/j.1365-294X.2005.02683.x [DOI] [PubMed] [Google Scholar]
- Sosef M. (1994). Refuge begonias: taxonomy, phylogeny and historical biogeography of Begonia sect. loasibegonia and sect. Scutobegonia in relation to glacial rain forest refugia in Africa. Agric. Univ. Wageningen Pap. 94, 1–306. [Google Scholar]
- Suksathan P., Gustafsson M. H., Borchsenius F. (2009). Phylogeny and generic delimitation of Asian Marantaceae. Bot. J. Linn. Soc. 159, 381–395 10.1111/j.1095-8339.2009.00949.x [DOI] [Google Scholar]
- Taberlet P., Fumagalli L., Wust-Saucy A. G., Cosson J. F. (1998). Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 7, 453–464. 10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- Tate J. A. B., Simpson B. (2003). Paraphyly of Tarasa (Malvaceae) and diverse origins of the polyploid species. Syst. Bot. 28, 723–737 10.1043/02-64.1 [DOI] [Google Scholar]
- Tutin C. E. G. (1998). Gorillas and their food plants in the lope reserve, Gabon, in Chorology, Taxonomy and Ecology of the Floras of Africa and Madagascar, eds Huxley C. R., Lock J. M., Cutler D. F. (Kew: Royal Botanical Gardens; ), 227–243. [Google Scholar]
- Tutin C. E. G., Fernandez M. (1993). Relationships between minimum temperature and fruit production in some tropical forest trees in Gabon. Am. J. Primatol. 30, 195–211 10.1002/ajp.1350300305 [DOI] [PubMed] [Google Scholar]
- Vande Weghe J. P. (2004). Forests of Central Africa. Tielt: Lannoo Publishers. [Google Scholar]
- Ward M., Dick C. W., Gribel R., Lowe A. J. (2005). To self, or not to self. A review of outcrossing and pollen-mediated gene flow in neotropical trees. Heredity. 95, 246–254, 10.1038/sj.hdy.6800712 [DOI] [PubMed] [Google Scholar]
- White F. (1979). The Guineo-Congolian region and its relationships to other phytochoria. Bull. Jard. Bot. Nat. Belg. 49, 11–55. [Google Scholar]
- White L., Abernethy K. (1997). A Guide to the Vegetation of the Lope Reserve Gabon. Gabon: Wildlife Conservation Society; ECOFAC. [Google Scholar]
- Williamson E. A, Tutin C. E. G., Rogers M. E., Fernandez M. (1990). Composition of the diet of lowland gorillas at lope in Gabon. Am. J. Primatol. 21, 265–277 10.1002/ajp.1350210403 [DOI] [PubMed] [Google Scholar]
- Willis K. J., McElwain J. C. (2002). The Evolution of Plants. Oxford: Oxford University Press. [Google Scholar]
- Zeng X., Michalski S. G., Fischer M., Durka W. (2012). Species diversity and population density affect genetic structure and gene dispersal in a subtropical understory shrub. J. Plant Ecol. 5, 270–278 10.1093/jpe/rtr029 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.