Summary
Porcine induced pluripotent stem cells (iPSCs) provide useful information for translational research. The quality of iPSCs can be assessed by their ability to differentiate into various cell types after chimera formation. However, analysis of chimera formation in pigs is a labor-intensive and costly process, necessitating a simple evaluation method for porcine iPSCs. Our previous study identified mouse embryonic stem cell (ESC)-specific hypomethylated loci (EShypo-T-DMRs), and, in this study, 36 genes selected from these were used to evaluate porcine iPSC lines. Based on the methylation profiles of the 36 genes, the iPSC line, Porco Rosso-4, was found closest to mouse pluripotent stem cells among 5 porcine iPSCs. Moreover, Porco Rosso-4 more efficiently contributed to the inner cell mass (ICM) of blastocysts than the iPSC line showing the lowest reprogramming of the 36 genes (Porco Rosso-622-14), indicating that the DNA methylation profile correlates with efficiency of ICM contribution. Furthermore, factors known to enhance iPSC quality (serum-free medium with PD0325901 and CHIR99021) improved the methylation status at the 36 genes. Thus, the DNA methylation profile of these 36 genes is a viable index for evaluation of porcine iPSCs. genesis 51:763–776. © 2013 Wiley Periodicals, Inc.
Keywords: epigenetics, induced pluripotent stem cells, translational research
INTRODUCTION
The use of induced pluripotent stem cells (iPSCs) is expected to dramatically accelerate advances in medical care (Okita and Yamanaka, 2011; Takahashi and Yamanaka, 2006; Takahashi et al., 2007). In particular, iPSCs may offer novel therapies for previously intractable conditions, and a wide range of possible applications has been investigated including exploration of the pathogenic mechanisms of refractory diseases and the development of new drugs (Ebert et al., 2009; Imaizumi et al., 2012; Inoue and Yamanaka, 2011), cell therapy (Montserrat et al., 2011; Zhou et al., 2011), production of organs and tissues (Kobayashi et al., 2010; Usui et al., 2012), and generation of germ cells (Hayashi et al., 2011, 2012).
Before iPSCs can be used for clinical applications, it is essential that appropriate experiments using animal models are carried out to ensure their effectiveness and safety. In addition to the use of standard laboratory animals such as rodents, investigation of larger animal species, with closer physiological resemblance to humans, will significantly benefit translational research. The pig is one such species and has many similarities in anatomy and physiology to humans (van der Spoel et al., 2011; Zhao and Prather, 2011); pigs have often been used in biomedical studies as a large experimental model, which can produce data that can be easily applied to humans (Lunney, 2007; Petters, 1994; Prather et al., 2003). Therefore, the generation and evaluation of porcine iPSCs will provide useful information that could help promote clinical application of human iPSCs (Ezashi et al., 2009; Fujishiro et al., 2013; Montserrat et al., 2011; West et al., 2010; Wu et al., 2009).
The pluripotency of porcine iPSCs can be evaluated by determining their ability to form chimeras (Fujishiro et al., 2013; West et al., 2010). However, the production of chimeric fetuses and piglets is a labor-intensive and costly process that requires embryo manipulation and transfer. Indeed, few studies have used chimera-forming ability as a means of confirming the pluripotency of porcine iPSCs (Fujishiro et al., 2013; West et al., 2010, 2011). Therefore, it is essential to develop new methods, either for evaluating the pluripotency of porcine iPSCs, or for pre-screening iPSC lines for use in chimera formation experiments.
Epigenetic regulation, including DNA methylation and histone modifications, is fundamental to tissue- and/or cell-type specific gene expression (Golob et al., 2008; Ikegami et al., 2009; Lieb et al., 2006; Shiota et al., 2004). There are a large number of tissue-dependent differentially methylated regions (T-DMRs) in the mammalian genome (Shiota et al., 2002; Yagi et al., 2008). The DNA methylation status of T-DMRs is determined during embryonic development, and the DNA methylation profile of T-DMRs is distinctive in each cell type (Sakamoto et al., 2007; Shiota et al., 2002). Mouse ESCs, which are known to be pluripotent stem cells, exhibit unique DNA methylation profiles at T-DMRs. Genes involved in the establishment and maintenance of the pluripotent state, including Oct3/4 (Pou5f1), are hypomethylated in mouse ESCs (Hattori et al., 2004; Imamura et al., 2006). Genome-wide DNA methylation analyses have identified several hundred mouse ESC-specifically hypomethylated T-DMRs (EShypo-T-DMRs; Sato et al., 2010). Furthermore, it has been shown that the DNA methylation profiles of EShypo-T-DMRs in mouse iPSCs with a high efficiency of chimera formation are similar to those of ESCs (Aoi et al., 2008; Sato et al., 2010). These results indicate that the DNA methylation profile of EShypo-T-DMRs provide a viable index for screening high-quality iPSCs.
We recently generated naïve-like porcine iPSC lines using the four Yamanaka factors (Oct3/4, Sox2, Klf4, and c-Myc; Fujishiro et al., 2013); these factors are also used to generate mouse iPSCs. These porcine iPSC lines exhibit LIF-dependent proliferation abilities similar to those of mouse ESCs/iPSCs (Fujishiro et al., 2013); however, the lines also show different characteristics.
In this study, we sought to determine whether the DNA methylation index for mouse EShypo-T-DMRs could be used to evaluate porcine iPSC lines. If this approach is feasible for the evaluation of porcine iPSCs, then the same strategy could be applied to quality enhancement of iPSCs from a wide range of domesticated animal species and ultimately for human iPSCs.
RESULTS
DNA Methylation Profile of 36 Genes Known to be Specifically Hypomethylated in Mouse ESCs in Porcine iPSCs
To analyze DNA methylation profiles of porcine iPSCs, we selected 36 genes that are hypomethylated specifically in mouse ESCs (EShypo-T-DMRs; Sato et al., 2010). The 36-gene set was categorized into three groups; those targeted by Oct3/4 (Oct3/4-targets); Klf4, Sox2, or c-Myc (KSM-targets); and genes, which are not targets of these four factors (non-targets; Fig. 1a). In the course of iPSC establishment, activation of target genes of the four Yamanaka factors is required after introduction into somatic cells. Among the four factors, Oct3/4 is of utmost importance since iPSC lines have not been established without Oct3/4 introduction to date. Thus, target genes of Oct3/4, such as Sall4 (Tsubooka et al., 2009), are thought to have crucial roles in iPSC establishment, and the Oct3/4 target genes (Oct3/4-targets) were separated from the target genes of the other three Yamanaka factors (KSM-targets). Although “non-targets” are genes that are not directly bound by the four Yamanaka factors, their methylation levels in mouse ESCs were lower than those in differentiated tissues/cells (Sato et al., 2010), suggesting that the DNA methylation statuses of non-targets can also be useful as an index for evaluation of porcine iPSCs. The classification of target genes was based on ChIP-seq data for several transcription factors, including the four Yamanaka factors (Chen et al., 2008). We initially selected 56 porcine genes orthologous to mouse genes with EShypo-T-DMRs, whose DNA methylation levels were low in mouse ESCs but 30% or more higher in differentiated tissues/cells (Sato et al., 2010). However, 16 of these were found to be hypomethylated in porcine somatic tissues, and further, four genes were hypermethylated in porcine blastocysts, consisting of pluripotent cells (Supporting Information Fig. 1). Therefore, these 20 genes were excluded, and the remaining 36 were used for DNA methylation analyses. Among these 36 genes, several are known to be hypomethylated in mouse ESCs, but to be in a poised state of transcriptional activation, rather than actively expressed. This type of epigenetic regulation has been reported in H3K4me3/H3K27me3-enriched bivalent regions (Meissner et al., 2008; Xu et al., 2007).
Figure 1.
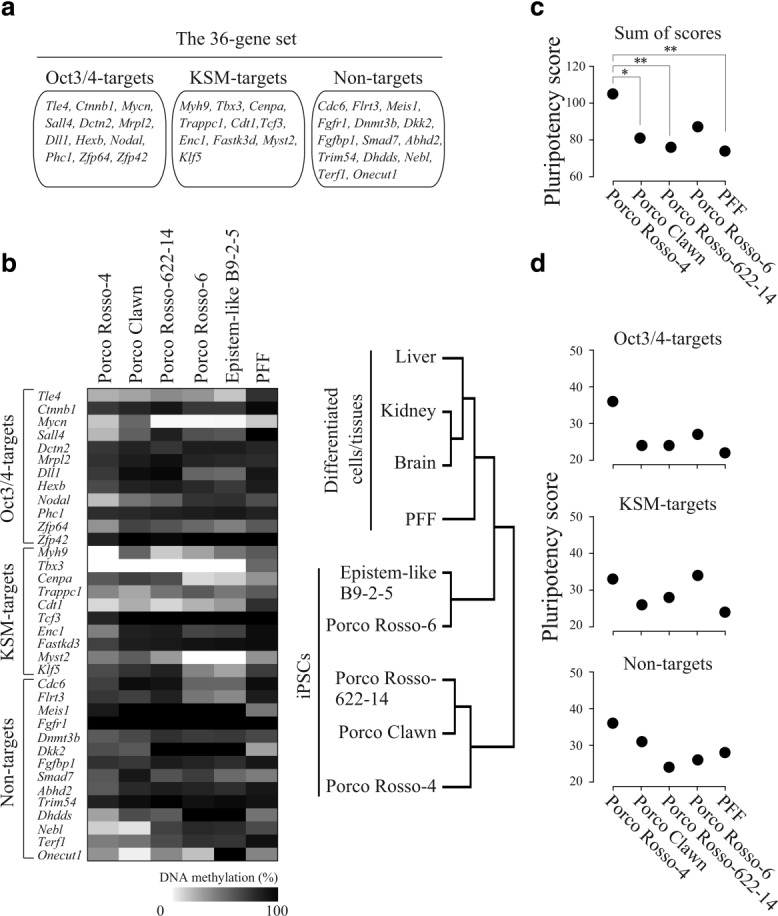
DNA methylation profiles of porcine iPSCs. (a) The 36-gene set analyzed for DNA methylation profiling of porcine iPSCs. The genes were retrieved from EShypo-T-DMRs previously determined in mouse ESCs, and were categorized into three groups: Oct3/4-targets, KSM-targets, and non-targets. (b) DNA methylation status of the 36-gene set in five iPSC lines (Porco Rosso-4, Epistem-like B9-2-5, Porco Rosso-6, Porco Rosso-622-14, and Porco Clawn) and porcine fetal fibroblast (PFF) by COBRA assay. The methylation level is represented as a heatmap (left panel). Based on the methylation level determined by COBRA assay, differentiated cell or tissues (brain, liver, kidney, and PFF) and five porcine iPSC lines were clustered according to the similarity of their DNA methylation profiles at the 36 selected genes using the Euclidean distance (right panel). (c) Sum of pluripotency scores of the 36 genes for porcine naïve-like iPSCs and PFF. Depending on percentage difference in methylation levels between blastocysts and iPSCs, pluripotency scores for each gene were classified into five categories: < 20%, 20–40%, 40–60%, 60–80%, or > 80%, with pluripotency scores of 5, 4, 3, 2, or 1, respectively. Total scores of the 36-gene set are plotted. Statistical comparison was performed by Wilcoxon test. *P < 0.05; **P < 0.01. (d) Pluripotency scores of porcine iPSCs and PFF for each of the three gene-groups. Total scores of the genes belonging to each group (Oct3/4-targets, KSM-targets, and non-targets) are plotted.
We performed DNA methylation analyses of the 36 selected genes in five porcine iPSC lines; Porco Rosso-4, −6, −622-14, Epistem-like B9-2-5, and Porco Clawn. Epistem-like B9-2-5 was cultured in typical non-mouse ESC/iPSC media containing bFGF (Fujishiro et al., 2013), and its colony shape and gene expression patterns were similar to those of mouse epiblast stem cells (EpiSCs; Brons et al., 2007; Tesar et al., 2007). The other four cell lines were cultured with porcine LIF, and their colony shape was similar to that of mouse ESCs (Fujishiro et al., 2013). DNA methylation levels of the 36 genes were examined by combined bisulfite restriction analysis (COBRA) assay (Xiong and Laird, 1997) in iPSCs and porcine fetal fibroblast (PFF) used to produce the iPSC lines (Fig. 1b, left panel). Hierarchical clustering of DNA methylation status was performed on the basis of the 36-gene set for somatic cells/tissues and iPSC lines (Fig. 1b, right panel). Somatic tissues (brain, liver, and kidney) and PFF clustered together, whereas the five iPSC lines clustered separately, indicating that the DNA methylation profile of the 36-gene set could distinguish between differentiated cells/tissues and iPSCs.
Next, we determined a “Pluripotency score” based on comparison of methylation levels of the 36 genes between blastocysts consisting of pluripotent cells and the four naïve-like iPSC lines. Since Epistem-like B9-2-5 is distinguishable from naïve-like iPSC lines based on the mouse EpiSC-like colony shape and bFGF-dependent proliferation (Fujishiro et al., 2013), Epistem-like B9-2-5 was excluded in the following experiments. Scoring was performed for each gene, and the sum of the scores of the 36 genes was plotted (Fig. 1c). In this way, high pluripotency scores are awarded to iPSC lines with DNA methylation profiles close to those of pluripotent cells. Among the four iPSC lines examined, Porco Rosso-4 showed the highest pluripotency score, which was statistically significant when compared with PFF, Porco Rosso-622-14, and Porco Clawn. We further examined the pluripotency score depending on the gene groups (Oct3/4-targets, KSM-targets, or non-targets; Fig. 1d). The Oct3/4-target genes had higher pluripotency scores in the Porco Rosso-4 cell line (36) than the other three cell lines and PFF (22–27). This tendency was also observed for non-target genes. By contrast, the score of KSM-target genes in Porco Rosso-4 was similar to that of Porco Rosso-6. Thus, among the four iPSC lines examined, the DNA methylation profile of Porco Rosso-4 is most similar to that of pluripotent cells. This was consistent with the results of the hierarchical clustering analysis, where the Porco Rosso-4 line was separate from the Epistem-like B9-2-5 line.
We confirmed the DNA methylation status of six gene loci (Ctnnb1, Sall4, Tle4, Dll1, Enc1, and Nebl), whose DNA methylation level was clearly different among the examined iPSC lines (Fig. 1b, left panel), by bisulfite sequencing. All 6 gene loci were hypermethylated in PFF and liver, whereas hypomethylated status was observed in blastocysts (Fig. 2). DNA methylation levels of the Oct3/4-target genes, Ctnnb1 and Sall4, were 24 and 2%, respectively in the Porco Rosso-4 line, which had the highest pluripotency score among the four iPSC lines. However, Porco Rosso-622-14, which had the lowest pluripotency score, exhibited hypermethylation at the Ctnnb1 (65%) and Sall4 (61%) loci. This indicates that Ctnnb1 and Sall4 are highly demethylated in Porco Rosso-4 but not in Porco Rosso-622-14 cells. At the other four gene loci, partial demethylation was also observed in Porco Rosso-4 but not Porco Rosso-622-14. These bisulfite sequencing results confirm that, in Porco Rosso-4 iPSC line, DNA methylation patterns of the 6 genes, we analyzed changed to the expected direction as pluripotent cells, and Oct3/4-target genes, Sall4 and Ctnnb1, especially underwent demethylation within the entire sequenced regions in the majority of the cell population.
Figure 2.
DNA methylation status of EShypo-T-DMRs in iPSCs (Porco Rosso-4 and Porco Rosso-622-14) analyzed by sodium bisulfite sequencing. Open and closed circles indicate unmethylated and methylated CpG dinucleotides, respectively. Arrowheads indicate CpG sites analyzed by COBRA assay. The methylation level (%) was based on the methylated CpGs/all examined CpGs.
Contribution of Porcine iPSCs to the Inner Cell Mass (ICM) of Blastocysts
Established iPSC lines are intended for use in transplantation and complementation experiments. Considering that the naïve-like iPSC lines do not differ greatly in terms of morphology and marker gene expression (Fujishiro et al., 2013), it is more appropriate to select candidate iPSC lines using different indices. We next investigated contribution of the iPSC lines to the ICM of blastocysts using the aggregation method (Fig. 3a). We performed two independent experiments (Exps. 1 and 2), and found that Porco Rosso-4 cells contributed better to the ICM compared with Porco Rosso-622-14 in both experiments (Fig. 3b). In Exps. 1 and 2, the number of embryos that exhibited ICM contribution of iPSCs statistically differed between Porco Rosso-4 (14/57, 24.6%) and Porco Rosso-622-14 (2/51, 3.9%; Fig. 3c). The sum of pluripotency scores (105) for the three gene groups (Oct3/4-targets, KSM-targets, and non-targets) for Porco Rosso-4 was higher than that of Porco Rosso-622-14 (76), indicating that a higher pluripotency score coincides with higher efficiency of incorporation of iPSCs into the ICM. Thus, the pluripotency score based on the DNA methylation status of the 36 genes provides a feasible index for evaluating porcine iPSCs.
Figure 3.
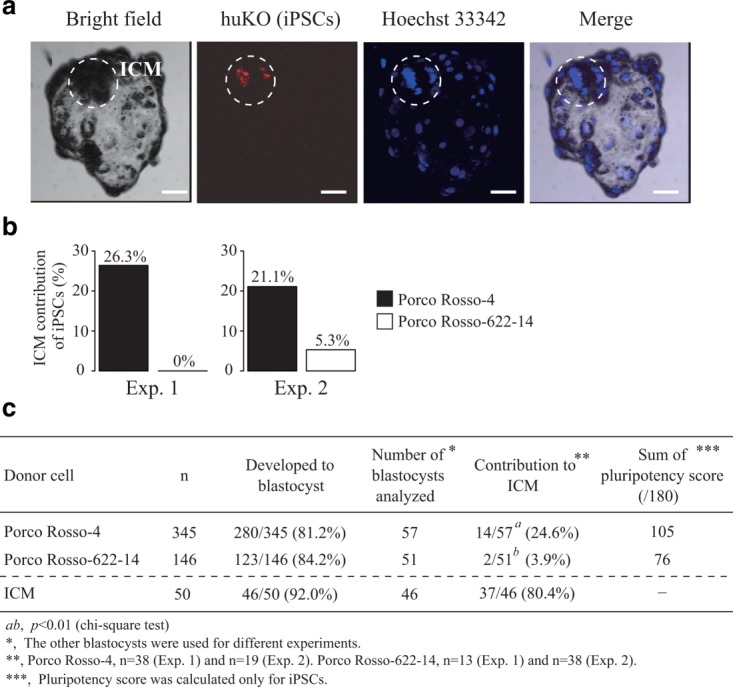
Contribution of iPSCs to the ICM of blastocysts. (a) Porcine iPSCs (Porco Rosso-4 and Porco Rosso-622-14) were aggregated with porcine parthenogenetic 4- to 8-cell-, or morula-stage embryos. After in vitro culture, contribution of iPSCs to blastocysts was analyzed by fluorescence of the transgene, humanized Kusabira-Orange (huKO). Scale bar = 50 µm. (b) The percentage of ICM contribution of Porco Rosso-4 and Porco Rosso-622-14. Aggregation experiments using porcine iPSCs (Porco Rosso-4 and Porco Rosso-622-14) were performed twice independently (Exps. 1 and 2). (c) Summary of the contribution of iPSCs to the ICM of blastocysts. Using the ICM cells dissociated from blastocysts as donor cells, most aggregated embryos developed into blastocysts, and donor ICM cells could contribute efficiently to the ICM of the host blastocyst, confirming the contribution of pluripotent cells to the ICM. Statistical comparison was performed by chi-square test.
DNA Methylation Analysis in huKO-Negative Cells
We previously reported that approximately 5% of the cell population in Porco Rosso-4 became negative for humanized Kusabira-Orange (huKO) fluorescence (huKO-negative cells), and their characteristics were closer to those of the expected pluripotent cells (Fujishiro et al., 2013). This implies that high-quality porcine iPSCs were enriched in huKO-negative fraction. To confirm whether the DNA methylation profile of EShypo-T-DMRs is a useful index for screening porcine iPSCs, we analyzed the DNA methylation status of huKO-negative cells. The huKO-negative cells in Porco Rosso-4 were collected by cell sorting, together with cells with high expression of huKO (huKO positive; Fig. 4a). EShypo-T-DMRs (Dll1 and Enc1) were analyzed by bisulfite sequencing, because approximately half of the sequenced clones exhibited obvious hypomethylation in Porco Rosso-4 (Figs. 2 and 4), suggesting that Porco Rosso-4 contains a certain proportion of the cells properly demethylated at these two loci. Based on the bisulfite sequence data (Fig. 4b), the percentage of the cells hypomethylated at these two loci in Porco Rosso-4 was calculated on the basis of the number of unmethylated CpGs in the sequenced clones before and after sorting (Fig. 4c). Before sorting, the percentages of hypomethylated cells at the Dll1 and Enc1 loci were estimated as 36% and 50%, respectively. In huKO-negative fraction, 67–80% of the cells were considered as hypomethylated, whereas the proportion of hypomethylated cells in the huKO-positive fraction were lower than that of the cells before sorting, indicating that huKO-negative cells were hypomethylated at these EShypo-T-DMRs as expected from the DNA methylation patterns in mouse ESCs. Thus, the DNA methylation profile based on the EShypo-T-DMRs reflects the characteristics of the expected cells.
Figure 4.
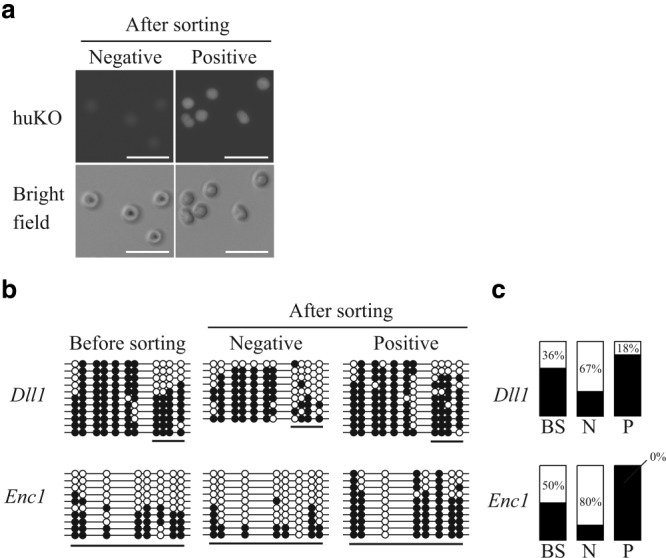
DNA methylation status of huKO-negative cells after sorting of Porco Rosso-4. (a) The huKO-negative and huKO-positive cells of Porco Rosso-4 iPSC line after sorting. Scale bar = 50 µm. (b) DNA methylation status of Dll1 and Enc1 before and after sorting (huKO-negative and huKO-positive). Open and closed circles indicate unmethylated and methylated CpG dinucleotides, respectively. (c) The percentage of (hypomethylated clones)/(all sequenced clones) in the underlined regions of Figure 4b. Hypomethylated clones were defined on the basis of the number of unmethylated CpGs underlined below the circles in each sequenced clone. At the Dll1 locus, the DNA methylation statuses of four CpGs were clearly different between the bulk Porco Rosso-4 line and PFF in Figure 2, whereas the other CpGs were almost fully methylated in all of the sequenced clones, indicating that the methylation statuses of the four selected CpGs in the underlined region were informative enough for evaluating the ratio of properly reprogrammed cells in huKO-negative and huKO-positive fractions. Thus, sequenced clones with three or more unmethylated CpGs in the four CpGs were designated as hypomethylated at the Dll1 locus. At the Enc1 locus, about half of the sequenced clones exhibited six or more unmethylated CpGs within the eight CpGs examined in the bulk Porco Rosso-4 line (Fig. 2). However, the other sequenced clones were almost fully methylated at the eight CpGs. Thus, sequenced clones exhibiting six or more unmethylated CpGs were designated as hypomethylated to strictly evaluate properly reprogrammed cells at the Enc1 locus. With respect to the other four EShypo-T-DMRs (Ctnnb1, Sall4, Tle4, and Nebl) analyzed in Figure 2, the DNA methylation statuses of hypomethylated CpGs in bulk Porco Rosso-4 cells did not differ between huKO-negative and huKO-positive cells (Supporting Information Fig. 2). Black and white bars indicate the ratio of hypermethylated and hypomethylated clones, respectively. BS, before sorting; N, huKO-negative cells; P, huKO-positive cells.
Improvement of Pluripotency Scores of Porcine iPSCs by SF + 2i Treatment
It is known that treatment with two signal-transduction inhibitors (2i) of PD0325901, an inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK), and CHIR99021, an inhibitor of GSK3β, is effective for establishment of a naïve state in iPSCs from rodent and human (Buehr et al., 2008; Hanna et al., 2009; Li et al., 2008). Additionally, 2i treatment has been shown to improve the characteristics of porcine iPSCs (Rodríguez et al., 2012). We examined whether the DNA methylation profile of the 36 selected genes is a useful index for evaluation of the changes in characteristics of porcine iPSCs after 2i treatment. We analyzed the DNA methylation status of Porco Clawn cultured in medium with FBS, or in serum-free (SF) or in SF+2i medium, by COBRA assay (Fig. 5a, left panel). Cells treated with 5-aza-2′-deoxycytidine (5-aza-dC), an inhibitor of DNA methyltransferase 1, were also analyzed for DNA methylation status.
Figure 5.
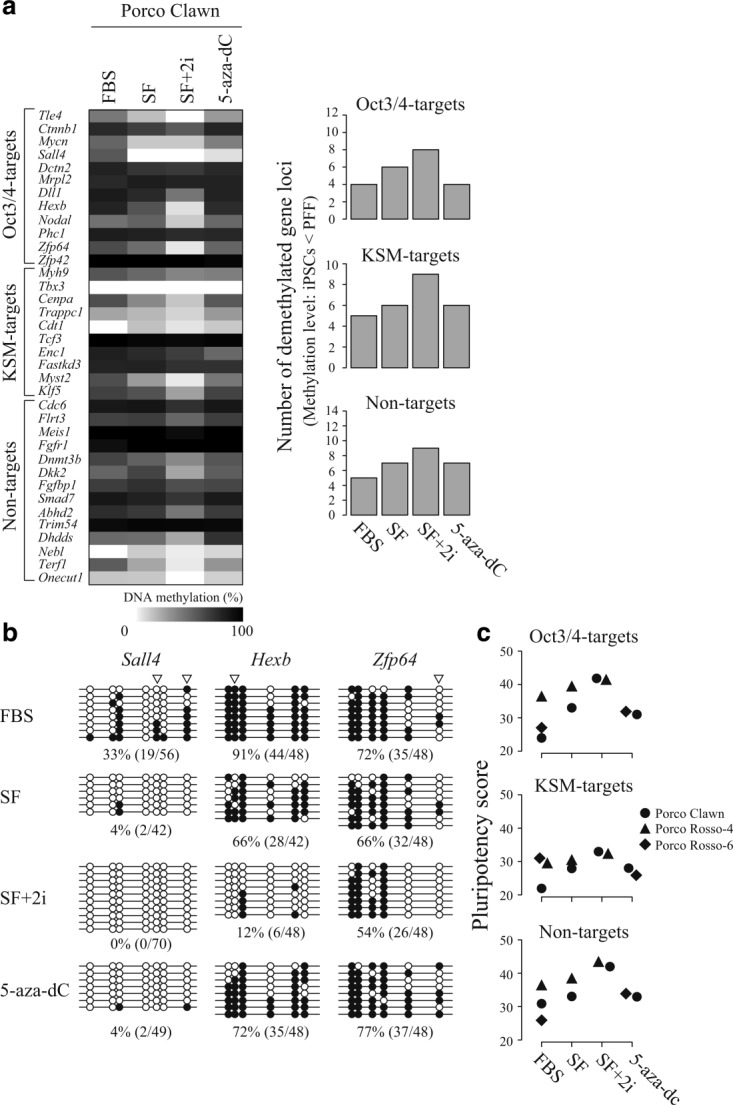
Improvement of DNA methylation status under different culture conditions. (a) DNA methylation status of Porco Clawn cells cultured in the presence of FBS, or in serum-free (SF), SF + 2i, or 5-aza-dC conditions was analyzed by COBRA assay; the methylation level is represented as a heatmap (left panel). The number of demethylated genes in each culture condition is shown in the right panel. (b) DNA methylation status of three Oct3/4-target genes (Sall4, Hexb, and Zfp64) in Porco Clawn cells cultured under different conditions analyzed by sodium bisulfite sequencing. Open and closed circles indicate unmethylated and methylated CpG dinucleotides, respectively. Arrowheads indicate CpG sites analyzed by COBRA assay. The methylation level (%) was calculated as methylated CpGs/all examined CpGs. (c) DNA methylation status in Porco Rosso-4 cells cultured under SF or SF + 2i conditions and in Porco Rosso-6 cultured in the presence of 5-aza-dC was analyzed by COBRA assay. Pluripotency scores of the iPSC lines cultured under each condition were plotted for the three gene-groups (Oct3/4-targets, KSM-targets, and non-targets).
In the course of mouse iPSC establishment, promoter regions of pluripotency-related genes, including EShypo-T-DMRs become demethylated (Okita et al., 2007; Sato et al., 2010; Takahashi and Yamanaka, 2006). In this study, we observed that porcine iPSCs cultured under SF conditions exhibited effective demethylation of the Oct3/4-target genes compared with FBS-cultured cells (Fig. 5a, right panel). In addition, the number of demethylated and reprogrammed genes was greatly increased under the SF+2i condition. These tendencies were also observed in the KSM-target and non-target gene groups, suggesting that SF+2i condition induced demethylation of the 36 selected genes. However, 5-aza-dC treatment was not effective, and the number of hypomethylated Oct3/4-target genes remained unchanged compared with FBS-cultured controls.
Improvement of the DNA methylation status of Porco Clawn cells by culture under SF+2i conditions was confirmed by sodium bisulfite sequencing (Fig. 5b). Sall4, Hexb, and Zfp64, all of which are Oct3/4-targets, were hypermethylated in the presence of FBS, whereas demethylation occurred under the SF+2i conditions. By contrast, demethylation at Hexb and Zfp64 was much less pronounced in the 5-aza-dC-treated cells (Fig. 5b). These results suggest that the SF+2i condition was the most effective of those tested for reprogramming of the 36-gene set. Demethylation also occurred in the SF+2i-treated cells at the Dll1 and Enc1 loci (Supporting Information Fig. 3a), whereas a portion of cells remained hypermethylated at the Dll1 locus (Supporting Information Fig. 3b). In addition, DNA methylation analysis revealed that a proportion of cells still needed to be reprogrammed at the Enc1 locus even after SF+2i treatment.
To examine whether SF+2i culture conditions are effective in improving pluripotency scores, we cultured Porco Clawn, in addition to several other iPSC lines, under the same conditions (FBS, SF, and SF+2i), and DNA methylation status was analyzed by COBRA assay (Fig. 5c). Of note, the pluripotency scores of all the three gene groups (Oct3/4-targets, KSM-targets, and non-targets) were increased in Porco Rosso-4 cells under SF+2i condition compared with FBS, or SF conditions. Thus, SF+2i treatment is effective for multiple porcine iPSC lines.
DISCUSSION
The porcine iPSCs analyzed in this study exhibited LIF-dependent self-renewal and a round morphology, similar to mouse ESCs (Fujishiro et al., 2013). The expression patterns of several pluripotency-related marker genes, including Oct3/4, were also similar among the iPSC lines. As it is intended that these iPSC lines will be used for applications such as chimera formation (Okita et al., 2007) and transplantation experiments (Kobayashi et al., 2010; Usui et al., 2012), it is important that they are able to contribute to the ICM of the blastocyst embryos. In this context, a simple index, other than morphology and marker gene expression, correlating with ICM contribution efficiency is required to allow pre-screening to determine which iPSC lines are appropriate for use in labor- and cost-intensive transplantation experiments. We previously reported that mouse iPSC lines with high chimera-forming ability (Aoi et al., 2008) have DNA methylation profiles close to those of ESCs (Sato et al., 2010). In this study, we focused on the DNA methylation profile of mouse EShypo-T-DMRs and determined a 36-gene set applicable to porcine iPSC lines. The pluripotency scores, based on the DNA methylation profile of the 36-gene set, were revealed to differ among iPSC lines that were indistinguishable by morphology or marker gene expression. Moreover, the iPSC line with the highest pluripotency score contributed to the ICM as assessed by a chimera formation experiment, whereas the iPSC line with the lowest score contributed much less efficiently to the ICM. Collectively, the DNA methylation profile of the 36-gene set derived from mouse EShypo-T-DMRs correlated with the ability to contribute to the ICM of blastocyst embryos.
In the course of mouse iPSC establishment, reprogramming for activation is important for genes such as endogenous Oct3/4 and target genes of Yamanaka factors (Oct3/4, Sox2, Klf4, and c-Myc) (Okita et al., 2007; Takahashi and Yamanaka, 2006). In this study, we categorized the 36-gene set into three groups (Oct3/4-targets, KSM-targets, and non-targets), and analyzed DNA methylation status in porcine iPSCs. Among the three groups, differences in pluripotency score were most apparent in the Oct3/4-target genes. The Oct3/4-target group contains genes that are important for the maintenance of pluripotency in stem cells. For example, Sall4 is an Oct3/4-target gene (Chen et al., 2008; Sakaki-Yumoto et al., 2006; Tsubooka et al., 2009) and was hypomethylated in the porcine iPSC line exhibiting both a high pluripotency score and efficient contribution to ICM. Differences in pluripotency scores among the examined iPSC lines were less significant in the KSM-target group than the Oct3/4-target group. Non-target genes can be thought of as downstream targets of the four Yamanaka factors, thereby non-target genes should be secondarily reprogrammed after activation of the primary targets of the Yamanaka factors. Thus, the pluripotency score of non-target genes was relatively high in iPSCs with high Oct3/4- and KSM-target scores, most likely indicating the activation of the primary target genes. Therefore, although pluripotency scores of Oct3/4-target genes can be considered as the most important index for evaluation of porcine iPSCs, the DNA methylation profiles of all three groups making up the 36 genes are required for evaluation with high accuracy.
For establishing mouse iPSCs, 2i treatment has been reported as effective for obtaining high-quality lines (Buehr et al., 2008; Hanna et al., 2009; Li et al., 2008). The 2i consists of MEK- and GSK3b-inhibitors, both of which prevent differentiation of mouse ESCs. In the porcine iPSC lines we examined, 2i-treatment effectively induced demethylation of the 36-gene set, with a consequent increase in pluripotency scores. Since an increased pluripotency score indicates cells with closer characteristics to those of pluripotent stem cells, such as mouse ESCs, the DNA methylation profile of the 36-gene set is useful for detection of improvements of porcine iPSC lines. In contrast, treatment of 5-aza-dC was less effective for improvement of pluripotency scores, implying that simple DNA demethylation by such compounds is insufficient. Taken together, DNA methylation profiling is useful for evaluation of characteristic changes of porcine iPSCs.
Reprogramming of endogenous pluripotency-related genes such as Oct3/4 is crucial for stably maintaining iPSCs. By contrast, the first report of mouse iPSCs (Fbx15 iPSCs) observed incomplete demethylation of the endogenous Oct3/4 region and the cells were not transmitted through the germline (Takahashi and Yamanaka, 2006), whereas superior germline-transmittable iPSC lines (Nanog iPSCs) showed complete demethylation of the endogenous pluripotency-related genes including Oct3/4 (Okita et al., 2007). We previously reported that the endogenous Oct3/4 promoter region was relatively hypermethylated in the same porcine iPSC lines that were examined in this study (Fujishiro et al., 2013). However, one iPSC line that we analyzed contained cells showing reprogrammed and hypomethylated status within the whole examined endogenous Oct3/4 locus, suggesting that a small population of the cells were activated at the endogenous Oct3/4 locus. In mouse ESCs, a 1.5-fold increase or two-fold decrease in endogenous Oct3/4 expression led to cellular differentiation (Niwa et al., 2000). Therefore, the appropriate expression level of endogenous Oct3/4 is important for maintenance of pluripotency. In the pig, however, no bona fide pluripotent stem cells, such as mouse ESCs, have been described to date; thus, the appropriate expression level of endogenous Oct3/4 remains to be determined. Based on the bisulfite sequencing data of endogenous Oct3/4 in our previous study (Fujishiro et al., 2013) and several Oct3/4-targets in this study, a proportion of the cells appear to be properly reprogrammed in the iPSC lines we examined. In addition, the finding that huKO-negative cells, which are likely closer to pluripotent cells (Fujishiro et al., 2013), exhibited hypomethylation of EShypo-T-DMRs compared with the bulk iPSC line further support the usefulness of DNA methylation profile. Thus, DNA methylation analysis can evaluate the ratio of properly reprogrammed cells in each iPSC line without reference cells such as ESCs in the case of mouse iPSCs.
The porcine iPSC lines examined in this study were likely heterozygous and contained both properly and improperly reprogrammed cells, even after SF/2i-treatment, as shown by the measurement of the DNA methylation patterns of several EShypo-T-DMRs. For aggregation experiments, one iPSC colony large enough to be picked up was dissociated into single cells and aggregated with early embryonic cells. Under the present culture conditions, properly reprogrammed cells such as huKO-negative cells, which we proved to have better characteristics as stem cells (Fujishiro et al., 2013), exhibited slower proliferation than improperly reprogrammed cells (data not shown). This implies that visible iPSC colonies, which can be used for aggregation experiments to analyze ICM contribution, contain substantial numbers of improperly reprogrammed cells due to higher proliferation potential than properly reprogrammed cells. Thus, the data demonstrating the ICM contribution of iPSCs in this study may have underestimated the ratio of properly reprogrammed cells that could be deduced from DNA methylation analysis. In this study, we identified several genes whose methylation profiles could be used to evaluate the ratio of properly reprogrammed cells. Thus, we will be able to identify appropriate culture conditions for the maintenance of properly reprogrammed cells by using the DNA methylation profiles of these genes.
In this study, we focused on the DNA methylation profile of the 36-gene set derived from EShypo-T-DMRs and established a new index for evaluation of porcine iPSCs correlating with their efficient contribution to the ICM of blastocysts. We provide proof-of-principle of the concept that mouse EShypo-T-DMR information is applicable to evaluate porcine iPSC lines. The gene set we used in this study is effective on validation of stemness under in vitro culture condition and in blastocyst stage embryos as one aspect of pluripotency. However, the other aspect of pluripotency to have broad lineage potential is also important but cannot be assessed by the gene set used in this study. We previously identified differentiated-tissue-specific hypomethylated loci (Tissuehypo-T-DMRs), which are related to certain specific somatic cell lineages and are hypermethylated in mouse ESCs (Sato et al., 2010). By selecting another porcine gene set from Tissuehypo-T-DMRs in addition to the 36-gene set examined in this study, we will be able to perform more strict evaluation of porcine iPSCs in terms of pluripotency of both stemness and broad lineage potential. Since our previous findings indicated that hundreds of mouse EShypo- and Tissuehypo-T-DMRs could be used for evaluation of mouse iPSCs, we anticipate deriving further information about EShypo- and Tissuehypo-T-DMRs in multiple porcine iPSC lines with higher accuracy using next generation sequencer to perform ultra-deep bisulfite sequencing.
In conclusion, information regarding DNA methylation derived from mouse EShypo-T-DMRs is a feasible index for evaluation of porcine iPSCs as a pre-screening tool, distinct from morphology and marker gene expression analysis. Evaluation of porcine iPSC lines by DNA methylation analysis is simple and time/cost-saving. The concept of evaluation of iPSCs by DNA methylation profile is readily applicable to other mammalian cells, including human iPSCs/ESCs, which cannot be evaluated by chimera formation for ethical reasons.
METHODS
Animal Care
All of the animal experiments in this study were approved by the Institutional Animal Care and Use Committee of Meiji University (IACUC-11-1).
Chemicals
Chemicals were purchased from the Sigma Chemical Co. (St. Louis, MO), unless otherwise indicated.
Porcine iPSCs
Establishment of porcine iPSCs was described previously (Fujishiro et al., 2013). Porcine iPSC lines, Porco Rosso-4, −6, −622-14, and Epistem-like B9-2-5, expressing humanized Kusabira-Orange (huKO) were derived from PFF of huKO-transgenic pigs (Matsunari et al., 2008). Porco Clawn iPSC line was derived from PFF of Clawn miniature pigs (Japan Farm, Kagoshima, Japan). All the iPSC lines were maintained in knockout DMEM medium (Invitrogen, Rockville, MD) containing 15% fetal bovine serum (FBS, Invitrogen) or knockout serum replacement (KSR, Invitrogen), 1% NEAA (Invitrogen), 1% glutamax-L (Invitrogen), 100 µM 2-mercaptoethanol (Invitrogen), 50 U/ml penicillin, 50 µg/ml streptomycin, 10 µM forskolin (Biomol, Farmingdale, NY), and 0.5% porcine LIF with or without 5 µM 5-aza-2′-deoxycytidine (5-aza-dC) for 10 days and two signal-transduction inhibitors (2i) of 0.5 µM PD0325901 and 3 µM CHIR99021 for three weeks on MMC-treated STO cells. huKO-negative and huKO-positive cells of Porco Rosso-4 were sorted by BD FACS Aria (Becton Dickinson, NJ).
COBRA Assay and Sodium Bisulfite Genomic Sequencing
DNA methylation analysis by COBRA assay was performed based on our previous results (Sato et al., 2010) focusing on 58 genes that were hypomethylated in mouse ESCs. Genomic DNA was purified using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), and digested with a restriction enzyme Hind III (Takara Bio, Shiga, Japan). Digested genomic DNA was purified with a QIAquick Gel Extraction Kit (Qiagen), and bisulfite reactions were performed using the EZ DNA Methylation-Direct Kit (Zymo Research, Irvine, CA). Bisulfite-treated DNA was amplified with BioTaq HS DNA polymerase (Bioline, London, UK) using specific gene primers (Supporting Information Table 1). Primer sequences are based on the porcine genome assembly, Sscrofa9.2 (November 2009 build) from the UCSC genome database. PCR was performed under the following conditions: 95°C, 10 min; 40 cycles of 95°C, 30 s; 60°C, 30 s; 72°C, 1 min; final extension 72°C, 2 min. Amplified PCR products were digested with Hpy CH4 IV (New England BioLabs, Beverly, MA) at 37°C for 3 h, or Taq I (Takara Bio) at 65°C for 3 h and analyzed by electrophoresis. DNA methylation levels were calculated using the formula: estimated methylation degree (%) = 100 × IC/(IC + IUC), where IC and IUC represent intensities of digested and undigested bands, respectively. The intensities of bands were determined using ImageJ software provided by the National Institutes of Health (http://rsb.info.nih.gov/ij/). COBRA assay was independently performed twice for all samples. PCR products were cloned into the pGEM T-Easy vector (Promega, Madison, WI), and 10 to 16 clones sequenced to determine DNA methylation status.
Preparation of Porcine Parthenogenetic Embryos
Porcine parthenogenetic embryos were generated as reported previously (Matsunari et al., 2008). Briefly, porcine ovaries were collected at local abattoirs. In vitro maturated oocytes with expanded cumulus cells were treated with 1 mg/ml of hyaluronidase dissolved in Tyrode’s lactose medium containing 10 mM HEPES and 0.3% (w/v) polyvinylpyrrolidone (HEPES-TL-PVP), and separated from the cumulus cells by gentle pipetting. Oocytes with evenly granulated ooplasm and an extruded first polar body were selected for subsequent experiments. Oocytes were transferred to an activation solution consisting of 0.3 M mannitol (Nacalai Tesque, Kyoto, Japan), 50 µM CaCl2, 100 µM MgCl2, and 0.01% (w/v) PVA, and activated by applying a single direct current pulse (150 V/mm, 100 µsec) using an electrical pulsing machine (LF201; Nepa Gene, Chiba, Japan). Activated oocytes were cultured and treated with 5 µg/ml cytochalasin B for 3 h to suppress extrusion of the second polar body followed by in vitro culture for up to 4 days to obtain parthenogenetic host embryos at 4 to 8-cell- and morula-stages. In vitro culture of parthenogenetic embryos was performed in porcine zygote medium-5 (PZM-5; Research Institute for the Functional Peptides, Yamagata, Japan) under paraffin oil (Kanto Chemical, Tokyo, Japan) in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2 at 38.5°C.
Production of Aggregated Embryos
Parthenogenetic embryos at 4- to 8-cell (day 3) or morula (day 4) stages were used as host embryos for aggregation of iPSCs. Host 4- to 8-cell embryos or morulae were decompacted by incubation in Ca2+/Mg2+-free Dulbecco phosphate-buffered saline (DPBS; Nissui, Tokyo, Japan) containing 0.1 mM EDTA-2Na and 0.01% (w/v) PVA for 15–20 min, followed by removal of the zona pellucidae by digestion with 0.25% (w/v) pronase in DPBS. Small clumps (∼30 cells) of iPSCs were isolated from the feeder layer and kept in a drop of 21 mM Hepes-buffered MEM (Invitrogen) supplemented with 5 mg/ml BSA (Sigma) (MEM-HEPES). Aggregation of iPSC clumps and host embryos was carried out using the micro-well method (Nagy and Rossant, 1993). Each clump of iPSCs was aggregated with host blastomeres isolated from two parthenogenetic embryos at 4- to 8-cell or morula stages.
Some of the parthenogenetic host embryos were aggregated with ICM clumps isolated from parthenogenetic blastocysts on day 6 by immunosurgery as described previously (Nagashima et al., 2004). Isolated ICM cells were labeled with fluorescent carbocyanine dye (DiI, Takara Bio) for 30 min, followed by thorough washing with MEM-HEPES.
Embryos produced by the aggregation method were cultured for 48–72 h in PZM-5. Blastocysts at day 6 were observed by confocal microscopy to analyze the incorporation of the donor cells into the ICM. Images of blastocysts placed in a drop of DPBS containing 5 µg/ml Hoechst 33342 were taken using a confocal fluorescence microscope (FV-1000; Olympus, Tokyo, Japan).
Acknowledgments
The authors acknowledge Dr. Masaki Nagaya for helpful discussions on this manuscript and Mr. Taisuke Matsuda for technical help.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure 1 Supplementary Information
Figure 2 Supplementary Information
Figure 3 Supplementary Information
Table 1 Supplementary Information
Literature cited
- Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, McLay R, Hall J, Ying QL, Smith A. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T, Telugu BP, Alexenko AP, Sachdev S, Sinha S, Roberts RM. Derivation of induced pluripotent stem cells from pig somatic cells. Proc Natl Acad Sci USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishiro SH, Nakano K, Mizukami Y, Azami T, Arai Y, Matsunari H, Ishino R, Nishimura T, Watanabe M, Abe T, Furukawa Y, Umeyama K, Yamanaka S, Ema M, Nagashima H, Hanazono Y. Generation of naive-like porcine-induced pluripotent stem cells capable of contributing to embryonic and fetal development. Stem Cells Dev. 2013;22:473–482. doi: 10.1089/scd.2012.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golob JL, Paige SL, Muskheli V, Pabon L, Murry CE. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev Dyn. 2008;237:1389–1398. doi: 10.1002/dvdy.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Mitalipova M, Cheng AW, Cassady JP, Staerk J, Carey BW, Lengner CJ, Foreman R, Love J, Gao Q, Kim J, Jaenisch R. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Nishino K, Ko YG, Hattori N, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Ikegami K, Ohgane J, Tanaka S, Yagi S, Shiota K. Interplay between DNA methylation, histone modification and chromatin remodeling in stem cells and during development. Int J Dev Biol. 2009;53:203–214. doi: 10.1387/ijdb.082741ki. [DOI] [PubMed] [Google Scholar]
- Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S, Takanashi M, Funayama M, Hirayama A, Soga T, Hishiki T, Suematsu M, Yagi T, Ito D, Kosakai A, Hayashi K, Shouji M, Nakanishi A, Suzuki N, Mizuno Y, Mizushima N, Amagai M, Uchiyama Y, Mochizuki H, Hattori N, Okano H. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Yamanaka S. The use of induced pluripotent stem cells in drug development. Clin Pharmacol Ther. 2011;89:655–661. doi: 10.1038/clpt.2011.38. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee YS, Usui J, Knisely AS, Hirabayashi M, Nakauchi H. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying QL. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Beck S, Bulyk ML, Farnham P, Hattori N, Henikoff S, Liu XS, Okumura K, Shiota K, Ushijima T, Greally JM. Applying whole-genome studies of epigenetic regulation to study human disease. Cytogenet Genome Res. 2006;114:1–15. doi: 10.1159/000091922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney JK. Advances in swine biomedical model genomics. Int J Biol Sci. 2007;3:179–184. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunari H, Onodera M, Tada N, Mochizuki H, Karasawa S, Haruyama E, Nakayama N, Saito H, Ueno S, Kurome M, Miyawaki A, Nagashima H. Transgenic-cloned pigs systemically expressing red fluorescent protein, Kusabira-Orange. Cloning Stem Cells. 2008;10:313–323. doi: 10.1089/clo.2008.0024. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montserrat N, Bahima EG, Batlle L, Häfner S, Rodrigues AM, González F, Izpisúa Belmonte JC. Generation of pig iPS cells: A model for cell therapy. J Cardiovasc Transl Res. 2011;4:121–130. doi: 10.1007/s12265-010-9233-3. [DOI] [PubMed] [Google Scholar]
- Nagashima H, Giannakis C, Ashman RJ, Nottle MB. Sex differentiation and germ cell production in chimeric pigs produced by inner cell mass injection into blastocysts. Biol Reprod. 2004;70:702–707. doi: 10.1095/biolreprod.103.022681. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J. Production of completely ES cell-derived fetuses. In: Joyner AL, editor. Gene targeting: a practical approch. UK: IRL Press; 1993. pp. 147–179. [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–2207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petters RM. Transgenic livestock as genetic models of human disease. Reprod Fertil Dev. 1994;6:643–645. doi: 10.1071/rd9940643. [DOI] [PubMed] [Google Scholar]
- Prather RS, Hawley RJ, Carter DB, Lai L, Greenstein JL. Transgenic swine for biomedicine and agriculture. Theriogenology. 2003;59:115–123. doi: 10.1016/s0093-691x(02)01263-3. [DOI] [PubMed] [Google Scholar]
- Rodríguez A, Allegrucci C, Alberio R. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS One. 2012;7:e49079. doi: 10.1371/journal.pone.0049079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Suzuki M, Abe T, Hosoyama T, Himeno E, Tanaka S, Greally JM, Hattori N, Yagi S, Shiota K. Cell type-specific methylation profiles occurring disproportionately in CpG-less regions that delineate developmental similarity. Genes Cells. 2007;10:1123–1132. doi: 10.1111/j.1365-2443.2007.01120.x. [DOI] [PubMed] [Google Scholar]
- Sato S, Yagi S, Arai Y, Hirabayashi K, Hattori N, Iwatani M, Okita K, Ohgane J, Tanaka S, Wakayama T, Yamanaka S, Shiota K. Genome-wide DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) residing in mouse pluripotent stem cells. Genes Cells. 2010;15:607–618. doi: 10.1111/j.1365-2443.2010.01404.x. [DOI] [PubMed] [Google Scholar]
- Shiota K. DNA methylation profiles of CpG islands for cellular differentiation and development in mammals. Cytogenet Genome Res. 2004;105:325–334. doi: 10.1159/000078205. [DOI] [PubMed] [Google Scholar]
- Shiota K, Kogo Y, Ohgane J, Imamura T, Urano A, Nishino K, Tanaka S, Hattori N. Epigenetic marks by DNA methylation specific to stem, germ and somatic cells in mice. Genes Cells. 2002;7:961–969. doi: 10.1046/j.1365-2443.2002.00574.x. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14:683–694. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- Usui J, Kobayashi T, Yamaguchi T, Knisely AS, Nishinakamura R, Nakauchi H. Generation of kidney from pluripotent stem cells via blastocyst complementation. Am J Pathol. 2012;180:2417–2426. doi: 10.1016/j.ajpath.2012.03.007. [DOI] [PubMed] [Google Scholar]
- van der Spoel TI, Jansen of Lorkeers SJ, Agostoni P, van Belle E, Gyöngyösi M, Sluijter JP, Cramer MJ, Doevendans PA, Chamuleau SA. Human relevance of pre-clinical studies in stem cell therapy: systematic review and meta-analysis of large animal models of ischaemic heart disease. Cardiovasc Res. 2011;91:649–658. doi: 10.1093/cvr/cvr113. [DOI] [PubMed] [Google Scholar]
- West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;8:1211–1220. doi: 10.1089/scd.2009.0458. [DOI] [PubMed] [Google Scholar]
- West FD, Uhl EW, Liu Y, Stowe H, Lu Y, Yu P, Gallegos-Cardenas A, Pratt SL, Stice SL. Chimeric pigs produced from induced pluripotent stem cells demonstrate germline transmission and no evidence of tumor formation in young pigs. Stem Cells. 2011;10:1640–1643. doi: 10.1002/stem.713. [DOI] [PubMed] [Google Scholar]
- Wu Z, Chen J, Ren J, Bao L, Liao J, Cui C, Rao L, Li H, Gu Y, Dai H, Zhu H, Teng X, Cheng L, Xiao L. Generation of pig induced pluripotent stem cells with a drug-inducible system. J Mol Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- Xu J, Pope SD, Jazirehi AR, Attema JL, Papathanasiou P, Watts JA, Zaret KS, Weissman IL, Smale ST. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:12377–12382. doi: 10.1073/pnas.0704579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Hattori N, Ohgane J, Tanaka S, Liu XS, Shiota K. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MT, Prather RS. The multi-potentiality of skin-derived stem cells in pigs. Theriogenology. 2011;75:1372–1380. doi: 10.1016/j.theriogenology.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang W, Liu Y, Fernandez de Castro J, Ezashi T, Telugu BP, Roberts RM, Kaplan HJ, Dean DC. Differentiation of induced pluripotent stem cells of swine into rod photoreceptors and their integration into the retina. Stem Cells. 2011;29:972–980. doi: 10.1002/stem.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1 Supplementary Information
Figure 2 Supplementary Information
Figure 3 Supplementary Information
Table 1 Supplementary Information



