Abstract
Translocations are an increasingly common tool in conservation. The maintenance of genetic diversity through translocation is critical for both the short- and long-term persistence of populations and species. However, the relative spatio-temporal impacts of translocations on neutral and functional genetic diversity, and how this affects genetic structure among the conserved populations overall, have received little investigation. We compared the impact of translocating different numbers of founders on both microsatellite and major histocompatibility complex (MHC) class I diversity over a 23-year period in the Seychelles warbler (Acrocephalus sechellensis). We found low and stable microsatellite and MHC diversity in the source population and evidence for only a limited loss of either type of diversity in the four new populations. However, we found evidence of significant, but low to moderate, genetic differentiation between populations, with those populations established with fewer founders clustering separately. Stochastic genetic capture (as opposed to subsequent drift) was the main determinant of translocated population diversity. Furthermore, a strong correlation between microsatellite and MHC differentiation suggested that neutral processes outweighed selection in shaping MHC diversity in the new populations. These data provide important insights into how to optimize the use of translocation as a conservation tool.
Keywords: conservation, differentiation, drift, genetic capture, genetic diversity, major histocompatibility complex, re-introduction
Introduction
The translocation of populations for species conservation and ecosystem restoration is an increasingly common conservation tool (Moritz 1999; Ewen et al. 2012; IUCN/SSC 2013). However, translocation success rates have been poor across many taxa, often for unknown reasons (Griffith et al. 1989; Wolf et al. 1996; Godefroid et al. 2011). Close monitoring of translocated populations is crucial if we are to understand the drivers of success and failure (Miller et al. 1999; Allendorf & Luikart 2007), but until recently such monitoring has often been absent or inadequate (Fischer & Lindenmayer 2000; Armstrong & Seddon 2008). Translocations have been particularly widely used in the conservation of oceanic island species. Such islands possess some of the most globally threatened and evolutionarily distinct taxa (Kier et al. 2009; Lee & Jetz 2011), contributing disproportionally to global biodiversity (Whittaker 1998). Further, the small size of many oceanic islands means that eradication of alien predators and restoration of native biota are achievable goals (Nogales et al. 2004; Howald et al. 2007), enhancing the prospects of successful translocations.
Many biotic and abiotic factors can influence translocation success, including genetic diversity (Sarrazin & Barbault 1996; Wolf et al. 1998; Seddon et al. 2007; Groombridge et al. 2012). Maintaining genetic diversity is one of the IUCN's three conservation priorities, and its role in species extinction risk is now largely accepted (Spielman et al. 2004; Frankham 2005; O'Grady et al. 2006). However, translocations typically involve small source populations and limited numbers of founders, resulting in founder effects (e.g. Taylor & Jamieson 2008; Cardoso et al. 2009). Furthermore, genetic drift is stronger in smaller populations, will erode genetic diversity (Kimura 1983) and will cause interpopulation divergence if populations are isolated (e.g. Brekke et al. 2011). Small, isolated populations also suffer inbreeding (Frankham et al. 2002) and inbreeding depression (Charlesworth & Charlesworth 1987). Purging of the mutational load may alleviate this over time (Crnokrak & Barrett 2002), but its effectiveness in wild populations is uncertain (Boakes et al. 2007). In the longer term, the loss of genetic diversity also reduces a population's ability to adapt to future challenges, that is its evolutionary potential (Franklin 1980; Franklin & Frankham 1998). Translocations can, therefore, have considerable long-lasting genetic impacts on populations and hence on entire species (Biebach & Keller 2009), and there is a clear need to integrate genetic considerations into translocation programmes (Jamieson & Lacy 2012).
Most studies on the genetic impacts of translocation investigate diversity at putatively neutral markers such as microsatellites (e.g. Larson et al. 2002; Taylor & Jamieson 2008; Brekke et al. 2011). Such variation may not correlate well with the genetic diversity important to the evolutionary potential of the new populations (Reed & Frankham 2001; Aguilar et al. 2004). It may be more relevant to directly assess variation at important functional loci, such as the major histocompatibility complex (MHC). These highly polymorphic loci encode molecules central to antigen recognition in the adaptive immune response of vertebrates (Hughes & Yeager 1998). This key role has made the MHC an attractive candidate for studying functional diversity in and among populations (reviewed in Bernatchez & Landry 2003; Sommer 2005; Spurgin & Richardson 2010). A few studies have compared the impact of translocation on neutral and functional diversity in wild populations (see Sutton et al. 2011; Bauer et al. 2013; Monzón-Argüello et al. 2013). Perhaps the best example to date is a study by Miller and Lambert (2004) which showed that drift outweighed selection in shaping MHC diversity in re-introduced populations of two bird species in New Zealand. However, the reasons for differences in loss of neutral and functional variation during the process of translocation itself remain unclear.
How many individuals to translocate is a difficult question to answer, although it is a central concern for conservationists. Guidelines recommend that ‘adequate’ numbers of founders should be taken (IUCN/SSC 2013), although this is rarely quantified (Tracy et al. 2011). The goal of translocation is to establish a ‘viable’ population (IUCN/SSC 2013), often defined by the ‘minimum viable population’ concept, with estimates ranging from 50 individuals for short-term persistence up to 5000 individuals to maintain evolutionary potential (Franklin 1980; Lande 1995; Franklin & Frankham 1998; Willi et al. 2006). However, translocations often use <50 founders owing to ecological, logistic and economic constraints (Komdeur 1994; Cardoso et al. 2009; Jamieson 2011; Tracy et al. 2011). Genetic data have only recently been incorporated into estimates of what constitutes ‘adequate’ founder sizes, even though the genetic consequences of small population size are directly relevant. Weeks et al. (2011) introduced the concept of ‘genetic capture’, where the minimum number of individuals to translocate is determined by the capture of ≥95% of source population genetic diversity. Models have also been developed to predict allele retention accounting for post-translocation parameters such as survival, population growth, carrying capacity, overlapping generations and/or specific mating systems (Brekke et al. 2011; Tracy et al. 2011; Weiser et al. 2012). Modelling approaches provide vital benchmarks for conservationists, but there are drawbacks. Importantly, these methods are largely based on the loss of neutral variation in the founding population. Drift may disproportionately reduce variation at functional loci if they are highly variable due to a history of balancing selection (Sutton et al. 2011). Therefore, such methods are only useful for functional diversity if the loci are selectively neutral at small population sizes (Tracy et al. 2011). Complementing modelling approaches with replicated empirical data on neutral and functional variation, in systems with adequate pre- and post-translocation sampling, should help inform best practice for future translocation programmes.
Here, we present data from a study spanning 23 years and involving four translocations of different founder sizes of the Seychelles warbler (Acrocephalus sechellensis), an isolated island species. The objective was to compare the spatio-temporal impact of translocation on neutral and functional diversity within and across populations. First, we characterize neutral and functional genetic diversity in the source population and how that has changed over time. Second, we compare predicted vs. observed levels of genetic capture across different founder sizes. Third, we quantify genetic diversity in the four new translocated populations, including how that compares with variation in the source population and if it changes over time. Fourth, we quantify levels of genetic differentiation among all the Seychelles warbler populations now in existence. Fifth, we estimate the effective population size for each population. We discuss the implications of our findings for the use of translocation in the conservation of this and other endangered species.
Materials and methods
Study populations
By the mid-20th century, the Seychelles warbler was on the verge of extinction due to habitat destruction and the introduction of invasive predators, with the last population of 26–50 individuals (Crook 1960; Spurgin et al. in review) existing on Cousin Island (4°20′S, 55°40′E, 0.29 km²). This population has been under intense study since 1986 (>96% individuals ringed since 1997, Richardson et al. 2001) as part of a long-term project (Komdeur 1992; Richardson et al. 2007; Barrett et al. 2013). Each year, birds are caught and unringed individuals are identified with a unique combination of coloured leg rings (herein referred to as catch-year samples). A ca. 25 μl blood sample is taken from every bird and stored in absolute ethanol. Four translocations have been undertaken as part of the species conservation plan (Richardson 2001), with Cousin as the source for each (Fig. S1, Supporting information). In brief, 29 birds were translocated to both Aride (4°12′S, 55°40′E, 0.68 km²) and Cousine (4°21′S, 55°39′E, 0.25 km²) in 1988 and 1990, respectively (Komdeur 1994). In 2004, 58 birds were translocated to Denis (3°48′S, 55°40′E, 1.42 km², Richardson et al. 2006) and in 2011, 59 birds to Frégate (4°35′S, 55°56′E, 2.19 km², Wright & Richardson 2012). For each translocation, founders were selected from across the whole source population based on body condition, age (avoiding very young or old birds), breeding experience and sex, to maximize the chances of population establishment. The translocations were undertaken blind in regard to genetic characteristics, although translocating known first-order relatives was avoided. Movement between populations is virtually absent, with rates of inter-island dispersal at only 0.1% (Komdeur et al. 2004). Complete sampling of the founding populations was conducted for Denis and Frégate, but the Aride and Cousine translocations were undertaken prior to routine blood sampling and so few founders were sampled (4 of 29 in each translocation). The following catch-year samples were used in this study: Cousin 1993, 2005, 2011; Aride 1993, 2005, 2011; Cousine 1997, 2005, 2011; Denis 2004, 2011; Frégate; 2011 (sample sizes in Table1).
Table 1.
Catch-year sampling regime and marker summary statistics for microsatellite and major histocompatibility complex (MHC) data across the five Seychelles warbler populations. Cousin is the source of all other populations. Abbreviations are number of individuals scored (N), expected heterozygosity (HE), total number of MHC alleles in the population (Alleles), mean MHC alleles per individual (MHC/ind), nucleotide diversity (Pi) and an index of allelic richness (Theta K)
| Island | Catch year | Microsatellites | MHC | ||||||
|---|---|---|---|---|---|---|---|---|---|
| N | HE | Allelic richness | N | Alleles | MHC/ind | Pi | Theta K | ||
| Cousin | 1993 | 49 | 0.49 | 2.96 | 52 | 10 | 3.94 | 19.76 | 2.05 |
| 2005 | 169 | 0.48 | 2.89 | 156 | 10 | 4.61 | 19.71 | 1.54 | |
| 2011 | 163 | 0.48 | 2.91 | 91 | 10 | 4.71 | 19.50 | 1.71 | |
| Aride | 1993 | 27 | 0.49 | 2.82 | 27 | 10 | 4.85 | 18.84 | 2.34 |
| 2005 | 30 | 0.47 | 2.84 | 30 | 10 | 4.67 | 18.68 | 2.30 | |
| 2011 | 29 | 0.45 | 2.81 | 29 | 10 | 4.93 | 18.97 | 2.28 | |
| Cousine | 1997 | 24 | 0.45 | 2.60 | 23 | 10 | 5.09 | 19.70 | 2.43 |
| 2005 | 29 | 0.42 | 2.59 | 29 | 9 | 4.03 | 19.13 | 2.10 | |
| 2011 | 30 | 0.45 | 2.70 | 30 | 10 | 4.17 | 20.16 | 2.05 | |
| Denis | 2004 | 58 | 0.50 | 2.95 | 56 | 10 | 4.77 | 19.69 | 1.91 |
| 2011 | 35 | 0.48 | 2.85 | 30 | 10 | 4.97 | 19.25 | 2.25 | |
| Frégate | 2011 | 59 | 0.49 | 2.90 | 58 | 10 | 4.55 | 19.52 | 1.92 |
Molecular and statistical analyses
Statistical analyses were performed using r version 2.15 (R Development Core Team 2012), unless stated. Samples were genotyped at 30 microsatellite loci following Spurgin et al. (in review). Rare alleles (frequency <0.01) were verified by two or more independent PCRs from different samples. We tested for deviations from Hardy–Weinberg equilibrium and linkage disequilibrium between loci using genepop version 4.1 (Raymond & Rousset 1995b) and for null alleles using cervus version 3.0 (Marshall et al. 1998). Genetic diversity in each catch-year sample on each island was quantified by calculating expected heterozygosity (HE) using arlequin version 3.5 (Excoffier & Lischer 2010). Allelic richness was quantified using a rarefaction approach in hp-rare version 1 (Kalinowski 2005) as sample size differences can bias the estimations (Leberg 2002).
Variation at exon 3 of MHC class I, which codes for the peptide-binding region involved in antigen recognition (Hughes & Yeager 1998), was screened using reference strand-mediated conformation analysis (RSCA) using the primers from Richardson & Westerdahl (2003), following the method of Worley et al. (2008). Each segregating RSCA variant corresponded to a unique 255-bp amino acid coding sequence (hereafter termed ‘allele’ for simplicity, Richardson & Westerdahl 2003). Ten MHC class I alleles have been detected in the Seychelles warbler, with individuals possessing 2–8 alleles each, suggesting that at least four class I loci are amplified (Richardson & Westerdahl 2003). Our primers were sited within exon 3. Consequently, we were not able to screen all the variation within this exon, and it is possible that some additional polymorphism exists (e.g. Llaurens et al. 2012). However, the amplicon includes all the codons of the peptide-binding region where we expect most variation to be found (Hughes & Yeager 1998). Further, to minimize the effect of this issue, we employed two primer sets which vary at the 3′ end where a known polymorphism occurs (see Richardson & Westerdahl 2003). Finally, any missed variation would not affect the main results or conclusions of the present study which seeks to address how the variation we have screened is captured across translocated populations. It is impossible at present to identify locus zygosity, due to homogeneity of alleles between multiple, duplicated loci within the MHC (Westerdahl 2007). Instead, we measured MHC diversity by calculating the total number of different alleles in each catch-year sample, the mean number of alleles per individual (MHC/ind), nucleotide diversity (Pi) and theta K (allelic richness) in arlequin by entering the nucleotide sequence and number of individuals carrying each allele as haplotype data, following Miller et al. (2010). This approach may overestimate rare alleles and underestimate common ones but is the best available (Ekblom et al. 2007).
Changes in genetic diversity over time were analysed using a randomization approach (Manly 1997). For each diversity measure, the data from the earliest and latest years of each population were pooled and randomly resampled with replacement 100 000 times. The P value was calculated as the proportion of times the difference between the means of the resampled data sets were equal to or greater than the observed difference between earliest and latest years. Differences in microsatellite variation between years were also tested using a global differentiation exact test (Raymond & Rousset 1995a) in arlequin, with a 30 000 step Markov chain. Seychelles warblers can live up to 17 years (Brouwer et al. 2010) and are routinely sampled multiple times throughout their life as part of this long-term study system (e.g. Barrett et al. 2013). To check for any effect of including the same individuals across catch-year samples, diversity measures were also compared for lay-year (year of hatching, estimated at first ringing) cohorts on Cousin. Patterns of diversity across lay years and catch years were qualitatively the same (data not shown) so only catch-year data are reported. This enabled us to use the largest sample sizes available and hence most accurate estimations of diversity for each year in our analyses. Differences in diversity between populations were assessed using the same randomization approach. As we observed no differences in variation within islands over time (see Study populations), for between-island comparisons, samples were pooled for each island to improve the accuracy of the rarefaction measures (i.e. allelic richness). Differences in mean MHC/ind between islands were tested using a Kruskal–Wallis test.
To model the expected genetic capture at each translocation, we constructed rarefaction curves of observed allelic richness using the 2011 Cousin sample (microsatellites; n = 163, MHC; n = 91) with 1000 repetitions. As we did not have source population diversity data prior to every translocation and diversity did not change over time within islands (see Study populations), we used the 2011 sample as a proxy for pretranslocation diversity at all four genetic capture events. A proportionally greater loss of genetic diversity is expected from microsatellite loci of higher initial diversity (Hedrick 1999), so microsatellite genetic capture was investigated using two models: all loci (n = 30) and ‘diverse loci’ only (n = 7), defined as possessing ≥4 alleles, a threshold that separates out the ca. 25% most polymorphic loci in our data set. As mean MHC/ind varied over time (see Study populations), a second MHC curve was constructed using the 1993 Cousin sample (n = 52) to check the accuracy of capture estimates for the earlier translocations to Aride (1988) and Cousine (1990). The rarefaction curves were then used to estimate the expected allelic richness captured during the translocations by taking the mean of the 1000 rarefaction repetitions for each number of translocated birds (Aride/Cousine 29, Denis 58, Frégate 59). To test model accuracy, we compared the observed microsatellite allelic richness captured for Denis and Frégate (where we had complete founder samples) with the distribution of expected values generated by rarefaction. A one-tailed P value was obtained by calculating the percentile of simulated data in which the observed mean was located.
Population structure within and among islands was tested by calculating microsatellite and MHC pairwise FST in arlequin across all twelve catch-year samples. We also calculated pairwise DEST (Jost 2008), using the r package MMOD (Winter 2012) for microsatellites and SPADE (Chao & Shen 2010) for MHC. The measures were strongly correlated for both microsatellites and MHC (both rM > 0.90, P < 0.001); hence, only FST is reported. The relationship between pairwise microsatellite and MHC FST values was assessed using a Mantel test. A Bayesian algorithm was also implemented in structure version 2.3 (Pritchard et al. 2000) to determine the most likely number of genetic clusters (K). As all islands have recent common ancestry, limited differentiation was expected, so samples from the latest sampling period (2011) were used along with a model of no admixture, prior information on sampling location and correlated allele frequencies, features which are suited to detecting subtle structure (Hubisz et al. 2009). We carried out four independent iterations of 500 000 repetitions with a burn-in of 20 000 at each clustering level for K = 1–5. We analysed the results using structure harvester (Earl & vonHoldt 2012), which implements the ad hoc ΔK test (Evanno et al. 2005), a more accurate estimate of the most likely number of clusters than assessing log probability alone. The structure results were visualized using distruct version 1.1 (Rosenberg 2004).
Estimates of the effective population size (Ne) of each population were obtained using two different methods: (i) We used the approximate Bayesian computation approach implemented in ONeSAMP (Tallmon et al. 2008), with Ne priors of 2–150 for each island. This method bases Ne on a single population sample (from 2011); (ii) We used a linkage disequilibrium approach in LDNE (Waples & Do 2008) with a random mating model to account for complex patterns of breeding in this species (Komdeur 1992; Richardson et al. 2001). The lowest allele frequency allowed was 0.02 to address the trade-off between estimate precision and bias (Waples & Do 2008), although bias should be minimal given the comparatively low polymorphism observed in the microsatellites.
Results
Microsatellite genotypes were compiled for 658 individuals and MHC genotypes for 581 individuals across the five populations. All populations were in Hardy–Weinberg equilibrium at all microsatellite loci. Linkage disequilibrium was detected in nine different loci pairs across the five islands after sequential Bonferroni correction (26 of 4933 pairwise comparisons significant). However, inconsistency of patterns between and within populations suggests they are not truly linked. Null allele frequency estimates were also inconsistent across years within each island, with no locus frequency (F) ≥0.1 more than twice except for locus Ase3 (F ≥ 0.1 in four catch-year samples, Table S1, Supporting information). Excluding the Ase3 locus did not qualitatively alter the results (data not shown). All 30 loci were retained in the final analyses.
Microsatellite diversity in the source population on Cousin had an overall mean (± SE) HE of 0.49 ± 0.005 and allelic richness of 2.92 ± 0.01. Neither measure differed across years (randomization tests, HE: P = 0.67; allelic richness: P = 0.85) and the global exact test found no overall differentiation across years (P > 0.99). Mean MHC/ind increased from 1993 (3.93 ± 0.19) to 2011 (4.71 ± 0.16, P = 0.003). Full diversity estimates are given in Table1.
Resampling from the source population, the genetic capture model estimated that 95% (3.07 ± 0.01) of the overall microsatellite allelic richness would be captured with the translocation of 59 individuals, but only 87% (4.86 ± 0.01) of allelic richness for diverse loci. Virtually identical values were observed for 58 individuals (all loci: 3.06 ± 0.01, diverse loci: 4.86 ± 0.01). There was no difference between expected and observed allelic richness translocated to Frégate (all loci: P = 0.36, diverse loci: P = 0.37, Fig.1a) or Denis (all loci: P = 0.35, diverse loci: P = 0.35), indicating a good fit of the genetic capture models. It is therefore reasonable to assume that the rarefaction curve estimates of genetic capture for 29 individuals (all loci = 91%, 2.94 ± 0.10; diverse loci = 85%, 4.45 ± 0.10) provide an accurate representation of genetic diversity initially captured from the source population during the translocations to Cousine and Aride. Across all 30 loci, we observed a loss of eight and ten alleles in the Denis and Frégate translocations, respectively. On Denis, all losses were of rare alleles (frequency < 0.01 in source population) from diverse loci, and on Frégate, two alleles were also lost from less diverse loci, including the biallelic Pte24-CEST locus, leading to fixation of a single allele in this population. The lack of founder genotypes or complete population sampling for Aride and Cousine means we could not directly determine the exact roles of genetic capture vs. subsequent drift in allele loss for these two translocations.
Figure 1.
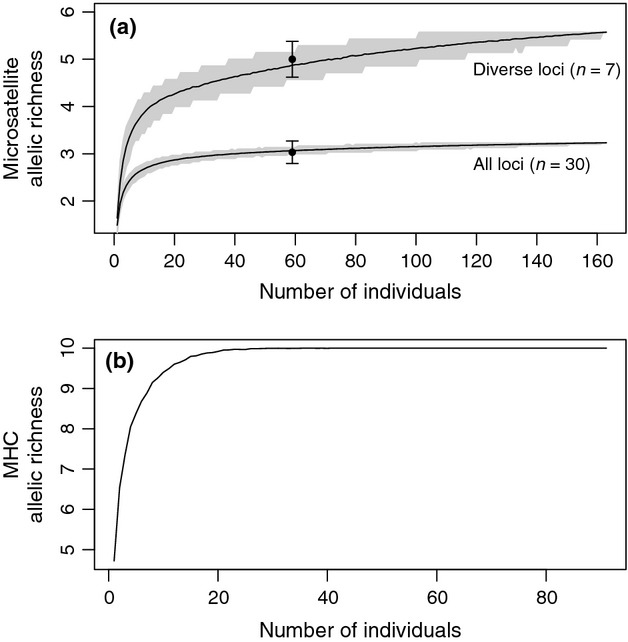
Rarefaction curves constructed from 1000 replicates of the genetic variation observed on Cousin Island in 2011. (a) Microsatellite allelic richness (n = 163). Diverse loci have ≥4 alleles, and the shaded areas lie between the 5–95th percentiles. Observed mean allelic richness of Frégate founders (n = 59) given as points with SE bars, (b) major histocompatibility complex variation (n = 91).
The MHC rarefaction curves estimated that ca. 20 (2011 sample) to ca. 25 (1993 sample) individuals would be required for complete sampling of known class I variation (Fig.1b). All MHC class I alleles were subsequently found in the Denis and Frégate founders at translocation, and in Aride and Cousine in subsequent catch-year samples (Table1).
Diversity estimates for translocated populations are given in Table1. Although some microsatellite alleles were lost in the translocation process, no significant differences in either microsatellite HE or allelic richness across either the full suite of loci or diverse loci alone were detected between islands (pooled samples, all P > 0.20, Fig.2). There were also no differences in microsatellite diversity over time within each population (all P > 0.30). There was no difference in mean MHC/ind (H = 6.03, P = 0.20) between islands (pooled samples). Similarly, there were no differences in MHC/ind across time within each translocated population (all P > 0.07).
Figure 2.
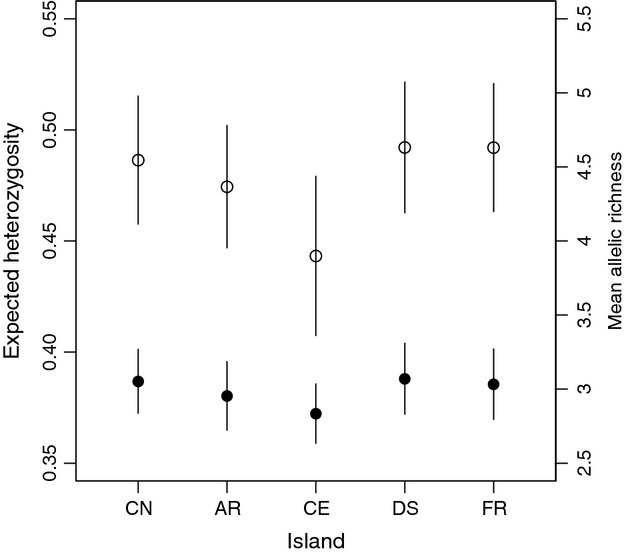
Genetic diversity and differentiation between populations of the Seychelles warbler; HE (white points) and rarefied allelic richness (black points) across all microsatellite loci. Error bars given are SE. CN = Cousin, AR = Aride, CE = Cousine, DS = Denis, FR = Frégate.
Microsatellite pairwise FST analyses revealed moderate differentiation between the populations on Aride and Cousine (2011, FST = 0.08, P < 0.001). Subtle differentiation was also detected between all other pairwise comparisons (2011, FST = 0.01–0.05, all P < 0.001) except for between Cousin and Frégate (P = 0.73). The MHC FST analyses also revealed subtle differentiation between Cousine and: Aride (2011, FST = 0.02, P = 0.002), Denis (2011, FST = 0.02, P = 0.005) and Cousin (2011, FST = 0.01, P = 0.03). Pairwise FST are given in Table S2 (Supporting information). A positive correlation was found between microsatellite and MHC FST (rM = 0.69, P < 0.001, Fig.3). The structure analysis identified two genetically distinct clusters across the five populations (Fig. S2, Supporting information). When visualized, the islands of Cousin, Denis and Frégate contained a mixture of both clusters, but clear segregation of the clusters was observed in the Aride and Cousine populations (Fig.4).
Figure 3.
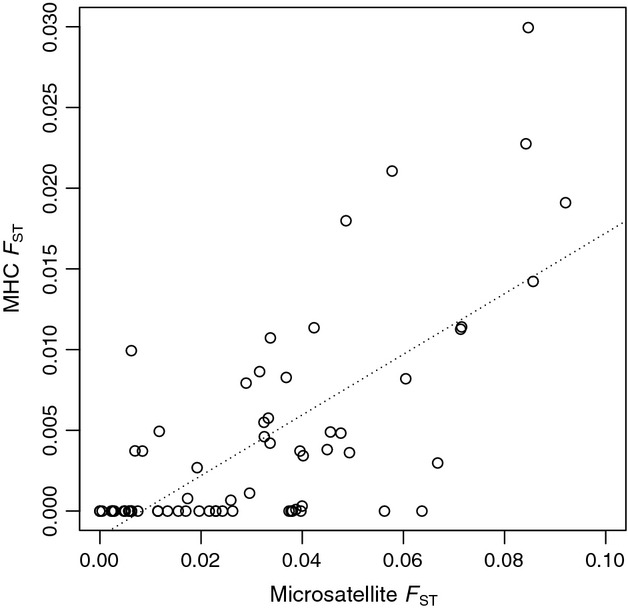
Comparison of microsatellite and major histocompatibility complex FST differentiation across catch-year samples between populations of the Seychelles warbler. Mantel test r = 0.69, P < 0.001 (using all island and catch-year pairwise comparisons).
Figure 4.

STRUCTURE plot of genetic clustering across the five Seychelles warbler populations in 2011; Cousin (CN), n = 163. Aride, n = 29. CE, n = 30. Denis, n = 35. Frégate, n = 59. Two clusters represented by gray and white bars.
Estimates of Ne varied between methods (Table2), but both methods estimated Cousin to have the largest Ne and Cousine to have the smallest Ne.
Table 2.
Effective population size estimates for five Seychelles warbler populations using two methods on samples from 2011. Medians are given with credible limits for ONeSAMP and 95% confidence intervals for LDNE
| Island | ONeSAMP | LDNE |
|---|---|---|
| Cousin | 35 (31–42) | 68 (59–82) |
| Aride | 24 (21–31) | 39 (26–70) |
| Cousine | 23 (21–28) | 22 (16–34) |
| Denis | 28 (25–34) | 36 (26–61) |
| Frégate | 26 (23–30) | 54 (38–85) |
Discussion
The Seychelles warbler, with its translocation history and long-term sampling regime, presents a useful case study of the spatio-temporal impact of conservation translocations on neutral and functional genetic diversity. We found low and temporally stable microsatellite diversity, and low MHC diversity in the source population on Cousin. Our rarefaction models predicted allele retention accurately, and genetic capture (as opposed to subsequent drift) appeared to be the main determinant of translocated population diversity. A small number of rare alleles from neutral markers were lost during translocation, but no statistically significant loss of either neutral or functional diversity was detected in any of the four new populations. Further, diversity in these new populations remained stable over time. Importantly, however, the populations established with a lower number of founders (Aride and Cousine) were central to the low to moderate levels of genetic differentiation observed among populations, indicating that translocations produced subtle changes in allele frequencies, if not in levels of diversity per se.
Conservationists have been advised to capture ≥95% of the source population's genetic diversity during translocations to limit any bottleneck effects caused by the translocation process (Weeks et al. 2011). Our rarefaction models show that translocations to Denis and Frégate (58 and 59 birds, respectively) captured ca. 95% of neutral diversity (87% for diverse loci), but the earlier translocations of 29 birds to Aride and Cousine only captured ca. 91% (85% for diverse loci). In line with this, we found a slight, albeit nonsignificant, decrease in both allelic richness and heterozygosity in the Aride and Cousine populations (Fig.2). We would expect a more pronounced effect of loss on allelic richness measures than heterozygosity (Allendorf 1986), but we see no significant difference. A similar study by Taylor and Jamieson (2008) reported little subsequent loss of genetic variation in serial translocations of saddlebacks (Philesturnus carunculatus) involving even smaller numbers of founders (lowest n = 16), although the source population also had lower initial variation. Our result supports their suggestion that long-term population history (i.e. previous severe bottlenecks) may negate any contemporary bottleneck effects caused by small numbers of founders. This is simply because if all but the commonest alleles are already lost, there is little variation left to lose in subsequent bottlenecks. However, this will only be true for the most extreme cases of genetically depauperate populations (as in the saddlebacks). The Seychelles warbler is severely bottlenecked, and yet we still find some loss of variation during translocations, indicating that care – and suitably high numbers of founders – must be taken to avoid further genetic impoverishment of translocated populations. Furthermore, statistical and biological significance may not concur, as rare alleles can play important roles in evolution (Allendorf 1986). Indeed, another approach to conserving genetic diversity in translocations is to maximize the probability of retaining rare alleles in the founding population (Weiser et al. 2012). This would require much larger founder sizes – in our case, our rarefaction curves suggest that to capture 99% of variation (thus capturing the majority of rare alleles), we would have needed to translocate in excess of 130 individuals. However, when planning translocations, conservation managers are inevitably faced with balancing genetic factors against other considerations (namely logistics, expense and restricted source populations), and the best genetic approach may not be possible. Where neutral markers are a good proxy for functional diversity (as our study suggests can be the case in bottlenecked populations), the development of predictive models of neutral diversity loss makes it possible to compare the genetic consequences of different management options prior to translocation (Weiser et al. 2013) – a practice that we would encourage wherever possible.
Disentangling the impact of genetic capture, its associated founder effects, and subsequent drift on genetic diversity is generally not possible, requiring virtually complete sampling of a population before, during and, for extended periods, after the translocation event. Complete microsatellite genotypes of all founders on Denis and Frégate enabled us to identify genetic capture as the main determinant of diversity in the translocated populations, and to pinpoint alleles lost specifically through genetic capture. All but two of the alleles that were lost were rare (frequency < 0.01) in the source population, in line with theoretical expectations (Allendorf 1986; Stockwell et al., 1996). The fixation of an allele at locus Pte24-CEST on Frégate further demonstrates the stochastic nature of genetic capture. This locus became fixed in the largest founding population, whilst remaining biallelic across three other translocations of smaller founder numbers. Our data demonstrate that the loss of rare alleles in the founding populations is due to incomplete genetic capture. Additionally, there is little evidence for significant changes in diversity over time, suggesting that drift subsequent to the translocations has had little effect on these populations. This may partly be explained by the rapid population growth observed in the translocated populations, which will have limited the effects of drift (Nei et al. 1975).
The rarefaction models for MHC diversity suggested that ca. 20–25 Seychelles warblers would be required to capture all the known source MHC variation, which was achieved in all translocations. MHC copy number variation may exist in these populations, but it is difficult to separate from the variance in number of alleles shared across duplicated loci (e.g. Eimes et al. 2011). It may be therefore logical to translocate individuals with the largest number of different alleles, irrespective of whether this was the result of copy number variation or across-loci heterozygosity, as this would maximize the MHC variation in the founding population.
The Seychelles warbler possesses a depauperate MHC diversity compared with other Acrocephalus species, with only ten class I alleles observed (Hansson & Richardson 2005) and no class II variation detected (Hutchings 2009). Pathogen-mediated balancing selection is believed to maintain MHC diversity in large populations (Doherty & Zinkernagel 1975; Slade & McCallum 1992; Spurgin & Richardson 2010). However, with decreasing population size, the effects of drift are more severe, and selection needs to be stronger to maintain diversity (Kimura 1983). Studies have shown that neutral processes outweigh selection in shaping MHC diversity in small, bottlenecked and/or isolated populations (e.g. Seddon & Ellegren 2004; Miller et al. 2010; Agudo et al. 2011; Sutton et al. 2011), although instances of selection acting to maintain MHC diversity have been documented (e.g. Aguilar et al. 2004; Oliver & Piertney 2012). Here, we find that the pattern of microsatellite and MHC differentiation is highly positively correlated (Fig.3), suggesting that any MHC-based selection that occurs in the Seychelles warbler (Richardson et al. 2005; Brouwer et al. 2010) is not strong enough to override the effect of neutral processes during translocations. Instead, our data support the conclusion that demographic processes shape both neutral and functional diversity in a similar way. In our case, it seems that the stochastic process of genetic capture of small founder numbers has been the main driver of differentiation between translocated Seychelles warbler populations (Fig.4). Although our analysis clearly suggests k = 2 genetic clusters across the populations, it can be interpreted as three ‘groupings’: the heterogeneous populations of Cousin, Denis and Frégate forming one group and Aride and Cousine each forming another, with the latter two diverging in opposite directions to one another due to their smaller numbers of founders.
Overall, there appears less variation in the MHC than observed at the microsatellites. Admittedly, these particularly variable microsatellites were originally selected from a larger panel of markers to resolve parentage, which may explain their sensitivity in detecting population differentiation, where MHC was less suited. An obvious but key point is that the observable impact of translocations on genetic diversity is wholly dependent on the variability of the loci used in the study. Careful choice of good, highly polymorphic markers is therefore important to enable higher resolution in studies investigating genetic variation loss.
Defining a ‘successful’ translocation is difficult and can lead to misinterpretation regarding long-term possibility of failure and, potentially, to inadequate future conservation effort (Griffith et al. 1989; Fischer & Lindenmayer 2000). Two biologically relevant aspects on which success can be judged are persistence and resilience (Fischer & Lindenmayer 2000). Many avian translocation studies report high mortality during/immediately following release and often complete failure of populations to become established (e.g. Brekke et al. 2011; Jamieson 2011; White et al. 2012). The Seychelles warbler programme is therefore unusual, with no mortality occurring during any of the four translocations (Komdeur 1994; Richardson et al. 2006; Wright & Richardson 2012). No loss of variation is observed subsequently in the translocated populations (after the loss due to the initial genetic capture). This indicates that survival is high, that reproductive representation of founders is balanced and that the rapid population growth observed during establishment has limited the severity of the founder bottleneck. Current census population estimates for each island are Cousin = 320, Aride = 1850, Cousine = 210, Denis = 300 and Frégate = 80. The populations on Cousin, Aride and Cousine are at carrying capacity (DSR, pers. obs.), with differences in the area and quality of habitat on the different islands responsible for the variation in island capacity. Denis and Frégate are also still in the early stages of post-translocation population growth. The translocations can therefore be considered extremely successful in the short-medium term. The long-term genetic resilience of a population will depend on the functional variation present. Other bottlenecked species, such as southern elephant seals (Mirounga leonina), Chatham Island black robins (Petroica traversi), cheetah (Acinonyx jubatus) and falcons (Falco spp.), survive despite extremely low MHC variability (Slade 1992; Miller & Lambert 2004; Castro-Prieto et al. 2011; Gangoso et al. 2012). Decreased pathogen exposure within such populations may partly explain their apparent viability (Slade 1992; Miller & Lambert 2004), and depauperate parasite loads are found in the Seychelles warbler (Hutchings 2009). However, whether the remaining MHC diversity provides adequate resilience against any novel pathogens that may enter these populations in the future is unknown.
The variability in effective population size (Ne) estimates between the methods means caution should be exercised with interpretations. Both the severity and duration of bottleneck events will affect estimates of Ne (Nei et al. 1975; Frankham 1995). Although the bottlenecks were relatively severe (29 and 58 of 59), all founders were sourced from an already bottlenecked population (Spurgin et al. in review). Further, each population has experienced rapid growth over many generations (with the exception of Frégate, only established at the end of 2011), which would limit further reduction in Ne. The general scale and order of Ne estimates across populations therefore appear logical in relation to known history, as well as founder and census population sizes. Our results show that while the translocations have clearly been very successful in massively expanding the overall Seychelles warbler population and range without significant extra loss of diversity, most of the estimates of Ne for our populations fall below any recommended minimum viable sizes (50–5000, Lande 1995; Franklin & Frankham 1998). This and the low levels of diversity at both neutral and functional loci mean that concerns regarding the evolutionary potential of this species still cannot be discounted. Given these results and the evidence that some genetic divergence exists, assisted gene flow between translocated populations may be required in the future to maintain all the Seychelles warblers as one large, undifferentiated and hence more viable population. The general environmental conditions are similar between islands (indeed host islands were selected based on their similarity to Cousin); but we cannot fully discount the possibility of different selection pressures across populations due to factors we have not been able to assess. It may therefore be logical to check for adaptive differences between populations before undertaking assisted gene flow as this could cause outbreeding depression, an important consideration for translocation projects in general (Edmands 2007).
Conclusion
The demographic history of the Seychelles warbler is typical of many endangered species (Hudson et al. 2000; Robertson et al. 2009; Grueber & Jamieson 2011; Bristol et al. 2013), and our results should therefore be of general applicability. Questions have been raised about inferring functional variation based solely on any one given locus, that is the MHC (Acevedo-Whitehouse & Cunningham 2006; Radwan et al. 2010). Future work on other important immune genes, such as toll-like receptors (e.g. Grueber et al. 2013), individual variation in gene copy number (e.g. Eimes et al. 2011) and broader genomic studies (Angeloni et al. 2012), will help our understanding of how and why genetic variation influences population persistence. What role candidate genes vs. genomic approaches will play in conservation programmes will also undoubtedly be an important avenue of research. However, conservation biology is a crisis discipline (Soulé 1985) and requires efficient, evidence-based decision-making. From a practical perspective, in the case of the Seychelles warbler, investigating MHC variation as an example of functional genetic diversity provided no extra information (above and beyond that of the microsatellite data) to help estimate the numbers of founders required. Evidence is now accumulating for drift outweighing selection in small populations and that maintaining maximal genetic diversity is vital in species conservation. Based on this, we suggest that in bottlenecked, isolated species, such as the Seychelles warbler, genetic decisions on the number of founders to use in translocations could be made most efficiently and cost-effectively based on neutral diversity measures. However, this depends on the demographic history of the organism. It may be that this is an acceptable course of action only in bottlenecked, genetically depauperate populations, where drift has largely overridden selection. In our study, a handful (e.g. five) of the most polymorphic microsatellite loci would have allowed us to accurately determine the required number of founders to be translocated and to monitor the genetic diversity of the resulting populations. Further, using a suite of markers means that decisions are not made on a single functional region at the potential expense of other important regions (Radwan et al. 2010). Lastly, as next-generation sequencing technologies become cheaper and hence more accessible to conservation programmes, it should be possible to provide conservation managers with increasingly accurate information on genome-wide variation within and between populations to aid vital evidence-based decision-making (Angeloni et al. 2012). The results presented here add to the growing evidence base on the impact and use of translocations as an important conservation tool and will, we hope, help inform translocation practices and the conservation of other endangered species.
Acknowledgments
We thank Nature Seychelles for allowing us to work on Cousin Island and the proprietors of Aride, Cousine, Denis and Frégate for allowing access. We thank M. van der Velde, M. Mannarelli, J. Gill and numerous colleagues in the field and laboratory for help with the project. RSCA work was performed at NERC Biomolecular Analysis Facility Sheffield. DJW was supported by a UK Natural Environment Research Council CASE PhD Studentship with BirdLife International. The Seychelles Bureau of Standards and Department of Environment gave permission for sampling and fieldwork.
D.S.R. and D.J.W. designed the study. The long-term fieldwork and translocations were managed by D.S.R. and J.K. The molecular analyses were undertaken in the laboratories of D.S.R., J.K. and T.B. D.J.W. and L.G.S. analysed the data. D.J.W., L.G.S. and D.S.R. drafted the manuscript, with input from N.J.C., T.B. and J.K. All authors edited and agreed to the manuscript.
Data accessibility
Individual microsatellite and MHC genotypes with catch year and island have been deposited in the DRYAD data repository (doi:10.5061/dryad.596k1).
Supporting Information
Additional supporting information may be found in the online version of this article.
Map of the inner Seychelles archipelago with location, date and number of founding individuals for the four Seychelles warbler translocations.
Graph generated by STRUCTURE HARVESTER (Earl & vonHoldt 2012), displaying the change in ΔK against number of clusters (K) calculated following the method of Evanno et al. (2005), highlighting that K = 2 is the most likely number of genetic clusters across five island populations of Seychelles warbler.
Null allele estimates for each of 30 microsatellite loci across each catch-year sample period for five island populations of Seychelles warblers where CN=Cousin, AR=Aride, CE=Cousine, DS=Denis and FR=Frégate. Estimates >0.1 highlighted in bold type.
Pairwise FST between each population sample across five island populations of Seychelles warblers. Microsatellites (lower) and major histocompatibility complex (upper) data.
References
- Acevedo-Whitehouse K, Cunningham AA. Is MHC enough for understanding wildlife immunogenetics? Trends in Ecology & Evolution. 2006;21:433–438. doi: 10.1016/j.tree.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Agudo R, Alcaide M, Rico C, et al. Major histocompatibility complex variation in insular populations of the Egyptian vulture: inferences about the roles of genetic drift and selection. Molecular Ecology. 2011;20:2329–2340. doi: 10.1111/j.1365-294X.2011.05107.x. [DOI] [PubMed] [Google Scholar]
- Aguilar A, Roemer G, Debenham S, et al. High MHC diversity maintained by balancing selection in an otherwise genetically monomorphic mammal. Proceedings of the National Academy of Sciences. 2004;101:3490–3494. doi: 10.1073/pnas.0306582101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf F. Genetic drift and the loss of alleles vs. heterozygosity. Zoo Biology. 1986;5:181–190. [Google Scholar]
- Allendorf F, Luikart G. Conservation and the Genetics of Populations. London, U.K: Blackwell Publishing; 2007. [Google Scholar]
- Angeloni F, Wagemaker N, Vergeer P, Ouborg J. Genomic toolboxes for conservation biologists. Evolutionary Applications. 2012;5:130–143. doi: 10.1111/j.1752-4571.2011.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong DP, Seddon PJ. Directions in reintroduction biology. Trends in Ecology & Evolution. 2008;23:20–25. doi: 10.1016/j.tree.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Barrett ELB, Burke TA, Hammers M, Komdeur J, Richardson DS. Telomere length and dynamics predict mortality in a wild longitudinal study. Molecular Ecology. 2013;22:249–259. doi: 10.1111/mec.12110. [DOI] [PubMed] [Google Scholar]
- Bauer MM, Miller MM, Briles WE, Reed KM. Genetic variation at the mhc in a population of introduced wild Turkeys. Animal Biotechnology. 2013;24:210–228. doi: 10.1080/10495398.2013.767267. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? Journal of Evolutionary Biology. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Biebach I, Keller LF. A strong genetic footprint of the re-introduction history of Alpine ibex (Capra ibex ibex) Molecular Ecology. 2009;18:5046–5058. doi: 10.1111/j.1365-294X.2009.04420.x. [DOI] [PubMed] [Google Scholar]
- Boakes EH, Wang J, Amos W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity. 2007;98:172–182. doi: 10.1038/sj.hdy.6800923. [DOI] [PubMed] [Google Scholar]
- Brekke P, Bennett PM, Santure AW, Ewen JG. High genetic diversity in the remnant island population of hihi and the genetic consequences of re-introduction. Molecular Ecology. 2011;20:29–45. doi: 10.1111/j.1365-294X.2010.04923.x. [DOI] [PubMed] [Google Scholar]
- Bristol RM, Tucker R, Dawson DA, et al. Comparison of historical bottleneck effects and genetic consequences of re-introduction in a critically endangered island passerine. Molecular Ecology. 2013;22:4644–4662. doi: 10.1111/mec.12429. [DOI] [PubMed] [Google Scholar]
- Brouwer L, Barr I, Van de Pol M, et al. MHC-dependent survival in a wild population: evidence for hidden genetic benefits gained through extra-pair fertilizations. Molecular Ecology. 2010;19:3444–3455. doi: 10.1111/j.1365-294X.2010.04750.x. [DOI] [PubMed] [Google Scholar]
- Cardoso M, Eldridge M, Oakwood M, et al. Effects of founder events on the genetic variation of translocated island populations: implications for conservation management of the northern quoll. Conservation Genetics. 2009;10:1719–1733. [Google Scholar]
- Castro-Prieto A, Wachter B, Sommer S. Cheetah paradigm revisited: MHC diversity in the world's largest free-ranging population. Molecular Biology and Evolution. 2011;28:1455–1468. doi: 10.1093/molbev/msq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Shen T-J. Program SPADE (Species Prediction And Diversity Estimation) 2010. Available at: http://chao.stat.nthu.edu.tw.
- Charlesworth D, Charlesworth B. Inbreeding depression and its evolutionary consequences. Annual Review of Ecology and Systematics. 1987;18:237–268. [Google Scholar]
- Crnokrak P, Barrett SCH. Perspective: purging the genetic load: a review of the experimental evidence. Evolution. 2002;56:2347–2358. doi: 10.1111/j.0014-3820.2002.tb00160.x. [DOI] [PubMed] [Google Scholar]
- Crook J. The present status of certain rare land birds of the Seychelles islands. Seychelles Government Bulletin. 1960.
- Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Earl D, vonHoldt B. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation management. Molecular Ecology. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Eimes JA, Bollmer JL, Whittingham LA, Johnson JA, Van Oosterhout C, Dunn PO. Rapid loss of MHC class II variation in a bottlenecked population is explained by drift and loss of copy number variation. Journal of Evolutionary Biology. 2011;24:1847–1856. doi: 10.1111/j.1420-9101.2011.02311.x. [DOI] [PubMed] [Google Scholar]
- Ekblom R, Saether SA, Jacobsson P, et al. Spatial pattern of MHC class II variation in the great snipe (Gallinago media) Molecular Ecology. 2007;16:1439–1451. doi: 10.1111/j.1365-294X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Ewen JG, Armstrong DP, Parker KA, Seddon PJ. Reintroduction Biology: Integrating Science and Management. Oxford: Wiley-Blackwell; 2012. [Google Scholar]
- Excoffier L, Lischer HEL. ARLEQUIN suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fischer J, Lindenmayer DB. An assessment of the published results of animal relocations. Biological Conservation. 2000;96:1–11. [Google Scholar]
- Frankham R. Effective population-size adult-population size ratios in wildlife - a review. Genetical Research. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Frankham R. Genetics and extinction. Biological Conservation. 2005;126:131–140. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Franklin I. Evolutionary change in small populations. In: Soulé ME, Wilcox BA, editors. Conservation Biology: An Evolutionary-Ecological Perspective. Sunderland, MA: Sinauer Associates; 1980. pp. 135–150. [Google Scholar]
- Franklin I, Frankham R. How large must populations be to retain evolutionary potential? Animal Conservation. 1998;1:69–70. [Google Scholar]
- Gangoso L, Alcaide M, Grande JM, et al. Colonizing the world in spite of reduced MHC variation. Journal of Evolutionary Biology. 2012;25:1438–1447. doi: 10.1111/j.1420-9101.2012.02529.x. [DOI] [PubMed] [Google Scholar]
- Godefroid S, Piazza C, Rossi G, et al. How successful are plant species reintroductions? Biological Conservation. 2011;144:672–682. [Google Scholar]
- Griffith B, Scott JM, Carpenter JW, Reed C. Translocation as a species conservation tool: status and strategy. Science. 1989;245:477–480. doi: 10.1126/science.245.4917.477. [DOI] [PubMed] [Google Scholar]
- Groombridge JJ, Raisin C, Bristol R, Richardson DS. Genetic Consequences of Reintroductions and Insights from Population History. In: Ewen JG, Armstrong DP, Parker KA, Seddon JM, editors. Reintroduction Biology: Integrating Science and Management. Oxford: Wiley-Blackwell; 2012. pp. 395–440. [Google Scholar]
- Grueber CE, Jamieson IG. Low genetic diversity and small population size of Takahe Porphyrio hochstetteri on European arrival in New Zealand. Ibis. 2011;153:384–394. [Google Scholar]
- Grueber CE, Wallis GP, Jamieson IG. Genetic drift outweighs natural selection at toll-like receptor (TLR) immunity loci in a re-introduced population of a threatened species. Molecular Ecology. 2013;22:4470–4482. doi: 10.1111/mec.12404. [DOI] [PubMed] [Google Scholar]
- Hansson B, Richardson DS. Genetic variation in two endangered Acrocephalus species compared to a widespread congener: estimates based on functional and random loci. Animal Conservation. 2005;8:83–90. [Google Scholar]
- Hedrick PW. Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution. 1999;53:313–318. doi: 10.1111/j.1558-5646.1999.tb03767.x. [DOI] [PubMed] [Google Scholar]
- Howald G, Donlan CJ, Galvan JP, et al. Invasive rodent eradication on islands. Conservation Biology. 2007;21:1258–1268. doi: 10.1111/j.1523-1739.2007.00755.x. [DOI] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson QJ, Wilkins RJ, Waas JR, Hogg ID. Low genetic variability in small populations of New Zealand kokako Callaeas cinerea wilsoni. Biological Conservation. 2000;96:105–112. [Google Scholar]
- Hughes AL, Yeager M. Natural selection at major histocompatibility complex loci of vertebrates. Annual Review of Genetics. 1998;32:415–435. doi: 10.1146/annurev.genet.32.1.415. [DOI] [PubMed] [Google Scholar]
- Hutchings K. Parasite-mediated selection in an island endemic, the Seychelles warbler. (Acrocephalus sechellensis), University of East Anglia; 2009. [Google Scholar]
- IUCN/SSC. Guidelines for Reintroductions and Other Conservation Translocations. Switzerland: Version 1.0 IUCN Species Survival Commission, Gland; 2013. [Google Scholar]
- Jamieson IG. Founder effects, inbreeding, and loss of genetic diversity in four avian reintroduction programs. Conservation Biology. 2011;25:115–123. doi: 10.1111/j.1523-1739.2010.01574.x. [DOI] [PubMed] [Google Scholar]
- Jamieson IG, Lacy RC. Managing Genetic Issues in Reintroduction Biology. In: Ewen JG, Armstrong DP, Parker KA, Seddon JM, editors. Reintroduction Biology: Integrating Science and Management. Oxford: Wiley-Blackwell; 2012. pp. 441–475. [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Molecular Ecology. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes. 2005;5:187–189. [Google Scholar]
- Kier G, Kreft H, Lee TM, et al. A global assessment of endemism and species richness across island and mainland regions. Proceedings of the National Academy of Sciences. 2009;106:9322–9327. doi: 10.1073/pnas.0810306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Komdeur J. Importance of habitat saturation and territory quality for evolution of cooperative breeding in the Seychelles warbler. Nature. 1992;358:493–495. [Google Scholar]
- Komdeur J. Conserving the Seychelles Warbler Acrocephalus sechellensis by translocation from Cousin Island to the Islands of Aride and Cousine. Biological Conservation. 1994;67:143–152. [Google Scholar]
- Komdeur J, Piersma T, Kraaijeveld K, Kraaijeveld-Smit F, Richardson DS. Why Seychelles Warblers fail to recolonize nearby islands: unwilling or unable to fly there? Ibis. 2004;146:298–302. [Google Scholar]
- Lande R. Mutation and Conservation. Conservation Biology. 1995;9:782–791. [Google Scholar]
- Larson S, Jameson R, Bodkin J, Staedler M, Bentzen P. Microsatellite DNA and mitochondrial DNA variation in remnant and translocated sea otter (Enhydra lutris) populations. Journal of Mammalogy. 2002;83:893–906. [Google Scholar]
- Leberg P. Estimating allelic richness: effects of sample size and bottlenecks. Molecular Ecology. 2002;11:2445–2449. doi: 10.1046/j.1365-294x.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- Lee TM, Jetz W. Unravelling the structure of species extinction risk for predictive conservation science. Proceedings of the Royal Society B. 2011;278:1329–1338. doi: 10.1098/rspb.2010.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llaurens V, McMullan M, Van Oosterhout C. Cryptic MHC polymorphism revealed but not explained by selection on the class IIB peptide-binding region. Molecular biology and evolution. 2012;29:1631–1644. doi: 10.1093/molbev/mss012. [DOI] [PubMed] [Google Scholar]
- Manly BFJ. Randomization, Bootstrap and Monte Carlo Methods in Biology. London: Chapman and Hall; 1997. [Google Scholar]
- Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Statistical confidence for likelihood-based paternity inference in natural populations. Molecular Ecology. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- Miller HC, Lambert DM. Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae) Molecular Ecology. 2004;13:3709–3721. doi: 10.1111/j.1365-294X.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- Miller B, Ralls K, Reading RP, Scott JM, Estes J. Biological and technical considerations of carnivore translocation: a review. Animal Conservation. 1999;2:59–68. [Google Scholar]
- Miller HC, Allendorf F, Daugherty CH. Genetic diversity and differentiation at MHC genes in island populations of tuatara (Sphenodon spp.) Molecular Ecology. 2010;19:3894–3908. doi: 10.1111/j.1365-294X.2010.04771.x. [DOI] [PubMed] [Google Scholar]
- Monzón-Argüello C, Garcia de Leaniz C, Gajardo G, Consuegra S. Less can be more: loss of MHC functional diversity can reflect adaptation to novel conditions during fish invasions. Ecology and Evolution. 2013;3:3359–3368. doi: 10.1002/ece3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C. Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas. 1999;130:217–228. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. Bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Nogales M, Martin A, Tershy BR, et al. A review of feral cat eradication on islands. Conservation Biology. 2004;18:310–319. [Google Scholar]
- O'Grady JJ, Brook BW, Reed DH, et al. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biological Conservation. 2006;133:42–51. [Google Scholar]
- Oliver MK, Piertney SB. Selection maintains MHC diversity through a natural population bottleneck. Molecular Biology and Evolution. 2012;29:1713–1720. doi: 10.1093/molbev/mss063. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. 2012.
- Radwan J, Biedrzycka A, Babik W. Does reduced MHC diversity decrease viability of vertebrate populations? Biological Conservation. 2010;143:537–544. doi: 10.1016/j.biocon.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995a;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995b;86:248–249. [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Richardson DS. Species Conservation Assessment and Action Plan: Seychelles warbler (Timerl Dezil) Norwich: Nature Seychelles, Roche Caiman & University of East Anglia; 2001. [Google Scholar]
- Richardson DS, Westerdahl H. MHC diversity in two Acrocephalus species: the outbred Great reed warbler and the inbred Seychelles warbler. Molecular Ecology. 2003;12:3523–3529. doi: 10.1046/j.1365-294x.2003.02005.x. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Jury FL, Blaakmeer K, Komdeur J, Burke T. Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis) Molecular Ecology. 2001;10:2263–2273. doi: 10.1046/j.0962-1083.2001.01355.x. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Komdeur J, Burke T, von Schantz T. MHC-based patterns of social and extra-pair mate choice in the Seychelles warbler. Proceedings of the Royal Society B. 2005;272:759–767. doi: 10.1098/rspb.2004.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DS, Bristol R, Shah NJ. Translocation of the Seychelles warbler Acrocephalus sechellensis to establish a new population on Denis Island, Seychelles. Conservation Evidence. 2006;3:54–57. [Google Scholar]
- Richardson DS, Burke T, Komdeur J. Grandparent helpers: the adaptive significance of older, postdominant helpers in the Seychelles warbler. Evolution. 2007;61:2790–2800. doi: 10.1111/j.1558-5646.2007.00222.x. [DOI] [PubMed] [Google Scholar]
- Robertson BC, Frauenfelder N, Eason DK, Elliott G, Moorhouse R. Thirty polymorphic microsatellite loci from the critically endangered kakapo (Strigops habroptilus) Molecular Ecology Resources. 2009;9:664–666. doi: 10.1111/j.1755-0998.2008.02506.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Sarrazin F, Barbault R. Reintroduction: challenges and lessons for basic ecology. Trends in Ecology & Evolution. 1996;11:474–478. doi: 10.1016/0169-5347(96)20092-8. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ellegren H. A temporal analysis shows major histocompatibility complex loci in the Scandinavian wolf population are consistent with neutral evolution. Proceedings of the Royal Society B. 2004;271:2283–2291. doi: 10.1098/rspb.2004.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon PJ, Armstrong DP, Maloney RF. Developing the science of reintroduction biology. Conservation Biology. 2007;21:303–312. doi: 10.1111/j.1523-1739.2006.00627.x. [DOI] [PubMed] [Google Scholar]
- Slade RW. Limited MHC polymorphism in the southern elephant seal - implications for MHC evolution and marine mammal population biology. Proceedings of the Royal Society B. 1992;249:163–171. doi: 10.1098/rspb.1992.0099. [DOI] [PubMed] [Google Scholar]
- Slade RW, McCallum HI. Overdominant vs frequency-dependent selection at MHC loci. Genetics. 1992;132:861–862. doi: 10.1093/genetics/132.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Frontiers in Zoology. 2005;2:16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulé ME. What is conservation biology? BioScience. 1985;35:727–734. [Google Scholar]
- Spielman D, Brook BW, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proceedings of the National Academy of Sciences. 2004;101:15261–15264. doi: 10.1073/pnas.0403809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proceedings of the Royal Society B. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgin LG, Wright DJ, Van der Velde M, et al. Revealing the demographic history of an endangered species provides evolutionary context and informs conservation
- Stockwell CA, Mulvey M, Vinyard GL. Translocations and the preservation of allelic diversity. Conservation Biology. 1996;10:1133–1141. [Google Scholar]
- Sutton JT, Nakagawa S, Robertson BC, Jamieson IG. Disentangling the roles of natural selection and genetic drift in shaping variation at MHC immunity genes. Molecular Ecology. 2011;20:4408–4420. doi: 10.1111/j.1365-294X.2011.05292.x. [DOI] [PubMed] [Google Scholar]
- Tallmon DA, Koyuk A, Luikart G, Beaumont MA. ONeSAMP: a program to estimate effective population size using approximate Bayesian computation. Molecular Ecology Resources. 2008;8:299–301. doi: 10.1111/j.1471-8286.2007.01997.x. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Jamieson IG. No evidence for loss of genetic variation following sequential translocations in extant populations of a genetically depauperate species. Molecular Ecology. 2008;17:545–556. doi: 10.1111/j.1365-294X.2007.03591.x. [DOI] [PubMed] [Google Scholar]
- Tracy LN, Wallis GP, Efford MG, Jamieson IG. Preserving genetic diversity in threatened species reintroductions: how many individuals should be released? Animal Conservation. 2011;14:439–446. [Google Scholar]
- Waples RS, Do CHI. LDNE: a program for estimating effective population size from data on linkage disequilibrium. Molecular Ecology Resources. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Weeks AR, Sgro CM, Young AG, et al. Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evolutionary Applications. 2011;4:709–725. doi: 10.1111/j.1752-4571.2011.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser EL, Grueber CE, Jamieson IG. AlleleRetain: a program to assess management options for conserving allelic diversity in small, isolated populations. Molecular Ecology Resources. 2012;12:1161–1167. doi: 10.1111/j.1755-0998.2012.03176.x. [DOI] [PubMed] [Google Scholar]
- Weiser EL, Grueber CE, Jamieson IG. Simulating retention of rare alleles in small populations to assess management options for species with different life histories. Conservation Biology. 2013;27:335–344. doi: 10.1111/cobi.12011. [DOI] [PubMed] [Google Scholar]
- Westerdahl H. Passerine MHC: genetic variation and disease resistance in the wild. Journal of Ornithology. 2007;148:469–477. [Google Scholar]
- White TH, Collar NJ, Moorhouse RJ, et al. Psittacine reintroductions: common denominators of success. Biological Conservation. 2012;148:106–115. [Google Scholar]
- Whittaker RJ. Island Biogeography: Ecology, Evolution and Conservation. Oxford: Oxford University Press; 1998. [Google Scholar]
- Willi Y, Van Buskirk J, Hoffmann AA. Limits to the adaptive potential of small populations. Annual Review of Ecology, Evolution, and Systematics. 2006;37:433–458. [Google Scholar]
- Winter DJ. MMOD: an R library for the calculation of population differentiation statistics. Molecular Ecology Resources. 2012;12:1158–1160. doi: 10.1111/j.1755-0998.2012.03174.x. [DOI] [PubMed] [Google Scholar]
- Wolf CM, Griffith B, Reed C, Temple SA. Avian and mammalian translocations: update and reanalysis of 1987 survey data. Conservation Biology. 1996;10:1142–1154. [Google Scholar]
- Wolf CM, Garland T, Griffith B. Predictors of avian and mammalian translocation success: reanalysis with phylogenetically independent contrasts. Biological Conservation. 1998;86:243–255. [Google Scholar]
- Worley K, Gillingham M, Jensen P, et al. Single locus typing of MHC class I and class II B loci in a population of red jungle fowl. Immunogenetics. 2008;60:233–247. doi: 10.1007/s00251-008-0288-0. [DOI] [PubMed] [Google Scholar]
- Wright DJ, Richardson DS. The Translocation of Seychelles warblers (Acrocephalus sechellensis) to Frégate Island. Norwich: University of East Anglia; 2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Map of the inner Seychelles archipelago with location, date and number of founding individuals for the four Seychelles warbler translocations.
Graph generated by STRUCTURE HARVESTER (Earl & vonHoldt 2012), displaying the change in ΔK against number of clusters (K) calculated following the method of Evanno et al. (2005), highlighting that K = 2 is the most likely number of genetic clusters across five island populations of Seychelles warbler.
Null allele estimates for each of 30 microsatellite loci across each catch-year sample period for five island populations of Seychelles warblers where CN=Cousin, AR=Aride, CE=Cousine, DS=Denis and FR=Frégate. Estimates >0.1 highlighted in bold type.
Pairwise FST between each population sample across five island populations of Seychelles warblers. Microsatellites (lower) and major histocompatibility complex (upper) data.
Data Availability Statement
Individual microsatellite and MHC genotypes with catch year and island have been deposited in the DRYAD data repository (doi:10.5061/dryad.596k1).


