Abstract
Vacuolar (V)-ATPase is a proton-translocating enzyme that acidifies cellular compartments for various functions such as receptor-mediated endocytosis, intracellular trafficking and protein degradation. Previous studies in Dermacentor variabilis chronically infected with Rickettsia montanensis have identified V-ATPase as one of the tick-derived molecules transcribed in response to rickettsial infection. To examine the role of the tick V-ATPase in tick–Rickettsia interactions, a full-length 2887-bp cDNA (2532-bp open reading frame) clone corresponding to the transcript of the V0 domain subunit a of D. variabilis V-ATPase (DvVATPaseV0a) gene encoding an 843 amino acid protein with an estimated molecular weight of ∼96 kDa was isolated from D. variabilis. Amino acid sequence analysis of DvVATPaseV0a showed the highest similarity to VATPaseV0a from Ixodes scapularis. A potential N-glycosylation site and eight putative transmembrane segments were identified in the sequence. Western blot analysis of tick tissues probed with polyclonal antibody raised against recombinant DvVATPaseV0a revealed the expression of V-ATPase in the tick ovary. Transcriptional profiles of DvVATPaseV0a demonstrated a greater mRNA expression in the tick ovary, compared with the midgut and salivary glands; however, the mRNA level in each of these tick tissues remained unchanged after infection with R. montanensis for 1 h. V-ATPase inhibition bioassays resulted in a significant decrease in the ability of R. montanensis to invade tick cells in vitro, suggesting a role of V-ATPase in rickettsial infection of tick cells. Characterization of tick-derived molecules involved in rickettsial infection is essential for a thorough understanding of rickettsial transmission within tick populations and the ecology of tick-borne rickettsial diseases.
Keywords: Dermacentor variabilis, Rickettsia montanensis, vacuolar-ATPase
Introduction
Ticks are important vectors of human pathogens, including bacteria, viruses and parasites (Sonenshine, 2005). Among the bacterial pathogens transmitted by ticks are Anaplasma spp., Ehrlichia spp., Francisella tularensis, and the obligate intracellular spotted fever group (SFG) Rickettsia. Acquisition of SFG Rickettsia by ticks can be either through an infectious bloodmeal from a vertebrate host or acquired via transovarial transmission. Although the kinetics of the infection process are not well-defined, transmission results in maintenance of SFG Rickettsia among tick populations. The tick-derived molecules associated with sustained transmission are relatively unknown, but characterization of their function may lead to novel points of intervention for tick-borne rickettsial diseases.
At the cellular level, based primarily on studies in vertebrate hosts, the process of rickettsial infection includes induced endocytosis and phagosomal escape to facilitate intracytoplasmic living of Rickettsia (Walker & Ismail, 2008). Host-derived molecules essential for rickettsial invasion include KU70 (Martinez et al., 2005) and, in ticks, histone H2B is associated with rickettsial entry of tick cells in vitro (Thepparit et al., 2010); however, the specific molecular processes by which Rickettsia invade tick cells is yet to be defined. In order to understand the mechanisms of rickettsial survival in the arthropod, previous studies have used molecular techniques such as differential display and subtractive hybridization-PCR to identify several Dermacentor variabilis-derived molecules thought to be associated with tick response to rickettsial infection (Macaluso et al., 2003; Mulenga et al., 2003). Among the putatively identified molecules differentially transcribed in response to rickettsial infection is vacuolar-ATPase (V-ATPase).
The structure and function of V-ATPase consists of multiple subunits separated into the membrane-associated V0 and water-soluble V1 domains. The V0 complex contains the proton-translocating pore and the V1 complex uses ATP to drive proton translocation across the membrane (Nishi & Forgac, 2002). The subunit composition of V-ATPase domains is variable; for example, the Saccharomyces cerevisiae V0 domain consists of six different subunits and the V1 domain is composed of eight different subunits (Kane, 2006; Forgac, 2007). A similar V1 domain is present in the midgut of the tobacco hornworm, Manduca sexta, with a V0 domain containing only four unique subunits (Merzendorfer et al., 2000). Despite the variability in subunits that compromise the V0 and V1 domains, the enzyme uses ATP hydrolysis to acidify compartments for receptor-mediated endocytosis, intracellular trafficking, and protein degradation (Kane, 2006; Forgac, 2007; Wieczorek et al., 2009).
Several bacterial pathogens require host cell V-ATPase to facilitate intracellular living. For example, in Listeria monocytogenes, phagosomal escape relies on acidification of the phagosome mediated by V-ATPase (Portnoy et al., 1992). V-ATPase is also suspected to have a role in the invasion of tick cells by Anaplasma marginale (Kocan et al., 2009). Although acidification of the vacuole is not definitively required for SFG Rickettsia infection (Welch et al., 2012), clathrin-coated pits that concentrate V-ATPases are probably an integral part of host cell invasion (Munderloh & Kurtti, 1995); however, the role of tick V-ATPase and the influence of SFG Rickettsia on this molecule remains to be elucidated. Based on the observed differential transcription of V-ATPase in SFG Rickettsia-infected ticks, we report here the molecular characterization of the full-length transcript and functional V-ATPase inhibition bioassays to further delineate the interaction between SFG Rickettsia and their tick hosts.
Results
Cloning and sequence analysis of DvVATPaseV0a
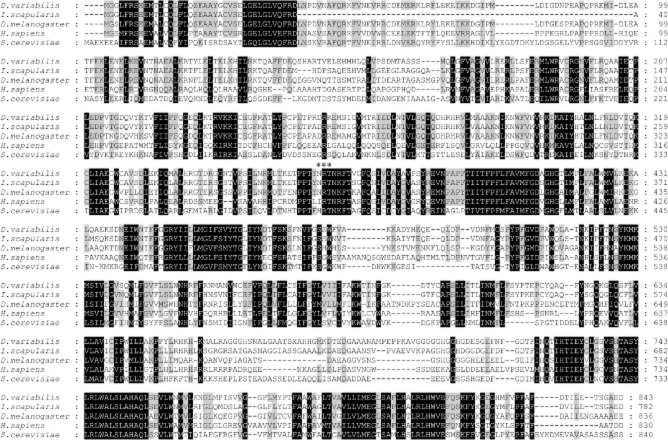
A full-length cDNA clone corresponding to the D. variabilis transcript of the V-ATPase V0 subunit a (DvVATPaseV0a) was isolated using the gene-specific primers designed from differential display PCR (Macaluso et al., 2003). Using MacVector, sequence analysis revealed a 2887-bp full-length cDNA with a 2532-bp open reading frame (ORF; GenBank accession number HM185485). The deduced amino acid sequence comprised 843 residues with an estimated molecular weight of 96 kDa. Amino acid sequence analysis of DvVATPaseV0a using a web-based multiple sequence alignment, multiple sequence comparison by log-expectation (muscle), showed 77, 66, 44 and 40% identity to VATPaseV0a from Ixodes scapularis, Drosophila melanogaster, Homo sapiens, and S. cerevisiae, respectively, as shown in Fig. 1. When analysed using the NetNGlyc 1.0 Server, an Asn-Xaa-Ser/Thr (asparagine-any amino acid-serine/threonine) sequon contained a potential N-glycosylation site at the asparagine (N358). Similarly to other organisms, DvVATPaseV0a possesses eight segment transmembrane portions predicted by TopPred software (Fig. 2).
Figure 1.
Multiple sequence alignment of VATPaseV0a amino acid sequences. muscle software was used to create a sequence alignment and calculate percent identity of VATPaseV0a from Dermacentor variabilis (GenBank accession number HM185485), Ixodes scapularis (GenBank accession number XP002414796), Drosophila melanogaster (GenBank accession number NP733274), Homo sapiens (GenBank accession number NP006010), and Saccharomyces cerevisiae (GenBank accession number NP014913.3). Identical and similar amino acids are highlighted in black and grey, respectively. The figure was created using GeneDoc software. Asterisks represent Asn-Xaa-Ser/Thr sequon in which asparagine (N358) residue was predicted to be glycosylated using NetNGlyc 1.0 Server.
Figure 2.
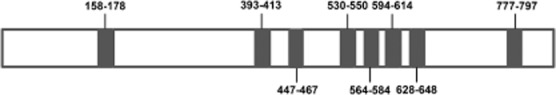
Schematic diagram representing the putative transmembrane regions of DvVATPaseV0a protein. Transmembrane segments of DvVATPaseV0a were predicted using TopPred software. The start and end amino acid positions of each domain are indicated. Putative transmembrane domains are shaded in black.
Expression of recombinant DvVATPaseV0a, antibody generation, and detection of VATPaseV0a in tick tissues
Full-length recombinant (r)DvVATPaseV0a was produced as a hexahistidine (6xHis) fusion protein using the baculovirus expression system in Sf9 insect cells. The size of the recombinant protein produced corresponded to an estimated molecular weight analysis of full-length cDNA sequence ∼96 kDa. A polyclonal antibody against rDvVATPaseV0a was generated in BALB/c mice. Western blotting analysis showed the rDvVATPaseV0a polyclonal antibody specifically recognizing three rDvVATPaseV0a protein bands, but not the His-tagged protein standard. The occurrence of multiple protein bands, after purifying and concentrating, may result from self-binding in the more concentrated protein preparation. To exclude the possibility of non-rDvVATPaseV0a protein contamination, the ∼96 kDa of purified rDvVATPaseV0a was excised from the gel and subjected to sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. The results, again, showed three bands appearing in the same positions as in the first purification, confirming that self-binding occurred in the concentrated protein preparation (data not shown).
Transcriptional analysis of the putative DvVATPaseV0a identified mRNA expression in tick salivary glands, midgut and ovary (Macaluso et al., 2003). To assess DvVATPaseV0a expression in tick tissues, protein extracts of salivary glands, midgut, and ovary were assessed by Western blot using antibodies to rDvVATPaseV0a. The result of the Western blot analysis shown in Fig. 3 revealed a detectable DvVATPaseV0a protein band in the ovary. The apparent molecular mass of this detected band was a little bigger than a 96-kDa band of the rDvVATPaseV0a which reflected the presence of post-translational modification on the protein in tick ovary. Although the same amount of protein was loaded (30 μg), no band was detected in the midgut and salivary glands, indicating a much lower amount of DvVATPaseV0a in the midgut and salivary glands.
Figure 3.
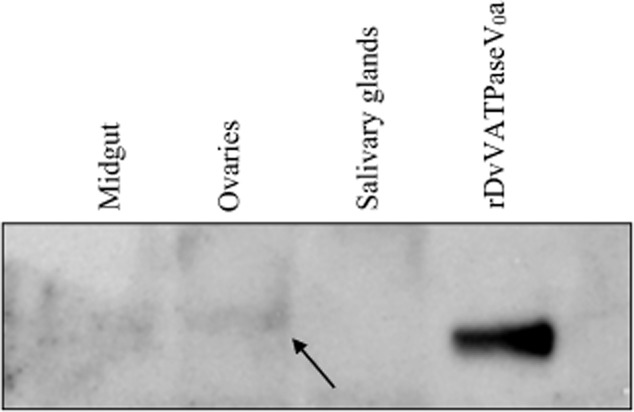
Detection of VATPaseV0a in the midgut, ovary and salivary glands from Dermacentor variabilis using rDvVATPaseV0a polyclonal antibody. Tick tissues, including midgut, ovary and salivary glands, were dissected from the female unfed ticks and the protein was extracted. Thirty micrograms of total proteins from each tissue were then analysed by SDS-PAGE followed by Western immunoblot using anti-rDvVATPaseV0a polyclonal antibody. The arrow indicates a DvVATPaseV0a protein band detected in tick ovary.
Expression of VATPaseV0a mRNA in backless Dermacentor variabilis
To determine a transcriptional profile of VATPaseV0a in D. variabilis tissues (midgut, ovary and salivary glands) in response to an early stage of rickettsial infection, backless ticks were generated and exposed to Rickettsia montanensis. After 1 h, the tissues were dissected from ticks, extracted for total RNA and measured for DvVATPaseV0a mRNA level by quantitative reverse transcription (qRT)-PCR. As shown in Fig. 4, mRNA was detectable in all tick tissues and in both unexposed and R. montanensis-exposed ticks. Correlating with the protein expression pattern, DvVATPaseV0a mRNA was expressed more highly in the ovary than in the other tissues, with a significant difference in unexposed ovaries compared with in the midgut (P = 0.0154) and salivary glands (P = 0.0155); however, in all tissues the mRNA level remained unchanged in R. montanensis-exposed ticks compared with the unexposed control.
Figure 4.
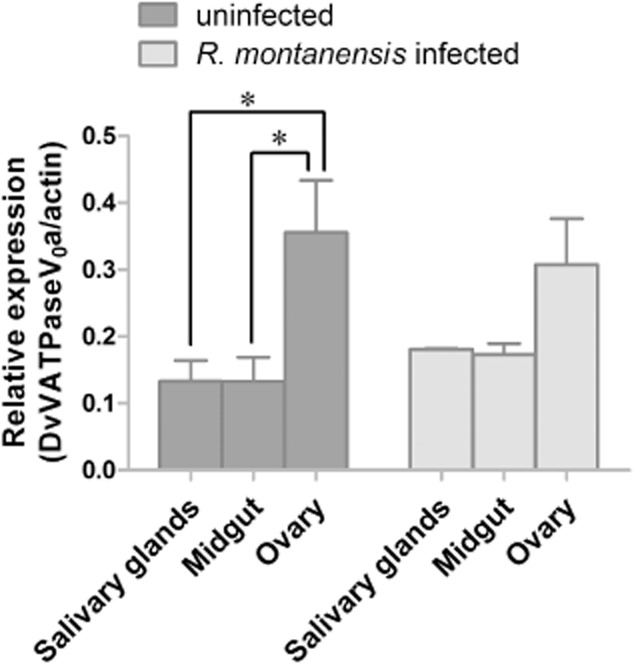
Transcriptional profile of VATPaseV0a in Dermacentor variabilis tissues. Backless ticks were generated by taking off the dorsal cuticle and were exposed to Rickettsia montanensis for 1 h. The tick tissues (midgut, ovary and salivary glands) were then dissected out and extracted for total RNA. The level of DvVATPaseV0a mRNA was measured by quantitative reverse transcription-PCR and normalized to tick actin mRNA. Data shown are mean relative expression from two independent experiments. Error bar represents sem values and *P ≤ 0.05 was considered significantly different. P values of 0.0154 and 0.0155 represent uninfected ovary compared with midgut and salivary glands, respectively.
Involvement of tick V-ATPase in Rickettsia montanensis infection
To assess the function of tick V-ATPase in response to R. montanensis infection, V-ATPase inhibition assays were performed in the D. variabilis-derived cell line DVE1. Tick cells were treated for 2 h with either 5, 0.5, or 0.05 μM of V-ATPase inhibitor, bafilomycin A1, prior to infection with R. montanensis at a multiplicity of infection (MOI) of 10. After 1 h, removal of R. montanensis from the cells occurred before washing cells twice with phosphate-buffered saline (PBS), followed by low-speed centrifugation to exclude the possibility of collecting extracellular rickettsiae. Genomic DNA (gDNA) was then extracted from the cells and the percentage of rickettsial infection in comparison with control cells was assessed by quantitative PCR (qPCR). As shown in Fig. 5, inhibition of V-ATPase in DVE1 cells reduced percent relative invasion compared with the untreated control by 27% at 5 μM (P = 0.0005) and 0.5 μM (P = 0.0005) and by 13% at 0.05 μM (P = 0.0877).
Figure 5.
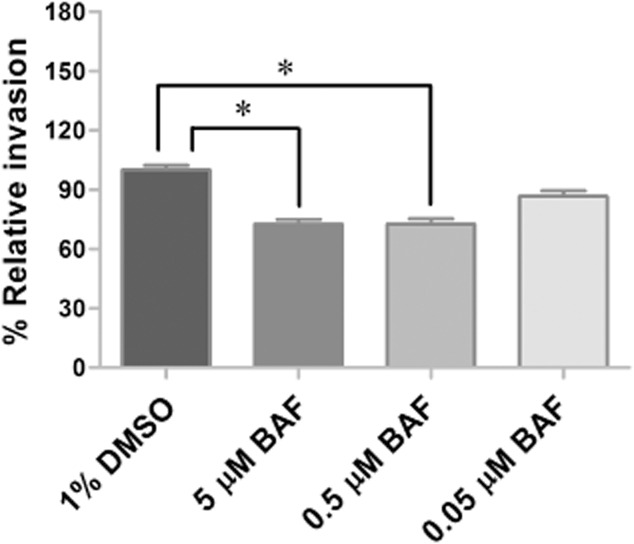
Effect of V-ATPase inhibitor on Rickettsia montanensis infection of DVE1 cells. DVE1 cells were treated for 2 h with bafilomycin A1 (BAF) prior to infection with R. montanensis at a multiplicity of infection of 10. After 1 h, Rickettsia was removed. The cells were washed twice with phosphate-buffered saline and collected by low-speed centrifugation. Genomic DNA was then isolated and percent relative invasion was assessed by quantitative PCR. Data shown are mean percent relative invasion from two independent experiments. Error bar represents sem values. The asterisks mark significant difference from untreated control cells (*P = 0.0005).
Discussion
The present study provides novel sequence and functional analyses for the VATPaseV0a from D. variabilis. A full-length cDNA of DvVATPaseV0a was isolated and multiple sequence alignment showed highest similarity in amino acid sequences to other arthropod species, including the available sequence from another ixodid tick, I. scapularis. The composition of V-ATPase is known to vary across species, yet enzymatic activity appears conserved, independent of subunit variations (Kane, 2006; Forgac, 2007; Wieczorek et al., 2009). Amino acid sequence analysis revealed transmembrane segments and also a potential site for N-glycosylation which is consistent with the studies of coated vesicle (Adachi et al., 1990) and yeast (Ryan et al., 2008) VATPaseV0a, in which the proteins were illustrated as transmembrane glycoproteins. Nevertheless, further investigation is needed to confirm the presence of N-glycosylation of tick-derived VATPaseV0a.
V-ATPase is ubiquitously found in and is responsible for acidification of a variety of intracellular organelles in eukaryotic cells, such as endosomes and lysosomes. The enzyme is essential for several biological processes, e.g. protein sorting, protein processing and degradation, coupled transport of small-molecule, receptor-mediated endocytosis, and receptor recycling (V-ATPase; Nishi & Forgac, 2002). To further characterize tick, the protein was expressed, and the polyclonal antibody raised against rDvVATPaseV0a was then used to confirm the presence of V-ATPase in tick tissues. The results showed a detectable protein band with a molecular weight slightly greater than the predicted 96 kDa, reflecting post-translational modification only in the tick ovary. Previous characterization of the C subunit of V-ATPase from Amblyomma americanum embryos and salivary glands identified a role for V-ATPase in salivary fluid secretion (water balance), but V-ATPase was not essential to the process (McSwain et al., 1997); thus, it is interesting to investigate alternate roles for V-ATPase at the molecular and functional level. The detection of DvVATPaseV0a protein in the tick ovary but not in the midgut and salivary glands corresponds to the observed transcriptional pattern (both in unexposed and Rickettsia-exposed ticks) in which the mRNA in the tick ovary was expressed at a higher level compared with other tissues. The inability to detect V-ATPase in the salivary glands and midgut, combined with abundant expression in the ovary might signify an essential role for this molecule in the ovary. The increased expression of V-ATPase in the tick ovary might be associated with receptor-mediated endocytosis and protein transport, the two processes that are important for ovulation and embryogenesis in which V-ATPase is involved. In many organisms, such as Caenorhabditis elegans (Grant & Hirsh, 1999), D. melanogaster (Schonbaum et al., 1995), Aedes aegypti (Sappington et al., 2008), and also D. variabilis (Mitchell et al., 2007), vitellogenin, a yolk precursor protein, is taken up through the receptor-mediated endocytosis pathway during oocyte growth. Studies using RNA interference in C. elegans showed that V-ATPase is required for ovulation and oogenesis. Specifically, the inhibition of C. elegans V1 subunit C and V0 subunit a (Oka & Futai, 2000; Oka et al., 2001) induced embryonic lethality. Moreover, studies of wnt/β-catenin signalling, a cascade important for numerous biological functions including embryonic development, revealed an involvement of V-ATPase in acidification of the pathway (Cruciat et al., 2010). Taken together, these findings suggest that V-ATPase is physiologically important in the ovary, but the mechanisms by which V-ATPase functions in the tick ovary needs to be further elucidated.
As tick tissues including the salivary glands, midgut and ovary are essential for both horizontal and vertical transmission of SFG Rickettsia (Munderloh & Kurtti, 1995), the role of V-ATPase was then investigated during R. montanensis invasion of D. variabilis. Using a backless tick bioassay, no difference of DvVATPaseV0a transcript levels were observed between unexposed and Rickettsia-exposed ticks. These results differ from previous analyses in which ticks chronically infected with R. montanensis had upregulated V-ATPase transcription compared with R. montanensis-uninfected control ticks (Macaluso et al., 2003). Comparison between the studies is difficult because of the point of assessment of infection in the current study (1 h) differs from chronically infected ticks. It is interesting to speculate that increased V-ATPase in chronically infected ticks, which maintain rickettsiae vertically through transovarial transmission, might be associated with increased vitellogenin processing or oocyte production; thus chronic infection is associated with an increased fitness advantage for an infected tick.
To better characterize the role of V-ATPase in rickettsial infection of tick cells, the D. variabilis-derived tick cell line, DVE1, was incorporated into V-ATPase inhibition bioassays. Bafilomycin A1 is a well characterized inhibitor for V-ATPase, and previous studies have shown that V0 domain subunits a (Crider et al., 1994; Zhang et al., 1994; Wang et al., 2005) and c (Rautiala et al., 1993; Bowman & Bowman, 2002; Bowman et al., 2006) are targets of the inhibitor. In ticks, the V-ATPase in salivary glands was impaired by bafilomycin A1 (McSwain et al., 1997). In the bioassay developed in the present study, a significant dose-dependent decrease in percent relative invasion was observed in cells treated with V-ATPase inhibitor compared with the untreated control. Because the role of V-ATPase was examined in the early stage of infection, it is possible that V-ATPase might play a role in receptor-mediated endocytosis, a process by which Rickettsia enter the cells (Munderloh & Kurtti, 1995). In addition to intracellular compartments, V-ATPase in clathrin-coated vesicles is present at the cell surface and is involved in receptor-mediated endocytosis and functions in recycling receptors (Forgac, 2000). An alternative mechanism by which rickettsial infection might be dependent on V-ATPase is related to the rearrangement of host cytoskeletal components. For example, V-ATPase V1 subunits B (Holliday et al., 2000) and C (Vitavska et al., 2003) bind to actin filament (F-actin); and, in M. sexta V-ATPase subunit C binds not only to F-actin but also to a monomeric G-actin (Vitavska et al., 2005). The biological function of V-ATPase and F-actin interaction was speculated to be involved in the organization of actin dynamics (Wieczorek et al., 2009). As previously characterized in vertebrate cells, SFG Rickettsia manipulate host actin in order to invade host cells (Dramsi & Cossart, 1998; Gouin et al., 2004). While, the mechanisms by which tick V-ATPase functions in response to rickettsial infection, including actin rearrangement, is unknown; the current data suggest the possibility that V-ATPase is involved in rickettsial infection of tick cells via a receptor-mediated endocytosis process.
Several bacterial pathogens use V-ATPase as a mechanism for invasion and survival in host cells. In L. monocytogenes, V-ATPase is required for the activation of the pore-forming haemolysin, essential for phagosomal escape (Portnoy et al., 1992). A dual role for V-ATPase in Chlamydia infection favours bacterial infection and defends the host cell against infection through autophagy (Yasir et al., 2011). With respect to tick-borne bacterial pathogens, V-ATPase is thought to play a role in F. tularensis phagosomal escape (Chong et al., 2008; Santic et al., 2008); however, the essentiality of V-ATPase is debatable (Clemens et al., 2009). Similarly to the R. montanensis and D. variabilis model, V-ATPase was upregulated in D. variabilis exposed to A. marginale (de la Fuente et al., 2007) and a role for the molecule in cell invasion and pathogen development was identified (Kocan et al., 2009). The essential role of V-ATPase in tick physiology, both feeding and oogenesis, make this molecule a viable target for exploitation by rickettsiae, and may provide insight into the maintenance of SFG Rickettsia in tick populations. Further studies to investigate the mechanism(s) by which Rickettsia manipulate tick V-ATPase to facilitate either horizontal or vertical transmission are required.
Experimental procedures
Tick dissection and tissue preparation
As previously described by Macaluso et al. (2001), Rickettsia-free D. variabilis originally derived at Old Dominion University were routinely maintained at Louisiana State University. After feeding for 3–5 days, unmated female ticks were removed from host animals, washed twice with 70% ethanol and rinsed with distilled water. Tick tissues, including midgut, ovary, and salivary glands, were removed from the ticks and washed in sterile PBS (0.137 mM NaCl, 2.7 mM KCl and 8 mM Na2HPO4, pH 7.4) or 0.1% diethyl pyrocarbonate (DEPC)-treated water. Ovaries were transferred into microtubes containing RNAlater (Ambion, Austin, TX, USA) for RNA extraction. For protein preparation, all organs were placed in protease inhibitor cocktail (PIC, Roche, Indianapolis, IN, USA). Tissues were immediately processed or stored at −80 °C until used for extraction.
Protein and mRNA extraction from tick tissues
Tick tissue from at least five D. variabilis were rinsed with DEPC-treated water before total RNA and mRNA were extracted from using the NucleoSpin RNAII and NucleoTrap mRNA Mini kits (Clontech, Mountain View, CA, USA), respectively, according to the manufacturer's instructions. For protein extraction, the tissue samples were thawed on ice for 15 min and washed once with PBS supplemented with PIC. The tissues were then lysed by adding 100 μl of lysis buffer (100 mN NaH2PO4, 10 mM Tris-Cl, 8 M urea, pH 8.0), homogenizing with plastic pestle for 15 min and sonicating in a bath sonicator (Crest Ultrasonics, Trenton, NJ, USA) for 10 min. The lysate was centrifuged at 16 000 × g for 10 min at 4 °C. The supernatant was then transferred to 1.7-ml centrifuge tubes. The protein solution was diluted two-fold and the protein concentration was measured by DC protein assay (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. Then, 30 μg of protein from the midgut, ovary and salivary glands was analysed using SDS-PAGE and Western immunoblot.
Cloning of the tick V-ATPase V0 subunit a full-length cDNA
The full-length cDNA for DvVATPaseV0a was generated using the SMART RACE cDNA Amplification Kit (Clontech) according to the manufacturer's protocol. Briefly, 1 μg of mRNA extracted from ovary was used to create 3′ and 5′ rapid amplification of cDNA ends-ready cDNA. Gene-specific primers designed from a partial sequence of DvVATPaseV0a gene (Macaluso et al., 2003) were used to amplify the 3′ and 5′ end fragments. PCR products were cloned into TOPO TA cloning vectors (Invitrogen, Carlsbad, CA, USA) and the plasmids were isolated using the SV miniprep kit (Promega, Madison, WI, USA) according to the manufacturers' protocols. The cloned plasmid inserts were sequenced by the dye terminator method on a 373 automated fluorescence sequencing system (Applied Biosystems, Carlsbad, CA, USA) at the University of Maryland, Baltimore. For DNA sequence analysis, the MacVector software program (Accelrys, San Diego, CA, USA) was used. Similarity comparison of DNA sequence was carried out against the protein database in GenBank using BlastX. Amino acid sequence analyses were conducted using web-based software suites. muscle was used to create a sequence alignment file and calculated percent identity (pairwise alignment). The alignment output was created using GeneDoc software. The transmembrane regions were predicted using TopPred. A potential N-glycosylation site was obtained using NetNGlyc 1.0 Server.
Construction of plasmid and expression of rDvVATPaseV0a
The Baculovirus Expression System with Gateway Technology (Invitrogen) was used to express rDvVATPaseV0a according to manufacturer's protocol. Briefly, the ORF of DvVATPaseV0a was subcloned into the pENTR/D-TOPO entry vector (Invitrogen) and then transferred into the cloning cassette of the pDEST10 vector (N-terminal His fusion vector, Invitrogen). The cloned ORF DvVATPaseV0a plasmid was transformed into DH10Bac E. coli (Invitrogen), which contains the baculovirus shuttle vector (bacmid), to produce a recombinant bacmid harbouring DvVATPaseV0a. The white colonies of recombinant bacmid DNA in a background of blue colonies containing the unaltered parent bacmid were selected and isolated from small-scale selective (50 μg/ml kanamycin, 7 μg/ml Gentamycin, 10 μg/ml tetracycline, 100 μg/ml Bluo-gal, and 40 μg/ml isopropyl-beta-D-thiogalactopyranoside) medium. These recombinant baculoviruses (1 μg) were used to transfect 9 × 105 Sf9 insect cells (Gibco, Carlsbad, CA, USA) using Cellfectin reagent (Invitrogen) according to the manufacturer's recommendation. The transfected cells were maintained at 27 °C and the culture medium containing the recombinant baculoviruses was collected at 7 days post-transfection as a primary viral stock (P1).
The P1 viral stock (40 μl) was used to infect fresh Sf9 (2 × 106) cells and incubated at 27 °C for 4 days. The medium containing baculoviruses was then collected and designated as P2 viral stock. The P2 stock was diluted tenfold from 10−3 to 10−8 and the viral titer was determined by endpoint dilution as described by O'Reilly et al. (1994). To express the protein, Sf9 cells were infected with rDvVATPaseV0a baculoviruses at MOI of 1 and maintained at 27 °C for 4 days. The cells were then harvested, washed once with PBS and kept at −20 °C until used for protein purification.
Purification of DvVATPaseV0a from polyacrylamide gel
The infected cells were resuspended in lysis buffer and stirred at room temperature for 1 h. The lysate was centrifuged at 16 000 × g for 30 min and the supernatant was removed. The pellet was then added with 1x NuPAGE LDS Sample Buffer (Invitrogen) supplemented with 100 mM dithiothreitol (DTT, Fisher Scientific, Pittsburgh, PA, USA) and loaded onto NuPAGE 4–12% Bis-Tris Zoom gels (Invitrogen). The gels were then negatively stained with E-zinc Reversible Stain Kit (Pierce, Rockford, IL, USA) according to the manufacturer's instructions. The unstained band of rDvVATPaseV0a was then excised from the gel and the residual stain from the edges of the excised gel pieces was erased by soaking them in Tris-glycine buffer (25 mM Tris and 192 mM glycine). The excised gel pieces were crushed using a clean pestle and added with 0.5 ml elution buffer (50 mM Tris-HCl, 150 mM NaCl, and 0.1 mM EDTA; pH 7.5). The mixture was then incubated in a rotary shaker at 30 °C overnight and the protein was collected after centrifugation.
Protein identification
To confirm peptide sequences of purified rDvVATPaseV0a protein, SDS-PAGE coupled with mass spectrophotometry analysis was performed as described previously by Sunyakumthorn et al. (2008). Briefly, the protein band was excised using the Proteome Works Spot Cutter (Bio-Rad), and a MassPrep Station (Waters, Milford, MA, USA) was used as the digestion robot. The peptides were then extracted from the gel plugs and separated by liquid chromatography using an Atlantis dC18 column (75 μm by 100 mm; Waters/Micromass, Milford, MA, USA). For analysis, a Q-Tof (quadrupole time-of-flight) Micro (Waters/Micromass) hybrid mass spectrometer was used and electrospray analysis (positive mode) was performed. ProteinLynx Global Server, version 2.0 (Waters/Micromass) was used for data acquisition and analysis. Database comparative analysis with an online Mascot (Matrix Science, Boston, MA, USA) tandem mass spectrophotometry ion search against the NCBInr/Proteobacteria database was carried out. The resultant peaks of the ∼96-kDa band matched the V-ATPase protein.
Preparation of a polyclonal antibody specific for rDvVATPaseV0a
Polyclonal antibodies to purified rDvVATPaseV0a were generated in BALB/c mice as described by Mulenga et al. (2003). Briefly, three mice were immunized subcutaneously with rDvVATPaseV0a protein (∼30 μg protein per mouse) emulsified with equal volume of TiterMax Gold adjuvant (Sigma-Aldrich, St. Louis, MO, USA), followed by two more injections (∼60–80 μg protein per mouse) at 2-week intervals. Antiserum with reactivity toward rDvVATPaseV0a was collected 2 weeks after the third injection and stored at 4 °C for subsequent studies.
SDS-PAGE and Western blot analysis
Proteins were separated by NuPAGE 4–12% Bis-Tris gel (Invitrogen) and electronically transferred to polyvinylidene difluoride membrane (Bio-Rad). The membrane was blocked with 5% (w/v) skim milk in tris buffer saline (TBS)-Tween (T) buffer (20 mM Tris-HCl, 500 mM NaCl and 0.1% (v/v) Tween-20; pH 7.5) at room temperature for 1 h and incubated with either 1: 5000 dilution of anti-6xHis monoclonal antibody (Clontech) or a 1 : 200–500 dilution of polyclonal anti-rDvVATPaseV0a in TBS-T for 2 h. After three washes, the membrane was incubated with horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Pierce) diluted 1: 20 000 at room temperature for 1 h and then washed three times with TBS-T. The bound antibody complexes were detected using an enhanced chemiluminescent system (Pierce).
Cell culture
Sf9 cells (Gibco) were cultured in SF900 II serum-free medium (Gibco) supplemented with penicillin/streptomycin (50 U/ml and 50 μg/ml, respectively, Gibco). DVE1 cells (derived from D. variabilis) were grown in L15C medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Hyclone, Waltham, MA, USA), 5% tryptose phosphate broth (Difco, Sparks, MD, USA), 0.1% lipoprotein-cholesterol concentrate (MP Biomedicals, Santa Ana, CA, USA), 6 mM HEPES solution (Sigma-Aldrich), and 0.06% sodium bicarbonate (Sigma-Aldrich). Vero cells were grown in DMEM high glucose (Invitrogen) containing 5% fetal bovine serum (Hyclone). For all cell lines, conditioned medium was replaced with new medium once a week. Sf9 and DVE1 cells were passaged (1 : 5 and 1 : 3, respectively) approximately every 2 and 4 weeks, respectively. Vero cells were subcultured (1 : 6 or 1 : 12) every 1–2 weeks with 0.05% trypsin-EDTA (Invitrogen). Sf9 and DVE1 cells were grown in a humidified incubator at 27 and 32 °C, respectively. Vero cells were maintained in a humidified 5% CO2 incubator at 34 °C.
Rickettsia culture and purification
Rickettsia montanensis were maintained in Vero cells as described by Sunyakumthorn et al. (2012). The isolation of R. montanensis from infected Vero cells was carried out as previously described by Weiss (1973) with minor modification. Briefly, Rickettsia-infected cells were detached by scraping and lysed by vortexing with sterile 3-mm borosilicate glass beads for 5 min (Sigma-Aldrich). The cell lysate was then transferred aseptically to 15-ml centrifuge tubes and centrifuged at 4 °C, 275 × g for 3 min to pellet cellular debris. The supernatant was transferred to a 10-ml syringe and filtered through a 2-μm syringe filter. For all bioassays, the number of Rickettsia was determined by counting Rickettsia stained with a LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Carlsbad, CA, USA) in a Petroff-Hausser bacterial counting chamber (Hausser Scientific, Horsham, PA, USA) and examined using a Leica microscope (Buffalo Grove, IL, USA; Kurtti et al., 2005).
Expression of VATPaseV0a mRNA in backless Dermacentor variabilis
The backless D. variabilis was used as a model to study an expression of DvVATPaseV0a mRNA in tick tissues (midgut, ovary, salivary glands) in response to R. montanensis infection using a modified protocol of Bell (1980), as previously described by Sunyakumthorn et al. (2012). Briefly, six unfed female ticks were washed with 70% ethanol followed by 1% benzalkonium chloride solution and rinsed three times with sterile water. The ticks were air-dried and afterwards the mouthparts and the legs were excised. The ticks were then cut along the perimeter of alloscutum and the dorsal cuticle was removed. The backless ticks were individually transferred onto 96-well plate containing complete L15B medium and incubated at 34 °C. After 24 h, three ticks were infected with R. montanensis (2.4 × 108 per tick) for 1 h while the other three were kept in the medium without exposure to Rickettsia. Tick tissues were collected by removing and pooling the tissues from three different ticks. The tissues were placed in RNAlater and the samples were kept at −20 °C until used for RNA extraction. The experiments were performed independently twice.
Total RNA isolation and relative quantitative reverse transcription-PCR
Total RNA from uninfected- and R. montanensis-infected tick tissues was extracted using an RNeasy Mini Kit (Qiagen, Germantown, MD, USA), treated with TURBO DNase (Ambion), and purified using an RNA cleanup kit (Zymo Research, Irvine, CA, USA) as described in the manufacturers' manuals. First-strand cDNA was synthesized from 40 ng total RNA using iScript reverse transcription kit (Bio-Rad) according to the manufacturer's instructions. All qPCR reactions were performed in 96-well plates in a 35-μl volume containing 100 nM of each primer (5′-CTCCTGGCCGTGATTTGTAT- 3′ and 5′-GCTGCTCCGTCCTCTGTATC- 3′ for V-ATPase, 5′-CTCGTTCTTGGGAATGGAAG-3′ and 5′-CTTGATCTTCATGGTGGAAG G- 3′ for actin), DNase/RNase-free water, 2 μl of cDNA template (samples) or water (negative control) and 2X iTaq SYBR Green Supermix with ROX (Bio-Rad). The mixtures were then aliquoted in triplicate 10-μl reactions onto 384-well plates and run on an ABI 7900HT unit (Applied Biosystems, Carlsbad, CA, USA) at Louisiana State University, School of Veterinary Medicine. No RT reaction (water was added instead of reverse transcriptase) was performed to confirm an absence of gDNA. Results were analysed using an ABI 7900HT sequence detection system (sds v2.3) software. Data are presented as the percent difference in threshold cycle (CT) value (ΔCT = CT Actin − CT V-ATPase).
DvV-ATPase inhibition assay
DVE1 cells (1 × 105) were seeded onto 96-well plates (Greiner Bio-One, Monroe, NC, USA) and incubated at 32 °C for 48 h. The cells were treated with 5, 0.5, and 0.05 μm of the V-ATPase inhibitor, bafilomycin A1 (EMD Millipore, Billerica, MA,USA), or medium containing 0.1% dimethhyl sulphoxide (inhibitor vehicle control) for 2 h. R. montanensis was then inoculated onto the treated cells at MOI of 10 and the plate was centrifuged at 700 × g for 2 min to facilitate the binding of Rickettsia to host cells. After 1 h, Rickettsia was removed and the cells were added with 150 μl PBS. The samples were centrifuged at 275 × g for 4 min to collect only infected host cells. After removal of supernatant, the cell pellet was washed with 1 ml PBS and centrifuged at 275 × g for 4 min. The samples were stored at −20 °C until used for gDNA isolation. According to the manufacturer's instructions, gDNA was extracted from the samples using DNeasy Blood & Tissue Kit (Qiagen). Genomic DNA was eluted in 35 μl DNase/RNase free water. The number of Rickettsia and tick cells were then quantified by qPCR. The experiments were performed in quadruplicate for each concentration of the inhibitor used and the results were the combination of two independent experiments.
Construction of a standard reference plasmid for qPCR
A standard reference plasmid containing portions of R. montanensis ompB (RmOmpB) and D. variabilis calreticulin (DvCRT) genes was generated and used to create a standard curve in qPCR assays as described by Reif et al. (2008). Briefly, fragments of DvCRT (132 bp) and RmOmpB (106 bp) were amplified using CRTDv321FxbaI (5′-AAAAAATCTAGAAGGAGAAAAGCAAGGGACTG-3′)/CRTDv452R (5′CAATGTTCTGCTCGTGCTTG-3′) and OmpBRm2832F (5′GCGGTGGTGTTCCTAATAC-3′)/OmpBRm2937RxbaI (5′-AAAAAATCTAGACCTAAGTTGTTATAGTCTGTAGTG-3′) primer pairs, respectively. The amplicons were then digested with XbaI (New England BioLabs, Ipswich, MA, USA) and ligated together. The ligation product was amplified using OmpBRm2832F (5′-GCGGTGGTGTTCCTAATAC-3′) and CRTDv452R (5′-CAATGTTCTGCTCGTGCTTG-3′) primers, cloned into pCR4-TOPO vector (Invitrogen) and sequenced.
Quantification of Rickettsia and tick cells by qPCR
To quantify copies of R. montanensis and DVE1 genes in samples, probe-based qPCRs were performed as described by Thepparit et al. (2011). Briefly, serial dilutions of plasmids containing RmOmpB and DvCRT genes were used to create a standard curve. qPCR was performed using 0.3 μM each DvCRT_TYE665 (TYE665/5′-TGGAGAAGGGCTCGAACTTGGC-3′/IAbRQSp) and RmOmpB_HEX (HEX/5′-CGGGGCAAAGATGCTAGCGCTTCACAGTTA CCCCG-3′/IABk FQ) probes and 0.2 μM each CRTDv321F (5′-AGGAGAAAAGCAAGGGACTG-3′), CRTDv452R (5′CAATGTTCTGCTCGTGCTTG-3′), OmpBRm2832F (5′-GCGGTGG TGTTCCTAATAC-3′) and OmpBRm2937R (5′- CCTAAGTTGTTATAGTCTGTAGTG-3′) primers. Percent relative invasion was calculated by comparing the ratios of rickettsial gene to tick gene between treated and untreated control groups.
Statistical analysis
anova was conducted using the sas statistical package (Version 9.3) GLM procedure. For mRNA level measurement in backless ticks, the relative gene expression was analysed using a two-way interaction (rickettsial infection and tick tissues). Pairwise t-tests of least-square means were used to examine the interaction effects of relative mRNA expression of DvVATPaseV0a between unexposed and Rickettsia-exposed ticks. For V-ATPase inhibition assays, the same tests were used to elucidate a role of the V-ATPase inhibitor, bafilomycin A1, on rickettsial invasion of tick cells. P-values of ≤0.05 were considered to indicate statistical significance.
Acknowledgments
We thank Drs Timothy Kurtti and Ulrike Munderloh for the DVE1 cells. We also thank Jacqueline Macaluso for helpful comments. This research was supported by the National Institutes of Health AI077784 (K.R.M.) and AI43006 (A.F.A.). This work was also part of N. Petchampai's doctoral dissertation.
References
- Adachi I, Puopolo K, Marquez-Sterling N, Arai H, Forgac M. Dissociation, crosslinking and glycosylation of the coated vesicle proton pump. J Biol Chem. 1990;265:967–973. [PubMed] [Google Scholar]
- Bell LJ. Organ culture of Rhipicephalus appendiculatus with maturation of Theileria parva in tick salivary glands in vitro. Acta Trop. 1980;37:319–325. [PubMed] [Google Scholar]
- Bowman BJ, Bowman EJ. Mutations in subunit c of the vacuolar ATPase confer resistance to bafilomycin and identify a conserved antibiotic binding site. J Biol Chem. 2002;277:3965–3972. doi: 10.1074/jbc.M109756200. [DOI] [PubMed] [Google Scholar]
- Bowman BJ, McCall ME, Baertsch R, Bowman EJ. A model for the proteolipid ring and bafilomycin/concanamycin-binding site in the vacuolar ATPase of Neurospora crassa. J Biol Chem. 2006;281:31885–31893. doi: 10.1074/jbc.M605532200. [DOI] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, Klose KE, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, Horwitz MA. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect Immun. 2009;77:1757–1773. doi: 10.1128/IAI.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crider BP, Xie XS, Stone DK. Bafilomycin inhibits proton flow through the H+ channel of vacuolar proton pumps. J Biol Chem. 1994;269:17379–17381. [PubMed] [Google Scholar]
- Cruciat CM, Ohkawara B, Acebron SP, Karaulanov E, Reinhard C, Ingelfinger D, et al. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327:459–463. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Cossart P. Intracellular pathogens and the actin cytoskeleton. Annu Rev Cell Dev Biol. 1998;14:137–166. doi: 10.1146/annurev.cellbio.14.1.137. [DOI] [PubMed] [Google Scholar]
- Forgac M. Structure, mechanism and regulation of the clathrin-coated vesicle and yeast vacuolar H(+)-ATPases. J Exp Biol. 2000;203(Pt 1):71–80. doi: 10.1242/jeb.203.1.71. [DOI] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- de la Fuente J, Blouin EF, Manzano-Roman R, Naranjo V, Almazán C, Pérez de la Lastra JM, et al. Functional genomic studies of tick cells in response to infection with the cattle pathogen, Anaplasma marginale. Genomics. 2007;90:712–722. doi: 10.1016/j.ygeno.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- Grant B, Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, et al. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem. 2000;275:32331–32337. doi: 10.1074/jbc.M004795200. [DOI] [PubMed] [Google Scholar]
- Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocan KM, Zivkovic Z, Blouin EF, Naranjo V, Almazán C, Mitra R, et al. Silencing of genes involved in Anaplasma marginale-tick interactions affects the pathogen developmental cycle in Dermacentor variabilis. BMC Dev Biol. 2009;16:9–42. doi: 10.1186/1471-213X-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtti TJ, Simser JA, Baldridge GD, Palmer AT, Munderloh UG. Factors influencing in vitro infectivity and growth of Rickettsia peacockii (Rickettsiales: Rickettsiaceae), an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni (Acari, Ixodidae) J Invertebr Pathol. 2005;90:177–186. doi: 10.1016/j.jip.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. Infection and transovarial transmission of rickettsiae in Dermacentor variabilis ticks acquired by artificial feeding. Vector Borne Zoonotic Dis. 2001;1:45–53. doi: 10.1089/153036601750137660. [DOI] [PubMed] [Google Scholar]
- Macaluso KR, Mulenga A, Simser JA, Azad AF. Differential expression of genes in uninfected and Rickettsia-infected Dermacentor variabilis ticks as assessed by differential-display PCR. Infect Immun. 2003;71:6165–6170. doi: 10.1128/IAI.71.11.6165-6170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Seveau S, Veiga E, Matsuyama S, Cossart P. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123:1013–1023. doi: 10.1016/j.cell.2005.08.046. [DOI] [PubMed] [Google Scholar]
- McSwain JL, Luo C, de Silva GA, Palmer MJ, Tucker JS, Sauer JR, et al. Cloning and sequence of a gene for a homologue of the C subunit of the V-ATPase from the salivary gland of the tick Amblyomma americanum (L) Insect Mol Biol. 1997;6:67–76. doi: 10.1046/j.1365-2583.1997.00158.x. [DOI] [PubMed] [Google Scholar]
- Merzendorfer H, Reineke S, Zhao XF, Jacobmeier B, Harvey WR, Wieczorek H. The multigene family of the tobacco hornworm V-ATPase: novel subunits a, C, D, H, and putative isoforms. Biochim Biophys Acta. 2000;1467:369–379. doi: 10.1016/s0005-2736(00)00233-9. [DOI] [PubMed] [Google Scholar]
- Mitchell RD, 3rd, Ross E, Osgood C, Sonenshine DE, Donohue KV, Khalil SM, et al. Molecular characterization, tissue-specific expression and RNAi knockdown of the first vitellogenin receptor from a tick. Insect Biochem Mol Biol. 2007;37:375–388. doi: 10.1016/j.ibmb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Mulenga A, Macaluso KR, Simser JA, Azad AF. Dynamics of Rickettsia-tick interactions: identification and characterization of differentially expressed mRNAs in uninfected and infected Dermacentor variabilis. Insect Mol Biol. 2003;12:185–193. doi: 10.1046/j.1365-2583.2003.00400.x. [DOI] [PubMed] [Google Scholar]
- Munderloh UG, Kurtti TJ. Cellular and molecular interrelationships between ticks and prokaryotic tick-borne pathogens. Annu Rev Entomol. 1995;40:221–243. doi: 10.1146/annurev.en.40.010195.001253. [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases-nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- O'Reilly DR, Miller LM, Luckow VA. Baculovirus Expression Vectors; A Laboratory Manual. New York: Oxford University Press; 1994. [Google Scholar]
- Oka T, Futai M. Requirement of V-ATPase for ovulation and embryogenesis in Caenorhabditis elegans. J Biol Chem. 2000;275:29556–29561. doi: 10.1074/jbc.M002756200. [DOI] [PubMed] [Google Scholar]
- Oka T, Toyomura T, Honjo K, Wada Y, Futai M. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem. 2001;276:33079–33085. doi: 10.1074/jbc.M101652200. [DOI] [PubMed] [Google Scholar]
- Portnoy DA, Chakraborty T, Goebel W, Cossart P. Molecular determinants of Listeria monocytogenes pathogenesis. Infect Immun. 1992;60:1263–1267. doi: 10.1128/iai.60.4.1263-1267.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautiala TJ, Koskinen AM, Väänänen HK. Purification of vacuolar ATPase with bafilomycin C1 affinity chromatography. Biochem Biophys Res Commun. 1993;194:50–56. doi: 10.1006/bbrc.1993.1783. [DOI] [PubMed] [Google Scholar]
- Reif KE, Stout RW, Henry GC, Foil LD, Macaluso KR. Prevalence and infection load dynamics of Rickettsia felis in actively feeding cat fleas. PLoS ONE. 2008;3:e2805. doi: 10.1371/journal.pone.0002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M, Graham LA, Stevens TH. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol Biol Cell. 2008;19:5131–5142. doi: 10.1091/mbc.E08-06-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sappington TW, Hays AR, Raikhel AS. Mosquito vitellogenin receptor: purification, developmental and biochemical characterization. Insect Biochem Mol Biol. 1995;25:807–817. doi: 10.1016/0965-1748(95)00016-o. [DOI] [PubMed] [Google Scholar]
- Schonbaum CP, Lee S, Mahowald AP. The Drosophila yolkless gene encodes a vitellogenin receptor belonging to the low density lipoprotein receptor superfamily. Proc Natl Acad Sci U S A. 1995;92:1485–1489. doi: 10.1073/pnas.92.5.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine DE. The biology of tick vectors of human disease. In: Goodman JL, Dennis DT, Sonenshine DE, editors. Tick-Borne Diseases of Humans. Washington, DC: ASM Press; 2005. pp. 12–36. [Google Scholar]
- Sunyakumthorn P, Bourchookarn A, Pornwiroon W, David C, Barker SA, Macaluso KR. Characterization and growth of polymorphic Rickettsia felis in a tick cell line. Appl Environ Microbiol. 2008;74:3151–3158. doi: 10.1128/AEM.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyakumthorn P, Petchampai N, Kearney MT, Sonenshine DE, Macaluso KR. Molecular characterization and tissue-specific gene expression of Dermacentor variabilis α-catenin in response to rickettsial infection. Insect Mol Biol. 2012;21:197–204. doi: 10.1111/j.1365-2583.2011.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepparit C, Bourchookarn A, Petchampai N, Barker SA, Macaluso KR. Interaction of Rickettsia felis with histone H2B facilitates the infection of a tick cell line. Microbiology. 2010;156(Pt 9):2855–2863. doi: 10.1099/mic.0.041400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thepparit C, Sunyakumthorn P, Guillotte ML, Popov VL, Foil LD, Macaluso KR. Isolation of a Rickettsial pathogen from a non-hematophagous arthropod. PLoS ONE. 2011;6:e16396. doi: 10.1371/journal.pone.0016396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitavska O, Wieczorek H, Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem. 2003;278:18499–18505. doi: 10.1074/jbc.M212844200. [DOI] [PubMed] [Google Scholar]
- Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6:375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- Wang Y, Inoue T, Forgac M. Subunit a of the yeast V-ATPase participates in binding of bafilomycin. J Biol Chem. 2005;280:40481–40488. doi: 10.1074/jbc.M509106200. [DOI] [PubMed] [Google Scholar]
- Weiss E. Growth and physiology of rickettsiae. Bacteriol Rev. 1973;37:259–283. doi: 10.1128/br.37.3.259-283.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Reed SCO, Haglund CM. Establishing intracellular infection: escape from the phagosome and intracellular colonization (Rickettsiaceae. In: Palmer GH, Azad AF, editors. Intracellular Pathogens II: Rickettsiales. Washington, DC: ASM Press; 2012. pp. 243–269. [Google Scholar]
- Wieczorek H, Beyenbach KW, Huss M, Vitavska O. Vacuolar-type proton pumps in insect epithelia. J Exp Biol. 2009;212(Pt11):1611–1619. doi: 10.1242/jeb.030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasir M, Pachikara ND, Bao X, Pan Z, Fan H. Regulation of Chlamydial infection by host autophagy and vacuolar ATPase-bearing organelles. Infect Immun. 2011;79:4019–4028. doi: 10.1128/IAI.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Feng Y, Forgac M. Proton conduction and bafilomycin binding by the V0 domain of the coated vesicle V-ATPase. J Biol Chem. 1994;269:23518–23523. [PubMed] [Google Scholar]