Abstract
N-myc belongs to the myc proto-oncogene family, which is involved in numerous cellular processes such as proliferation, growth, apoptosis, and differentiation. Conditional deletion of N-myc in the mouse nervous system disrupted brain development, indicating that N-myc plays an essential role during neural development. How the development of the olfactory epithelium and neurogenesis within are affected by the loss of N-myc has, however, not been determined. To address these issues, we examined an N-mycFoxg1Cre conditional mouse line, in which N-myc is depleted in the olfactory epithelium. First changes in N-myc mutants were detected at E11.5, with reduced proliferation and neurogenesis in a slightly smaller olfactory epithelium. The phenotype was more pronounced at E13.5, with a complete lack of Hes5-positive progenitor cells, decreased proliferation, and neurogenesis. In addition, stereological analyses revealed reduced cell size of post-mitotic neurons in the olfactory epithelium, which contributed to a smaller olfactory pit. Furthermore, we observed diminished proliferation and neurogenesis also in the vomeronasal organ, which likewise was reduced in size. In addition, the generation of gonadotropin-releasing hormone neurons was severely reduced in N-myc mutants. Thus, diminished neurogenesis and proliferation in combination with smaller neurons might explain the morphological defects in the N-myc depleted olfactory structures. Moreover, our results suggest an important role for N-myc in regulating ongoing neurogenesis, in part by maintaining the Hes5-positive progenitor pool. In summary, our results provide evidence that N-myc deficiency in the olfactory epithelium progressively diminishes proliferation and neurogenesis with negative consequences at structural and cellular levels. © 2013 The Authors. Developmental Neurobiology Published by Wiley Periodicals, Inc. Develop Neurobiol 74: 643–656, 2014
Keywords: neurogenesis, N-myc, olfactory epithelium, vomeronasal organ, mouse
INTRODUCTION
The myc family of proto-oncogenes consists of c-myc, N-myc, and L-myc, three related genes involved in diverse biological processes such as proliferation, differentiation, and apoptosis [reviewed in (Henriksson and Luscher, 1996; Facchini and Penn, 1998; Eilers and Eisenman, 2008)]. Mice deficient in c-myc or N-myc die at about embryonic day 10 (E10) or E12, respectively (Charron et al., 1992; Trumpp et al., 2001), whereas L-myc deficiency has no lethal phenotype (Hatton et al., 1996). N-myc-deficient embryos display delayed development of organs, which normally express high levels of N-myc, such as the heart, lung, and gut (Charron et al., 1992). Interestingly, these developmental defects in N-myc mutants occurred despite the compensatory increase of c-myc expression (Stanton et al., 1992), which underlines the essential functions of N-myc during development. Conditional deletion of N-myc in neuronal progenitor cells prevented the early lethal phenotype and uncovered N-myc as crucial factor during development of the nervous system (Knoepfler et al., 2002; Dominguez-Frutos et al., 2011; Kopecky et al., 2011). N-myc-deficient mice exhibited abnormal behavior in correlation with a twofold reduction in brain mass, including severe defects in the cerebellum (Knoepfler et al., 2002). In addition, conditional deletion of N-myc in the otic placode severely affects inner ear development, including perturbed morphology and disorganized neuronal innervation (Dominguez-Frutos et al., 2011; Kopecky et al., 2011).
As part of the peripheral nervous system, the olfactory placode gives rise to the olfactory epithelium (also known as main olfactory epithelium) and the vomeronasal organ. In general, olfactory sensory neurons in the olfactory epithelium transmit odor sensation, while the vomeronasal organ detects pheromones and both structures project axons to discrete target regions of the olfactory bulb. In addition, gonadotropin-releasing hormone (GnRH) neurons have been suggested to originate from the epithelia of the vomeronasal organ and medial wall of the olfactory pit (Schwanzel-Fukuda and Pfaff, 1989; Wray et al., 1989). The GnRH neurons migrate in association with olfactory epithelial- and vomeronasal organ-derived axons towards the hypothalamus in the forebrain (Wray et al., 1994; Norgren et al., 1995; Yoshida et al., 1995). The production of GnRH in the hypothalamus controls the reproductive system in vertebrates by stimulating the release of gonadotrophins from the anterior pituitary, which affects the gonadal functions [reviewed in (Wray, 2010)]. To date, no correlation between N-myc and GnRH neurons has been reported.
The olfactory epithelium is one of the few regions in the nervous system, where neurogenesis persists throughout life to generate olfactory sensory neurons. Therefore, it serves as a useful model system to study regulatory processes during embryonic and adult neurogenesis (Cau et al., 1997; Kawauchi et al., 2009; Tucker et al. 2010; Fletcher et al., 2011; Maier et al., 2011; Packard et al., 2011). Neurogenesis in the olfactory epithelium begins already at olfactory placodal stages in both mouse and chick, and the sensory neuronal lineage contains stem-like cells, neuronal precursor cells at different maturity stages and post-mitotic olfactory neurons (Kawauchi et al., 2005; Maier and Gunhaga, 2009; Wei et al., 2013). The different cell types can be defined by the expression of specific molecular markers; stem-like progenitor cells express the basic helix-loop-helix repressor gene Hes5 (Cau et al., 2000; Maier and Gunhaga, 2009), the immediate neuronal precursor cells are defined by the expression of Neurogenin1 (Ngn1) (Cau et al., 2002; Maier and Gunhaga, 2009), and cells committed to leave the cell cycle express the terminal neuronal differentiation marker NeuroD1 (Cau et al., 2002). All post-mitotic neurons express the general neuronal markers HuC/D (Fornaro 2003) and Tuj1 (Wei et al., 2013), while a subset of the neurons express the LIM-homeodomain transcription factor Lhx2 (Hirota and Mombaerts, 2004; Kolterud et al., 2004). However, the potential impact of myc proto-oncogenes on the development of the olfactory epithelium and neurogenesis therein has not been addressed.
In this study, we have analyzed the influence of N-myc on the development of the olfactory epithelium and neurogenesis within, using a recently described conditional mouse line, where N-myc deficiency is restricted to Foxg1-positive cells (Dominguez-Frutos et al., 2011). The expression of Foxg1 (also known as brain factor-1) is detected already at the initiation of neurogenesis in the olfactory placode of mice at E9.5 (Xuan et al., 1995), and also at later stages of olfactory development (Kawauchi et al., 2009). Our results show a progressive effect of N-myc deficiency displaying reduced proliferation, neurogenesis, and disturbed morphogenesis in the mouse olfactory epithelium from E10.5 until E13.5. At E13.5, the population of stem-like progenitors is depleted and proliferation and the generation of neurons are reduced, resulting in a severely reduced olfactory epithelium. Thus, N-myc is an essential factor for ongoing neurogenesis and proper development of the olfactory sensory epithelium.
METHODS
Transgenic Mouse Embryos
A recently described conditional N-myc transgenic mouse line (Dominguez-Frutos et al., 2011) was used. Briefly, the N-mycFoxg1Cre line was generated by crossing N-mycflox/flox mice (Knoepfler et al., 2002) with a mouse strain carrying a Cre recombinase under control of the Foxg1 locus (Hebert and McConnell, 2000). N-mycFoxg1Cre (hereafter referred to as N-myc−/−) mutants display Foxg1-mediated loxP recombination in the telencephalon and discrete head structures including the olfactory epithelium. Less than 10% of homozygous N-myc mutants are obtained from heterozygous crosses as previously discussed in (Dominguez-Frutos et al., 2011). Both, the N-myc and Foxg1 gene loci are localized on the same chromosome, which leads to the heterozygous loss of the Foxg1 coding region in Foxg1-N-myc mutants (Hebert and McConnell, 2000). This could explain the breeding anomaly of the N-myc−/− strain in respect to the Mendelian inheritance pattern. The generation and genotyping of the N-myc−/− mouse line was performed as previously described (Dominguez-Frutos et al., 2011). The embryos were fixed in 4% PFA at 4°C for 3–6 hours, cryoprotected in 30% sucrose at 4°C overnight (ON), embedded in TissueTek (Gibco, Stockholm, Sweden), frozen and stored at −80°C. The use of N-myc−/− mutant and control mice was approved by the Committee on the Welfare of Experimental Laboratory Animals of the University of Valladolid.
In Situ RNA Hybridization and Immunohistochemistry
In situ RNA hybridization was performed essentially as previously described (Wilkinson and Nieto, 1993) on transversal consecutive sections (10 µm) of the entire olfactory epithelium. Applied mouse digoxigenin-labeled probes were as follows: c-myc (Kapeli and Hurlin, 2011), Hes1 (Apelqvist et al., 1999), Hes5 (Machold et al., 2007), Ngn1 (gift from G. Fishell), NeuroD1 (Cau et al., 1997), N-myc (Potvin et al., 2010), and Notch1 (Stump et al., 2002). Immunohistochemistry was performed using standard protocols. Briefly, sections were blocked in 10% fetal calf serum at room temperature (RT) and primary antibodies were incubated at 4°C ON. Antibodies used were as follows; monoclonal mouse antibodies: anti-HuC/D (1:200, Molecular Probes, Göteborg, Sweden) and anti-neuronal class III Beta-Tubulin (Tuj1) (1:500, Covance, USA), polyclonal rabbit antibodies: anti-phospho-Histone H3 (1:500, Millipore, Solna, Sweden), anti-Lhx2 (1:4000, gift from Thomas M. Jessell), anti-cleaved Caspase3 (1:1000, Cell Signaling, Stockholm, Sweden), and anti-GnRH (1:1000, Fisher, Göteborg, Sweden). Sections were incubated with the appropriate Alexa Fluor secondary antibodies (1:400, Molecular Probes, Göteborg, Sweden) for 1 hour at RT and nuclei were stained using DAPI (1:600, Sigma, Stockholm, Sweden). Slides were mounted with fluorescent or Glycergel mounting medium (Dako, Stockholm, Sweden).
Stereology
The size of HuC/D+ (HuC/D-positive) neurons was analyzed using the unbiased estimation of the volume of particles (Gundersen, 1986). Images of the olfactory epithelium and vomeronasal organ were taken using a 0.75 numerical aperture lens on a Nikon Eclipse E800 microscope, equipped with a CCD camera connected to a PC (Nikon Imaging Software NIS-Elements). Images of HuC/D and DAPI staining were merged and processed with Photoshop CS2 software (Adobe) and the diameter of 25–30 neurons per structure and hemisphere was measured using ImageJ software (http://rsb.info.nih.gov/ij/). The unbiased volume of HuC/D+ neurons was calculated with the following estimation; π/3 times of the diameter length l0 raised to the third power (Gundersen, 1986).
Statistical Analysis and Imaging
The quantification of Caspase3, HuC/D, Lhx2, pHH3, and GnRH immunopositive cells as well as Hes5, Ngn1, and NeuroD1 in situ positive cells were performed using a 0.75 numerical aperture lens on a Nikon Eclipse E800 microscope. At all stages used in this study, the left and right hemispheres were analyzed separately and the mean values used for statistics. In addition, at E11.5 and E13.5, the medial and lateral parts of the olfactory epithelium were analyzed separately. All quantitative data of cell numbers and cell size were compared between age-matched N-myc−/− mice and control littermates. To consider the reduced morphology of the olfactory epithelium and vomeronasal organ in N-myc−/− embryos, the total number of cells was determined by counting the number of DAPI-positive nuclei. All data from the cell counting were corrected to the total cell number by structure and hemisphere. Quantification and image generation was performed using a Nikon Eclipse E800 microscope for simultaneous Epi-fluorescence/DIC observations, equipped with a CCD camera connected to a PC (Nikon Imaging Software NIS-Elements). Images were processed using Photoshop CS2 (Adobe). The graphs represent the mean number or mean ± SEM if not stated otherwise. Significant effects were confirmed by Student's t test, with p values of <0.05 (*), < 0.01 (**), < 0.0001 (***) accepted as statistically significant.
RESULTS
N-myc is Expressed at the Onset of Neurogenesis in the Olfactory Placode
To examine whether N-myc is expressed at the onset of neurogenesis in the olfactory placode, we analyzed the expression of N-myc and various markers of the sensory neuronal lineage in E9.5 wild-type mouse embryos. N-myc expression was scattered throughout the olfactory placode at E9.5, and consistent with the onset of neurogenesis cells in the olfactory placode also expressed Hes5, Ngn1, NeuroD1, HuC/D, and Tuj1 (Fig. 1). However, Lhx2+ post-mitotic neurons were not generated at E9.5 (Fig. 1). Proliferative cells were detected in the olfactory placode indicated by expression of phosphorylated Histone H3 (pHH3), a marker for mitotic cells (Sholl-Franco et al., 2010) (Fig. 1). Thus, the transcription factor N-myc is expressed at the onset of neurogenesis in the olfactory placode of mice.
Figure 1.
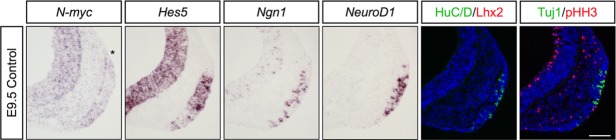
N-myc is expressed at E9.5, at the onset of neurogenesis, in the olfactory placode. At E9.5, cells in the mouse olfactory placode express the neurogenic markers N-myc, Hes5, Ngn1, and NeuroD1. HuC/D+ post-mitotic neurons, Tuj1+ neurons, and pHH3+ proliferative cells are detectable in the olfactory placode, but not Lhx2+ post-mitotic neurons. The borders of the olfactory placode are indicated by asterisks. Scale bar: 100 µm.
Neurogenesis in the Olfactory Epithelium is not Dependent on N-myc at E10.5
To study the influence of N-myc on early neurogenesis in the olfactory epithelium, we analyzed E10.5 N-myc−/− mice and their control littermates. As expected, N-myc expression was present in the olfactory epithelium of control embryos, and was completely absent in N-myc−/− embryos at E10.5 [Fig. 2(A)]. However, no differences in the generation of Hes5+ stem-like progenitors and Ngn1+ neuronal precursors were detectable between N-myc−/− and control mice [Fig. 2(A)]. Double labeling with HuC/D and Lhx2 revealed the onset of Lhx2 expression in a subset of post-mitotic neurons in the olfactory epithelium at E10.5 [Fig. 2(A)]. The generation of Lhx2+ neurons, Tuj1+ neurons was unaffected in N-myc mutants, and the quantitative analysis of HuC/D+ neurons and proliferative pHH3+ cells revealed no differences between N-myc−/− mice and their control littermates [Fig. 2(A–C)]. These data indicate that at E10.5, early neurogenesis in the olfactory epithelium is not affected by N-myc deficiency.
Figure 2.
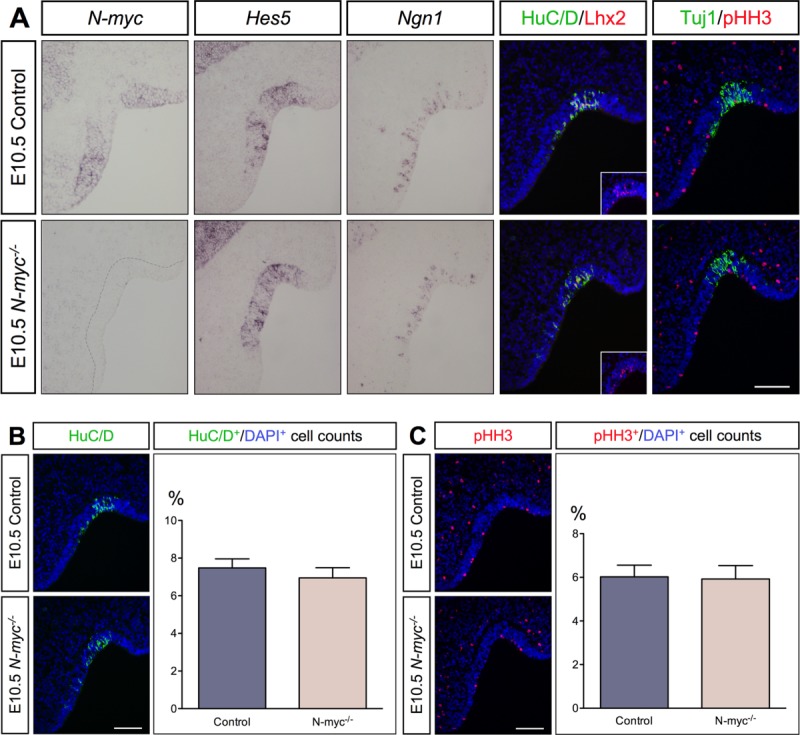
At E10.5, the development of the olfactory epithelium is not dependent on N-myc. (A) The expression of N-myc is completely missing in the olfactory epithelium of E10.5 N-myc mutants, whereas the generation of Hes5+ stem-like progenitors and Ngn1+ neuronal precursors are not different between both genotypes. The generation of Lhx2+, HuC/D+, and Tuj1+ post-mitotic neurons are not different between mutants and controls. The onset of Lhx2 expression is indicated in the insets. (B,C) N-myc deficiency did not alter the number of HuC/D+ post-mitotic neurons (B) or proliferation, indicated by pHH3+ staining (C). Statistical analysis of the cell counts in comparison to the total cell number in E10.5 control embryos (n = 5) and N-myc−/− animals (n = 4) (HuC/D p = 0.5296, pHH3 p = 0.9104). Error bars represent ±SEM, Student's t test. Scale bars: 100 µm.
To evaluate whether a possible up-regulation of c-myc expression could compensate for the loss of N-myc activity at this early stage, we analyzed c-myc expression in the olfactory epithelium at E10.5. However, no difference in c-myc expression was detected in the olfactory epithelium of N-myc−/− mice at E10.5 compared to control embryos (Supporting Information Fig. 1).
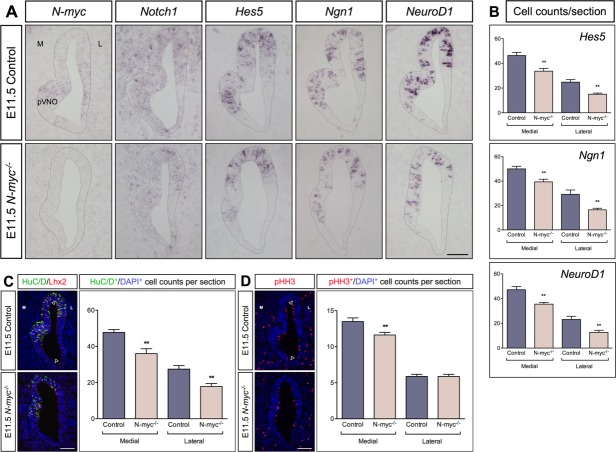
Altered Neurogenesis and Morphological Defects Observed in N-myc−/− Mice at E11.5
Next, we examined the olfactory epithelium in N-myc−/− mice and their control littermates at E11.5. Consistent with the results from E10.5, N-myc expression in the olfactory epithelium was only detected in control mice, but not in E11.5 N-myc−/− embryos [Fig. 3(A)]. In addition, the generation of Hes5+ stem-like progenitors, Ngn1+ neural precursors, and NeuroD1+ neuronal differentiation was reduced in the olfactory epithelium of N-myc−/− mutants [Fig. 3(A)].
Figure 3.
Altered proliferation, neurogenesis, and morphology in the olfactory epithelium at E11.5. (A) N-myc expression is completely missing in the olfactory epithelium of E11.5 N-myc−/− mice, Notch1 expression is down-regulated and the generation of Hes5+ progenitors, Ngn1+ neuronal precursors, and NeuroD1+ cells are decreased compared to controls. At this stage, the morphology of the olfactory epithelium is disturbed, displaying a less prominent area of the prospective vomeronasal organ (pVNO) in mutant mice. (C,D) The number of HuC/D+ and Lhx2+ post-mitotic neurons (C), and the number of pHH3+ mitotic cells are reduced (D). The borders between the medial (M) and lateral (L) part of the olfactory epithelium are indicated by arrowheads. (B–D) Statistical analysis of the cell counts in comparison to the total cell number defined by DAPI in the E11.5 olfactory epithelium of controls (n = 7) and N-myc−/− embryos (n = 6) for Hes5 (control versus N-myc−/− medial **p = 0.0041, and lateral **p = 0.0013), Ngn1 (control versus N-myc−/− medial **p = 0.0060, and lateral **p = 0.0071), NeuroD1 (control versus N-myc−/− medial **p = 0.0023, and lateral **p = 0.0062), HuC/D (control versus N-myc−/− medial **p = 0.0025, and lateral **p = 0.0037), and pHH3 (control versus N-myc−/− medial, n = 8, **p = 0.0096). Error bars represent ±SEM, Student's t test. Scale bars: 100 = µm.
Interestingly, the olfactory epithelium was smaller in N-myc−/− embryos, exhibiting a less prominent prospective vomeronasal organ in comparison to control animals [Fig. 3(A)]. This morphological defect was not caused by increased apoptosis as Caspase3 expression was not changed (data not shown). In contrast, the number of HuC/D+ post-mitotic neurons was reduced in both the medial and lateral part of the N-myc-deficient olfactory epithelium [Fig. 3(B)]. In addition, the number of Lhx2+ neurons was also reduced in the medial and lateral part of the olfactory epithelium in N-myc−/− embryos, to a similar extent as HuC/D+ neurons (Supporting Information Fig. S2). Moreover, proliferation indicated by pHH3 expression was also reduced, however, only in the medial part of the N-myc-deficient olfactory epithelium [Fig. 3(C)]. Double labeling with HuC/D and Lhx2 indicated that approximately 90% of HuC/D+ post-mitotic neurons also expressed Lhx2 in both N-myc−/− and wild-type embryos [Fig. 3(B)].
Both proliferation and Hes5 expression has been shown to be regulated by Notch activity (Ohtsuka et al., 1999; Basak and Taylor, 2007). Subsequently, we also examined the expression of Notch1 in the olfactory epithelium. At E11.5, Notch1 expression was clearly reduced in the olfactory epithelium of N-myc−/− mice [Fig. 3(A)], indicating that N-myc activity is required for Notch activity and proliferation in the olfactory epithelium. Thus, at E11.5 loss of N-myc reduces Notch activity, proliferation, and ongoing neurogenesis, resulting in a smaller olfactory epithelium.
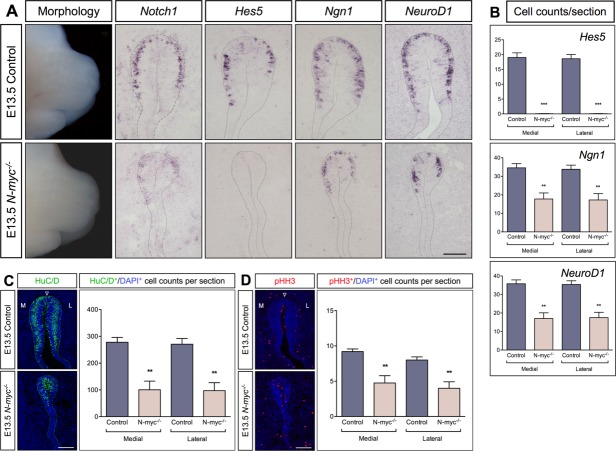
Progressive Effects on Olfactory Neurogenesis in N-myc−/− Mutants
To cover a later phase of olfactory neurogenesis, we investigated E13.5 mouse embryos, where the vomeronasal organ has separated from the olfactory epithelium. Despite the normal appearance of the mouse head, especially the nose, the olfactory epithelium was severely reduced in size in E13.5 N-myc−/− mutants [Fig. 4(A)]. Important to note is that apoptosis was not the cause for the smaller olfactory epithelium, since the number of Caspase3+ cells was similar in N-myc−/− compared to wild-type mice (Supporting Information Fig. S3A). In contrast, the number of pHH3+ mitotic cells was reduced in both the medial and lateral part of the N-myc-deficient olfactory epithelium [Fig. 4(C)]. Thus, the differential loss in proliferation between the medial and lateral parts observed in the olfactory epithelium of N-myc mutants at E11.5 were no longer detectable at E13.5
Figure 4.
Progressive reductions of proliferation, neurogenesis, and morphology in the olfactory epithelium at E13.5. (A) The nose is indistinguishable between N-myc mutants and control embryos. However, Notch1 expression was reduced, the generation of Hes5+ progenitors was completely diminished, and the olfactory epithelium was malformed and severely reduced in size in N-myc mutants. Moreover, the generation of Ngn1+ neuronal precursors, and NeuroD1+ cells decreased in N-myc mutants. (C,D) The number of HuC/D+ post-mitotic neurons (C) and proliferative pHH3+ cells (D) are reduced in the medial (M) and lateral (L) part of the olfactory epithelium. (B–D) Statistical analysis of the cell counts in comparison to the total cell number in the olfactory epithelium of E13.5 controls (n = 5) and N-myc mutants (n = 4) for Hes5 (control versus N-myc−/− medial ***p ≤ 0.0001, and lateral ***p ≤ 0.0001), Ngn1 (control versus N-myc−/− medial **p = 0.0032, and lateral **p = 0.0040), NeuroD1 (control versus N-myc−/− medial **p = 0.0012, and lateral **p = 0.0011), HuC/D (control versus N-myc−/− medial **p = 0.0015, and lateral **p = 0.0018), and pHH3 (control versus N-myc−/− medial **p = 0.0030, and lateral **p = 0.0040). Error bars represent ±SEM, Student's t test. Scale bars: 100 µm.
Interestingly, at E13.5 the generation of Hes5+ stem-like progenitor cells was completely lost [Fig. 4(A)], without any compensatory up-regulation of Hes1 (Supporting Information Fig. S3B). Consistently, Notch1 expression was diminished in N-myc-deficient mice [Fig. 4(A)]. Furthermore, the generation of Ngn1+ neural precursors and NeuroD1+ terminally differentiated neurons appeared to be reduced in N-myc−/− embryos [Fig. 4(A)]. Consistently, the number of HuC/D+ neurons was reduced in both the medial and lateral part of the N-myc-deficient olfactory epithelium [Fig. 4(B)]. Our data at E13.5 indicate a progressive effect of N-myc deficiency, in which decreased proliferation, reduced Notch activity and depletion of the Hes5+ progenitor cells results in a significantly smaller olfactory pit and suppressed neurogenesis.
Development of the Vomeronasal Organ and GnRH Neurons are Dependent on N-myc
At E11.5, the vomeronasal organ is part of the olfactory epithelium, but separates from it around E13 to develop independently. The severe effect of N-myc deficiency on the E13.5 olfactory epithelium guided us to also analyze the vomeronasal organ in these embryos. We detected a much smaller vomeronasal organ in N-myc−/− mutants compared to control littermates (Fig. 5). The decreased size of the vomeronasal organ was not due to an increase in apoptosis, as measured by the presence of Caspase3+ cells (data not shown). The generation of Ngn1+ neural precursors and terminal neuronal differentiation indicated by NeuroD1 was strongly reduced in N-myc-deficient mice [Fig. 5(A)]. Furthermore, the number of HuC/D+ post-mitotic neurons was reduced in both the sensory and non-sensory part of the vomeronasal organ [Fig. 5(B)], whereas a reduction in proliferative pHH3+ cells was restricted to the sensory portion of the vomeronasal organ [Fig. 5(C)]. Although, Hes5+ stem-like progenitors were absent [Fig. 5(A)], Notch1 expression appeared to be less affected by N-myc deficiency in the vomeronasal organ compared to the olfactory epithelium [Fig. 5(A)]. No compensatory up-regulation of Hes1 expression was observed in N-myc mutants (Supporting Information Fig. S3B).
Figure 5.
Development of the vomeronasal organ is dependent on N-myc. (A) The vomeronasal organ (VNO) is clearly smaller in N-myc-deficient mice compared to control animals. The expression of Notch1 is mildly down-regulated, whereas the generation of Hes5+ stem-like progenitors is completely absent, and Ngn1+ neuronal precursors and NeuroD1+ neurons are reduced in the N-myc-deficient VNO. The medial (M) and lateral (L) area of the VNO are indicated. (C,D) The number of HuC/D+ post-mitotic neurons is reduced in the sensory (SE) and non-sensory (NSE) part (C), while pHH3+ proliferative cells are only reduced in the SE-VNO (D). (B–D) Statistical analysis of the total cell counts in the VNO of E13.5 controls (n = 5) and N-myc mutants (n = 4) for Hes5 (control versus N-myc−/− SE-VNO ***p ≤ 0.0001), Ngn1 (control versus N-myc−/− SE-VNO ***p = 0.0002), NeuroD1 (control versus N-myc−/− SE-VNO ***p = 0.0003, and NSE-VNO ***p ≤ 0.0001), HuC/D (control versus N-myc−/− SE-VNO **p = 0.0028, and NSE-VNO ***p = 0.0003) (C), and pHH3 (control versus N-myc−/− SE-VNO **p = 0.0052) (D). Error bars represent ±SEM, Student's t test. Scale bars: 100 µm.
It has been suggested that GnRH neurons originate from the epithelia of the vomeronasal organ and medial wall of the olfactory pit (Schwanzel-Fukuda and Pfaff, 1989; Wray et al., 1989). Since both of these structures are severely reduced in size in the N-myc mutants [Figs. 4(A) and 5(A)], we analyzed whether the generation of GnRH neurons were disturbed in N-myc mutants at E13.5. Our results show an approximate 85% reduction of GnRH neurons in N-myc mutants at E13.5 compared to wild-type embryos (Fig. 6). These results indicate that N-myc plays an important role for the development of the vomeronasal organ including the generation of GnRH neurons.
Figure 6.
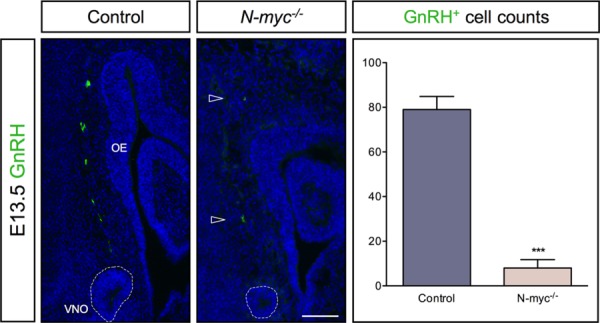
Severely reduced numbers of GnRH neurons in -myc-deficient mice. At E13.5, the numbers of GnRH neurons are severely reduced (∼85%) (arrowheads) in N-myc-deficient mice (n = 3) compared to wild type embryos (n = 4) (***p = 0.0002). The olfactory epithelium (OE) and vomeronasal organ (VNO) are indicated. Error bars represent ±SEM, Student's t test. Scale bar: 100 µm.
N-myc is Important for Neurons to Maintain Their Cell Size
To further examine the smaller olfactory pit and vomeronasal organ in N-myc-deficient mice, we analyzed the size of single cells in these structures. HuC/D staining is mainly restricted to the cytoplasm of post-mitotic neurons (Fornaro et al., 2003), which are representative for a large subpopulation of cells in the olfactory epithelium. Therefore, HuC/D+ neurons provided a sufficient number of cells for stereological analysis in the affected olfactory epithelium of N-myc−/− mutants. At E10.5, the size of HuC/D+ neurons was not different between N-myc−/− and control embryos [Fig. 7(A)]. However, at E11.5 HuC/D+ neurons were about 18% smaller in the olfactory epithelium of N-myc−/− mutants, with the smallest neurons located in the lateral part [Fig. 7(B)]. At E13.5, these neurons showed a size reduction of approximately 24% in N-myc-deficient mice throughout the olfactory epithelium [Fig. 7(C)].
Figure 7.
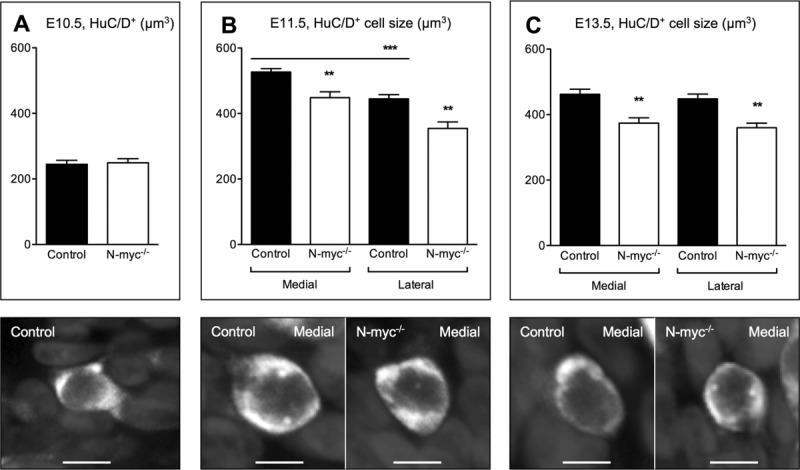
N-myc-deficient neurons are smaller in the olfactory epithelium. Stereological analysis of the size of HuC/D+ neurons in the olfactory epithelium at E10.5 (control n = 5, N-myc−/− n = 4), and in the medial and lateral part at E11.5 and E13.5 (E11.5: control n = 7, N-myc−/− n = 6; E13.5: control n = 5, N-myc−/− n = 4). (A) No difference in cell size between E10.5 controls and N-myc mutants. (B) At E11.5, HuC/D+ neurons are smaller in the lateral part (medial control versus lateral control ***p = 0.0003), and N-myc-deficient neurons are smaller in both the medial (**p = 0.0022) and lateral part (**p = 0.0025) compared to controls. Note that HuC/D+ neurons doubled their size between E10.5 and E11.5. (C) At E13.5, the size of HuC/D+ neurons is similar between the lateral and medial part of controls, but smaller in the medial (**p = 0.0058) and lateral part (**p = 0.0037) of N-myc mutants compared to controls. Error bars represent ±SEM, Student's t test. Scale bars: 5 µm.
It is worth to note that in wild-type mice, HuC/D+ neurons almost doubled their size between E10.5 and E11.5 especially in the medial part [Fig. 7(A,B)], suggesting a period of high cellular activity (Pena et al., 2001). At E13.5, HuC/D+ neurons in the olfactory epithelium were similar in size in the medial and lateral part [Fig. 7(C)]. Reduction in the size of HuC/D+ neurons from E11.5 to E13.5 were restricted to the medial part, which suggests a more uniform activity of neurons in the entire olfactory epithelium at E13.5 [Fig. 7(B,C)]. Consistently, we also detected smaller HuC/D+ neurons in the separated vomeronasal organ of E13.5 N-myc−/− mutants (data not shown). Thus, reduced neurogenesis and proliferation in combination with smaller HuC/D+ neurons might explain the progressive reduction in size of the olfactory epithelium and vomeronasal organ in N-myc-deficient embryos. In summary, our results indicate that N-myc acts as an important factor to maintain cellular activity, proliferation, ongoing neurogenesis, and proper morphogenesis of the olfactory epithelium and vomeronasal organ, including the generation of GnRH neurons.
DISCUSSION
Neurogenesis is the process by which neurons are generated from neural stem cells and progenitors. A few structures in the nervous system maintain neurogenesis throughout life, including the olfactory epithelium belonging to the peripheral nervous system. Neurogenesis in the olfactory epithelium occurs in an ordered manner and specific cell types in the neuronal lineage can be identified by distinct markers (Cau et al., 2002; Beites et al., 2005; Murdoch and Roskams, 2007; Maier and Gunhaga, 2009). In addition, the olfactory epithelium has the potential to recover almost completely after injury [reviewed in (Schwob, 2002)], which makes this structure a valuable model system to study regulatory mechanisms of neurogenesis. Despite expanding knowledge about the control of olfactory neurogenesis (Duggan et al., 2008; Tucker et al., 2010; Gokoffski et al., 2011; Maier et al., 2011; Packard et al., 2011), little has been known how members of the myc family influence the development of and neurogenesis in the olfactory epithelium. Now our results provide evidence that N-myc is required for normal development of the olfactory epithelium to maintain proliferation, neurogenesis and subsequent morphogenesis of the olfactory pit and the vomeronasal organ.
Our results show that although N-myc is expressed already at E9.5 in the olfactory placode, at the initiation of olfactory neurogenesis, the first changes in N-myc-deficient mice are observed somewhat later, at E11.5. At this stage, N-myc−/− embryos exhibit reduced neurogenesis in the entire olfactory epithelium, whereas decreased proliferation is only detected in the medial part. These results suggest that the critical role of N-myc in the olfactory epithelium is restricted to stages of established neurogenesis. Our finding that N-myc is required to maintain proliferation and neurogenesis is in agreement with previous studies suggesting that N-myc regulates proliferation and differentiation in the brain, inner ear, and retina (Knoepfler et al., 2002; Martins et al., 2008; Dominguez-Frutos et al., 2011; Kopecky et al., 2011). Consistent with our results, data from the LaMantia lab has also shown that at E11.5 rapid proliferation occurs in the medial part of the olfactory epithelium, while proliferation in the lateral part proceeds in a slow and symmetric manner (Tucker et al., 2010). Interestingly, the rapidly dividing precursor cells in the medial domain of the olfactory epithelium are suggested to give rise to olfactory sensory neurons, vomeronasal neurons, and GnRH neurons (Tucker et al., 2010). Based on results that high levels of N-myc expression are present in rapidly cycling progenitors and low levels of N-myc in more slowly replicating cells, it has been suggested that N-myc regulates the cell cycle in neuronal progenitors (Knoepfler et al., 2002; Wey et al., 2010; Dominguez-Frutos et al., 2011). Thus, it is possible that the higher levels of N-myc expression detected in the medial part of the olfactory epithelium define the rapidly dividing neurogenic precursor cells. Furthermore, this might explain why reduced proliferation was only observed in the medial part of the N-myc-deficient olfactory epithelium.
At E11.5, the vomeronasal organ is part of the medial olfactory epithelium, and it is not until around E13 that the vomeronasal organ separates and develops independently. In addition, GnRH neurons are first detected at E11 in the medial wall of the developing olfactory epithelium, and later in the separated vomeronasal organ (Schwanzel-Fukuda and Pfaff, 1989; Wray et al., 1989). Moreover, it has been shown that GnRH neurons migrate in association with olfactory epithelial- and vomeronasal organ-derived axons towards the hypothalamus in the forebrain (Wray et al., 1994; Norgren et al., 1995; Yoshida et al., 1995). Our analysis of N-myc−/− embryos show that already at E11.5, the medial olfactory epithelium is smaller and exhibits a less prominent prospective vomeronasal organ, and at E13.5, the vomeronasal organ is severely reduced in size. Consistently, our results provide evidence that the generation of GnRH neurons are decreased by approximately 85% in E13.5 N-myc-deficient embryos. A clinical aspect of a reduced number of GnRH neurons in the hypothalamus is the human Kallmann syndrome (MacColl et al., 2002; Balasubramanian et al., 2010). Patients with Kallmann syndrome suffer from reproductive dysfunction, specifically hypogonadotropic hypogonadism and sometimes also from anosmia (loss of smell). Two genes, KAL and KAL2 (fibroblast growth factor 1 receptor) have been coupled to a small percentage of Kallmann syndrome cases (Franco et al., 1991; Legouis et al., 1991; Dode et al., 2003). Consequently, the majority of patients with Kallmann's syndrome have mutations in unknown genes, among which our current study has identified N-myc as a candidate gene. The embryonic origin of GnRH neuron progenitors has been debated, apart from the olfactory epithelium the adenohypophyseal placode and neural crest cells have also been suggested as possible origins for GnRH neurons in zebrafish (Whitlock et al., 2003). However, in mutant mice with either missing or disrupted anterior pituitaries, GnRH neurons develop normally in association with the vomeronasal organ (Metz and Wray, 2010). Regardless of the origin of the GnRH neurons, our results indicate that a normal development of the medial wall of the olfactory pit and the vomeronasal organ is critical for the generation of GnRH neurons. Whether the reduction of GnRH neurons in N-myc−/− embryos are a direct effect caused by the loss of N-myc activity, or a secondary effect due to a disturbed development of the medial wall of the olfactory epithelium and the vomeronasal organ remains to be determined.
Our results show a progressive decrease in proliferation and Notch1 expression in the N-myc-deficient olfactory epithelium, and by E13.5 the Hes5+ proliferative pool of cells is completely lost. Consistently, our data also show a progressive reduction in the generation of neurons. Moreover, although the structure of the nose was indistinguishable between wild-type and mutant mice, the morphology of the olfactory epithelium was malformed, and both the olfactory epithelium and the vomeronasal organ were much smaller in size in N-myc−/− mice. Our results indicate that the reduction in size of the olfactory structures is not due to increased cell death, but rather caused by decreased proliferation. In addition, our stereological analysis of a distinct population of cells indicated that the generated HuC/D+ neurons are smaller in N-myc mutants. However, since the stereological analysis was restricted to HuC/D+ neurons, other cell populations might be also affected. Thus, our results suggest that the reduction of proliferation and neurogenesis, in combination with smaller cell size is responsible for the severe atrophy in both the olfactory epithelium and vomeronasal organ observed in N-myc-deficient mice. This hypothesis is in agreement with findings that myc genes can activate the expression of several genes that are involved in the regulation of cell size, protein synthesis, and growth (Iritani and Eisenman, 1999; Coller et al., 2000; Boon et al., 2001). Consistently, a previous study suggested that a decrease in cell size might explain the smaller brain of N-myc-deficient mice (Knoepfler et al., 2002).
While N-myc has been shown to be widely expressed in the nervous system, c-myc expression is confined to other tissues and organs (Stanton et al., 1992). Interestingly, increased c-myc expression was detected at E10.5 in the neuroepithelium of N-myc-deficient mice (Stanton et al., 1992), indicating a compensatory feed-back loop of myc genes in the nervous system. However, the up-regulation of c-myc expression was not sufficient to attenuate the severe phenotype of N-myc deletion (Stanton et al., 1992). Our data now show that the loss of N-myc in the mouse olfactory epithelium does not stimulate an up-regulation of c-myc levels. On the other hand, N-myc has been shown to rescue the essential role of c-myc during embryonic development and compensate most of its functions (Malynn et al., 2000). In conclusion, our data provide evidence that N-myc is an essential factor for ongoing proliferation and neurogenesis in the olfactory epithelium, and for proper morphogenesis of the olfactory pit and vomeronasal organ.
Acknowledgments
The authors would like to thank Robert Eisenman for providing N-myc mutants and Iris López-Hernandez for genotyping of mice. They also thank the following persons for kindly providing plasmids; H. Edlund (Hes1), G. Fishell (Hes5, Ngn1), F. Guillemot (NeuroD1), P. Hurlin (c-myc), and V. Taylor (Notch1). They are grateful to members of the Gunhaga lab for helpful discussions.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe, de Angelis M, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Dwyer A, Seminara SB, Pitteloud N, Kaiser UB, Crowley WF., Jr Human GnRH deficiency: A unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology. 2010;92:81–99. doi: 10.1159/000314193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak O, Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Exp Cell Res. 2005;306:309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- Dominguez-Frutos E, Lopez-Hernandez I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, et al. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. J Neurosci. 2011;31:7178–7189. doi: 10.1523/JNEUROSCI.0785-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan CD, DeMaria S, Baudhuin A, Stafford D, Ngai J. Foxg1 is required for development of the vertebrate olfactory system. J Neurosci. 2008;28:5229–5239. doi: 10.1523/JNEUROSCI.1134-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: Recent discoveries lead to new insights. FASEB J. 1998;12:633–651. [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J. p63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72:748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornaro M, Geuna S, Fasolo A, Giacobini-Robecchi MG. HuC/D confocal imaging points to olfactory migratory cells as the first cell population that expresses a post-mitotic neuronal phenotype in the chick embryo. Neuroscience. 2003;122:123–128. doi: 10.1016/j.neuroscience.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- Gokoffski KK, Wu HH, Beites CL, Kim J, Kim EJ, Matzuk MM, Johnson JE, et al. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development. 2011;138:4131–4142. doi: 10.1242/dev.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen HJ. Stereology of arbitrary particles. A review of unbiased number and size estimators and the presentation of some new ones, in memory of William R. Thompson. J Microsc. 1986;143:3–45. [PubMed] [Google Scholar]
- Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, et al. Expression and activity of L-Myc in normal mouse development. Mol Cell Biol. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Henriksson M, Luscher B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv Cancer Res. 1996;68:109–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- Hirota J, Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc Natl Acad Sci USA. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeli K, Hurlin PJ. Differential regulation of N-Myc and c-Myc synthesis, degradation, and transcriptional activity by the Ras/mitogen-activated protein kinase pathway. J Biol Chem. 2011;286:38498–38508. doi: 10.1074/jbc.M111.276675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Kim J, Santos R, Wu HH, Lander AD, Calof AL. Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development. 2009;136:1453–1464. doi: 10.1242/dev.034967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolterud A, Alenius M, Carlsson L, Bohm S. The Lim homeobox gene Lhx2 is required for olfactory sensory neuron identity. Development. 2004;131:5319–5326. doi: 10.1242/dev.01416. [DOI] [PubMed] [Google Scholar]
- Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Dev Dyn. 2011;240:1373–1390. doi: 10.1002/dvdy.22620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouis R, Hardelin JP, Levilliers J, Claverie JM, Compain S, Wunderle V, Millasseau P, et al. The candidate gene for the X-linked Kallmann syndrome encodes a protein related to adhesion molecules. Cell. 1991;67:423–435. doi: 10.1016/0092-8674(91)90193-3. [DOI] [PubMed] [Google Scholar]
- MacColl G, Quinton R, Bouloux PM. GnRH neuronal development: Insights into hypogonadotrophic hypogonadism. Trends Endocrinol Metab. 2002;13:112–118. doi: 10.1016/s1043-2760(01)00545-8. [DOI] [PubMed] [Google Scholar]
- Machold RP, Kittell DJ, Fishell GJ. Antagonism between notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2007;2:5. doi: 10.1186/1749-8104-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier E, Gunhaga L. Dynamic expression of neurogenic markers in the developing chick olfactory epithelium. Dev Dyn. 2009;238:1617–1625. doi: 10.1002/dvdy.21966. [DOI] [PubMed] [Google Scholar]
- Maier E, Nord H, von Hofsten J, Gunhaga L. A balance of BMP and notch activity regulates neurogenesis and olfactory nerve formation. PLoS One. 2011;6:e17379. doi: 10.1371/journal.pone.0017379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O'Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- Martins RA, Zindy F, Donovan S, Zhang J, Pounds S, Wey A, Knoepfler PS, et al. N-myc coordinates retinal growth with eye size during mouse development. Genes Dev. 2008;22:179–193. doi: 10.1101/gad.1608008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz H, Wray S. Use of mutant mouse lines to investigate origin of gonadotropin-releasing hormone-1 neurons: Lineage independent of the adenohypophysis. Endocrinology. 2010;151:766–773. doi: 10.1210/en.2009-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, Roskams AJ. Olfactory epithelium progenitors: Insights from transgenic mice and in vitro biology. J Mol Histol. 2007;38:581–599. doi: 10.1007/s10735-007-9141-2. [DOI] [PubMed] [Google Scholar]
- Norgren RB, Jr, Gao C, Ji Y, Fritzsch B. Tangential migration of luteinizing hormone-releasing hormone (LHRH) neurons in the medial telencephalon in association with transient axons extending from the olfactory nerve. Neurosci Lett. 1995;202:9–12. doi: 10.1016/0304-3940(95)12210-9. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Giel-Moloney M, Leiter A, Schwob JE. Progenitor cell capacity of NeuroD1-expressing globose basal cells in the mouse olfactory epithelium. J Comp Neurol. 2011;519:3580–3596. doi: 10.1002/cne.22726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol. 2001;430:250–263. doi: 10.1002/1096-9861(20010205)430:2<250::aid-cne1029>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Potvin E, Beuret L, Cadrin-Girard JF, Carter M, Roy S, Tremblay M, Charron J. Cooperative action of multiple cis-acting elements is required for N-myc expression in branchial arches: Specific contribution of GATA3. Mol Cell Biol. 2010;30:5348–5363. doi: 10.1128/MCB.00353-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269:33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- Sholl-Franco A, Fragel-Madeira L, Macama Ada C, Linden R, Ventura AL. ATP controls cell cycle and induces proliferation in the mouse developing retina. Int J Dev Neurosci. 2010;28:63–73. doi: 10.1016/j.ijdevneu.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- Stump G, Durrer A, Klein AL, Lutolf S, Suter U, Taylor V. Notch1 and its ligands Delta-like and Jagged are expressed and active in distinct cell populations in the postnatal mouse brain. Mech Dev. 2002;114:153–159. doi: 10.1016/s0925-4773(02)00043-6. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Tucker ES, Lehtinen MK, Maynard T, Zirlinger M, Dulac C, Rawson N, Pevny L, et al. Proliferative and transcriptional identity of distinct classes of neural precursors in the mammalian olfactory epithelium. Development. 2010;137:2471–2481. doi: 10.1242/dev.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Lang MF, Jiang X. Calretinin is expressed in the intermediate cells during olfactory receptor neuron development. Neurosci Lett. 2013;542:42–46. doi: 10.1016/j.neulet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9:537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock KE, Wolf CD, Boyce ML. Gonadotropin-releasing hormone (GnRH) cells arise from cranial neural crest and adenohypophyseal regions of the neural plate in the zebrafish, Danio rerio. Dev Biol. 2003;257:140–152. doi: 10.1016/s0012-1606(03)00039-3. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Wray S. From nose to brain: Development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010;22:743–753. doi: 10.1111/j.1365-2826.2010.02034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci U S A. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Key S, Qualls R, Fueshko SM. A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev Biol. 1994;166:349–354. doi: 10.1006/dbio.1994.1320. [DOI] [PubMed] [Google Scholar]
- Xuan S, Baptista CA, Balas G, Tao W, Soares VC, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Tobet SA, Crandall JE, Jimenez TP, Schwarting GA. The migration of luteinizing hormone-releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J Neurosci. 1995;15:7769–7777. doi: 10.1523/JNEUROSCI.15-12-07769.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.