Summary
Constitutive Centromere Associated Network (CCAN) proteins, particularly CENP-C, CENP-T and the CENP-H/-I complex, mechanically link CENP-A-containing centromeric chromatin within the inner kinetochore to outer kinetochore proteins, like the Ndc80 complex, that bind kinetochore microtubules. Accuracy of chromosome segregation depends critically upon Aurora B phosphorylation of Ndc80/Hec1. To determine how CCAN protein architecture mechanically constrains intrakinetochore stretch between CENP-A and Ndc80/Hec1 for proper Ndc80/Hec1 phosphorylation, we used super-resolution fluorescence microscopy and selective protein depletion. We found that at bi-oriented chromosomes in late prometaphase cells, CENP-T is stretched ~16 nm to the inner end of Ndc80/Hec1, much less than expected for full-length CENP-T. Depletion of various CCAN linker proteins induced hyper-intrakinetochore stretch (an additional 20-60 nm) with corresponding significant decreases in Aurora B phosphorylation of Ndc80/Hec1. Thus, proper intrakinetochore stretch is required for normal kinetochore function and depends critically on all the CCAN mechanical linkers to the Ndc80 complex.
Introduction
The core of the inner kinetochore in human cells includes CENP-A and a group of 16 chromatin-proximal proteins termed the constitutive centromere-associated network (CCAN). CENP-A, a modified histone H3, is the epigenetic marker for the centromere site for kinetochore assembly. CCAN proteins include the CENP-H/I complex (CENP-H/-I/-K/-L/-M/-N), CENP-O class protein (COMA complex (CENP-O/-P/-Q/-U(50)), CENP-R), CENP-T/W complex (CENP-T/-W/-S/-X) and CENP-C. CENP-A and the CCAN proteins are conserved from yeast to humans (excluding CENP-M and CENP-R; these proteins are not identified in yeast) (Biggins, 2013; Perpelescu and Fukagawa, 2011). CENP-C and CENP-T are known to have a crucial role for mechanically linking centromeric chromatin within the inner kinetochore to the kinetochore microtubule (kMT) attachment proteins of the outer kinetochore like the Ndc80 complex and other protein members of the highly conserved KMN network (Knl1, Zwint-1; Mis12, Dsn1, Nsl1, Nnf1; and Ndc80/Hec1, Nuf2, Spc24, Spc25) (Rago and Cheeseman, 2013). The Ndc80 complex, a 57 nm long hetero-tetramer comprising a Spc24-Spc25 dimer bound to a Ndc80/Hec1-Nuf2 dimer, is the major kMT binding unit (Cheeseman and Desai, 2008; Ciferri et al., 2008; Wei et al., 2007). The N-terminus of CENP-T binds directly to Spc24/Spc25 at the inner end of the Ndc80 complex, while the N-terminus of CENP-C binds to the Mis12 complex, which in combination with KNL1 can also bind Spc24/Spc25 (Malvezzi et al., 2013; Nishino et al., 2013; Przewloka et al., 2011; Screpanti et al., 2011). The primary MT binding domains of the Ndc80 complex are at the N-terminus of Ncd80/Hec1, which is located at the outer end of the Ndc80 complex.
Because both CENP-T and CENP-C have long extendable disordered domains (Suzuki et al, 2011), their linkage to the KMN network is proposed to play a major role in “intrakinetochore stretch” of kinetochores of bi-oriented chromosomes produced by kMT formation. The intrakinetochore stretch that has an important role in silencing the spindle assembly checkpoint (SAC) has been measured by the increased separation between the mean centroids of different colored fluorescent labels, one on CENP-A and the other on inner-end of one of the KMN network proteins, either the Ndc80 complex (Maresca and Salmon, 2009), or the Mis12 complex (Uchida et al., 2009). Intrakinetochore stretch also may play an important role in the prometaphase correction mechanism for errors in kMT attachment that produce aneuploidy, the gain or loss of chromosomes that can lead to birth defects during development or cancer in tissue cells (Bakhoum et al., 2009; Silkworth and Cimini, 2012). Error correction is dependent on Aurora B kinase phosphorylation of kMT attachment proteins, especially the primary MT binding site of Ndc80/Hec1 at its outer N-terminal end. Phosphorylation weakens binding of the Ndc80 complex to MTs in vitro and causes destabilization of kMT attachments in vivo and prevents silencing of SAC. Conversely, dephosphorylation in prometaphase causes hyper stabilization of kMT attachment and induces errors in sister chromosome segregation in anaphase (Guimaraes et al., 2008; Liu et al., 2009; Welburn et al., 2010). A gradient in the level of Aurora B phosphorylation is reported to decrease with distance towards the outer kinetochore from its source within the inner centromere (Lampson and Cheeseman, 2011; Liu et al., 2009; Tanaka, 2013). As a consequence, the amount of intrakinetochore stretch allowed by the CCAN protein linkers to the Ndc80 complex may provide important positional control of the level of Aurora B phosphorylation so that errors are destabilized in prometaphase while proper attachments persist. If so, hyper-intrakinetochore stretch should produce abnormally lower than normal phosphorylation at the primary Ndc80/Hec1 MT binding domain.
This study addresses several unresolved questions about CCAN protein architecture important for understanding the molecular basis of intrakinetochore stretch and its control of outer kinetochore protein function such as phosphorylation of the MT binding domain of Ndc80. Here, we use a two-color super-resolution fluorescence microscopy method to achieve nanometer scale measurements (variance typically 3-7 nm) for the mean positions along the axis of kMTs for the CCAN protein network within human kinetochores at metaphase relative to the mean positions of the protein components of the KMN network (Varma et al., 2013; Wan et al., 2009). These measurements revealed that the mean positions of most CCAN proteins, except CENP-C and CENP-A, are localized close to the periphery of centromeric chromatin and extend toward the outer kinetochore. CENP-T is stretched about 19 nm on average between its C-terminal DNA binding domain and its N-terminal Spc24/Spc25 binding domain. Surprisingly, the majority of CENP-C is not bound to the Mis12 complex. Intrakinetochore stretch was constant independent of 2 fold changes in the stretch of the centromere between sister kinetochores indicating that above a low tension, intrakinetochore stretch is constrained. We used protein depletion assays and nm-scale distance measurements to determine for bi-oriented chromosomes in late prometaphase whether the three major CCAN linkers CENP-T, CENP-H/I and CENP-C constrain intrakinetochore stretch for bi-oriented chromosomes between the mean positions of CENP-A and the MT binding site on the Ndc80/Hec1. We used antibodies to Aurora B phosphorylation sites within the primary MT binding domain of the Ndc80/Hec1 to test if hyper-intrakinetochore stretch changes Ndc80/Hec1 phosphorylation. We find that all three linkers are required to prevent in late prometaphase hyper-intrakinetochore stretch that correlates with a significant reduction in the Aurora B phosphorylation of the Ndc80 complex, a reduction known to produce unequal segregation of the genome (DeLuca et al., 2006).
RESULTS
CCAN Protein Architecture at Metaphase HeLa Kinetochores
We used the Delta method (Varma et al., 2013; Wan et al., 2009) to measure mean separation along the kMT axis at nm-scale accuracy between the centroids of different colored pairs of fluorescent labels (see Methods) for cells fixed in paraformaldehyde and labeled with site specific antibodies. We have shown previously that our fixation and antibody labeling procedures yield the same Delta values as obtained from live cell measurements of red and green pairs of fluorescent protein markers for kinetochore proteins (Varma et al., 2013).
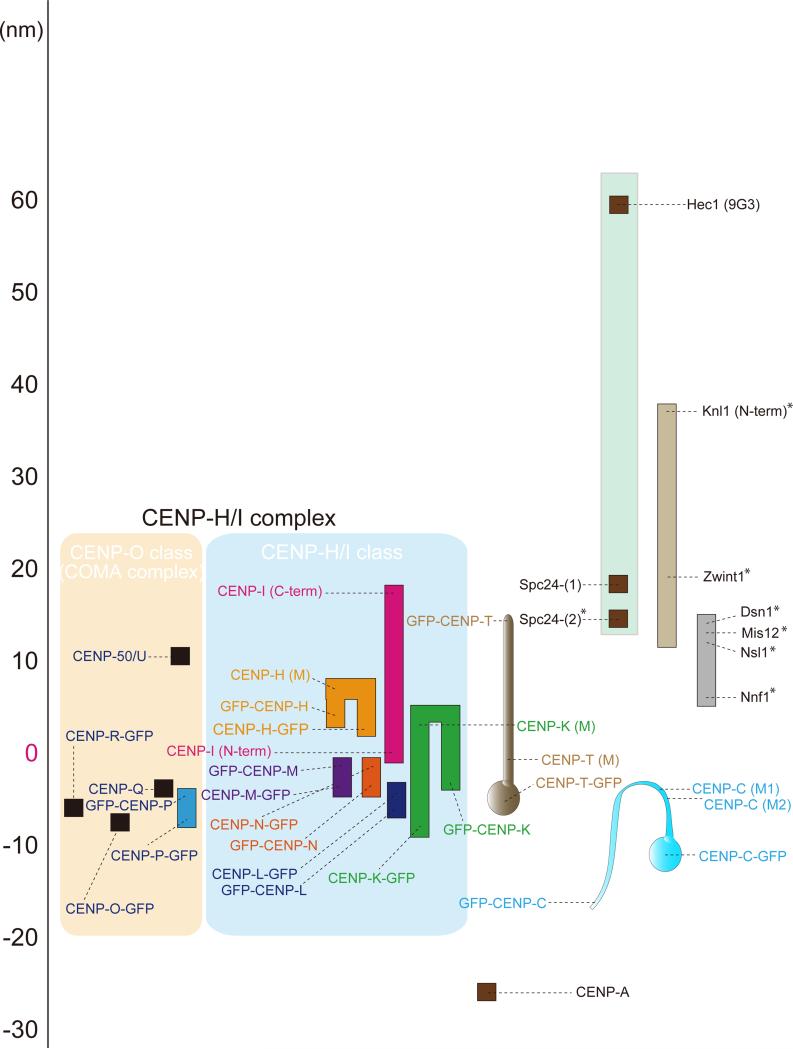
Figure 1 shows a nm-scale map of protein position calculated from Delta measurements between the protein epitopes indicated and the Ndc80/Hec1 (9G3 antibody), the primary kMT binding end of the Ndc80 complex. The mean Delta values for the CCAN protein labels had a standard deviation of 5-8 nm for an average of >100 measured sister-kinetochore pairs (Table S1 and Figures S1-S2). In the large map in Figure 1, we have set the position of the N-terminus of CENP-I to zero nm along the kMT axis as an indication of the peripheral surface of the centromere. This makes label positions towards the outer kinetochore positive and towards the inner kinetochore negative. Note, the mean position of the plus-ends of kMTs is not known precisely, but it is close to the N-terminus of CENP-I in cold treated metaphase PtK1 cells that lack any centromere tension (Wan et al., 2009) and inside of the outer plate of HeLa cells, which is the mean location of immunogold labeled Hec1(9G3) (DeLuca et al., 2005). Also, there are three known reasons why the end-to-end length from Ndc80/Hec1(9G3) to Spc24 measured in Figure 1 and previous studies (Wan et al., 2009, Varma et al., 2013) is 40~45 nm along the kMT axis instead of the 57 nm full length of the Ndc80 complex (Wei et al., 2005). First, the measured length between 9G3-Spc24 does not include globular domains of both Ndc80-Nuf2, which are about 3 nm long. Second, Ndc80/Hec1 complex has a loop domain in the middle of its alpha-helical coiled coil domain that allows bending of the Ndc80 complex (Wang et al., 2009). Finally, the Ndc80 complex projects at an angle from the MT wall, based on electron microscopy images of Ndc80/Nuf2 bound to MTs in vitro (Ciferri et al., 2008; Wang et al., 2008).
Figure 1.
Nanometer (nm) scale map of CCAN protein positions along the sister kinetochore axis relative to the KMN network proteins in human kinetochores at metaphase. The map was constructed from Delta measurements relative to Ndc80/Hec1(9G3) (Table S1 and Figures S1-S2). Scale on the far-left is set equal to zero at the position of the N-terminus of CENP-I centroid. Positive values are outward (towards the spindle microtubules), while negative values are inward (towards the centromere interior). Colored boxes indicate kinetochore proteins. Black dots indicate the mean Delta values that have been corrected for kinetochore tilt (Wan et al., 2009). The S.D. was typically 3-7 nm and, n > = 100 kinetochores averaged (Table S1). *Mean values obtained from Wan et al., 2009 and Varma et al., 2013.
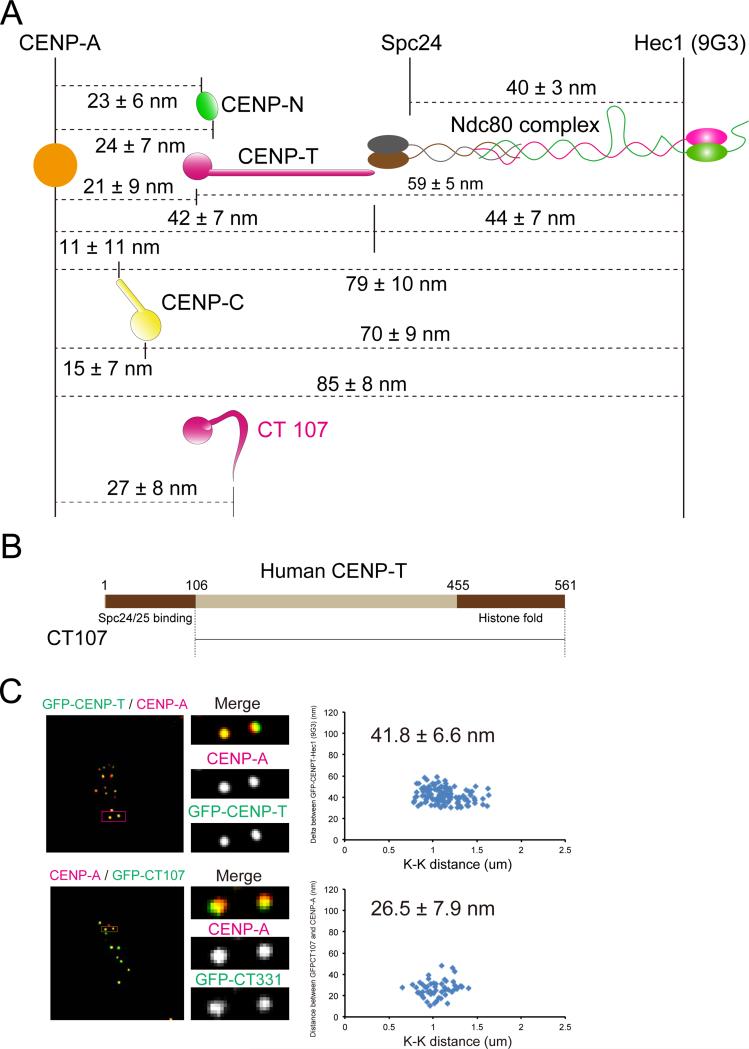
To test the accuracy of the map in Figure 1, we measured separations directly between CENP-A (using a CENP-A antibody) or Hec1(9G3) and key centromeric DNA binding proteins (CENP-N, CENP-T, and CENP-C) (Figures 2A and S2). The mean separation distances measured in Figure 2A are almost identical (within 1-2 nm in most cases) to those obtained from the map in Figure 1.
Figure 2.
Nanometer (nm) separations measured between mean centroid positions of key CCAN proteins in the linkage between CENP-A chromatin and the Ndc80 complex at metaphase. (A) Summary of separation measurements between the DNA binding domains of CENP-T, CENP-C, and CENP-N, within human kinetochores at metaphase relative to fluorescent labels on CENP-A and the Ndc80/Hec1 complex obtained from Delta analysis in Figure S2. Included are mean positions of the two ends of CT 107, which lacks the N-terminal of CENP-T that binds Spc24/25, as well as the positions of the two ends of normal CENP-T. Note that the mean position of the N-terminus of CT107 is close to the DNA binding C-terminal of CENP-T and not close to Spc24/Spc25. (B) Schematic representation of human CENP-T. The N-terminal region (1-106 for CENP-T) binds to Spc24/25 globular domain. The C-terminal region (455-561) is a histone fold domain for DNA binding. (C) Two-color immunofluorescence (left) and Delta analysis (right) in GFP-CENP-T (upper) and GFP-CT 107 (lower) expressed in CENP-T depleted cells.
CENP-H/I essential components
The CENP-H/I complex is essential for chromosome segregation and also has a crucial role for CENP-A incorporation and inner kinetochore conformation (Cheeseman et al., 2008; Foltz et al., 2006; Okada et al., 2006). CENP-H/-I/-K/-L/-M/-N are tightly bound to each other (Okada et al., 2006). The mean position of CENP-N, a CENP-A chromatin binding protein, was near the zero position of the N-terminus of CENP-I, about 23 nm outside of the mean position of CENP-A within the centromere (Figures 1 and S2B). Unexpectedly, we found that the mean position of the C-terminus of CENP-I localized ~18 nm outside the N-terminus of CENP-I and close to Spc24 at the inner end of the Ndc80 complex. The C-terminus, N-terminus and middle of CENP-H were found to localize within the extended CENP-I protein (Figures 1 and S1B). The average positions of both N-terminus and C-terminus of CENP-M localized slightly inward (around 3-4 nm) from zero (Figures 1 and S1C). The N-terminus and C-terminus of CENP-K localized 5 and 3 nm inside of zero while middle of CENP-K localized outward (towards the pole) less than 5 nm (Figures 1 and S1D). CENP-L localized slightly inward from CENP-M and CENP-N and overlapped with CENP-K (Figures 1 and S1E). This data indicate that the CENP-H/-I complex appears bound to the periphery of CENP-A chromatin and extends outward, with CENP-I extending beyond the outer end of the Mis12 complex and overlapping the inner ends of the Ndc80 and Knl1 complexes.
CENP-O class proteins (COMA complex)
CENP-O class proteins were first identified as the COMA complex (Ctf19, Okp1, Mcm21, and Ame1) in S. Cerevisiae, where they have an essential function for kinetochore assembly and cell viability (De Wulf et al., 2003). The vertebrate CENP-O class proteins (CENP-O/-P/-Q/-U(50)/-R) are part of the CENP-H/-I sub-complex but they are not essential for cell viability. Depletion of CENP-O class proteins does not affect any other CCAN kinetochore localizations including the CENP-H/I essential components (Hori et al., 2008). However, CENP-O class proteins have an important role for faithful chromosome segregation (Hori et al., 2008; Toso et al., 2009). CENP-U/50 localized 10 nm outward from the zero position (Figures 1 and S1H). CENP-O, -P, -Q, and -R overlapped each other and localized slightly inward compared to zero (Figures 1 and S1I-L).
CENP-T complex
CENP-T is one of the key inner kinetochore components that recruit the KMN network proteins (Gascoigne et al., 2011; Hori et al., 2013; Malvezzi et al., 2013). The C-terminus of CENP-T, which makes a hetero-tetramer with CENP-W/-S/-X using a histone fold-like structure, is essential for centromeric DNA binding (Amano et al., 2009; Nishino et al., 2012). The mean position of CENP-T C-terminus was ~6 nm inward from the zero position indicating that CENP-T C-terminus predominantly binds CENP-A-containing chromatin at the periphery of the centromere (Figures 1, 2A and S2C).
The N-terminus of CENP-T (Figure 2B) is reported to directly wrap around the Spc24/25 globular domain of the Ndc80 complex that faces the centromere and CENP-T is a major contributor to kinetochore localization of the Ndc80 complex (Malvezzi et al., 2013; Nishino et al., 2013). We found the mean position of the N-terminus of CENP-T localized to the exact same position as Spc24 (Figures 1, 2A and S2C) as predicted by CENP-T-Spc24/25 binding data in vitro (Malvezzi et al., 2013; Nishino et al., 2013). We also measured the mean separation of the N-terminus and C-terminus of CENP-T relative to CENP-A mean position (Figures 2A and S2C) and these values gave the same results as measurements relative to Ndc80/Hec1(9G3). The mean position of antibodies to the middle of CENP-T localized between the N-terminus and C-terminus of CENP-T indicating the major axis of CENP-T is extended parallel to the kMT axis at metaphase (Figures 1, 2A, and S1F). The N-terminus of a CENP-T truncated mutant CT107 (107-530aa), which lacks the Spc24/25 binding site (1-106 aa), didn't extend toward the outer kinetochore (Figures 2A-C). Thus, binding of the N-terminal end of CENP-T to Spc24/25 (Nishino et al., 2013) is essential for CENP-T extension from the inner kinetochore into the outer kinetochore.
CENP-C
CENP-C also has a critical role in recruiting the KMN network proteins (Przewloka et al., 2011; Screpanti et al., 2011). CENP-C has multiple DNA binding sites (Carroll et al., 2010; Kato et al., 2013; Tomkiel et al., 1994; Yang et al., 1996) within the C-terminal half of the protein. To confirm centromere-targeting domains of CENP-C, we examined localization of three CENP-C truncated mutants, CC426 (426-943 aa), CC690 (690-943 aa), and CC759 (759-943 aa) (Figure S3A). Both CC426 and CC690 clearly localized to the kinetochore throughout the cell cycle, but only a minor population of CC759 localized to the kinetochore (Figures S3B-C).
The mean position of DNA binding domain of CENP-C (CC426) relative to CENP-I N-terminus (zero position, Figure 1) increased with protein expression level of CENP-C (Figure S3D). As a result, for our Figure 1 and 2A measurements, we used stable cell lines expressing at about wild-type levels CENP-C with GFP fused to either the C- or N-terminus and resistant to RNAi of the endogenous CENP-C. We confirmed that GFP-CENP-C rescued the abnormal phenotype produced by depletion of endogenous CENP-C (see Methods). For these cells, the mean position of the C-terminus of CENP-C was localized 12 nm inward from the zero position (Figures 1, 2A and S2D), well within the depth of the CENP-A chromatin that extends inward and has a mean position 23 nm from the periphery of the centromere (Figure 1 and 2A). The N-terminus of CENP-C is known to play a major role in recruiting the Mis12 complex to the kinetochore (Przewloka et al., 2011; Screpanti et al., 2011). Interestingly, the mean position of the N-terminus of GFP-CENP-C localized ~16 nm inward from the centromere surface into the CENP-A chromatin (Figure 1 and 2A). This mean position for the N-terminus of CENP-C was also measured in previous live-cell imaging studies in HeLa cells and Ptk1 cells (Dumont et al., 2012; Varma et al., 2013). These results indicate that only a small minority of CENP-C N-terminus binds Mis12, while the majority remains within the CENP-A chromatin.
Intrakinetochore Stretch of CCAN Proteins Relative to the Primary MT Binding Domain of Ndc80/Hec1 Is Not Sensitive to Inter-kinetochore Centromere Stretch at Metaphase
Intrakinetochore stretch at metaphase is thought to be important for silencing SAC activity, stabilizing kMT attachment, and faithful chromosome segregation (Maresca and Salmon, 2010). The long disordered regions at the N-terminus of CENP-C and CENP-T are likely very floppy when not under tension (Maresca and Salmon, 2009; Suzuki et al., 2011). In our study, none of the protein separations for control bi-oriented chromosomes at late prometaphase, when the majority of chromosomes are bi-oriented at equator and few chromosomes are not bi-oriented, or metaphase exhibited a large positive-slope in plots of Delta verses inter-kinetochore distance (K-K) (Figures 3A and S1-S2). For example, at late prometaphase Delta measurements for endogenous CENP-A to Hec1(9G3) had a slope of 0.6 nm/um of centromere stretch that occurs during metaphase chromosome oscillations between the poles (Figure 3A). These data showed for bi-oriented chromosomes that both the mean deformation of CENP-A-containing chromatin and the CCAN protein linkage between the inner and outer kinetochore is constant and insensitive to the changes in tension from the inter-kinetochore stretch.
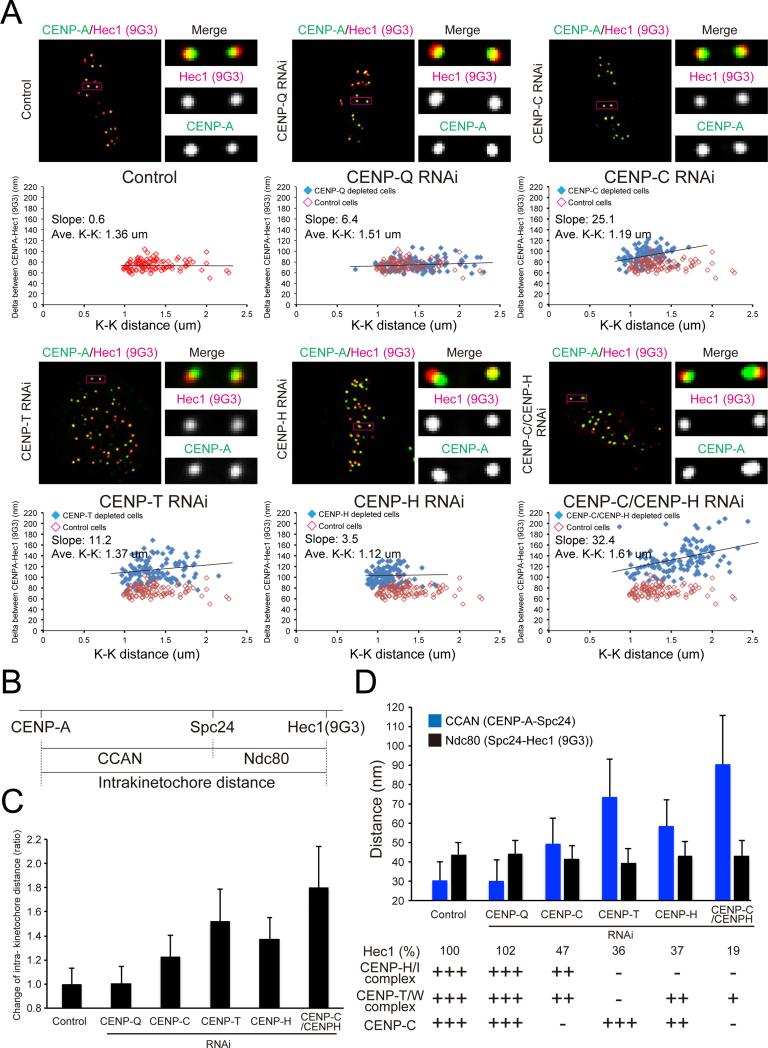
Figure 3.
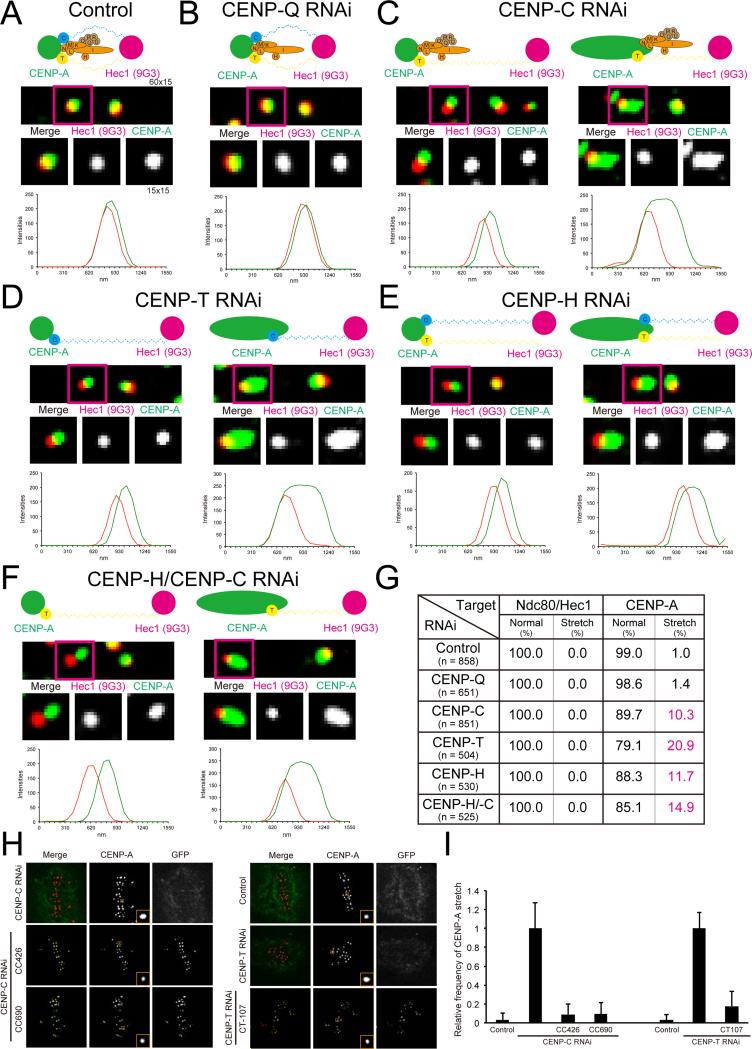
CENP-T, CENP-C and CENP-H/I complex are needed to limit intrakinetochore stretch of bi-oriented chromosomes in late prometaphase. (A) Immunofluorescence of both CENP-A and Ndc80/Hec1 (9G3) at kinetochores in control cells or CENP-Q RNAi cells or CENP-C RNAi cells or CENP-T RNAi cells or CENP-H RNAi cells or CENP-C/CENP-H RNAi cells (upper). Distribution of Delta mean separation between CENP-A and Hec1 (9G3) in each cell (lower). (B) Diagram showing that total intrakinetochore distance between CENP-A and Ndc80/Hec1 (9G3) is the sum of the separation between CENP-A to the inner end of the Ndc80 complex (Spc24/Spc25) and length of the Ndc80 complex. (C) Intrakinetochore distance between CENP-A and Ndc80/Hec1(9G3) for control and experimental cells from mean Delta values in Figure 3A) normalized by the value for controls. (D) Intrakinetochore stretch of CCAN domain (CENP-A and Spc24) contributes to intrakinetochore stretch but length of the Ndc80 complex remains constant (separation between Spc24 to Ndc80/Hec1 (9G3)) in experimental cells compared to controls. All images in Figure 3 are corrected chromatic aberration.
In our initial study (Wan et al., 2009), we found that mean position of GFPCENP-A was 20 nm inside of the mean position measured here for endogenous CENP-A and showed significant oscillation dependent intrakinetochore stretch (~30 nm/um). We found that GFP-CENP-A was over-incorporated into kinetochores ~4-fold compared to endogenous CENP-A (Figure S4A). This comparison suggests that the chromatin occupied by most of GFP-CENP-A is much more compliant than the chromatin occupied by endogenous CENP-A and CCAN proteins within the inner kinetochore.
Hyper-Intrakinetochore Stretch Occurs for Selective Depletion of CENP-T, CENP-H/-I Sub-Complex or CENP-C for bi-oriented chromosomes in late prometaphase
A previous study has shown CENP-T protein can potentially extend up to 90 nm in vitro as assayed by Atomic Force Microscopy (Suzuki et al., 2011). For unattached kinetochores, the end-to-end axial length of CENP-T is near zero (Suzuki et al., 2011). However, here we show that on average CENP-T is extended for bi-oriented chromosomes by only ~19 nm at metaphase and ~16 nm at late prometaphase along the kMT axis from its C-terminal DNA binding domain to its N-terminal Spc24/Spc25 binding domain (Figures 1 and 2A). Also, this linkage is stiff to tension generated by oscillations in K-K centromere stretch (Figures S1F and S2C). These data suggest that the kinetochore normally has a mechanism to inhibit hyper-intrakinetochore stretch at metaphase.
To test the above hypothesis, we measured changes in the mean separation of CENP-A and Hec1(9G3) for bi-oriented chromosomes at the spindle equator in late prometaphase, for cells selectively depleted of CENP-Q, CENP-C, CENP-T, CENP-H/-I or CENP-C and CENP-H/-I complex proteins by RNAi. Every CCAN RNAi in this study exhibited more than 95 % loss of target proteins on metaphase kinetochore (Figure S4B). In addition, we analyzed Ndc80, CENP-T, CENP-H and CENP-C intensities in each RNAi treated cell (Figure S4B). CENP-T, CENP-C, CENP-H, and CENP-H/-C RNAi induced prometaphase arrest (data not shown). The results agree with previous reports (Gascoigne et al., 2011; Hori et al., 2008; Okada et al., 2006).
Compared to control, depletion of CENP-Q did not affect intrakinetochore stretch (Figures 3A-C). Depletion of CENP-C, CENP-T, CENP-H (removes whole CENP-H/-I complex, Figure S4B) or CENP-H plus CENP-C RNAi produced an increase in the separation of CENP-A to Hec1(9G3) of ~20%, 50%, 40% and 80% respectively (Figures 3A-D).
To test if changes in the mean length of the Ndc80 complex along the kMT axis contributed to increased CENP-A to Hec1(9G3) separation, we measured the distance between Spc24 and Hec1(9G3) for each condition (Figure 3B). There was no difference in the mean Spc24 to Hec1(9G3) separation between control cells and the various CCAN protein depleted cells (Figures 3D and S4C-D). Therefore, changes in intrakinetochore stretch in late prometaphase and metaphase are produced by changes in the mean separation between CENP-A and the Spc24/Spc25 end of the Ndc80 complex.
We obtained the amount of CCAN-dependent stretch in our protein depletion assay (Figure 3D) by subtracting the mean axial length of the Ndc80 complex from the measured mean separations of CENP-A and Hec1(9G3) (Figure 3C). There was no difference in the mean separation of CENP-A and Spc24 between the control and CENP-Q depleted cells, which retained the CENP-T complex, CENP-C, and CENP-H/I complex (Figure 3D). Depleting the CENP-H/-I complex, which leaves both CENP-T and CENP-C at kinetochores, caused a 2 fold CCAN-dependent stretch (from ~30 to 60 nm; Figure 3D). For CENP-T depleted cells, which still retained CENP-C but not the CENP-H/-I complex, the CCAN dependent stretch increased further (from ~30 to 75 nm; Figure 3D). The CCAN-dependent stretch in CENP-C plus CENP-H doubly depleted cells, in which CENP-T still exists, was the largest (from ~ 30 to 90 nm; Figure 3D). These results suggest that CENP-T or CENP-C can extend much more than in normal metaphase cells, but that the whole CCAN proteins, including the CENP-H/-I complex, prevents hyper-stretching of either CENP-T or CENP-C.
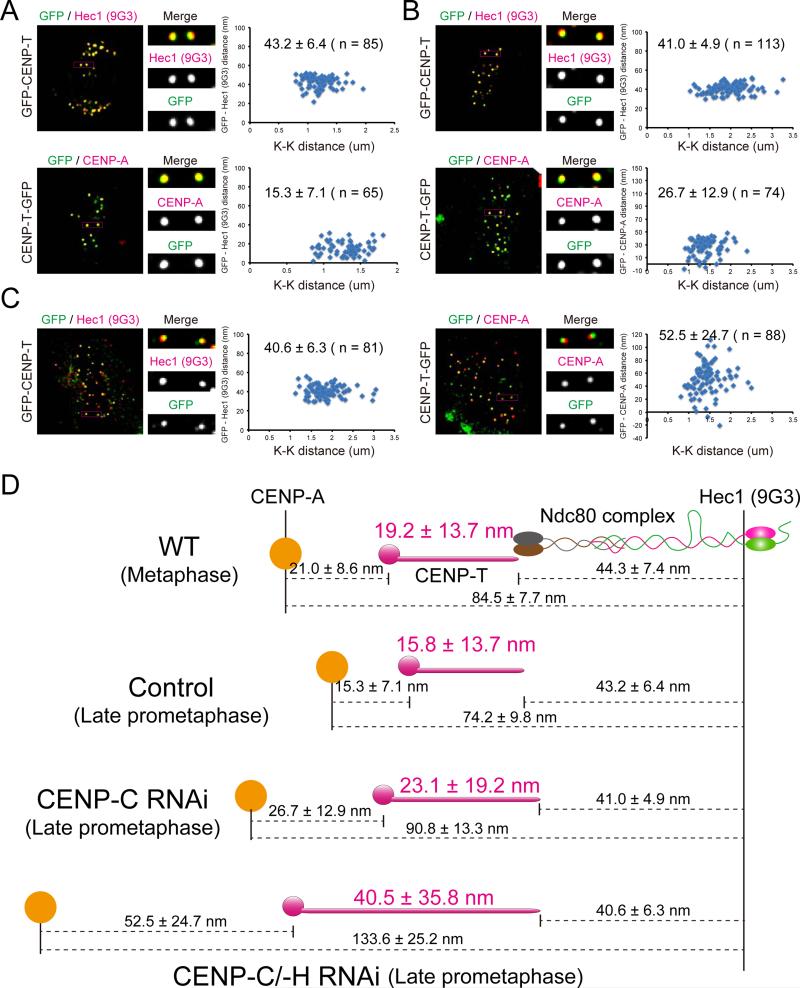
To determine the relative contributions of CENP-A chromatin stretch and CENP-T stretch to hyper-intrakinetochore stretch, we obtained measurements of the mean position of both ends of CENP-T relative to CENP-A and the Ndc80 complex (Figures 4A-D). Depletion of CENP-C or CENP-C/-H increased the length of CENP-T in late prometaphase from the control value of ~16 nm to ~23 nm or to ~41 nm respectively while the separation distance between the mean positions of CENP-A and the C-terminus of CENP-T increased from the control value of ~15 nm to ~27 nm and to ~53 nm respectively (Figures 4A-D). These results indicate that depletion of CENP-C or CENP-C/-H not only causes increased intrakinetochore stretch of CENP-T, but also equally affects the structure and compliance of CENP-A chromatin.
Figure 4.
Both CENP-A chromatin stretch and CENP-T stretch contribute to intrakinetochore stretch. Kinetochore immunofluorescence of both GFP and Ndc80/Hec1(9G3) or GFP and CENP-A in GFP-CENP-T or CENP-T-GFP stably expressed cell (A) treated with CENP-C RNAi (B) or CENP-C/-H RNAi (C). Corresponding Delta analysis (right). (D) Summary of mean separation measurements between CENP-A and C-terminus of CENP-T and the C-terminus of CENP-T to Spc24/Spc25 for changes in intrakinetochore stretch between normal late prometaphase and metaphase, and for changes in late prometaphase induced by depletion of CENP-C and CENP-C/-H.
CCAN proteins contribute to CENP-A chromatin compliance and compaction
In CENP-Q, CENP-T, CENP-C, and CENP-C/-H depleted cells the slope of the Delta verses K-K distance for CENP-A to 9G3 increase from near zero for controls to 6, 11, 25 and 32 nm per um change in inter-kinetochore centromere distance respectively (Figure 3A). The axial length of the Ndc80 complex exhibits small values for the slope indicating that it remains constant (Figures S4C-D). These data indicate that CENP-C has a major role and CENP-T has a minor role in controlling the compliance of the linkage between the centroid of CENP-A and the Spc24/Spc25 end of the Ndc80 complex.
We also found that CCAN proteins contribute to compaction of CENP-A-containing chromatin for bi-oriented chromosomes. In control and CENP-Q depleted late prometaphase cells, most fluorescent kinetochore spots both for CENP-A and Ndc80(9G3) were elliptical with the long axis perpendicular to the kMT axis (Figures 5A, 5G, and see S5 for criteria). In CENP-H, CENP-T, CENP-C, or CENP-C/CENP-H depleted cells in late prometaphase, all Ndc80/Hec1(9G3) and around 80% of CENP-A fluorescent kinetochore spots of bi-oriented chromosomes had the shape of controls, but the CENP-A and Ndc80 spots were much more separated from each other (Figures 5C-F, left, and 5G). These images were used to obtain the Delta measurements in Figures 3, 4 and S4. About 20 % of kinetochores exhibited CENP-A kinetochore fluorescence elongated about 3 fold over normal in the K-K axis direction and it was not possible to obtain accurate measurements of the centroid of CENP-A chromatin for these kinetochores by our Gaussian 3D fitting methods (Wan et al., 2009; Figures 5C-F, right, and 5G). This CENP-A chromatin “de-compaction” appears tension sensitive as it did not occur for unattached kinetochores of unaligned chromosomes in the various CCAN depleted cells. It was very rare that both sister kinetochores exhibited this de-compaction and the data in Figure 5G are typical of all cells. To test the hypothesis that CCAN DNA binding proteins, such as CENP-T/W/S/X, CENP-C, and CENP-N regulate CENP-A compaction during chromosome segregation, we examined the function of CENP-T or CENP-C DNA binding regions (Figure 5H). Expression of CENP-T and CENP-C DNA binding regions, such as CT107, CC426, and CC690, prevented, by an unknown mechanism, the de-compaction of CENP-A chromatin resulting from either CENP-T or CENP-C depletion (Figure 5I). Overexpression of the DNA binding domains of CENP-C was not healthy for the cells as it promoted abnormal chromosome segregation (mini chromosomes) and apoptosis (Figures S3E-F). Nevertheless, taken together these results show that CENP-T, CENP-C, and CENP-H/I complex have a critical role for the compaction of CENP-A-containing chromatin in bi-oriented chromosomes.
Figure 5.
CCAN regulates compaction of CENP-A containing chromatin during mitosis. (A) Two-color immunofluorescence of kinetochore CENP-A and Ndc80/Hec1(9G3) in control cells (top) with high magnification image of indicated region (middle), and line scan (bottom) for (A) Control; (B) CENP-Q depleted cells; (C) CENP-C depleted cells; (D) CENP-T depleted cells; (E) CENP-H depleted cells; and (F) CENP-C/CENP-H depleted cells. (G) The percentage of punctate spot or stretched spot of CENP-A and Ndc80/Hec1(9G3) immunofluorescence in control and experimental cells in (A) to (F). (H) Two-color immunofluorescence of CENP-A and GFP in control cells, CENP-C depleted cells, CENP-C depleted cells expressing GFP-CC-426 or GFP-CC-690, and CENP-T depleted cells expressing GFP-CC-426 or GFP-CT-107. (I) The relative frequency of CENP-A containing chromatin expansion for cells in (H).
Hyper-Intrakinetochore stretch is correlated with a reduction in the level of phosphorylation of the Ndc80/Hec1 N-terminus
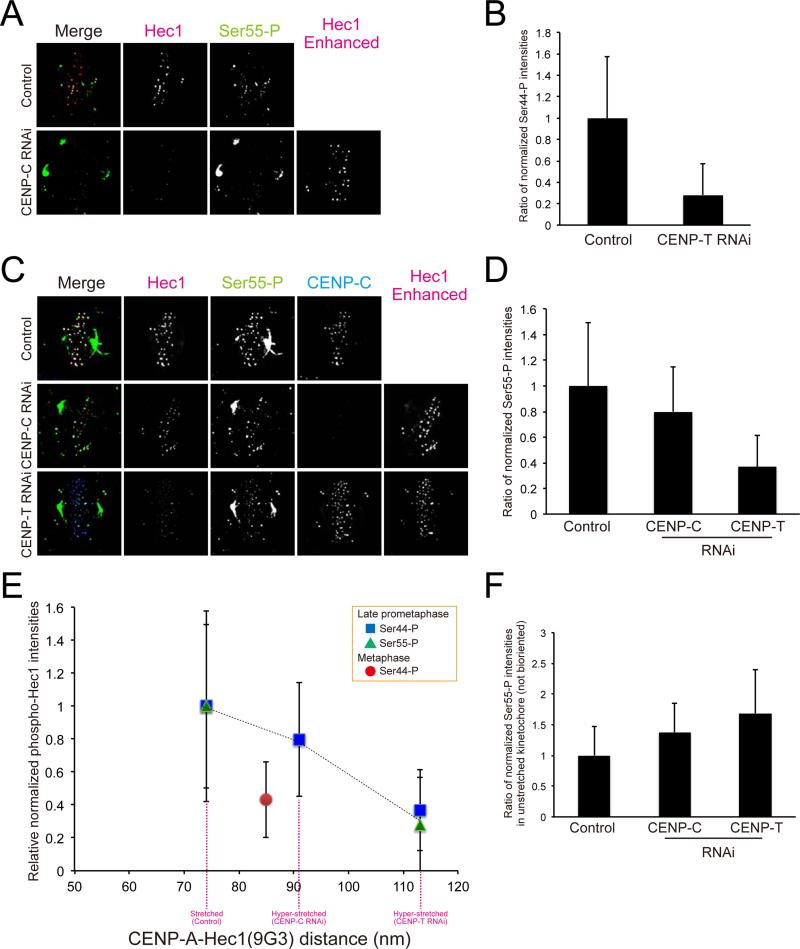
To test if hyper-intrakinetochore stretch reduces the level of Aurora B phosphorylation of the Ndc80/Hec1 in late prometaphase, we used two antibodies that have been previously shown to specifically recognize Aurora B phosphorylation epitopes at the N-terminus of Ndc80/Hec1: S44-P and S55-P (DeLuca et al., 2011; Figures 6A-D). Phosphorylation of these epitopes has important roles in controlling destabilization of kMT attachment in prometaphase (DeLuca et al., 2011). We measured integrated fluorescence intensities at kinetochores of bi-oriented chromosomes labeled with these antibodies in control, CENP-C, or CENP-T depleted cells and normalized by the value for Ndc80/Hec1(9G3) at the same kinetochore (Figures 6B and 6D). This normalization was required because both CENP-C and CENP-T RNAi induced reduction of Ndc80/Hec1 at kinetochores to 47% and 36% of the control level (Figure 3D and S4B). The phosphorylation of Ndc80/Hec1 decreased substantially with the extent of hyper-intrakinetochore stretch for kinetochores of bi-oriented chromosomes in late prometaphase: 20% and 65% reductions occurred compared to control cells for the 15 nm and 45 nm hyper-stretch produced by CENP-C and CENP-T RNAi (Figure 6E). In contrast, the phosphorylation per Ndc80 complex at kinetochores of unaligned chromosomes (no hyper-kinetochore stretch) was somewhat higher than controls with depletion of CENP-T or CENP-C (Figure 6F). As a control, we measured fluorescence for kinetochores labeled with an antibody specific for an Aurora B phosphorylated epitope on Knl1 (Welburn et al., 2010). There was a drop in the level of phosphorylation with intrakinetochore stretch for CENP-C and CENP-T depleted cells as occurs for S44-P and S55-P of Hec1(Ndc80) (Figures S6A-B), but the decrease was not as large perhaps because Knl1 is located more than 25 nm inward from Hec1(9G3) (Wan et al., 2009). These results indicate that the level of Aurora B phosphorylation for the N-terminus of Ndc80/Hec1 decreases rapidly for distances greater than the normal amount of intrakinetochore stretch of Ndc80/Hec1(9G3) away from CENP-A.
Figure 6.
Hyper-intrakinetochore stretch reduces the level of phosphorylation of the Ndc80/Hec1 N-terminus. (A) Two-color immunofluorescence for Ndc80/Hec1(9G3) and Ser44-P at kinetochores in control or CENP-T depleted cells. (B) Integrated fluorescence intensity of Ser44-P at kinetochores normalized by the value for Ndc80/Hec1(9G3) on bi-oriented kinetochores in control or CENP-T depleted cells. (C) Two-color immunofluorescence forNdc80/Hec1(9G3) and Ser55-P at kinetochores in CENP-C depleted or CENP-T depleted cells. (D) Integrated fluorescence intensity of Ser55-P at kinetochores normalized by the value for Ndc80/Hec1(9G3) on bi-oriented kinetochores in control, CENP-C depleted, and CENP-T depleted cells. (E) Data from (B) and (D) plotted as a function of the separation between CENP-A to Ndc80/Hec1. (F) Integrated fluorescence intensity of Ser55-P at kinetochores normalized by the value for Ndc80/Hec1(9G3) on unaligned kinetochores in control, CENP-C depleted, and CENP-T depleted cells. The enhanced Hec1(9G3) images are added to mark kinetochore positions in (A) and (B).
To test if the reduction of Ndc80/Hec1 phosphorylation that accompanies hyper-intrakinetochore stretch of bi-oriented chromosomes in late prometaphase induces stable kMT attachment, we measured kMT intensities in cold treated CENP-T or CENP-C/-H depleted cells (Figure S6C). In CENP-C or CENP-T depleted cells, the intensity of kMT fluorescence normalized by the level of Ndc80/Hec1 was slightly greater than for kinetochores in control cells (Figure S6D; p = 0.057, p < 0.01 (t-test), respectively). We also examined the level of Mad1 at kinetochores as Mad1 depletes with kMT attachment (Musacchio and Salmon, 2007). Mad1 at kinetochores of bi-oriented chromosomes in late prometaphase cells decreased as a function of hyper-intrakinetochore stretch as expected for enhanced stabilization of kMT attachment by lower levels of phosphorylation of Ndc80/Hec1 N-terminus (Figure S6E). We conclude that intrakinetochore stretch must be constrained to the normal amount for bi-oriented kinetochores in late prometaphase to prevent loss of the Aurora B phosphorylation needed to destabilize kMT attachment for error correction in prometaphase (DeLuca et al., 2006; DeLuca et al., 2011; Foley et al., 2011)
A major drop in phosphorylation of the Ndc80/Hec1 complex by Aurora B at metaphase is not predicted by intrakinetochore stretch
There is a major increase (>2 fold) in the stability of kMT attachment to kinetochores of bi-oriented chromosomes between prometaphase and metaphase (Kabeche and Compton, 2013). There is a corresponding substantial decrease in the level of Aurora B phosphorylation for the N-terminus of Ndc80/Hec1 at kinetochores of bi-oriented chromosomes (Figure 6E; DeLuca et al., 2011). The 10 nm increase in intrakinetochore stretch from late prometaphase to metaphase does not appear to play a major role in either of these changes (Figure 6E).
Discussion
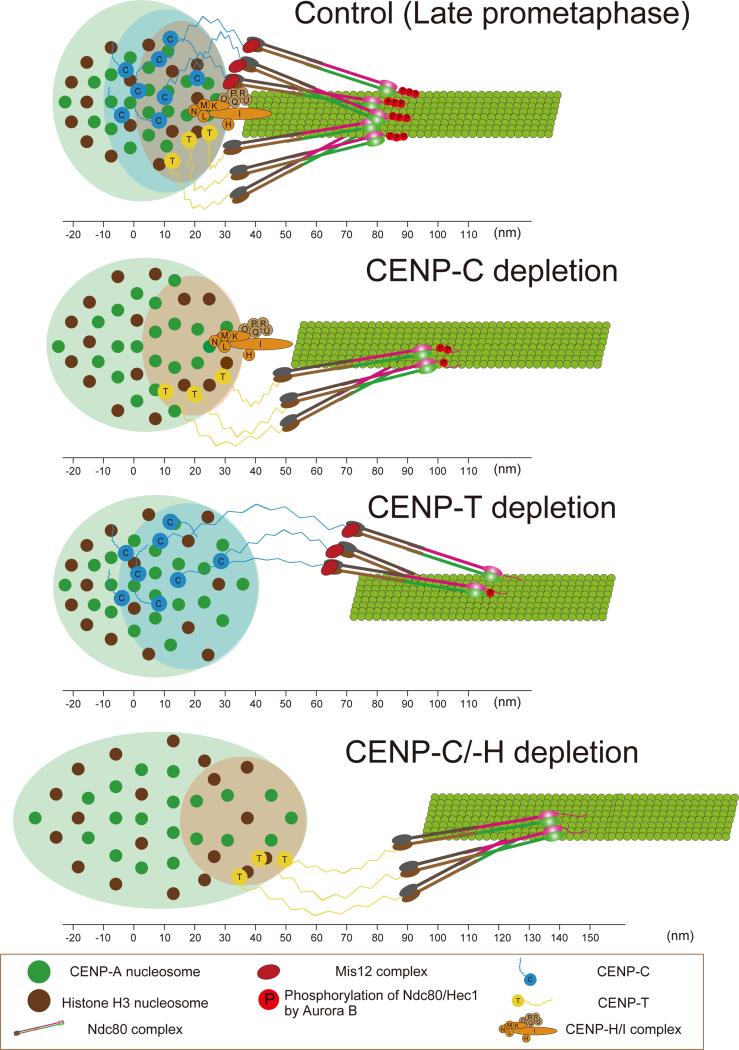
In Figure 7, we summarize how CCAN proteins organize the mechanical linkage between CENP-A-containing chromatin and KMN network proteins within human kinetochores of bioriented chromosomes in late prometaphase and metaphase (Figure 7, Top) and in late-prometaphase following depletion of CENP-C, CENP-T or CENP-C/H (Figure 7, bottom 3 panels). We found that most CCANs, except CENP-A and CENP-C, are localized close to the periphery of centromeric chromatin. The great majority of CENP-T extends from its C-terminal chromatin-binding domain to overlap at its N-terminus with Spc24/Spc25, and the great majority of CENP-I also extends into the outer kinetochore, similar to the extension of CENP-T (Figures 1 and 2A). In contrast the mean positions of both ends of CENP-C are slightly deeper than the other DNA binding CCAN proteins except for CENP-A (Figures 1 and 2A). The N-terminus of CENP-C is known to play a major role in recruiting Mis12 complex to the kinetochore and known to bind Mis12 complex in vitro (Przewloka et al., 2011; Screpanti et al., 2011). Our measurements support a model where only a minor fraction of CENP-C actually interconnects the inner kinetochore with Mis12 complex, a result predicted by a previous study (Ribeiro et al., 2010).
Figure 7.
A model for inner kinetochore protein architecture and how phosphorylation and the N-terminal kMT binding domain of the Ndc80 complex depends on intrakinetochore stretch for bi-oriented centromeres late prometaphase and metaphase control cells (Top panel) and in late prometaphase for CENP-C depleted, CENP-T depleted, and CENP-C/-H depleted human cells (Bottom 3 panels) . From the periphery of the centromeric chromatin, the depth of CENP-A is greatest, with the DNA binding domains of CENP-C being slightly deeper than those of CENP-T and CENP-N. CENP-T/-W/-S/-X complex, CENP-C, and CENP-H/I complex all contribute to limiting the separation between the mean positions of CENP-A and the inner end of the Ndc80 complex to about 40 nm for bi-oriented centromeres in control cells. This separation is increased to, much less than is possible and contribute for kMT assembly regulated by Aurora B. The depletion of CENP-T or CENP-C induced hyper-intrakinetochore stretch and a reduction of phosphorylation of Hec1 by Aurora B.
Our data indicate that the intrakinetochore stretch of CENP-T in metaphase HeLa cells is constrained to be ~19 nm on average compared to a potential stretch of 90 nm. A length of ~19 nm in metaphase is similar to that measured for chicken DT40 cells (Suzuki et al., 2011). The amount of tension per CENP-T may be one of the constraint factors since the end-to-end separation for CENP-T in metaphase DT40 cells without centromere tension is near zero (Suzuki et al., 2011) and the mean position of the N-terminal of CENP-T is near the C-terminus when the Spc24/Spc25 binding domain is deleted (Figures 2A-C). The long stretch of unstructured amino acids within CENP-T that extends to near the N-terminus could act as an entropic spring (Gardner et al., 2013) whose stiffness limits stretch to ~19 nm at the metaphase tension supported by each CENP-T molecule. CENP-C also has an extended region of unstructured amino acids near its N-terminus, which could also act as an entropic spring connected to Mis12. This spring would act in parallel with the CENP-T spring, reducing the force on both CENP-T and CENP-C from centromeric tension dependent on their relative stiffness and numbers. In support of this model, we found that deleting CENP-C, CENP-T, or CENP-C/-H caused an increase in intrakinetochore stretch between the mean positions of CENP-A and Spc24/Spc25 (Figure 3D) as expected for fewer springs supporting the same force from centromere stretch between sister kinetochores. However we found for controls that normal intrakinetochore stretch is not sensitive to changes in centromere tension that are produced by oscillations of bi-oriented chromosomes back-and forth-across the spindle equator (Figure 3A). In contrast, the hyper-intrakinetochore stretch that occurs with depletion of CENP-C, CENP-T, or CENP-C/-H is sensitive to changes in centromere tension. This suggests that depletion of CENP-T, CENP-C or CENP-H releases crosslinking within the kinetochore that constrains intrakinetochore stretch to a level produced by low centromere tension and prevents further intrakinetochore stretch in response to higher levels of tension produced by further centromere stretch between sister kinetochores.
We also analyzed for late prometaphase cells, the relative contributions of Ndc80 complex, CENP-A chromatin stretch, and CENP-T stretch to intrakinetochore stretch (Figures 4A-D). The mean end-to-end length of Ndc80 complex remained constant, while increases in intrakinetochore stretch were contributed by increases in the mean separation of CENP-A to the Spc24/Spc25 end of the Ndc80 complex. It is still unclear why both CENP-A chromatin and CENP-T are slightly more stretched in metaphase, compared to late prometaphase, but cell cycle changes in chromatin structure and a slight increase the number of kMTs in metaphase could contribute. Surprisingly, CENP-A chromatin stretch contributed about equally with CENP-T stretch to increased intrakinetochore stretch for the small change between normal prometaphase and metaphase and for the much larger changes induced by depletion of CENP-C, CENP-T or CENP-H.
Our data indicate that CCAN proteins contribute not only to the mechanical linkage between their CENP-A-chromatin binding sites and the KMN network, but they also have other binding domains that contribute to the compliance and compaction of CENP-A-containing chromatin within the inner kinetochore (Figures 5A-I), which supports a previous observation (Ribeiro et al., 2010). CENP-C has multiple DNA binding domains (Figure S3) and also forms a homo-dimer, which indicates that CENP-C has multiple mechanisms for chromatin cross-linking (Kato et al., 2013). The DNA binding domain of CENP-T also binds CENP-W/-S/-X, which also has chromatin binding ability (Nishino et al., 2012), making chromatin cross-linking a possibility. Note that the potential chromatin cross-linking ability of CENP-C or CENP-T do not depend on either the Spc24/Spc25 binding domain of CENP-T or on the Mis12 binding domain of CENP-C.
Another unexpected finding is that the CENP-H/-I complex also makes a major contribution to limiting intrakinetochore stretch. Depletion of the CENP-H/-I complex doubled the mean separation of CENP-A to Spc24/Spc24 (Figure 3D) while maintaining the control stiffness of the linkage (Figure 3A). Depleting both CENP-H/-I and CENP-C caused the largest intrakinetochore stretch, where as depleting CENP-C alone produced a moderate increase in intrakinetochore stretch. In addition, we found that CENP-I extended from the inner kinetochore to overlap with the Spc24/Spc25 end of the Ndc80 complex, the Mis12 complex and the C-terminus of Knl1. We conclude from this data that the CENP-H/-I complex also makes a major contribution in constraining intrakinetochore stretch of CENP-T and CENP-C and predict that CENP-I C-terminus makes a major contribution in recruiting Ndc80 complex to kinetochores, perhaps through recruiting the whole KMN network.
Our data also show that the CCAN protein linkage between the inner and outer kinetochores is capable of resisting spindle forces at kinetochores and producing intrakinetochore stretch large enough upon kMT attachment to control silencing of the spindle checkpoint (Maresca and Salmon, 2009, 2010; Uchida et al., 2009), but also small enough so that the level of Ndc80/Hec1 phosphorylation is sufficiently high to allow frequent enough release of kMTs to correct errors in attachment in prometaphase. We show here that kinetochores exhibiting hyper-intrakinetochore stretch in late prometaphase also exhibit substantially reduced levels of Ndc80/Hec1 phosphorylation (Figures 6A-E). Low levels of Ndc80/Hec1 phosphorylation in prometaphase has been shown to significantly enhance errors in anaphase chromosome segregation (Cimini et al., 2006; DeLuca et al., 2006; DeLuca et al., 2011). Low levels of Ndc80/Hec1 phosphorylation stabilize Ndc80 complex binding to MTs (Cheeseman and Desai, 2008; Wei et al., 2007). This enhanced binding affinity for kMTs at low phosphorylation helps explain how less than 30% of the normal level of Ndc80 complexes at kinetochores in CENP-T and CENP-H/CENP-C depleted cells are able to bind stably to their kMTs (Figures S6C-D) and produce nearly normal inter-kinetochore centromere stretch of bi-oriented chromosomes (Figure 3A). In contrast, we have found that the major reduction in the level of Aurora B phosphorylation for the N-terminus of Ndc80/Hec1 between late prometaphase and metaphase is not accounted for by the 10 nm increase that occurs in intrakinetochore stretch. Instead, most of this reduction in phosphorylation is likely produced by cell cycle regulation of Aurora B kinase and PP1 and PP2A phosphatase activities at kinetochores that makes kMTs twice as stable at kinetochores of bi-oriented chromosomes in metaphase in comparison to prometaphase (Campbell and Desai, 2013; DeLuca et al., 2011; Foley et al., 2011; Kabeche and Compton, 2013).
Experimental Procedures
Cell culture and Transfections
HeLa Cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2. For transfection conditions of RNAi and plasmids, see the Supplemental Information.
Immunofluorescence
Cells were fixed by 3% PFA at 37°C. Fixed samples were permeabilized by 0.5% NP40 (Roche) or 0.5% Triton (SIGMA) in PHEM buffer (120mM Pipes, 50mM HEPES, 20mM EGTA, 4mM magnesium acetate, pH7.0) at 37oC, rinsed in 0.05% Triton / PHEM, and incubated in 0.5% BSA (SIGMA) with BGS (Boiled Goat Serum) or BDS (Boiled Donkey Serum) for 30 min at room temperature. Then samples were incubated for 1 h at 37°C with primary antibodies. Primary antibodies are described detail in Supplementary Information. Samples were stained using DAPI (0.1ug/ml) after incubation with secondary antibodies. Secondary antibodies we used were conjugated Alexa488, Rhodamine Red-X, or Cy5 (Jackson ImmunoResearch Labs). Samples were mounted using Prolong Antifade (Molecular Probes) or house made mounting media (20 mM Tris pH 8.0, 0.5% N-propyl gallate, 90% Glycerol) after post fixation was performed by 3% PFA. If we mounted using Prolong Antifade, we observed samples by LM after increasing appropriate refraction index. For staining of phospho-Ndc80/Hec1, we added phosphatase inhibitors (PhosSTOP (Roche) and Calyculin A (Cell Signaling)) to the 3% PFA fixation buffer and permeablization buffers.
Imaging
Imaging was performed as described by Varma et al., 2013. For details, see Supplemental Information.
Fluorescence Intensity Measurements
Measurements of kinetochore integrated fluorescence intensity minus background were performed as described by Lawrimore et al., 2011 as described in Supplemental Information.
Delta Analysis
For each kinetochore, 3D centroid positions were first measured for each fluorescent color by a 3D Gaussian fitting function (Wan et al., 2009). For each sister kinetochore pair, the centroids of one color were projected to the axis defined by the centroids of the other color, and the average separation of the projection distance between the signals of different colors for that pair (Delta) was then calculated to correct for chromatic aberration (Wan et al., 2009). Mean Delta measurements were corrected for tilt of the face of the kinetochore relative to the axis between sister kinetochores as described by Wan et al., 2009. In addition to antibodies selective for a specific protein domain, like the Ndc80/Hec1 (9G3) antibody that binds near the MT binding domain of the Ndc80 complex, we also expressed CCAN proteins in HeLa cells with GFP fused to either the N- or C-terminus, and measured the separation between antibodies to GFP and 9G3 or endogenous CENP-A. For mean position measurement of the N-terminus or C-terminus of CENP-T and the N-terminus or C-terminus of CENP-C, we established stable cell lines expressed GFP-fused, RNAi-resistance, CENP-T or CENP-C and measured mean position in RNAi treated cells that substantially depleted endogenous CENP-T or CENP-C. In this study, we measured several protein positions previously determined by Wan et al., 2009, and found nearly identical values.
Statistical Analysis
All data were expressed, as means ±SD. Statistical significance was determined using Student's t-test for comparison between two independent groups. P < 0.05 was considered statistical significant and 0.5 < P < 0.1 was considered marginally statistical significant.
Supplementary Material
Acknowledgements
We would like to thank Drs. Iain Cheeseman, Andrea Mussachio, Gordon Chan, Tim Yen, Todd Stukenberg, Julian Haase, Paul Maddox, Arshad Desai, and Dileep Varma for providing valuable regents and advice. This work supported by Uehara Memorial Foundation, Kazato research foundation, and Japan Society and Promotion of Science (A. Suzuki), R01GM088371 (J.G. DeLuca), and 5R37GM24364 (E.D. Salmon) from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution
A.S. performed entire experiments and analyzed the data. B.L.B and A.S. performed Figure S3B and S6 experiments. X.W. confirmed the delta analysis data. J.G.D. provided critical regents and suggestion. A.S. and E.D.S. designed all experiments and wrote the manuscripts.
References
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. The Journal of cell biology. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nature cell biology. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013;194:817–846. doi: 10.1534/genetics.112.145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CS, Desai A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 2013;497:118–121. doi: 10.1038/nature12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. The Journal of cell biology. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nature reviews. Molecular cell biology. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Molecular biology of the cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Current biology: CB. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes & development. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF. Heel and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Molecular biology of the cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Heel. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Heel phosphorylation control kinetochore-microtubule attachment stability during mitosis. Journal of cell science. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–358. doi: 10.1126/science.1221886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nature cell biology. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nature cell biology. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gardner KA, Moore DA, Erickson HP. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Molecular microbiology. 2013 doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Heel. Current biology: CB. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Okada M, Maenaka K, Fukagawa T. CENP-0 class proteins form a stable complex and are required for proper kinetochore function. Molecular biology of the cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. The Journal of cell biology. 2013;200:45–60. doi: 10.1083/jcb.201210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature. 2013;502:110–113. doi: 10.1038/nature12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends in cell biology. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi E, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. The EMBO journal. 2013;32:409–423. doi: 10.1038/emboj.2012.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. The Journal of cell biology. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. Journal of cell science. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature reviews. Molecular cell biology. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nishino T, Rago E, Hori T, Tomii K, Cheeseman IM, Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. The EMBO journal. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature cell biology. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Current biology: CB. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Rago F, Cheeseman IM. Review series: The functions and consequences of force at kinetochores. The Journal of cell biology. 2013;200:557–565. doi: 10.1083/jcb.201211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Misl2 complex joins the inner and outer kinetochore. Current biology: CB. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell division. 2012;7:19. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hori T, Nishino T, Usukura J, Miyagi A, Morikawa K, Fukagawa T. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. The Journal of cell biology. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K. Regulatory mechanisms of kinetochore-microtubule interaction in mitosis. Cellular and molecular life sciences : CMLS. 2013;70:559579. doi: 10.1007/s00018-012-1057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J, Cooke CA, Saitoh H, Bernat RL, Earnshaw WC. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. The Journal of cell biology. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. The Journal of cell biology. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KS, Takagaki K, Kumada K, Hirayama Y, Noda T, Hirota T. Kinetochore stretching inactivates the spindle assembly checkpoint. The Journal of cell biology. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D, Wan X, Cheerambathur D, Gassmann R, Suzuki A, Lawrimore J, Desai A, Salmon ED. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. The Journal of cell biology. 2013;202:735–746. doi: 10.1083/jcb.201304197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. Journal of molecular biology. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nature structural & molecular biology. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR, 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Molecular cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Molecular and cellular biology. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.