SUMMARY
Atg16L1 mediates the cellular degradative process of autophagy and is considered a critical regulator of inflammation based on its genetic association with inflammatory bowel disease. Here we find that Atg16L1 deficiency leads to an exacerbated graft-versus-host disease (GVHD) in a mouse model of allogeneic hematopoietic stem cell transplantation (allo-HSCT). Atg16L1-deficient allo-HSCT recipients with GVHD displayed increased T cell proliferation due to increased dendritic cell (DC) numbers and co-stimulatory molecule expression. Reduced autophagy within DCs was associated with lysosomal abnormalities and decreased amounts of A20, a negative regulator of DC activation. These results broaden the function of Atg16L1 and the autophagy pathway to include a role in limiting a DC-mediated response during inflammatory disease, such as GVHD.
INTRODUCTION
Autophagy (also referred to as macroautophagy) consists of the sequestration of cellular material including organelles and long-lived proteins within double-membrane vesicles termed autophagosomes, and subsequent targeting of this cargo to the lysosome for degradation and recycling (Yang and Klionsky, 2010). Although initially characterized as a method to maintain viability during periods of nutrient deprivation, a variety of immune functions have been attributed to this pathway including the degradation of intracellular pathogens, cytokine secretion, lymphocyte homeostasis, and antigen presentation (Deretic, 2012; Levine et al., 2011; Ma et al., 2013). Consistent with this critical role in immunity, a common polymorphism in ATG16L1 is associated with a higher incidence of Crohn’s disease, a major type of inflammatory bowel disease (IBD) (Rioux et al., 2007). Atg16L1 promotes the conjugation of the ubiquitin-like molecule LC3 to phosphatidyl-ethanolamine (PE), a step that is required for the proper formation of the autophagosome (Fujita et al., 2008). While complete deletion of the gene is lethal (Saitoh et al., 2008), we previously generated viable mice that have reduced Atg16l1 expression and autophagy in all tissues examined due to insertion of a gene-trap cassette in the Atg16l1 locus (Cadwell et al., 2009). These Atg16L1 hypomorph (Atg16L1HM) mice do not display obvious abnormalities in the absence of an infectious trigger, but they develop intestinal abnormalities upon norovirus infection (Cadwell et al., 2010). Additionally, we have found that Atg16L1HM mice display significantly increased resistance towards intestinal infection by Citrobacter rodentium that is mediated by an enhanced mucosal immune response (Marchiando et al., 2013). These findings suggest that Atg16L1 has a broad physiological role in regulating the nature and intensity of the immune response to various inflammatory insults.
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) can be a life-saving procedure for individuals with malignancies as well as non-malignant diseases such as genetic blood disorders and bone marrow failure (Jenq and van den Brink, 2010). Allo-HSCT involves the transfer of HSCs from the bone marrow, peripheral blood, or umbilical cord blood of a donor into the patient following a conditioning regimen, frequently irradiation and/or chemotherapy. A significant complication of this procedure occurs when alloreactive T cells derived from the donor attack healthy tissue and cause a multi-organ disease referred to as graft-versus-host-disease (GVHD). Inflammation caused by the conditioning regimen and subsequent activation of antigen presenting cells such as dendritic cells (DCs) contribute to donor T cell proliferation and migration to target organs. GVHD involving the gastrointestinal tract in particular is a major contributor to transplant-related morbidity and mortality. Intestinal GVHD shares certain similarities with IBD including genetic susceptibility conferred by mutations in NOD2, a Crohn’s disease gene that encodes a bacterial sensor (Henault et al., 2012; Holler et al., 2004; Landfried et al., 2010; Tanabe et al., 2011). We previously demonstrated that Nod2−/− mice have increased susceptibility to GVHD in a mouse model of allo-HSCT, further supporting the resemblance in pathophysiology of IBD and GVHD (Penack et al., 2009). Despite the emerging understanding of the immune functions of Atg16L1, and interest in pharmacologically targeting autophagy, the role of this protein in intestinal GVHD and allo-HSCT outcome has not been examined.
In this study, we define a critical role for Atg16L1 in reducing morbidity and mortality following allo-HSCT. Atg16L1HM allo-HSCT recipient mice develop worse GVHD with increased donor T cell proliferation and migration to the intestine. We further demonstrate that autophagy has a cell-intrinsic anti-inflammatory role in DCs that is critical for reducing T cell alloreactivity. The DC hyperactivity due to Atg16L1-deficiency was associated with accumulation of aberrant lysosomes and an increase in Laptm5, a pro-inflammatory lysosomal protein that enhances NF-κB signaling by inhibiting the ubiquitin-editing enzyme A20. These results establish a pivotal role for the autophagy pathway in limiting DC-mediated T cell alloreactivity in a systemic disease.
RESULTS
Atg16L1 deficiency increases morbidity and mortality in a mouse model of allo-HSCT
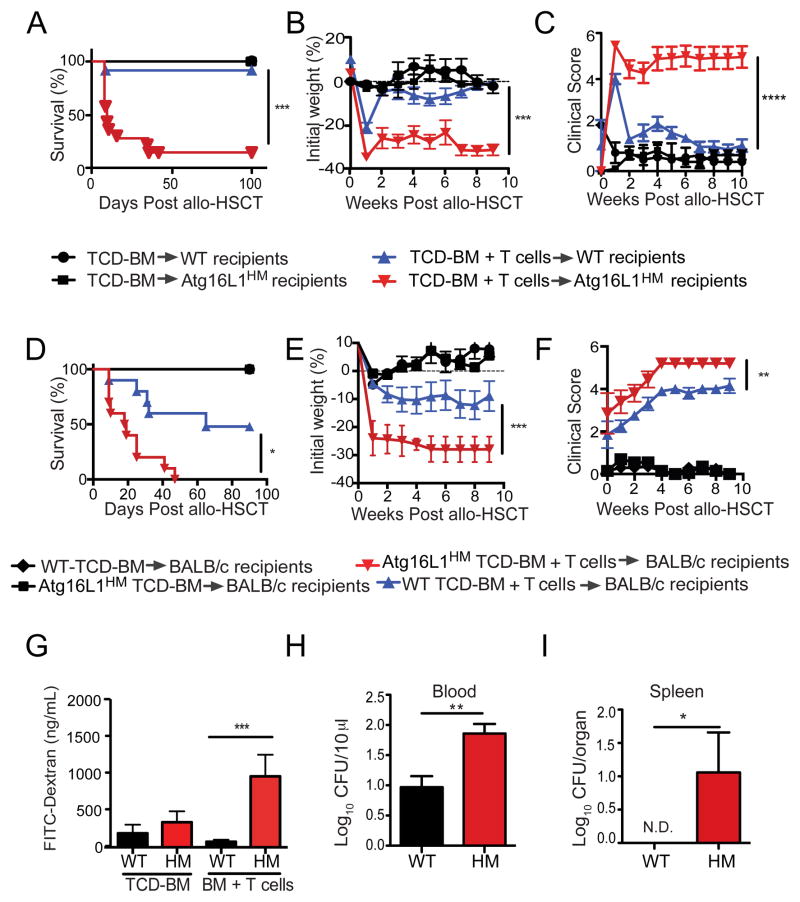
To investigate the role of Atg16L1 in allo-HSCT recipients, we utilized a major histocompatibility complex (MHC) disparate transplantation model in which T cell depleted (TCD) bone marrow (BM) cells with or without T cells from B10.BR donor mice were transplanted into lethally-irradiated C57BL/6 wild-type (WT) or Atg16L1HM recipient mice. Because Atg16L1HM mice are sensitive to the presence of infectious agents including common mouse pathogens, we initially examined WT and Atg16L1HM mice that are maintained in an animal facility free of murine norovirus (MNV), Helicobacter, and segmented filamentous bacteria (SFB). Compared to WT recipients, Atg16L1HM mice receiving TCD-BM and 2×106 T cells displayed increased lethality, weight loss, and clinical GVHD score (Figures 1A–C). This exacerbated GVHD is T cell-dependent since neither WT nor Atg16L1HM mice receiving TCD-BM without T cells showed signs of disease (Figures 1A–C). Also, the severity of GVHD was T cell dose-dependent. Transplantation of a lower number of T cells decreased weight loss and mortality, but GVHD was still significantly worse in Atg16L1HM compared to WT recipients (Figures S1A and S1B). The lack of certain pathogens and commensals commonly found in mouse facilities may explain the absence of lethality in WT recipients in these experiments. Indeed, we found that an increased number of WT mice succumbed to lethality when the experiment was repeated in a second animal facility. However, Atg16L1HM mice still displayed significantly diminished survival compared to their WT counterparts, indicating that the role of Atg16L1 in protection from GVHD is reproducible (Figure S1C). To investigate the role of Atg16L1 in donors, we utilized another common allo-HSCT model in which irradiated BALB/c mice receive the transplant from C57BL/6 donors. Again, we found an increase in morbidity and mortality in BALB/c recipients that received TCD-BM from Atg16L1HM mice compared to recipients of TCD-BM from WT mice, in a manner dependent on T cells (Figures 1D–F). Thus, Atg16L1 deficiency in either the donor or recipient leads to worse outcomes after an allo-HSCT.
Figure 1. Atg16L1 deficiency in donors and recipients increases morbidity and mortality in a mouse model of allo-HSCT.
(A–C) Lethally-irradiated C57BL/6 WT and Atg16L1HM recipient mice were transplanted with 5×106 TCD-BM cells with or without 2×106 splenic T cells from donor B10.BR mice and monitored for (A) survival, (B) weight loss, and (C) macroscopic signs of GVHD (see methods). n = 20 recipient mice/group. (D–F) Lethally-irradiated BALB/c recipient mice were transplanted with 5×106 TCD-BM cells from either C57BL/6 WT or Atg16L1HM mice with or without 1×106 T cells from donor WT mice and monitored for (D) survival, (E) weight loss, and (F) macroscopic signs of GVHD. n = 20 recipient mice/group. (G) Serum FITC-dextran concentration following oral gavage was measured in WT and Atg16L1HM mice on day 7 after irradiation followed by transfer with TCD-BM with or without T cells from B10.BR mice. n = 5–8 recipient mice/group. (H, I) Colony forming units (CFU) of bacteria present in (H) blood and (I) spleen harvested on day 7 from WT and Atg16L1HM mice receiving TCD-BM with T cells from B10.BR mice. N.D. not detected. n = 8 recipient mice/group. Data represent at least two independent experiments. Mean ± S.E.M. are shown in (B), (C), (E)–(I). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We took advantage of the consistently early onset of disease in Atg16L1HM recipient mice in the B10.BR→C57BL/6 model to further characterize the role of Atg16L1 during allo-HSCT. The pathophysiology of GVHD involves a feedforward amplification of the inflammation from the transplant procedure driven by inflammatory cytokines and tissue damage (Ferrara and Reddy, 2006). Upon screening a panel of cytokines and chemokines, we found that Atg16L1HM mice receiving TCD-BM and T cells had increased amounts of tumor necrosis factor (TNF-α), interleukin-12 (IL-12) p70, macrophage inflammatory protein α (MIP1α), and IL-7, and a decreased amount of IL-5 compared to similarly treated WT mice on day 7 post-transplantation (Figures S1D and S1E). Consistent with a critical role for cytokines in amplifying the inflammation, administration of anti-TNF-α antibodies prevented mortality in Atg16L1HM recipient mice (Figure S1F).
Allo-HSCT was also associated with inflammation of various target organs including the small intestine, colon, liver, and skin in both WT and Atg16L1HM recipient mice (Figures S2A–J). Although Atg16L1-deficiency did not lead to increases in scoring when analyzed by classical semi-quantitative histopathology parameters, this lack of obvious differences between genotypes could be a reflection of the rapid progression from disease onset to lethality. Therefore, as a more quantitative measure of intestinal damage, we examined intestinal permeability because compromised barrier function is one of the most serious complications associated with GVHD (Takatsuka et al., 2003). On day 7 after allo-HSCT transplanted with T cells, Atg16L1HM recipients of BM + T cells displayed significantly more intestinal permeability compared to WT recipients as determined by the presence of FITC-dextran in the serum following oral gavage (Figure 1G). This increase in permeability was not observed in mice receiving TCD-BM only or mice that were irradiated without transplantation (Figures 1G and S1G). As predicted by the loss of barrier function, Atg16L1HM mice receiving TCD-BM and T cells also had 10-fold higher numbers of bacteria in the spleen and blood on day 7 (Figures 1H and I). Therefore, Atg16L1 is critical for maintaining intestinal integrity during GVHD in allo-HSCT recipients.
Based on our observations in the mouse model, we examined the effect of the common T300A polymorphism in ATG16L1 associated with Crohn’s disease on allo-HSCT outcome in humans. We analyzed a cohort of 122 patients receiving an allograft from an HLA-identical sibling (Table S1). Individuals that succumbed to death following tumor-relapse were censored. The presence of ATG16L1T300A in either the donor or recipient was associated with increased non-relapse mortality (Figure S1H and I), although only the presence of the polymorphism in donors reached statistical significance in this small cohort. The distribution of the causes of treatment-related mortality were similar in all groups and were typical for the procedure; the most common cause of death were GVHD, with or without a secondary infection, and idiopathic pneumonia syndrome, which is a condition due to allogeneic T cell activity and frequently associated with GVHD. Although confirmation in a larger prospective study is needed, these data support a role for ATG16L1 in clinical allo-HSCT outcome.
Atg16L1 deficiency in allo-HSCT recipients leads to increased T cell proliferation and intestinal homing
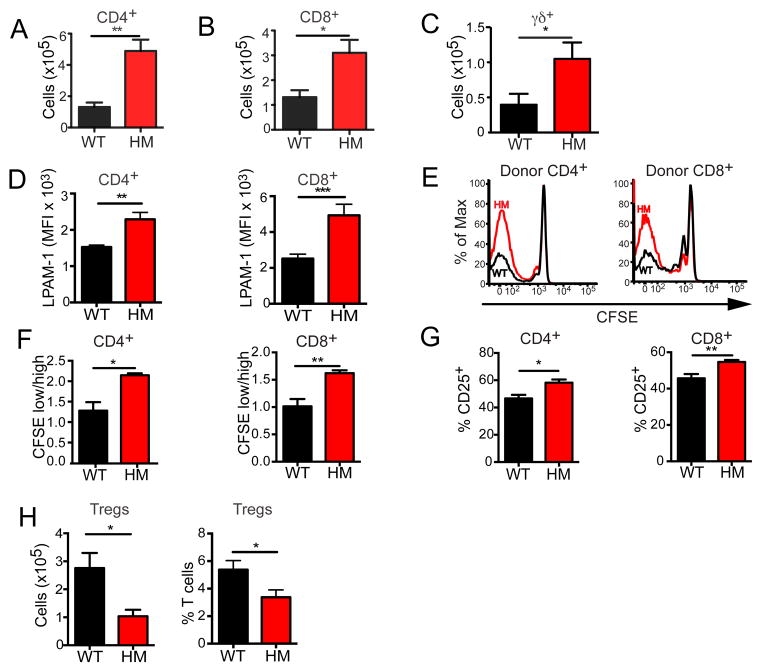
Given that the enhanced GVHD observed in Atg16L1-deficient mice is T cell-dependent, we examined the fate of T cells after transplantation. Flow cytometric analysis of intra-epithelial lymphocytes (IELs) indicated that Atg16L1HM recipients had significantly increased numbers of donor-derived CD4+, CD8+, and TCRγδ+ T cells in the small intestine on day 7 after allo-HSCT compared to WT recipients (Figures 2A–C). Consistent with the increased number of IELs, donor T cells harvested from the spleen of Atg16L1HM allo-HSCT recipient mice displayed increased expression of LPAM-1 (α4β7 integrin), a cell adhesion molecule responsible for T cell homing to the intestine during GVHD (Petrovic et al., 2004) (Figure 2D). Next, we measured alloreactive T cell proliferation by adoptive transfer of donor T cells labeled with the intravital fluorescent dye carboxyfluorescein succinimidyl ester (CFSE). A larger fraction of splenic donor CD4+ and CD8+ T cells displayed CFSE dilution and expressed CD25 (as a measure of activation) when recovered from the spleen after transfer into Atg16L1HM recipients compared to WT recipients, indicating that alloreactive T cells undergo increased proliferation and activation in Atg16L1-deficient allo-HSCT recipients (Figures 2E–G). We also observed a decrease in the number and relative proportion of FoxP3+ regulatory T cells in Atg16L1HM recipients compared to WT recipients (Figure 2H). These results indicate that Atg16L1-deficiency in allo-HSCT recipients leads to increased donor T cell alloactivation.
Figure 2. Donor T cells display increased proliferation and intestinal homing in Atg16L1-deficient recipients.
(A–D) Lethally-irradiated WT and Atg16L1HM recipients were transplanted with TCD-BM cells and T cells from B10.BR mice, and live H2-Kk+ donor T cells were analyzed by flow cytometry on day 7. IELs were harvested from the small intestines and the numbers of (A) CD4+, (B) CD8+, and (C) TCRγδ+ T cells were quantified. (D) Mean fluorescent intensity (MFI) of LPAM-1 cell surface expression on donor CD4+ and CD8+ T cells harvested from the spleen. n = 10 recipient mice/group. (E–G) Lethally-irradiated WT and Atg16L1HM recipients were transplanted with 107 CFSE-labeled splenic T cells from B10.BR donor mice, and recipient spleens were harvested 96 hrs after transfer for analysis by flow cytometry. (E) Representative histograms of CFSE dilution gated on donor CD4+ and CD8+ T cells and (F) ratios of proliferating to non-proliferating donor T cells are shown. (G) Quantification of the percentages of donor CD4+ and CD8+ T cells expressing CD25. n = 6 recipient mice/group. (H) Quantification by flow cytometry of the total number and percentages of CD4+CD25+Foxp3+ Tregs in the spleen from the recipient mice in (A–D). Mean ± S.E.M are shown in (A–D) and (F–H), and represent two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Atg16L1-deficient DCs have a higher activation status after allo-HSCT
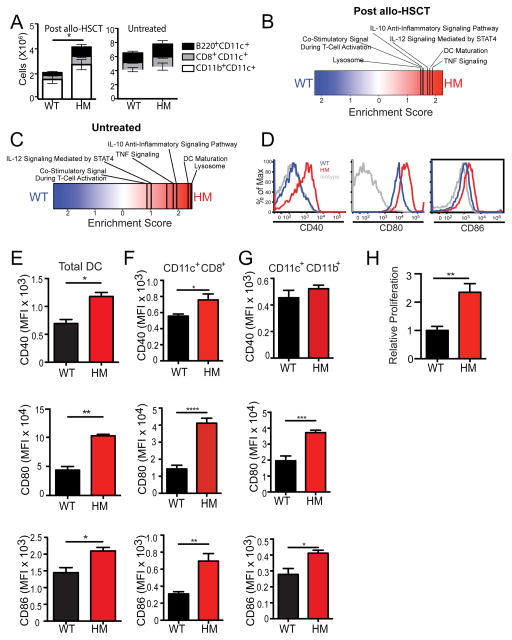
Our finding that Atg16L1 deficiency in either allo-HSCT donors or recipients confers susceptibility to exacerbated GVHD suggests that this protein is functioning within the hematopoietic compartment to increase T cell alloreactivity. CD11c+ antigen presenting cells, hereon referred to collectively as DCs, are considered to have a central role in promoting T cell alloreactivity after allo-HSCT (Hashimoto and Merad, 2011). Quantification of DCs revealed the presence of a higher number of recipient-derived DCs in the spleen of Atg16L1HM compared to WT recipients on day 7 after allo-HSCT (Figure 3A). At this time point, there were very few donor-derived DCs in the spleen, and their numbers were similar between genotypes. The increase in number of recipient-derived DCs observed in Atg16L1HM mice was not due to an expansion of a particular DC subset, and untreated ‘pre-transplant’ WT and Atg16L1HM had a similar number of splenic DCs (Figure 3A). To further examine these DCs, we performed an mRNA microarray analysis on CD11c+ cells from the spleens of WT and Atg16L1HM allo-HSCT recipients (Table S2). Consistent with the serum cytokine concentrations, Gene Set Enrichment Analysis (GSEA) indicated that DCs from Atg16L1HM mice preferentially expressed gene sets associated with TNFα, IL-12, and IL-10 signaling (Figures 3B and Table S2). A parallel TNF-α and IL-10 signature has been observed in human allo-HSCT recipients as well (E. Holler, unpublished observation). This analysis also revealed a transcriptional signature associated with DC maturation, co-stimulatory signaling, and lysosome function. Significance of microarray (SAM) analysis identified specific transcriptional differences (Z-score > 2) including decreased expression of autophagy genes Atg16l1 and Atg7, and increased expression of co-stimulatory molecules CD86 and CD80 in Atg16L1HM samples (Figure S3A). Pathway analysis of the genes that displayed increased expression in DCs from Atg16L1HM recipients yielded similar results as the GSEA results and indicated a significant enrichment in gene ontology (GO) terms such as programmed cell death, cytokine mediating signaling pathway, cellular response to stress, myeloid leukocyte differentiation, and others (Figure S3B). Thus, DCs harvested from Atg16L1-deficient allo-HSCT recipient mice have a hyperactive immune gene expression pattern.
Figure 3. DC activation is enhanced in Atg16L1-deficient allo-HSCT recipients.
(A) Lethally-irradiated WT and Atg16L1HM mice received TCD-BM and 2×106 T cells from B10.BR mice as in previous figures, and flow cytometry was used to quantify CD11c+B220+, CD11c+ CD8+, and CD11b+CD11c+ DC subsets in the spleen on day 7. Similar quantification was performed on splenic DCs from untreated mice. (B–C) Microarray analyses was performed on RNA extracted from purified splenic CD11c+ cells from either (B) allo-HSCT recipients on day 7 or (C) untreated mice as in (A). Enrichment score from GSEA shown. Extended analyses provided in Table S2. n = 3–4 biological replicates each. (D) Representative histograms of CD40, CD80, and CD86 cell surface expression on total CD11c+I-AE+ recipient DCs detected by flow cytometry on day 7 after allo-HSCT. (E–G) Quantification of CD40, CD80 and CD86 (MFI) from the flow cytometry analysis in (D) gated on (E) total CD11c+, (F) CD8+CD11c+, and (G) CD11b+ CD11c+ cells. (H) Purified CD11c+ cells from WT and Atg16L1HM recipient mice were co-cultured with B10.BR T cells at a 1:1 ratio for 72 hrs and proliferation was determined by 3H-thymidine incorporation. Bar graphs show mean ± S.E.M. and represent at least 5 mice/group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine if these transcriptional abnormalities preceded GVHD, we performed similar microarray analysis on untreated pre-transplant splenic CD11c+ cells harvested from WT and Atg16L1HM mice. GSEA and GO term analysis of enriched transcripts indicated that the above pathways were increased in DCs from untreated Atg16L1HM mice (Figures 3C, S3C–D, and Table S2). Under these conditions, transcriptional alterations associated with the lysosome were particularly prominent and included increased expression of autophagy related genes such as LC3 family members, lysosomal membrane protein-2 (LAMP-2), and LAMP-1, presumably representing a compensatory response (Figure S3D). These results reveal a potential abnormality in lysosomes in Atg16L1-deficient DCs, which is accompanied by enhanced immune activation that becomes more prominent following allo-HSCT.
To validate the microarray data, we examined cell surface expression of co-stimulatory molecules on splenic DCs by flow cytometry following allo-HSCT. Host-derived DCs from Atg16L1HM allo-HSCT recipients displayed increased expression of the classic co-stimulatory molecules CD40, CD80, and CD86 compared to DCs from WT recipients (Figures 3D and 3E). The relative increase in co-stimulatory molecule expression was particularly striking in splenic CD8+CD11c+ and CD11b+CD11c+ DC subsets (Figure 3F and 3G). However, DC subsets from the small intestinal lamina propria or mesenteric lymph nodes (MLNs) of WT and Atg16L1HM mice displayed similar co-stimulatory molecule expression (Figure S4A–F). A mixed lymphocyte reaction (MLR) assay has been extensively used to investigate GVHD since T cell alloreactivity in this in vitro co-culture system mirrors the extent and mechanism observed in vivo. Thus, we performed a MLR assay using B10.BR T cells and CD11c+ cells isolated from the spleen by bead purification on day 7 after allo-HSCT. DCs harvested from Atg16L1HM allo-HSCT recipients induced greater proliferation of allogeneic T cells compared to DCs harvested from WT recipients (Figure 3H). Thus, DCs in Atg16L1-deficient allo-HSCT recipients display increased activation and capacity to stimulate allogeneic T cell proliferation.
Atg16L1 functions within DCs to prevent hyperactivity
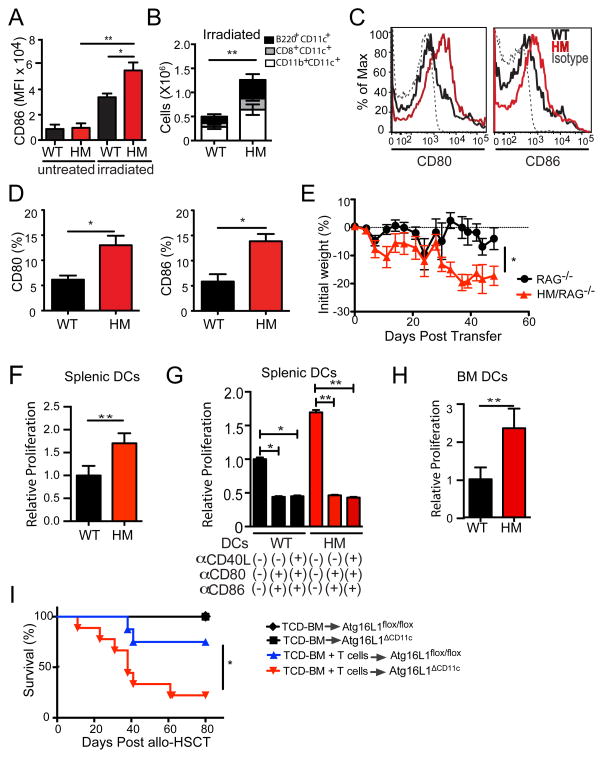
Because irradiation alone can induce upregulation of CD86 (Diab et al., 2013), we examined the cell surface expression of this co-stimulatory molecule in splenic DCs 24 hrs after irradiation. While irradiation induced a marked increased expression of CD86 in WT DCs, this increase was more pronounced in DCs from Atg16L1HM (Figure 4A). Spleens harvested from irradiated Atg16L1HM mice also had higher numbers of DCs compared to irradiated WT mice (Figure 4B). Remarkably, culturing bead-purified splenic CD11c+ DCs from naïve Atg16L1HM mice was sufficient to enhance CD80 and CD86 expression (Figure 4C and 4D). Thus, although irradiation and transplantation lead to the largest differences between DCs from WT and Atg16L1HM mice, these results suggest that Atg16L1-deficient DCs are more readily activated in general. Therefore, Atg16L1 may have a role in preventing excess alloreactivity in the absence of irradiation. To test this possibility, we performed an adoptive transfer of allogeneic (B10.BR) T cells into either Rag1−/− or Atg16L1HM mice that had been crossed to Rag1−/− mice (HM-Rag1−/−). Irradiation is not required for transferred T cells to reconstitute recipients in this model because Rag1−/− mice lack lymphocytes. As predicted, HM-Rag1−/− mice displayed significantly increased weight loss compared to Rag1−/− mice (Figure 4E). Thus, although conditioning with irradiation is a major contributor to the increased GVHD in Atg16L1HM recipients, Atg16L1-deficiency can also exacerbate alloreactivity without conditioning.
Figure 4. Atg16L1 functions within DCs to prevent hyperactivity.
(A) MFI of CD86 expression in CD11b+ splenic DCs analyzed by flow cytometry 24 hrs after WT and Atg16L1HM mice were lethally irradiated. (B) Quantification of indicated DC subsets by flow cytometry in the spleen of WT and Atg16L1HM mice 7 days after sub-lethal irradiation (550cGy). (C–D) CD11c+ splenic cells isolated from untreated WT or Atg16L1HM mice were cultured in media containing GM-CSF for 24 hrs, and CD80 and CD86 expression was analyzed by flow cytometry. n = 5/group (C) Representative histograms and (D) percentages of CD80+ and CD86+ cells are shown. (E) Rag1−/− or Atg16L1HM-Rag1−/− mice were injected i.v. with 2 × 106 allogeneic (B10.BR) T cells and monitored for weight loss. n = 8/group. (F–H) 3H-thymidine incorporation was measured in allogeneic B10.BR T cells co-cultured at a 1:1 ratio for 72 hrs with either (F) purified splenic DCs, (G) purified splenic DCs and 10μg/ml blocking antibodies against the indicated co-stimulatory molecules, or (H) BM-derived DCs (BMDCs). Relative proliferation was determined by normalizing data to the mean WT value. (I) Lethally-irradiated Atg16L1ΔCD11c recipient mice were transplanted with TCD-BM cells with or without 2 × 106 T cells from donor B10.BR WT mice and were monitored for survival. n = 8–10/group. Data in (A), (B), (D), (E), (F–H) represent mean ± S.E.M. Data are representative of at least two independent experiments. *P < 0.05, **P < 0.01.
To further test if Atg16L1-deficient DCs are intrinsically hyperactive, we performed MLR assays using splenic DCs isolated from untreated mice. Even in the absence of irradiation or transplantation, DCs harvested from Atg16L1HM mice induced increased alloreactive T cell proliferation compared to DCs from WT mice (Figure 4F). The addition of blocking antibodies against CD80, CD86, and CD40 inhibited T cell proliferation, demonstrating that the enhanced DC activity is dependent on co-stimulatory molecules (Figure 4G). Moreover, we found that BM-derived DCs (BMDCs) from Atg16L1HM mice were also able to induce increased alloreactive T cell proliferation (Figure 4H). These results indicate that Atg16L1 has a cell-intrinsic role in DCs that prevents excess alloreactive T cell proliferation in vitro. Next, we generated mice with a cell type-specific deletion of Atg16L1 in CD11c+ cells by breeding Atg16l1flox/flox mice with Cd11c-Cre mice (Atg16L1ΔCd11c) to examine the role of this gene in DCs in vivo (Figure S4G). Compared to Atg16l1flox/flox controls, Atg16l1ΔCd11c recipients displayed increased lethality and morbidity following allo-HSCT (Figures 4I and S4H). Atg16l1ΔCd11c c mice had increased numbers of DCs in the spleen compared to controls following irradiation, and culturing DCs from these mice led to increased CD80 expression as well (Figures S4I and S4J). Although Atg16L1-deficiency in other cell types is also likely contribute to allo-HSCT outcome, these results indicate that deletion in DCs is sufficient to confer exacerbated GVHD. These experiments collectively indicate that Atg16L1 function within the DC compartment has a critical role in preventing excess T cell alloreactivity in vitro and during GVHD.
Co-stimulatory molecule expression is associated with autophagy activity
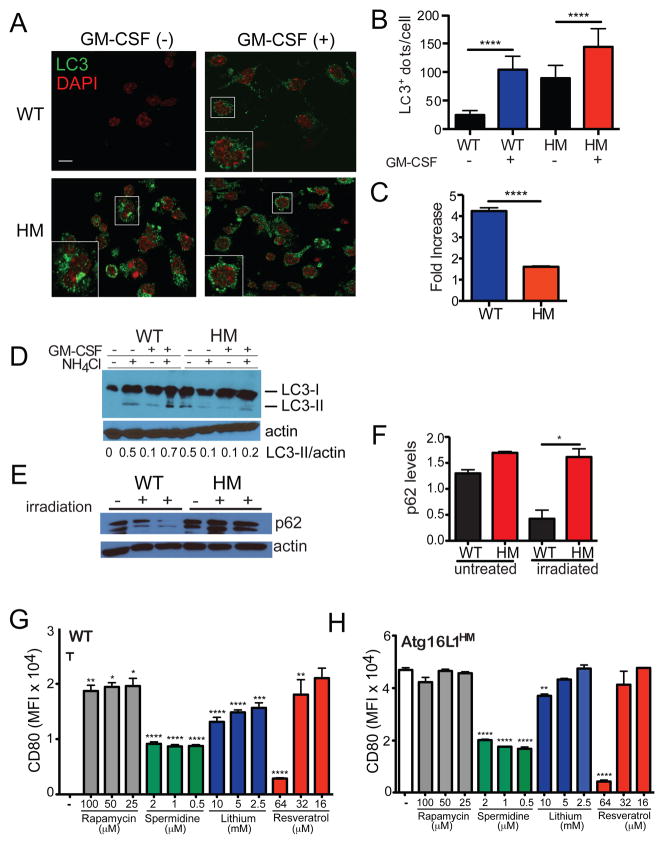
Granulocyte macrophage colony-stimulating factor (GM-CSF), a DC growth factor frequently added to culture media including in our assays, has been shown to induce autophagy (Zhang et al., 2012). We found that the presence of GM-CSF in the media of cultured splenic DCs harvested from WT mice led to a large increase in the number of LC3+ dots detected by immunofluorescence (IF), a marker of autophagy (Figures 5A–C). In contrast, DCs from Atg16L1HM mice displayed dysmorphic large LC3+ aggregates in resting conditions that were absent in WT cells, and there was a smaller increase in LC3+ structures upon GM-CSF treatment (Figures 5A–C). These results are consistent with the increase in autophagy-related transcripts observed in DCs harvested from untreated Atg16L1HM mice (Figure S3D). These results were also confirmed by Western blot analysis in which we detected more LC3 in DCs from Atg16L1HM mice under resting conditions (Figure 5D). While the addition of GM-CSF to DCs from WT mice led to an increase in the active form of LC3 (LC3-II) that was stabilized by the lysosomal inhibitor NH4Cl, only a small amount of LC3-II was detectable in DCs from Atg16L1HM mice after GM-CSF treatment (Figure 5D). We also confirmed these results during in vivo irradiation using the autophagy substrate p62 as a marker. Whole cell lysates of the spleen from irradiated WT mice displayed significantly reduced p62 expression compared to untreated controls, while samples from Atg16L1HM mice retained high amounts of p62 (Figure 5E and 5F). These results are consistent with a partial reduction in autophagy and failure to fully execute the pathway upon induction (He et al., 2013; Murthy et al., 2014).
Figure 5. Co-stimulatory molecule expression is associated with autophagy activity.
(A–C) CD11c+ splenic cells isolated from WT and Atg16L1HM mice were cultured with or without GM-CSF for 5 hrs and stained for LC3 and DAPI. (A) Representative immunofluorescence images, (B) Quantification of LC3+ dots per cell, and (C) fold increase in the number of LC3+ dots in DCs treated with GM-CSF relative to untreated cells are shown. (D) Whole cell lysates from DCs cultured as in (A) were analyzed by Western blot for LC3 and actin. Where indicated, cells received NH4Cl during the 5 hr culture to prevent degradation of LC3-II. Values displayed below represent the ratio of LC3-II over actin. Cells were pooled from 3 mice/genotype. (E, F) Western blot analysis and quantification of p62 relative to actin in the spleen harvested from WT and Atg16L1HM mice on day 7 after lethal irradiation. (G–H) Flow cytometric analysis of CD80 expression on CD11c+ splenic cells from (G) WT and (H) Atg16L1HM mice cultured in GM-CSF containing media overnight and the indicated concentrations of rapamycin, spermidine, lithium, or resveratrol. Autophagy inducing agents were added during the last 6–12 hrs prior to analysis. Bar graphs represent mean ± S.E.M, n = 3 mice/condition. *P < 0.05, **P < 0.01, *** P < 0.001, ****P < 0.0001.
Our findings suggest that it may be possible to reduce the amount of co-stimulatory molecule expression by inducing autophagy. We tested this possibility by culturing purified splenic DCs in the presence of a panel of known inducers of autophagy - rapamycin, lithium, spermadine, and resveratrol (Fleming et al., 2011). All of these agents reduced CD80 expression on WT DCs to various extents (Figure 5G). We next examined whether these autophagy inducers can reduce CD80 expression in Atg16L1HM DCs, which retain some degree of autophagy function. We found that addition of spermidine and resveratrol, the two agents that had the largest effect on WT DCs, led to reversal of the enhanced CD80 expression to amounts expressed in WT DCs (Figure 5H). Together with the above experiments, these results indicate that autophagy activity is inversely associated with DC hyperactivity.
Atg16L1 has been shown to prevent excess IL-1β production, most likely downstream of reactive oxygen species (ROS) produced by mitochondria that accumulate in the absence of autophagy (Saitoh et al., 2008). Thus, it is possible that the hyperactivity observed in Atg16L1-deficient DCs is mediated by enhanced cytokine or ROS production. However, antibody-mediated blockade of IL-1β or its receptor did not affect the increased capacity of Atg16L1HM DCs to stimulate alloreactive T cell proliferation (Figure S5A). Although an IL-12 signature was detected in Atg16L1-deficient DCs (Figure 3B and 3C), blocking this cytokine did not reduce DC hyperactivity either. Also, transfer of Atg16L1HM culture supernatant onto WT DCs did not transfer the increase in co-stimulatory molecule expression (Figure S5B). Additionally, we did not find differences in the amount of superoxide and hydroxy radicals when comparing WT and Atg16L1HM DCs (Figures S5C and S5D). These results suggest that Atg16L1-deficiency confers DC hyperactivity through a distinct mechanism.
Atg16L1 deficiency results in lysosomal abnormalities and decreased A20 expression
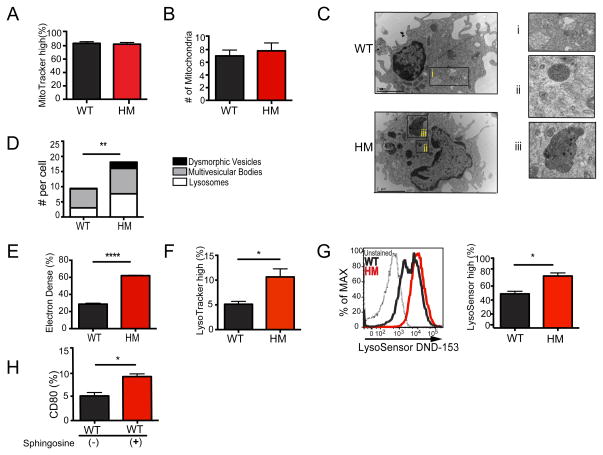
Base on the role of autophagy in membrane trafficking and organelles maintenance, we analyzed Atg16L1-deficient DCs for the presence of ultrastructural abnormalities by transmission electron microscopy (TEM). Instead of defects in mitochondria, we observed a striking increase in the number of electron dense lysosomal structures in cultured Atg16L1HM DCs compared to WT controls (Figure 6A–E). The accumulation of these lysosomal structures were not an artifact of culturing because we observed similar electron dense organelles in DCs isolated from untreated or irradiated mice without culturing (Figures S6A–D). In addition to an overall increase in lysosomes, we detected the accumulation of dysmorphic organelles that are morphologically similar to structures referred to as telolysosomes (Brunk and Terman, 2002). Accumulation of these vesicles has been associated with defective autophagy and decreased lysosomal activity (Levine and Kroemer, 2008). We corroborated these findings by flow cytometry. Atg16L1HM DCs displayed an increase in total lysosomal load as well as pH neutral dysfunctional lysosomes, which were detected by LysoTracker and LysoSensor Green staining respectively (Figures 6F and 6G). Also, we found that disrupting the lysosomal membrane with sphingosine (Ullio et al., 2012) was sufficient to increase CD80 expression in WT DCs (Figure 6H). These results indicate that hyperactivity in Atg16L1-deficient DCs is associated with decreased autophagy and defects in the lysosome compartment.
Figure 6. Atg16L1 deficiency results in lysosomal abnormalities.
(A–H) Splenic DCs isolated from WT and Atg16L1HM mice were cultured in GM-CSF containing media for 24 hours and analyzed by flow cytometry or TEM. (A) Percentage of cells displaying high MitoTracker incorporation by flow cytometry, (B) Quantification of mitochondria detected per cell by transmission electron microscopy (TEM). (C) Representative TEM images of splenic DCs (cultured in GM-CSF containing media for 24 hrs. i = multivesicular bodies, ii = lysosomes, and iii = dysmorphic vesicles. (D) Quantification of lysosomes, dysmorphic vesicles, and multivesicular bodies detected per cell by TEM. (E) Quantification of the percentage of vesicles detected in (C) that are electron dense. (F) Percentage of cells displaying high LysoTracker incorporation. (G) Representative histogram of LysoSensor DND-152 incorporation and quantification of the percentage of cells displaying high staining. (H) Flow cytometric analysis of CD80 expression on WT DCs cultured with or without 25μM Sphingosine for 3 hours. Bar graphs represent mean ± S.E.M, n = 3–6 mice/condition. Scale bar represents 10 μm for (A) and 2 μm for (F). *P < 0.05, **P < 0.01, ****P < 0.0001.
These findings are in concert with our microarray data indicating that Atg16L1-deficiency leads to increased lysosomal transcription. Of particular interest was an increase in Laptm5, a lysosomal transmembrane protein that promotes the degradation of several targets involved in immune signaling including the ubiquitin-editing enzyme A20 (Glowacka et al., 2012; Kawai et al., 2014; Ouchida et al., 2010; Ouchida et al., 2008). As a potent inhibitor of nuclear factor-kappaB (NF-κB) and mitogen activated protein kinase (MAPK) signaling, A20 is a key negative regulator of DC activity (Ganguly et al., 2013). Mice with DC-specific deletion of A20 display increases in DC numbers and co-stimulatory molecule expression, which causes enhanced T cell alloreactivity and systemic autoimmunity (Hammer et al., 2011; Kool et al., 2011). Therefore, an increase in Laptm5 due to Atg16L1-deficiency is predicted to downregulate A20, potentially explaining the increase in DC numbers and co-stimulatory molecule expression observed in Atg16L1HM mice. We first validated the microarray data by qRT-PCR on independent samples, and confirmed that Laptm5 expression is increased in Atg16L1HM DCs in the same conditions in which co-stimulatory molecule expression is increased (Figure S6E). Next, we found that A20 expression was decreased in Atg16L1HM DCs (Figure S6F and S6G). Also, we found that this decrease in A20 was associated with a corresponding increase in phosphorylated IκB and JNK, indicative of augmented NF-κB and MAPK signaling (Figure S6F and S6G). As CD80, CD86, and CD40 expression is regulated by these signaling pathways (Fong et al., 1996; Hinz et al., 2001; Zhao et al., 1996), these findings are consistent with the increased activation observed in Atg16L1HM DCs (Figure S6H).
DISCUSSION
Here we have shown that the autophagy gene Atg16l1 protects against exacerbated GVHD following allo-HSCT. In addition to significantly worse morbidity and mortality, Atg16L1HM allo-HSCT recipients displayed a compromised intestinal barrier that was associated with increased donor T cell proliferation, activation, and migration to the gastrointestinal tract. Atg16L1-deficiency in transplant donors also exacerbated GVHD in recipients, suggesting that this protein is functioning in a hematopoietic cell type during allo-HSCT. Several lines of evidence indicate that this increased GVHD and T cell alloreactivity is due to the effect of Atg16L1-deficiency in DCs. A higher number of recipient-derived DCs were recovered from Atg16L1HM recipient mice, and these DCs had a hyperactive gene expression profile. DCs from Atg16L1HM recipient mice also displayed elevated surface expression of co-stimulatory molecules known to have a role in acute GVHD and T cell alloreactivity. Moreover, DCs harvested directly from untreated Atg16L1HM mice as well as DCs differentiated from Atg16L1HM BM were able to stimulate increased donor T cell proliferation in vitro compared to their WT counterparts. Further, mice with cell type-specific deletion of Atg16L1 in DCs displayed worse allo-HSCT outcomes, and DCs harvested from these mice had increased co-stimulatory molecule expression. Our results are most consistent with a model in which Atg16L1-deficient DCs over-stimulate allogeneic T cells that proliferate and migrate to peripheral organs such as the intestine to cause damage. The ensuing disruption of the intestinal barrier most likely plays a critical role in accelerating disease through a feedforward loop of additional DC and T cell activation, which ultimately results in lethality.
In contrast to other situations in which autophagy supports antigen presentation by DCs (Cooney et al., 2010; Jagannath et al., 2009; Ma et al., 2012; Reed et al., 2013; Romao et al., 2013), we found a major immuno-suppressive function of Atg16L1 in this cell type. This discrepancy likely reflects the pathophysiology of GVHD, which does not necessarily rely on the proper trafficking of one particular antigen, and can be driven by either MHC class I or II molecules. In the context of our studies, Atg16L1-deficiency may prime the cells for hyperactivity in response to stress. Irradiation of Atg16L1HM mice or culturing DCs from these mice in conditioned media was sufficient to enhance co-stimulatory molecule expression. Also, Atg16L1-deficient DCs displayed alterations in gene expression that preceded transplantation, especially in transcripts associated with lysosomes. This finding is similar to previous studies that observed a compensatory increase in endo-lysosomal gene expression following autophagy inhibition in the intestinal epithelium (Cadwell et al., 2010; Patel et al., 2013). One of these genes increased in Atg16L1-deficient DCs was Laptm5, which has been shown to induce pro-inflammatory activity in macrophages by downregulating A20 (Glowacka et al., 2012). Consistent with other studies demonstrating a key role for A20 in preventing DC hyperactivity (Hammer et al., 2011; Kool et al., 2011), we find that increased Laptm5 in Atg16L1-deficient DCs is associated with a decrease in A20 and a corresponding increase in inflammatory signaling. How Laptm5 targets A20 for degradation is not known. Interestingly, Laptm5 has signaling functions in other immune cell types (Glowacka et al., 2012; Kawai et al., 2014; Ouchida et al., 2010; Ouchida et al., 2008), and overexpression is associated with accumulation of immature lysosomal vesicles, defective autophagy, and cell death in neuroblastomas (Inoue et al., 2009). Therefore, understanding the relationship between autophagy and this poorly understood lysosomal protein may be key to deciphering the complex relationship between lysosomes and inflammatory signaling in a variety of cell types.
These observations in a murine model of allo-HSCT may have important implications for assessing GVHD risk in humans. The IBD risk allele of ATG16L1 leads to a T300A mutation that destabilizes the protein product in the presence of TNF-α and metabolic stress by facilitating caspase-3-mediated cleavage (Lassen et al., 2014; Murthy et al., 2014). Our preliminary analysis of patients lacks the power to determine if the ATG16L1T300A allele is a true risk factor for GVHD due to other factors that can contribute to mortality following allo-HSCT. However, we were able to observe an association between this polymorphism and an adverse transplantation outcome even in a small cohort. Given the high frequency with which the ATG16L1T300A allele occurs in the human population, further studies (both retrospective and prospective) seem warranted to determine whether the presence of this allele in donor and/or host affects transplant outcome, in particular GVHD. Our mouse model also indicates that it will be important to examine the effect of the ATG16L1T300A allele on DCs, which is currently unclear (Strisciuglio et al., 2013; Wildenberg et al., 2012).
Finally, our findings provide experimental support for the concept that intestinal GVHD and IBD share underlying mechanisms responsible for the common pathophysiology observed between patients and animal models of each disease. In this context, it is notable that HM/Rag1−/− mice develop exaggerated disease upon transfer of allogeneic T cells in a model that resembles the T cell transfer colitis model frequently used in IBD studies. Our findings raise the possibility that therapeutically targeting DCs, potentially through manipulation of autophagy, could ameliorate intestinal barrier dysfunction in multiple disease settings.
EXPERIMENTAL PROCEDURES
Mice and bone marrow transplantation
Atg16L1HM and Atg16l1flox/flox mice have been described previously (Marchiando et al., 2013). WT refers to wild-type C57BL/6J mice initially purchased from Jackson Laboratory and bred onsite to generate animals for experimentation. BALB/c, B10.BR, and Cd11c-Cre mice were purchased from Jackson Laboratory. Atg16l1ΔCd11c mice and littermate Atg16l1flox/flox control mice used in experiments were generated from Atg16l1flox/flox and Atg16l1flox/flox Cd11c-Cre breeding pairs in which one parent was hemizygous for the Cre allele. Mice used in this study were 6–9 week old females at the outset of the experiment. Bone marrow (BM) transplantations were performed as previously described (Jagannath et al., 2009; Penack et al., 2010) with 1100 cGy (C57BL/6) or 850 cGy (BALB/c) split-dosed irradiation of recipients transplanted with 5×106 BM cell after T cell depletion with anti-Thy-1.2 and low-TOX-M rabbit complement (Cedarlane Laboratories). Donor T cells were prepared by harvesting splenocytes and enriching T cells by Miltenyi MACS purification of CD5. Recipient mice were monitored daily for survival and disease was quantified weekly for 5 clinical parameters (weight loss, hunched posture, activity, fur ruffling, and skin lesions) on a scale from 0 to 2. A clinical GVHD score was generated by the summation of the 5 criteria as previously described (Cooke et al., 1996). Last observation was carried forward for weight loss and clinical GVHD score for allo-HSCT experiments. All animal studies were performed according to approved protocols by the New York University School of Medicine and Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committees (IACUCs).
Flow cytometry and cell culture
All antibodies were obtained from BD Biosciences or Biolegend. CD11c+ cells were isolated from spleen by performing a collagenase and DNase digestion (Roche) at 37 °C for 15 min followed by MACS sorting with CD11c+ microbeads (Miltenyi). Cells were prepared for flow cytometry as previously described (Petrovic et al., 2004) and analyzed by LSR II flow cytometer (BD Biosciences) and FlowJo (TreeStar Software).
qRT-PCR and microarray gene-expression profiling
MACS purified CD11c+ cells from spleens of allo-HSCT recipients on day 7 or untreated mice were placed directly into TRIzol LS (Invitrogen). mRNA was isolated, amplified, and hybridized to an Affymetrix GeneChip (MOE430A). Affymetrix Power Tools software was used for processing and quantile normalization of fluorescence hybridization signals by the robust multichip averaging method (Irizarry et al., 2003). Transcripts were log2-normalized and average values for technical replicates were obtained for analysis of change in expression using Significance Analysis of Microarray (SAM). Differential gene expression was assessed using the Bioconductor package “siggenes” with a fold change cutoff of >2. Full results, including p-values and multiple-testing corrected q-values, are included in Supplementary Table 2. The Broad Institute GSEA software was used for gene-set enrichment analysis as previously described (Subramanian et al., 2005). Primers for Laptm5: Forward-GCGGTAAAGTGTCCTGTAGGTTC; Reverse- TCTTGACCACGCCGAACAGCAG:
Statistics
GraphPad Prism version 6 was used for statistical analysis. Differences between means were evaluated by two-tailed unpaired t test or one-way ANOVA and Holm-Sidak’s for experiments involving multiple comparisons. Survival was analyzed with the Mantel-Cox log-rank test. The means of the area under the curve was used to compare longitudinal weight loss and disease scores between conditions. Mann-Whitney test was used for analysis of intestinal permeability and bacterial translocation since data were not normally distributed.
Supplementary Material
Distribution of the ATG16L1 WT and T300A alleles in transplant donor and recipients. (B) Distribution of the causes of non-relapse mortality (NRM) in Figure S1. Major GVHD is defined as grade III or IV according to the Glucksberg criteria (Glucksberg et al., 1974). Although a cohort of an additional 69 patients receiving an allograft from unrelated donors was available during the period of this study, the effect of ATG16L1T300A on NRM could not be analyzed due to a high incidence of tumor-relapse in this group.
First tab shows complete gene list where fold change refers to Atg16L1HM over WT at steady state levels. Second tab shows GSEA analysis of leading edge genes from WT high and Atg16L1HM high gene sets. Third tab shows complete gene list where fold change refers to Atg16L1HM over WT after allo-HSCT. Fourth tab shows GSEA analysis of leading edge genes from Atg16L1HM high and WT allo-HSCT recipients high gene sets.
Supplemental Figure 1, related to Figure 1. Atg16L1 mutation increases morbidity and mortality after allo-HSCT with altered levels of pro-inflammatory cytokines. (A–B) Lethally-irradiated WT and Atg16L1HM recipients were transplanted with TCD-BM cells with and without a lower dose of splenic T cells (1×106) from donor B10.BR mice and monitored for (A) survival and (B) weight loss. n = 10 recipient mice/group. (C) WT mice in the B10.BR→B6 allo-HSCT model typically display more lethality than we observed in Figure 1A. The allo-HSCT from Figure 1A was repeated in a second animal facility (Memorial Sloan Kettering Cancer Center) to examine if the degree of lethality was facility-dependent. In contrast to the original experiment (performed in the ultra-clean animal room at New York University School of Medicine), WT recipients displayed substantial lethality. However, Atg16L1HM recipients remained significantly more susceptible. n = 10 recipient mice/group. (D–E) Cytokine and chemokine levels were measured by multiplex ELISA (Millipore) using serum from mice receiving the allo-HSCT described in Figure 1A–C on day 7 after the transplantation. Results were acquired with a Luminex 200 instrument and analyzed with xPONENT software. Cytokines displaying increases in Atg16L1HM allo-HSCT recipients are shown in C and those displaying decrease are shown in D. Untreated WT mice 6-weeks of age were used as additional controls to show baseline cytokine levels. No significant differences were seen in IL-1α, IL-1β, IL-2, IL-4, IL-15, IL-10, IL-13, IL-6, IL-17, IP-10, and RANTES when comparing B6 mice and Atg16L1HM receiving TCD-BM + T cells. n = 5–9 mice/group. (F) Survival of WT and Atg16L1HM treated with 500 μg of anti-TNFα or Isotype control (BioXcell) by i.p. injection on days −1 and 0, and every other day thereafter. Mice were transplanted with TCD-BM cells with 2×106 splenic T cells from donor B10.BR mice on day 0 as in Figure 1A–C. n = 10 mice/group. (G) Serum FITC-dextran concentration following oral gavage was measured in WT and Atg16L1HM mice on day 7 after lethal irradiation. (H–I) 122 donor-recipient pairs receiving allo-HSCT from HLA-identical sibling either at the University of Regensburg (n = 47, from May, 2000 until May, 2012) or at the University of Newcastle (n = 75, from January, 1993 until December, 2007) were included. Collection of samples for genotyping was approved by local ethical committees at both sites. DNA was typed for presence or absence of the ATG16L1T300A allele as described previously (Latiano et al., 2008). Results of genotyping were compared with major patient characteristics and outcome variables such as GVHD, non-relapse mortality (NRM), and overall survival using a SPSS based database and PASW statistic software for analysis. NRM and overall survival were calculated by Kaplan-Meier analysis, and Cox regression was used to compare risk factors in multivariate analysis. Patients receiving allo-HSCT from HLA-identical siblings were analyzed. DNA from patients and donors was isolated and typed for presence (heterozygous or homozygous) or absence of the ATG16L1T300A allele. The effect of the risk allele in donors and recipients were analyzed separately. (H) Rate of non-relapse mortality (NRM) in allo-HSCT recipients when the ATG16L1T300A allele is present versus absent in the donor. (I) Rate of NRM in allo-HSCT recipients when the ATG16L1T300A allele is present versus absent in the recipient. n = 6 mice/group. Mean ± S.E.M. are shown in (B), (D)–(E), and (G). *P < 0.05, **P < 0.01, ***P < 0.001.
Supplemental Figure 2, related to Figure 2. Histopathology of allo-HSCT recipients in the intestine and skin. (A–F) H&E-stained tissue sections from WT and Atg16L1HM allo-HSCT recipients (TCD-BM and 2×106 T cells as in Figure 1A–C) on day 7 were processed and examined for histopathology as previously described (Cadwell et al., 2010; Penack et al., 2009). Representative images of (A) small intestine and (B) skin, and blind quantification of GVHD histopathology using previous described parameters (Petrovic et al., 2004) shown for (C) small intestine, (D) colon, (E) liver, and (F) skin. Magnification: x10 for small intestines, x40 for skin. (G–J) Quantification of histopathology as described above on day 14-post allo-HSCT with a lower dose of T cells (1×106 T cells) to allow survival of mice. n = 6–8 mice/group. Mean ± S.E.M. are shown in (C–J).
Supplemental Figure 3, related to Figure 3. DCs from Atg16L1HM mice display an altered transcriptional profile. (A–D) Additional microarray analyses corresponding to the experiments described in Figure 3B and 3C. Heat-map presentation of gene-expression profiles after Significance Analysis of Microarray (SAM) (Z-score > 2) for (A) allo-HSCT recipients and (C) untreated mice. Complete list of genes can be found in Supplementary Table 2. Bar graphs in (B) and (D) represent p-values for biological processes (GO terms) displaying the most significant enrichment in Atg16L1HM allo-HSCT recipient and untreated Atg16L1HM mice compared to their respective WT controls. Enrichment in GO terms was determined by DAVID (Database for Annotation, Visualization and Integrated Discovery) using Z > 2 cutoff.
Supplemental Figure 4, related to Figure 4. Atg16L1 functions within DCs to prevent hyperactivity. (A–D) Flow cytometric analyses of small intestinal lamina propria cells from untreated or lethally (1100 cGy) irradiated WT and Atg16L1HM mice. CD3−B220−NK1.1−class-II+ CD11c+ cells were gated on either (A, B) CD11b−CD103+ or (C, D) CD11b+CD103+. MFI quantification shown for CD80 (A and C) and CD86 expression (B and D). No significant differences were noted when gating on percent cells that are CD80+ or CD86+, and there were no difference in cell numbers. (E–F) Similar flow cytometric analyses of mesenteric lymph nodes (MLNs) from untreated WT or Atg16L1HM mice. (G) Western blot comparing Atg16L1 levels (Sigma: A7356) in sorted splenic DCs from control (Atg16L1flox/flox) and Atg16L1ΔCD11c mice. (H) Clinical GVHD score of allo-HSCT recipient Atg16L1ΔCD11c mice from Figure 4I. (I) Quantification of indicated splenic DC subsets by flow cytometry from Atg16L1ΔCD11c and control mice 24 hours after lethal irradiation. n = 3 mice/group. (J) Flow cytometry analysis of CD80 expression on CD11c+ splenic cells isolated from Atg16L1ΔCD11c and control mice that were cultured for 24 hours. n = 3–4 mice/group. Mean ± S.E.M. are shown in (A)–(F) and (H)–(J). *P < 0.05.
Supplemental Figure 5, related to Figure 5. Increased co-stimulatory molecule expression on Atg16L1-deficient DCs is not transferrable. (A) 3H-thymidine incorporation of allogeneic B10.BR T cells co-cultured at a 1:1 ratio for 72 hours with DCs purified from the spleen of WT and Atg16L1HM mice, with or without neutralizing antibodies against IL-1β (5 μg/mL), anti-IL-1R1 (0.5 μg/mL), or anti-IL-12 (1 μg/mL) (R&D systems). (B) Flow cytometric analysis of CD80 expression on WT DCs cultured for 24 hours in supernatant collected from previously cultured Atg16L1HM DCs. (C, D) Flow cytometric analysis of CD11c+ DCs cultured with GM-CSF for 24 hours and stained for the ROS-detecting cell permeable dyes (C) dihydroethidium (DHE) and (D) 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA). n = 3 mice/condition. Bar graphs represent mean ± S.E.M and n = 3–6 mice/condition. ****P < 0.0001.
Supplemental Figure 6, related to Figure 6. Atg16L1 deficiency results in lysosomal abnormalities and decreased A20 expression. (A, B) Quantification of lysosomes, dysmorphic lysosomes, and multivesicular bodies detected per cell by TEM in splenic DCs harvested from (A) untreated mice or (B) mice 24 hrs after lethal irradiation. (C, D) Quantification of the percentage of vesicles detected per cell by TEM in splenic DCs harvested from (A) untreated mice or (B) mice 24 hrs after lethal irradiation. (E) qRT-PCR analysis of Laptm5 expression in splenic DCs from WT and Atg16L1HM mice cultured in GM-CSF containing media for 24 hrs. 5–7 mice/genotype. (F) Representative Western blot analyses of A20, phospho-IkB, phospho-JNK, and actin in whole cell lysates from DCs cultured as in (E). (G) Quantification of Western blot from (F). 6 mice/genotype. Bar graphs represent mean ± S.E.M, *P < 0.05, **P < 0.01, ****P < 0.0001. (H) Model figure: By inhibiting NF-κB and MAPK signaling, A20 prevents unregulated co-stimulatory molecule expression in DCs exposed to environmental stressors. In Atg16L1-deficient cells, decreased autophagy leads to accumulation of aberrant lysosomal structures and an accompanying lysosomal transcriptional response including enhanced expression of Laptm5. Laptm5 accumulation then mediates the degradation of A20 causing increased expression of the co-stimulatory molecules CD80, CD86, and CD40 that mediate enhanced T cell alloreactivity and subsequent GVHD.
HIGHLIGHTS.
Mice deficient in autophagy gene Atg16l1 are susceptible to graft-versus-host disease
Autophagy-deficient dendritic cells become hyperactive following an HSC transplant
Hyperactive dendritic cells stimulate donor T cells that damage the intestine
Acknowledgments
We would like to thank Smita Srivastava for assistance with culturing DCs, Michael Cammer for assistance with imaging, Fiangxia Liang for assistance with TEM, Herbert W. Virgin for Atg16L1flox/flox mice, Boris Reizis and Ana Maria Cuervo for advice, NYUSoM Flow Cytometry and Cell Sorting Center (5P30CA016087-33), and NYUSoM Microcopy Core supported by RR023704-01A1. This work was supported by US National Institute of Health (NIH) grants R01 DK093668 (K.C.), R01 HL069929 (M.v.d.B), P01 CA023766 (M.v.d.B.), Project 4 of P01-CA023766, R01 AI080455 (M.v.d.B.), AI100288 (M.v.d.B.), R01 AI101406 (M.v.d.B.), F32 HL115974 (V.M.H.), T32 AI007647 (V.M.H.), T32 AI100853 (K.M.), F30 DK098925 (K.M.), T32 GM0738 (K.M.); Dale F. Frey Award from the Damon Runyon Cancer Research Foundation (K.C.); The Uehara Memorial Foundation (Y.S.); Lymphoma Research Foundation PostDoctoral Fellowship Research Grant (Y.S.); U.S National Institute of Allergy and Infectious Diseases (NIAID Contract HHSN272200900059C) (M.v.d.B.); Swiss IBD Cohort Study: 3347CO-108792 (G.R.); The Lymphoma Foundation (M.v.d.B.); Susan and Peter Solomon Divisional Genomics Program (M.v.d.B.); and European Union’s Seventh Programme for research, technological development and demonstration grant agreement No [602587] (M.v.d.B.).
Footnotes
AUTHOR CONTRIBUTION
V.M.H., Y.S., K.C., and M.v.d.B. formulated the original hypothesis, designed the study, and analyzed results. V.M.H. and Y.S. performed the experiments. K.M., M.L.W., and N.V.S. provided technical assistance. K.S., G.R., A.M.D., and E.H. collected and analyzed human data. C.G.K.Z. performed the microarray analysis. C.L., A.C.F.M., G.M., and C.L. analyzed histology. S.M.L, J.D.W., and R.K.A. provided assistance with chemical inhibitors. V.M.H., Y.S., M.v.d.B. and K.C. wrote the manuscript, and all authors commented on the manuscript, data, and conclusions before submission.
COMPETEING FINANTIAL INTERESTS
The authors declare no competing financial interests.
Additional details are provided in the supplemental methods section.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Komatsu M, Virgin HWt, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke KR, Kobzik L, Martin TR, Brewer J, Delmonte J, Jr, Crawford JM, Ferrara JL. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88:3230–3239. [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy: an emerging immunological paradigm. J Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diab A, Jenq RR, Rizzuto GA, Cohen AD, Huggins DW, Merghoub T, Engelhorn ME, Guevara-Patino JA, Suh D, Hubbard-Lucey VM, et al. Enhanced responses to tumor immunization following total body irradiation are time-dependent. PLoS One. 2013;8:e82496. doi: 10.1371/journal.pone.0082496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara JL, Reddy P. Pathophysiology of graft-versus-host disease. Semin Hematol. 2006;43:3–10. doi: 10.1053/j.seminhematol.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- Fong TC, Wu Y, Kipps TJ. Identification of a promoter element that regulates tissue-specific expression of the human CD80 (B7.1) gene. J Immunol. 1996;157:4442–4450. [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D, Haak S, Sisirak V, Reizis B. The role of dendritic cells in autoimmunity. Nature reviews Immunology. 2013;13:566–577. doi: 10.1038/nri3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka WK, Alberts P, Ouchida R, Wang JY, Rotin D. LAPTM5 protein is a positive regulator of proinflammatory signaling pathways in macrophages. The Journal of biological chemistry. 2012;287:27691–27702. doi: 10.1074/jbc.M112.355917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nature immunology. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Merad M. Harnessing dendritic cells to improve allogeneic hematopoietic cell transplantation outcome. Semin Immunol. 2011;23:50–57. doi: 10.1016/j.smim.2011.01.005. [DOI] [PubMed] [Google Scholar]
- He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, et al. Beclin 2 Functions in Autophagy, Degradation of G Protein-Coupled Receptors, and Metabolism. Cell. 2013 doi: 10.1016/j.cell.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz M, Loser P, Mathas S, Krappmann D, Dorken B, Scheidereit C. Constitutive NF-kappaB maintains high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in Hodgkin/Reed-Sternberg cells. Blood. 2001;97:2798–2807. doi: 10.1182/blood.v97.9.2798. [DOI] [PubMed] [Google Scholar]
- Holler E, Rogler G, Herfarth H, Brenmoehl J, Wild PJ, Hahn J, Eissner G, Scholmerich J, Andreesen R. Both donor and recipient NOD2/CARD15 mutations associate with transplant-related mortality and GvHD following allogeneic stem cell transplantation. Blood. 2004;104:889–894. doi: 10.1182/blood-2003-10-3543. [DOI] [PubMed] [Google Scholar]
- Inoue J, Misawa A, Tanaka Y, Ichinose S, Sugino Y, Hosoi H, Sugimoto T, Imoto I, Inazawa J. Lysosomal-associated protein multispanning transmembrane 5 gene (LAPTM5) is associated with spontaneous regression of neuroblastomas. PLoS One. 2009;4:e7099. doi: 10.1371/journal.pone.0007099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–276. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- Jenq RR, van den Brink MR. Allogeneic haematopoietic stem cell transplantation: individualized stem cell and immune therapy of cancer. Nat Rev Cancer. 2010;10:213–221. doi: 10.1038/nrc2804. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Ouchida R, Yamasaki S, Dragone L, Tsubata T, Wang JY. LAPTM5 promotes lysosomal degradation of intracellular CD3zeta but not of cell surface CD3zeta. Immunology and cell biology. 2014 doi: 10.1038/icb.2014.18. [DOI] [PubMed] [Google Scholar]
- Kool M, van Loo G, Waelput W, De Prijck S, Muskens F, Sze M, van Praet J, Branco-Madeira F, Janssens S, Reizis B, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011;35:82–96. doi: 10.1016/j.immuni.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Landfried K, Bataille F, Rogler G, Brenmoehl J, Kosovac K, Wolff D, Hilgendorf I, Hahn J, Edinger M, Hoffmann P, et al. Recipient NOD2/CARD15 status affects cellular infiltrates in human intestinal graft-versus-host disease. Clin Exp Immunol. 2010;159:87–92. doi: 10.1111/j.1365-2249.2009.04049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741–7746. doi: 10.1073/pnas.1407001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. The Journal of biological chemistry. 2012;287:34149–34156. doi: 10.1074/jbc.M112.382812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–227. doi: 10.1016/j.immuni.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Marchiando AM, Ramanan D, Ding Y, Gomez LE, Hubbard-Lucey VM, Maurer K, Wang C, Ziel JW, van Rooijen N, Nunez G, et al. A Deficiency in the Autophagy Gene Atg16L1 Enhances Resistance to Enteric Bacterial Infection. Cell Host Microbe. 2013;14:216–224. doi: 10.1016/j.chom.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, van Lookeren Campagne M. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–462. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- Ouchida R, Kurosaki T, Wang JY. A role for lysosomal-associated protein transmembrane 5 in the negative regulation of surface B cell receptor levels and B cell activation. J Immunol. 2010;185:294–301. doi: 10.4049/jimmunol.1000371. [DOI] [PubMed] [Google Scholar]
- Ouchida R, Yamasaki S, Hikida M, Masuda K, Kawamura K, Wada A, Mochizuki S, Tagawa M, Sakamoto A, Hatano M, et al. A lysosomal protein negatively regulates surface T cell antigen receptor expression by promoting CD3zeta-chain degradation. Immunity. 2008;29:33–43. doi: 10.1016/j.immuni.2008.04.024. [DOI] [PubMed] [Google Scholar]
- Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. The EMBO journal. 2013;32:3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penack O, Holler E, van den Brink MR. Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115:1865–1872. doi: 10.1182/blood-2009-09-242784. [DOI] [PubMed] [Google Scholar]
- Penack O, Smith OM, Cunningham-Bussel A, Liu X, Rao U, Yim N, Na IK, Holland AM, Ghosh A, Lu SX, et al. NOD2 regulates hematopoietic cell function during graft-versus-host disease. The Journal of experimental medicine. 2009;206:2101–2110. doi: 10.1084/jem.20090623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Alpdogan O, Willis LM, Eng JM, Greenberg AS, Kappel BJ, Liu C, Murphy GJ, Heller G, van den Brink MR. LPAM (alpha 4 beta 7 integrin) is an important homing integrin on alloreactive T cells in the development of intestinal graft-versus-host disease. Blood. 2004;103:1542–1547. doi: 10.1182/blood-2003-03-0957. [DOI] [PubMed] [Google Scholar]
- Reed M, Morris SH, Jang S, Mukherjee S, Yue Z, Lukacs NW. Autophagy-Inducing Protein Beclin-1 in Dendritic Cells Regulates CD4 T Cell Responses and Disease Severity during Respiratory Syncytial Virus Infection. J Immunol. 2013 doi: 10.4049/jimmunol.1300477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, Ziegler U, Roesler J, Dengjel J, Reichenbach J, Munz C. Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing. The Journal of cell biology. 2013;203:757–766. doi: 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Strisciuglio C, Duijvestein M, Verhaar AP, Vos AC, van den Brink GR, Hommes DW, Wildenberg ME. Impaired autophagy leads to abnormal dendritic cell-epithelial cell interactions. J Crohns Colitis. 2013;7:534–541. doi: 10.1016/j.crohns.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka H, Iwasaki T, Okamoto T, Kakishita E. Intestinal graft-versus-host disease: mechanisms and management. Drugs. 2003;63:1–15. doi: 10.2165/00003495-200363010-00001. [DOI] [PubMed] [Google Scholar]
- Tanabe T, Yamaguchi N, Matsuda K, Yamazaki K, Takahashi S, Tojo A, Onizuka M, Eishi Y, Akiyama H, Ishikawa J, et al. Association analysis of the NOD2 gene with susceptibility to graft-versus-host disease in a Japanese population. Int J Hematol. 2011;93:771–778. doi: 10.1007/s12185-011-0860-5. [DOI] [PubMed] [Google Scholar]
- Ullio C, Casas J, Brunk UT, Sala G, Fabrias G, Ghidoni R, Bonelli G, Baccino FM, Autelli R. Sphingosine mediates TNFalpha-induced lysosomal membrane permeabilization and ensuing programmed cell death in hepatoma cells. J Lipid Res. 2012;53:1134–1143. doi: 10.1194/jlr.M022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg ME, Vos AC, Wolfkamp SC, Duijvestein M, Verhaar AP, Te Velde AA, van den Brink GR, Hommes DW. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology. 2012;142:1493–1503. e1496. doi: 10.1053/j.gastro.2012.02.034. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Freeman GJ, Gray GS, Nadler LM, Glimcher LH. A cell type-specific enhancer in the human B7.1 gene regulated by NF-kappaB. The Journal of experimental medicine. 1996;183:777–789. doi: 10.1084/jem.183.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the ATG16L1 WT and T300A alleles in transplant donor and recipients. (B) Distribution of the causes of non-relapse mortality (NRM) in Figure S1. Major GVHD is defined as grade III or IV according to the Glucksberg criteria (Glucksberg et al., 1974). Although a cohort of an additional 69 patients receiving an allograft from unrelated donors was available during the period of this study, the effect of ATG16L1T300A on NRM could not be analyzed due to a high incidence of tumor-relapse in this group.
First tab shows complete gene list where fold change refers to Atg16L1HM over WT at steady state levels. Second tab shows GSEA analysis of leading edge genes from WT high and Atg16L1HM high gene sets. Third tab shows complete gene list where fold change refers to Atg16L1HM over WT after allo-HSCT. Fourth tab shows GSEA analysis of leading edge genes from Atg16L1HM high and WT allo-HSCT recipients high gene sets.
Supplemental Figure 1, related to Figure 1. Atg16L1 mutation increases morbidity and mortality after allo-HSCT with altered levels of pro-inflammatory cytokines. (A–B) Lethally-irradiated WT and Atg16L1HM recipients were transplanted with TCD-BM cells with and without a lower dose of splenic T cells (1×106) from donor B10.BR mice and monitored for (A) survival and (B) weight loss. n = 10 recipient mice/group. (C) WT mice in the B10.BR→B6 allo-HSCT model typically display more lethality than we observed in Figure 1A. The allo-HSCT from Figure 1A was repeated in a second animal facility (Memorial Sloan Kettering Cancer Center) to examine if the degree of lethality was facility-dependent. In contrast to the original experiment (performed in the ultra-clean animal room at New York University School of Medicine), WT recipients displayed substantial lethality. However, Atg16L1HM recipients remained significantly more susceptible. n = 10 recipient mice/group. (D–E) Cytokine and chemokine levels were measured by multiplex ELISA (Millipore) using serum from mice receiving the allo-HSCT described in Figure 1A–C on day 7 after the transplantation. Results were acquired with a Luminex 200 instrument and analyzed with xPONENT software. Cytokines displaying increases in Atg16L1HM allo-HSCT recipients are shown in C and those displaying decrease are shown in D. Untreated WT mice 6-weeks of age were used as additional controls to show baseline cytokine levels. No significant differences were seen in IL-1α, IL-1β, IL-2, IL-4, IL-15, IL-10, IL-13, IL-6, IL-17, IP-10, and RANTES when comparing B6 mice and Atg16L1HM receiving TCD-BM + T cells. n = 5–9 mice/group. (F) Survival of WT and Atg16L1HM treated with 500 μg of anti-TNFα or Isotype control (BioXcell) by i.p. injection on days −1 and 0, and every other day thereafter. Mice were transplanted with TCD-BM cells with 2×106 splenic T cells from donor B10.BR mice on day 0 as in Figure 1A–C. n = 10 mice/group. (G) Serum FITC-dextran concentration following oral gavage was measured in WT and Atg16L1HM mice on day 7 after lethal irradiation. (H–I) 122 donor-recipient pairs receiving allo-HSCT from HLA-identical sibling either at the University of Regensburg (n = 47, from May, 2000 until May, 2012) or at the University of Newcastle (n = 75, from January, 1993 until December, 2007) were included. Collection of samples for genotyping was approved by local ethical committees at both sites. DNA was typed for presence or absence of the ATG16L1T300A allele as described previously (Latiano et al., 2008). Results of genotyping were compared with major patient characteristics and outcome variables such as GVHD, non-relapse mortality (NRM), and overall survival using a SPSS based database and PASW statistic software for analysis. NRM and overall survival were calculated by Kaplan-Meier analysis, and Cox regression was used to compare risk factors in multivariate analysis. Patients receiving allo-HSCT from HLA-identical siblings were analyzed. DNA from patients and donors was isolated and typed for presence (heterozygous or homozygous) or absence of the ATG16L1T300A allele. The effect of the risk allele in donors and recipients were analyzed separately. (H) Rate of non-relapse mortality (NRM) in allo-HSCT recipients when the ATG16L1T300A allele is present versus absent in the donor. (I) Rate of NRM in allo-HSCT recipients when the ATG16L1T300A allele is present versus absent in the recipient. n = 6 mice/group. Mean ± S.E.M. are shown in (B), (D)–(E), and (G). *P < 0.05, **P < 0.01, ***P < 0.001.
Supplemental Figure 2, related to Figure 2. Histopathology of allo-HSCT recipients in the intestine and skin. (A–F) H&E-stained tissue sections from WT and Atg16L1HM allo-HSCT recipients (TCD-BM and 2×106 T cells as in Figure 1A–C) on day 7 were processed and examined for histopathology as previously described (Cadwell et al., 2010; Penack et al., 2009). Representative images of (A) small intestine and (B) skin, and blind quantification of GVHD histopathology using previous described parameters (Petrovic et al., 2004) shown for (C) small intestine, (D) colon, (E) liver, and (F) skin. Magnification: x10 for small intestines, x40 for skin. (G–J) Quantification of histopathology as described above on day 14-post allo-HSCT with a lower dose of T cells (1×106 T cells) to allow survival of mice. n = 6–8 mice/group. Mean ± S.E.M. are shown in (C–J).
Supplemental Figure 3, related to Figure 3. DCs from Atg16L1HM mice display an altered transcriptional profile. (A–D) Additional microarray analyses corresponding to the experiments described in Figure 3B and 3C. Heat-map presentation of gene-expression profiles after Significance Analysis of Microarray (SAM) (Z-score > 2) for (A) allo-HSCT recipients and (C) untreated mice. Complete list of genes can be found in Supplementary Table 2. Bar graphs in (B) and (D) represent p-values for biological processes (GO terms) displaying the most significant enrichment in Atg16L1HM allo-HSCT recipient and untreated Atg16L1HM mice compared to their respective WT controls. Enrichment in GO terms was determined by DAVID (Database for Annotation, Visualization and Integrated Discovery) using Z > 2 cutoff.
Supplemental Figure 4, related to Figure 4. Atg16L1 functions within DCs to prevent hyperactivity. (A–D) Flow cytometric analyses of small intestinal lamina propria cells from untreated or lethally (1100 cGy) irradiated WT and Atg16L1HM mice. CD3−B220−NK1.1−class-II+ CD11c+ cells were gated on either (A, B) CD11b−CD103+ or (C, D) CD11b+CD103+. MFI quantification shown for CD80 (A and C) and CD86 expression (B and D). No significant differences were noted when gating on percent cells that are CD80+ or CD86+, and there were no difference in cell numbers. (E–F) Similar flow cytometric analyses of mesenteric lymph nodes (MLNs) from untreated WT or Atg16L1HM mice. (G) Western blot comparing Atg16L1 levels (Sigma: A7356) in sorted splenic DCs from control (Atg16L1flox/flox) and Atg16L1ΔCD11c mice. (H) Clinical GVHD score of allo-HSCT recipient Atg16L1ΔCD11c mice from Figure 4I. (I) Quantification of indicated splenic DC subsets by flow cytometry from Atg16L1ΔCD11c and control mice 24 hours after lethal irradiation. n = 3 mice/group. (J) Flow cytometry analysis of CD80 expression on CD11c+ splenic cells isolated from Atg16L1ΔCD11c and control mice that were cultured for 24 hours. n = 3–4 mice/group. Mean ± S.E.M. are shown in (A)–(F) and (H)–(J). *P < 0.05.
Supplemental Figure 5, related to Figure 5. Increased co-stimulatory molecule expression on Atg16L1-deficient DCs is not transferrable. (A) 3H-thymidine incorporation of allogeneic B10.BR T cells co-cultured at a 1:1 ratio for 72 hours with DCs purified from the spleen of WT and Atg16L1HM mice, with or without neutralizing antibodies against IL-1β (5 μg/mL), anti-IL-1R1 (0.5 μg/mL), or anti-IL-12 (1 μg/mL) (R&D systems). (B) Flow cytometric analysis of CD80 expression on WT DCs cultured for 24 hours in supernatant collected from previously cultured Atg16L1HM DCs. (C, D) Flow cytometric analysis of CD11c+ DCs cultured with GM-CSF for 24 hours and stained for the ROS-detecting cell permeable dyes (C) dihydroethidium (DHE) and (D) 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA). n = 3 mice/condition. Bar graphs represent mean ± S.E.M and n = 3–6 mice/condition. ****P < 0.0001.
Supplemental Figure 6, related to Figure 6. Atg16L1 deficiency results in lysosomal abnormalities and decreased A20 expression. (A, B) Quantification of lysosomes, dysmorphic lysosomes, and multivesicular bodies detected per cell by TEM in splenic DCs harvested from (A) untreated mice or (B) mice 24 hrs after lethal irradiation. (C, D) Quantification of the percentage of vesicles detected per cell by TEM in splenic DCs harvested from (A) untreated mice or (B) mice 24 hrs after lethal irradiation. (E) qRT-PCR analysis of Laptm5 expression in splenic DCs from WT and Atg16L1HM mice cultured in GM-CSF containing media for 24 hrs. 5–7 mice/genotype. (F) Representative Western blot analyses of A20, phospho-IkB, phospho-JNK, and actin in whole cell lysates from DCs cultured as in (E). (G) Quantification of Western blot from (F). 6 mice/genotype. Bar graphs represent mean ± S.E.M, *P < 0.05, **P < 0.01, ****P < 0.0001. (H) Model figure: By inhibiting NF-κB and MAPK signaling, A20 prevents unregulated co-stimulatory molecule expression in DCs exposed to environmental stressors. In Atg16L1-deficient cells, decreased autophagy leads to accumulation of aberrant lysosomal structures and an accompanying lysosomal transcriptional response including enhanced expression of Laptm5. Laptm5 accumulation then mediates the degradation of A20 causing increased expression of the co-stimulatory molecules CD80, CD86, and CD40 that mediate enhanced T cell alloreactivity and subsequent GVHD.