Abstract
Biological computation is a major area of focus in synthetic biology because it has the potential to enable a wide range of applications. Synthetic biologists have applied engineering concepts to biological systems in order to construct progressively more complex gene circuits capable of processing information in living cells. Here, we review the current state of computational genetic circuits and describe artificial gene circuits that perform digital and analog computation. We then discuss recent progress in designing gene circuits that exhibit memory, and how memory and computation have been integrated to yield more complex systems that can both process and record information. Finally, we suggest new directions for engineering biological circuits capable of computation.
Introduction
Cellular computation is a central enabler of many envisioned biological applications, including next-generation therapeutics, diagnostics, and biomanufacturing platforms. Scalable platforms for implementing biological computation are needed to realize engineered biological systems that have sophisticated sense-and-respond behaviors. Information processing in living cells involves integrating multiple inputs, performing computations on these signals, storing information in memory, and actuating outputs.
A significant body of literature has been published on the design of synthetic digital logic gates in living cells, where biological signals such as chemical concentrations are artificially thresholded to represent ‘0’ or ‘1’ (‘OFF’ or ‘ON’, respectively) states. However, there are challenges in creating genetic systems that mimic digital logic. Biological molecules do not generally exist at only two possible concentrations, but usually vary over a wide range of concentrations. In the absence of suitably sharp input-to-output transfer functions, this can lead to challenges in setting thresholds to define Boolean logic. Furthermore, achieving more complex computations with digital logic requires assembling together many simple digital logic units. However, unlike electronic systems in which transistors and other electronic components can be isolated on a substrate and physically wired together, biological components operate within compartments in which interactions between components are based on chemical specificity. Thus, it is challenging to construct large numbers of highly orthogonal parts in biological systems. As a result, digital logic can be difficult to scale in biological systems to achieve complex computations.
An alternative to digital computation is analog computation. While digital logic uses the states ‘0’ or ‘1’ to compute with, analog computation represents signals using the range of continuous values between these extremes and calculates mathematical functions on these signals using the laws that govern biochemical phenomena. Thus, analog computation enables complex computations to be performed without needing a large number of biological parts or resources by taking advantage of the underlying physics of the components. However, the limits and design rules behind analog circuits in biological systems are just starting to be understood. Here, we highlight multiple papers to illustrate key concepts behind biological computation, digital and analog processing, and memory. We refer the interested reader to excellent reviews for greater details1–3.
Digital-logic synthetic gene circuits
Early efforts to implement digital logic gates with synthetic gene circuits used both combinatorial approaches as well as rational design. For example, Guet et al. created a combinatorial library of gene networks based on the natural transcription factors (TFs) LacI, TetR, and lambda cI, as well as promoters responsive to these TFs4. Within this library were networks that were found to behave as NOR, NAND, and NOT-IF two-input logic gates. This work was followed by additional papers detailing the construction of digital logic gates using a variety of regulatory mechanisms, including regulation at transcriptional, translational and protein levels.
At the transcriptional layer, a large number of digital logic gates have been created. For example, Kramer et al5 used the combined action of both activating and repressing synthetic transcription factors (by leveraging the VP16 activation domain or the KRAB repression domain, respectively), in various combinations to build NOTIF, NOT-IF-IF, NAND, OR, NOR, and INVERTER gates. The NOR gate was a sequential transcription cascade, whereby in the absence of the inducer erythromycin (EM), a transcriptional activator bound to a promoter and induced expression of another transcriptional activator whose ability to induce the `output' reporter gene could be repressed by pristinamycin (PI). When neither small molecules (EM or PI) were present, the reporter was expressed (output = 1). If EM, PI, or both were present, then expression of the reporter was repressed (output = 0), thus forming a gate with NOR logic.
AND gates at the transcriptional level have been built by relying on mechanisms other than activation or repression by inducible transcription factors. Anderson et al6 used a T7 polymerase containing two amber stop codons in conjunction with the amber suppressor tRNA, supD. One promoter of the gate drove expression of the T7 polymerase, while the other drove expression of supD. If only the input signal to express the T7 polymerase was given, then translation of the T7 polymerase stopped prematurely at one of the amber stop codons. However, when the input to express the supD amber suppressor tRNA was also present, translation occurred through the amber stop codons, and full length, functional T7 polymerase was produced. T7 was then used to drive output gene expression from a T7-dependent promoter, thus forming an AND gate.
Lohmueller et al7 used inteins to create AND gates at the transcriptional level in mammalian cells. Inteins are self-splicing protein domains - if present within a protein, they will be removed, leaving the two adjacent domains fused together. The authors used split-inteins, a variation on standard inteins which, when fused to separate protein fragments, are able to splice the two fragments together to form a single protein. To build an AND gate, each half of a Zinc Finger (ZF) domain was fused to one half of a split-intein. The C-terminal half of the ZF was fused to a transcriptional activator VP64 domain. Expression of either half of the ZF led to minimal output gene expression. However, upon co-expression of both halves of the ZF, the split-inteins spliced the proteins together, forming a complete ZF DNA-binding domain fused to a VP64 activation domain. The ZF-VP64 protein was then used to drive expression of an output reporter. Construction of a NAND gate was achieved simply by replacing the activating VP64 domain with a repressive KRAB domain. The approach of split-inteins was subsequently extended to build 3-input AND gates8.
As an alternative to transcriptional regulation, regulation at the RNA level with RNA interference (RNAi) has been used for the construction of logic-based circuits. Rinaudo et al9 built circuits that behaved as either Conjunctive Normal Form (CNF) or Disjunctive Normal Form (DNF) evaluators, which are standardized strategies for representing logic functions that are also readily amenable to manipulation by automated algorithms. In this paper, the siRNAs that formed the `inputs’ to the logic circuits were added exogenously. Subsequent work10 used RNAi to create a variety of 2- and 3-input logic gates, but produced the siRNA inputs to these gates within cells using transcriptional control, a situation more amenable to subsequent integration of these circuits with other synthetic gene networks as well as endogenous signals. Xie et al11 applied these advancements to construct a complex, RNAi-based classifier circuit, that could classify, based on the levels of 5 different native miRNA species in a 5-input AND gate, whether a cell was in a certain `cancerous' state as defined by its miRNA profile. By using the classifier to control the expression of an apoptosis-promoting protein (hBax), the circuit was shown to selectively kill cancerous HeLa cells, with some degree of false positives and false negatives using non-cancerous HEK293 cells as a comparison.
Win et al12 took another approach for constructing logic gates at the RNA level. They engineered synthetic ribozymes, where the catalytic cleavage action of the ribozyme (which takes place in an `actuator' module of the RNA structure) could be controlled by a small-molecule binding to a separate `sensor’ module of the RNA device. The `sensor' and `actuator' modules were connected by a short RNA stem called the `transmitter' module. When the small molecule was bound to the sensor module, it was energetically favorable for the transmitter module to hybridize with the actuator module in such a way that the ribozyme’s catalytic activity was prevented. Without small-molecule binding to the sensor, the transmitter did not hybridize to the actuator module, thus allowing ribozyme cleavage to occur. By placing these synthetic ribozymes in the 3'UTR of a reporter gene, the expression or `output’ of the gene could be controlled. By combining synthetic ribozymes in various combinations, AND, NOR, NAND, and OR logic gates could be achieved. Performance-wise, both the AND and NOR gates were not particularly ideal representations of a digital function. In both, the [1, 0] and [0, 1] input states both displayed non-negligible levels of output, when ideally there should be minimal output gene expression. This more graded response is likely due to the additive nature of using ribozymes in certain configurations, as opposed to the implementation of Boolean AND gates using the T7 polymerase/supD6 and split-intein7,8 systems, where the two inputs are not additive but rather are both necessary for function. Other approaches to logic gate construction have used orthogonal ribosomes13 and allosteric switching of protein conformation14. As protein dynamics operate on very short timescales, using switchable protein conformations may allow for substantially higher switching speeds (i.e. from 0 to 1 or 1 to 0) than can be achieved through transcriptional or translational regulation14.
Sophisticated electronic systems are not formed from single logic gates, but multiple logic gates connected or `layered' in various configurations. Building on the success of single logic gates, synthetic biologists are now beginning to create larger genetic `programs' through the interconnection of multiple logic gates. Using a combination of transcriptional and translational control, Auslander et al15 built a 2-input N-IMPLY logic gate (where the output is 1 only in the presence of one of the two inputs) in mammalian cells. By connecting up two N-IMPLY gates, they created an XOR gate. Going further, they added an additional N-IMPLY logic gate to this XOR to create a half-subtractor circuit, capable of subtracting two bits from each other. By instead adding an AND gate to the XOR, they created a half-adder circuit which was capable of adding two bits together.
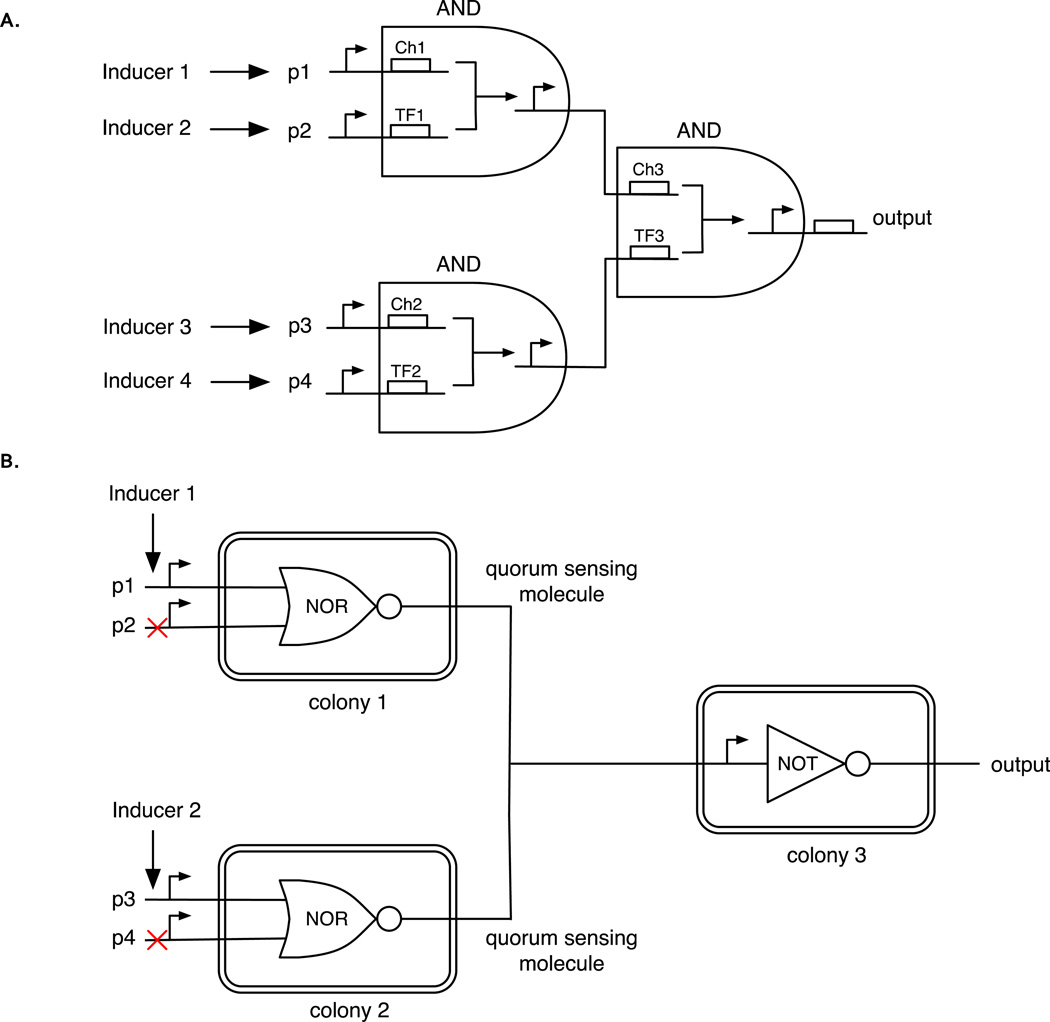
Moon et al16 connected two and three 2-input AND gates together to create single 3- and 4-input AND gates, respectively, in bacteria. The 4-input AND gate (Figure 1A) is the largest (in terms of the number of regulatory proteins used) synthetic gene circuit constructed to date. Notably, Moon et al16 considered the possible issue of delays when layering genetic logic gates. In published logic-circuit designs, there exists no clock to synchronize the progression of the signals (e.g., the processes of transcription, translation, protein binding etc.) through the regulatory layers. Therefore, any differential delay in the passing of various signals through the layers can cause different signals to reach downstream layers at different times, thus potentially resulting in incorrect inputs to the downstream layers at any given time and resulting in a `fault'. A mathematical model allowed the authors to derive a term that dictated whether or not a fault would occur between two layers of their circuits, which was largely dependent on the degradation rates of the regulatory proteins. No faults were observed in the 4-input or the more susceptible 3-input AND gates, although the authors pointed out that faults are likely to become a more prominent issue in biological logic gates as the number of regulatory layers increases.
Figure 1.
A. A 4-input genetic AND gate formed by coupling the output of two 2-input genetic AND gates into the input of a third 2-input genetic AND gate. Inducers 1, 2, 3, and 4 induce expression from promoters p1, p2, p3, and p4 respectively. Each AND gate comprises a transcription factor (e.g. TF1) and a corresponding chaperone (i.e. Ch1). Both are required together to activate transcription from the target promoter. The output of each of the left pair of genetic AND gates is used to induce expression from promoters in the right genetic AND gate. The output is only ‘1’ when the inducers are all present. B. A genetic AND gate formed from 3 different E. coli colonies connected via cell-cell communication modules. The cells of colony 1 all possess an identical genetic NOR gate. Colony 2 contains a genetic NOR gate that is different from the gate in colony 1. Colony 3 contains a genetic NOT gate. Inducers 1 and 2 induce expression from promoters p1 and p3, respectively. Promoters p2 and p4 are not induced in the operation of the AND gate. The output from colonies 1 and 2 is a diffusible quorum-sensing molecule that serves as the input to the promoter of the NOT gate. Thus, the output is only ‘1’ when inducers 1 and 2 are present. Figure 1A is adapted from Moon et al16.
An alternative approach to interconnecting logic gates in single cells has been to `wire’ gates between different cells of a population. For example, Tabor et al.17 used AHL (a diffusible quorum-sensing molecule that can be used for transmitting signals from one cell to another) to connect logic gates between E. coli cells. The authors used a light-repressible promoter to control expression of LuxI, an enzyme that produces AHL. The logic gate in each cell performed the following logic: IF (NOT light), THEN generate AHL; IF (AHL AND NOT (NOT light)), THEN generate pigment. Thus, cells in the dark produced AHL (as luxI expression was not repressed), but cells in the light would not. AHL from dark region diffused partially into the light region. Cells which were exposed to light and also sensed AHL from neighboring dark cells activated expression of lacZ and produce a dark pigment, thus defining the edge between the light and dark regions. The population therefore acted as an `edge detector’.
Two studies by Regot et al18 and Tamsir et al19 took this approach a step further. By creating cells that contained different logic gates that could again be interconnected with diffusible signaling molecules, it was possible to construct larger circuits on a population level, as opposed to within a single cell. One advantage of this approach is that the same genetic components can be re-used in different cells to construct different logic gates, thus helping to circumvent the limited number of wellcharacterized components that are available for building gene circuits. Tamsir et al19 connected together various versions of NOR, OR and NOT gates (with different inputs and outputs) to create all 16 2-input logic gates with populations of E. coli (Figure 1B shows the design of the 2-input AND gate). Specifically, they used spatially separated colonies on an agar surface, where each colony contained a single logic gate and links between colonies were established through diffusible quorum-sensing molecules. Regot et al18 followed a similar approach, but in a mixed population of yeast with cellular communication based on yeast pheromones (alpha-factors from S. cerevisiae and C. albicans). Using this strategy, they constructed all 2-input logic functions, a 2-to-1 multiplexer, and a half-adder circuit.
Analog synthetic gene circuits
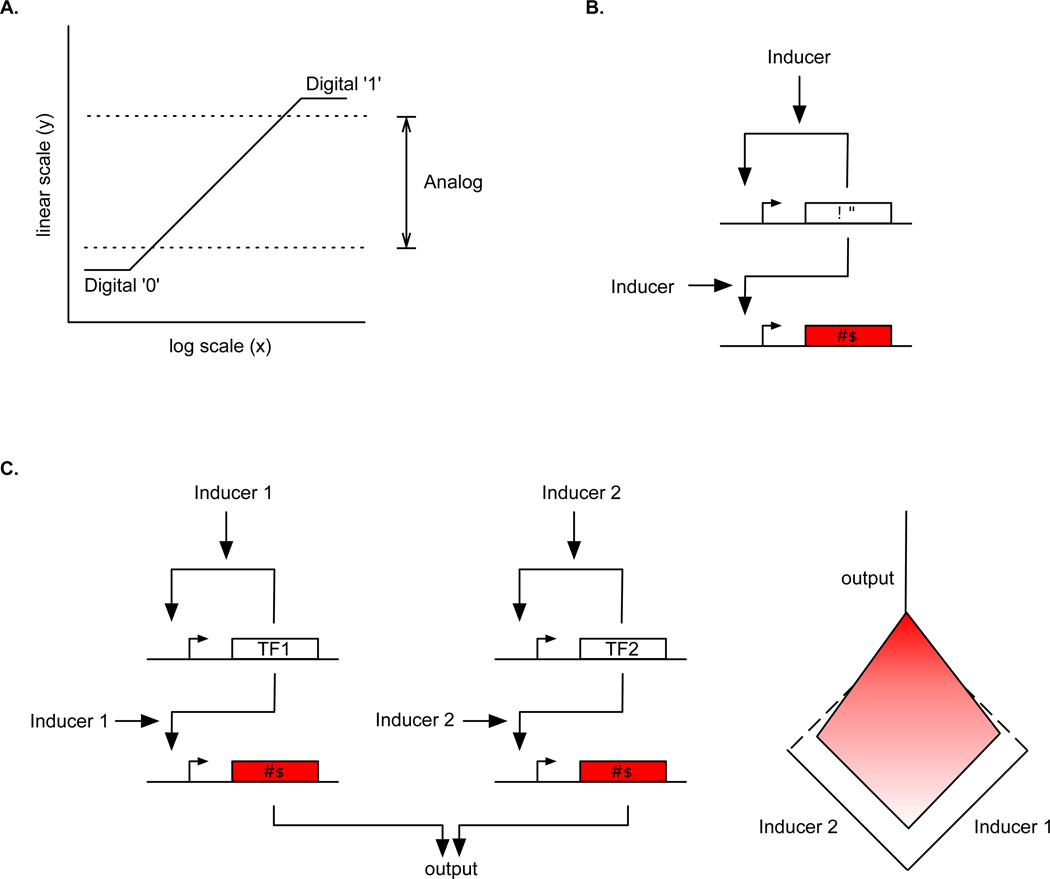
Analog processing represents an alternative to the conventional digital approach for cellular computation but has only recently begun to be explored in experimental settings. Daniel et al20 constructed synthetic genetic circuits that perform analog computation using small-molecule inducers to represent continuous input values and proteins to represent continuous output values. The authors started by taking a open-loop circuit with an inducible transcriptional activator (i.e., AraC) that displayed a sigmoidal, Hill-function-type input-output transfer function and engineering it to have a wide dynamic range. This was done to enable a greater range over which the input inducer (i.e., arabinose) concentration could be used to represent signals for analog computation. This was achieved by introducing a positive-feedback loop into the circuit such that the addition of inducer generated additional transcriptional activator proteins (Figure 2B). Furthermore, the authors placed the positive-feedback loop circuit on a low-copy plasmid and engineered a high-copy plasmid to contain extra promoters for the AraC transcriptional activator (i.e., the PBAD promoter driving expression of the mCherry fluorescent protein). These two effects thus delayed the saturation of the input-output transfer function with the addition of inducer and increased the input dynamic range. When the input-output transfer function was plotted on a log-linear graph, it transitioned almost linearly between low and high output levels. Thus, over this range, the circuit exhibited an output fluorescence which was a scaled logarithmic transformation of the input inducer concentration.
Figure 2.
A. Digital versus analog computation. Digital logic uses the extremes of an input-output transfer curve to represent ‘0’ and ‘1’ signals, while the analog paradigm uses the entire range in between the extremes for computation based on mathematical laws which determine how the output value(s) depends on the input value(s). B. Circuit design of the positive-feedback-and-shunt circuit, which implements a scaled positive-logarithmic transformation of the input inducer concentration to the output reporter (RP) protein concentration. The transcription factor (‘TF’) induces its own expression via a positive- feedback loop located on a low-copy plasmid and that of a reporter gene (‘RP’) from a high-copy plasmid. C. Two orthogonal versions of the circuit in B are used to make a genetic adder circuit. Each circuit uses a different inducible TF (TF1 and TF2) and inducer (Inducer 1 and Inducer 2). The output is the same for both circuits, and thus the net output is the sum of the outputs from both positive-logarithm circuits. The graph is an approximation of the result from Daniel et al20, which shows the additive nature of the analog circuit (x and y axes are in log scale, whereas the z axis is in linear scale).
This same motif was applied to another class of transcriptional activator proteins (i.e., LuxR), resulting in two wide-dynamic-range circuits which implemented positive-slope logarithms. By combining the two circuits, which accept two different inducer inputs but produce the same output protein, the authors created a circuit capable of addition since the total output is sum of the outputs from the individual circuits (Figure 2C). The authors also modified one of the positive-logarithm circuits to produce a transfer function with a wide-dynamic-range negative slope instead of a positive slope, thus implementing a negative logarithm. By combining a positive-logarithm and negative-logarithm circuit together, circuits that could perform subtraction as well as division were created. Division is a useful operation because it enables one to determine the relative ratio between two inputs, and many ratios between different chemical species are relevant to controlling biological pathways. Finally, using just two transcription factors, the authors created a feedback circuit which displayed a power-law relationship between its input and output.
An advantage of analog computation is that circuits can be built with far fewer components than their digital counterparts but achieve complex computational functions, by using the inherent behaviors of the underlying components. Leveraging analog computation can therefore help to reduce the size of genetic circuits and alleviate the burden on cellular resources. However, as intrinsic noise in chemical systems scales as the square-root of the molecular count21,22, analog systems are more susceptible to noise than digital systems when working with lower molecule numbers. Thus, the tradeoff between accuracy and resources must be carefully considered during system design.
Synthetic gene circuits exhibiting memory
The computational circuits described so far have outputs that are transient - if the inputs are removed, the outputs will not be sustained over a long timescale. Just as in electronic computing and neural processing, the development of sophisticated genetic programs will require that intermediate results (or `system states') and outputs are stored in cellular memories for downstream use.
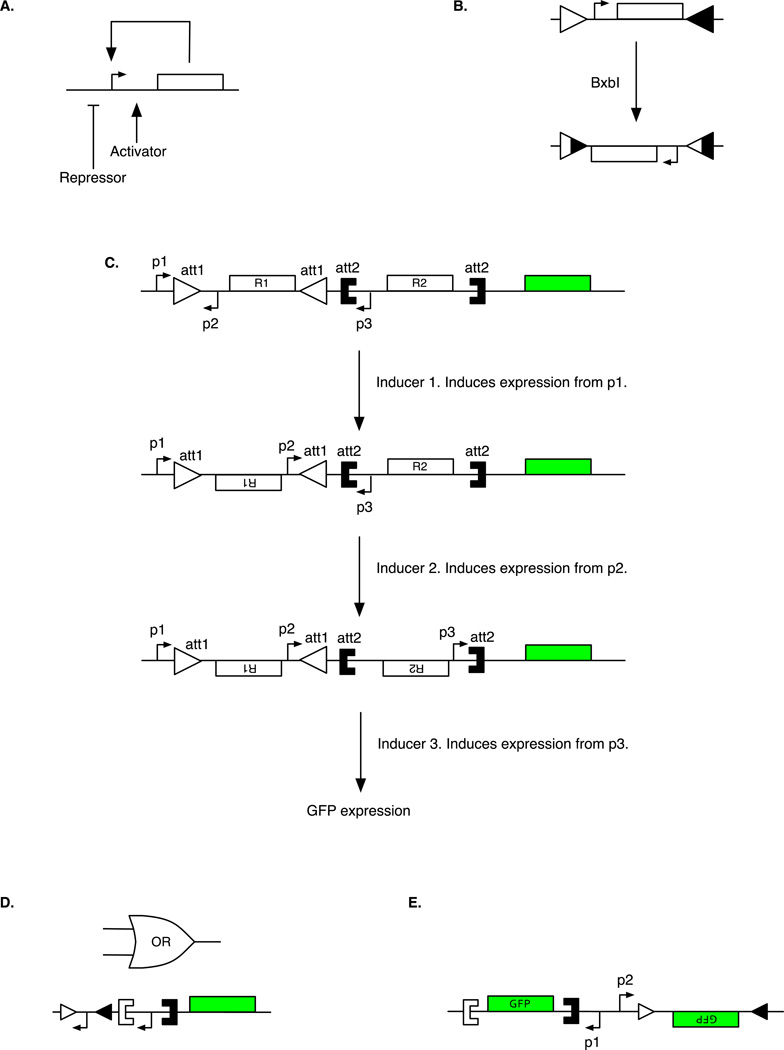
The toggle switch23 exhibited bistable behavior and was an early example of a synthetic gene circuit displaying what can be thought of as memory. A substantial body of subsequent work has focused on using bistable switches in memory-based circuits24–31. However, in contrast to the original toggle switch, which relied on negative feedback, many of these subsequent circuits have used positive feedback. Specifically, these circuits have involved a single gene whose protein product activates its own expression (Figure 3A). At low protein levels, the rate of expression from the gene is too low to overcome the degradation rate of the protein product, and the protein concentration is stably maintained at a low level. At high protein levels, the rate of expression is high enough to balance the degradation rate of the protein, allowing the protein concentration to be stably maintained at a high level. In order to switch the positive-feedback network between these two stable states, another input is required. This can either be one that activates the level of gene expression, pushing the system from the low to the high stable state29–32, or one that represses gene expression, pulling the system down from the high to the low stable state26,27.
Figure 3.
Strategies for building memory, counters, and integrated logic and memory in living cells. A. Design of a positive-feedback loop circuit where a transcription factor activates its own transcription. To control switching, the promoter is also controlled by an external activator or repressor input. B. In an example of recombinase-mediated memory, the Bxb1 recombinase recognizes the att sites denoted as triangles and inverts the intervening DNA, forming two new att sites which can no longer be recognized by Bxb1 for further inversion. C. Multiple-inducer 3-counter built by cascading recombinase-invertible memory units. Addition of Inducer 1 induces expression from promoter p1, producing recombinase R1 which flips DNA located between the att1 sites (triangles). This inverts promoter p2 so it is now capable of expressing recombinase R2. Subsequent addition of Inducer 2 induces expression from promoter p2, producing recombinase R2, which flips DNA between the att2 sites (square brackets). This inverts promoter p3 so it is now capable of expressing GFP upon addition of Inducer 3. D. Genetic OR gate based on recombinase logic. Triangles denote one pair of recombinase-recognition sites recognized by Bxb1 recombinase while square brackets denote another pair of recombinase-recognition sites recognized by phiC31 recombinase. E. Digital-to-analog converters can be implemented using multiple recombinase-invertible units within a single cell, each of which activates expression of a common output gene with a different expression level when flipped by their respective inputs. Thus, digital combinations of inputs lead to programmable levels of analog gene expression inside of living cells.
For example, by using a DNA-damage-responsive promoter to drive expression of a TF that serves as the activating input into the positive-feedback circuit, Burrill and Silver29 were able to detect and `remember' artificially induced DNA damage. This memory (i.e., the state of the circuit) was still maintained in >50% of cells after 6.5 days. Further work by Burrill et al31 created a variant of this memory network which could instead be switched by exposure to hypoxia (using an input promoter that is activated by HIF-1, a TF stabilized by hypoxia). The stability of the memory after induction by hypoxia was on the order of the previous DNA-damage-responsive circuit. The stated goal of such circuits was to track and record the history of cellular experiences and states within a population.
A parallel approach to implementing circuits capable of memory is to use recombinases for DNA-encoded memory. Recombinases are enzymes capable of recombining DNA located between specific sites known as `attachment' (or att) sites. Recombinases have traditionally been used in biotechnology for efficient cloning33 and genomic integration34. However, by placing pairs of the att recombinase-recognition sites on the same strand of DNA, recombinases can invert (`flip') or excise the intervening region of DNA (Figure 3B), depending on the relative orientation of the two sites. Recombinase-based inversion can be used to implement unidirectional recombination, in which the target DNA is flipped only once, after which two new att sites are formed which are no longer recognized by the recombinases, as well as bidirectional recombination in which the target DNA is continuously flipped. Although bidirectional recombinases have been used in gene circuits35,36, the continual flipping needs to be addressed in order to implement stable memory circuits. Alternatively, the stability of DNA flipped by unidirectional recombinases makes them ideal candidates for constructing memory circuits.
While transcription-based gene circuits can be knocked out of their stable state by random perturbations or unpredictable cellular influences (for instance, Ajo- Franklin et al.28 reported that their positive-feedback-based bistable circuit was sensitive to the growth rate of the cells), DNA which is reconfigured by recombinases is highly stable, even persisting after cell death. However, the restriction of being able to flip only once with unidirectional recombinases may be a limitation when designing certain types of circuits. Bonnet et al37 showed it was possible to overcome this limitation. Expressing the unidirectional recombinase Bxb1 resulted in a one-way flip of DNA, but co-expressing Bxb1 with a complementary Recombinase Directionality Factor (RDF) resulted in a reset of the DNA back into the original configuration. Although conceptually simple, a practical demonstration was difficult and required substantial optimization. The fundamental problem was that in the presence of the RDF, the recombinase action becomes bidirectional, since either the recombinase on its own or the recombinase-excisionase complex is able to flip DNA, each in a different direction. By optimizing the stoichiometry of the recombinase and the RDF, as well as the RDF degradation rate, the authors were eventually able to find a combination that allowed multiple rounds of controlled bidirectional flipping, with the majority of the population of cells flipping their DNA at each step. Nonetheless, this controlled bi-directionality must be demonstrated to be reliable and easily repeatable before it is likely to be used in place of a bistable genetic circuit (e.g. the toggle-switch).
Friedland and Lu et al36 used memory to create genetic circuits that could count up to three input events with two complementary approaches. The first exploited delays in a transcriptional regulatory cascade, with each input pulse of arabinose being short enough to only advance the cascade by roughly one step. The second approach used recombinase-invertible units called Single Invertase Memory Modules (SIMMs) that were arrayed into a cascade such that each pulse of an input inducer would generate the expression of a different recombinase, which would flip a specific stretch of DNA and advance the cascade by one step. This flipping resulted in the inversion of a promoter into the correct orientation for expressing the downstream recombinase gene, which was contained in the next SIMM, with the next input pulse. This approach enabled the creation of a counter that could count three inputs of the same inducer as well as a counter capable of recording multiple inducer events (Figure 3C). In the latter case, the recombinase-based counter could uniquely determine the sequence of the three input events, and thus implemented a sequence detector as well as a counter. In comparing the transcriptional-based and recombinase- based counters, the authors found that the transcriptional-based counter demonstrated a faster response to pulses, making it suited to counting events on the time scale of cell division. Inversion by recombinases takes place on the order of hours, and so the recombinase-based counter was more suited to counting events on longer time scales.
The two approaches, bistable circuits or recombinase-based circuits, both have advantages and disadvantages. Bistable transcriptional circuits are more amenable to reliable bidirectional switching, but each stable state is susceptible to perturbations. Furthermore, transcriptional circuits are unable to maintain memory after cell death. In contrast, controlled bidirectional switching with recombinases is not yet as reliable, but each stable state is highly stable, even after cell death. Depending on the desired application, synthetic biologists now have a suite of technologies that they can draw upon for implementing memory.
Integrating Logic and Memory
In order to realize more sophisticated genetic circuitry capable of implementing computations that contain state (i.e., stored information), both logic and memory must be integrated such that the results of logical operations can be stored and operated upon at later times. One step in this direction was undertaken by Lou et al38 when they coupled a NOR logic gate to a bistable switch which acted as a `memory module'. This allowed them to create a `push-on push-off' switch in which the same sequential input (ultraviolet light) caused the system to alternate between expressing GFP or RFP, and the state of the system (i.e., whether GFP or RFP is expressed) was `held' in the memory until the next pulse. The optimized circuit was able to switch back and forth three times, although with each subsequent switching event, progressively less cells successfully switched.
Taking another approach, recent studies have used recombinases to simultaneously perform logic and memory operations. Siuti et al39 created all 16 2-input logic gates using the unidirectional recombinases Bxb1 and phiC31 in combination with gene regulatory elements such as promoters, terminators, and output genes. For example, an OR gate can be constructed by having two inverted promoters in tandem, one surrounded by recombinase-recognition sites for Bxb1 and the other by sites for phiC31, upstream of an output gene (Figure 3D). When either Bxb1 or phiC31 is expressed by orthogonal inducer inputs, one of the promoters flips and can then drive downstream gene expression. A similar approach can be adopted to create all other logic functions. Since this approach involves flipping DNA elements, the history of inputs is stably recorded in the DNA of progeny cells and is reflected in the state of the circuits. For example, the OR gate described above has state associated with it and implements a circuit where the output is 1 if any of the two inputs were ever 1. These gates thus enable sequential logic, in contrast with memory-less combinatorial logic (for example, a memory-less combinatorial OR gate has an output that is 1 only if any one of the inputs are 1 at given time). While Siuti et al39 created logic gates based on flipping promoters, terminators, and output genes, the majority of the logic gates constructed by Bonnet et al40 relied on flipping only terminators. For example, an AND gate comprised two tandem unidirectional terminators present between a promoter and a reporter gene, where each terminator was flanked by recombination sites recognized by one of two different unidirectional recombinases (Bxb1 and TP901-1). For the promoter to express the output gene, both recombinases had to be expressed in order to flip the terminators into their inverted positions.
Siuti et al39 showed that it is possible to use recombinase-encoded circuits to implement digital-to-analog conversion41, in which digital combinations of inputs are translated into different analog levels of gene expression inside of the cell. By using two independent recombinases to orthogonally flip two promoters of different strengths that regulated two genes which encoded the same protein, each of the 4 possible digital combinations of promoter flipping resulted in 4 different protein levels (Figure 3E). Finally, by exploiting of the stability of recombinase-flipped DNA, Bonnet et al40 were able to use M13 phage to pass logic gates between cells, a process that would be difficult to achieve with other non-DNA encoded approaches.
Discussion
Genetic digital logic gates have been the focus of intensive study over the last decade, and recent work has made inroads into building larger computational circuits based on these gates. New strategies for creating orthogonal parts libraries7,42–45, modularizing genetic circuits46 and assembling large DNA constructs will contribute to the future scaling of digital circuits in living cells. Analog circuits have been a more recent development and will likely see further investigation as researchers attempt to develop more complex analog circuitry and alternative circuit topologies. As new circuit architectures continue to be described and refined, the field will need to develop standardized strategies for measuring and comparing the relative advantages and disadvantages of various cellular computing approaches.
For example, there is an important trade-off between the effects of noise on synthetic circuits and the effects of synthetic circuits on cellular resources47. At low molecule numbers, circuits are more susceptible to noise, but will impose a lower burden on host cells. Alternatively, at high molecule numbers, circuits can be less susceptible to noise, but will place a greater burden on host cells. As circuits which implement biological computations increase in complexity, it is unlikely they can be solely based on digital logic due to their burden on host cells and challenges in identifying sufficient numbers of orthogonal digital parts. Thus, important decisions in the design process for synthetic circuits will be when and how to combine digital and analog processing to balance this trade-off. In our opinion, hybrid computation, in which analog processing is used for the majority of the computations and digital logic is used to set thresholds, make decisions, and mitigate noise, may be a useful compromise between the two approaches, although this design strategy has not yet been used extensively in synthetic biology. Furthermore, integrating memory into these circuits will be another step towards increased sophistication.
Ultimately, the optimal integration of digital logic, analog computation, and memory circuits will enable a powerful next generation of genetic circuits. Although it is unlikely that biological computation will compete with silicon-based computing strategies in the short term, gene circuits that compute will be useful for biotechnology, biomedical engineering, and basic biology studies. For example, one can envision the design of synthetic gene circuits that can sense intracellular and extracellular signals in a biological process and optimize the production of a heterologous pathway or protein in response48. In addition, artificial sensors could be engineered into cells which are then used to detect signals in the human gut and respond by producing the appropriate therapeutics. Molecular recorders or ‘ticker tapes’ could be designed into human cells and used to study the sequence, timing, and frequency of cellular events involved in basic biological processes, such as development. In all these scenarios, the ability to sense, process, and record information is of central importance and will continue to drive innovation in the field of synthetic biology.
Highlights.
Cellular computation will enable applications envisioned by synthetic biologists.
Digital cellular circuits can perform increasingly complex logic functions.
Recent analog cellular circuits calculate math functions using just a few parts.
Memory devices can be based on bistable circuits or recombinase operations.
Integrating digital, analog and memory circuits will enable `next-gen’ circuits.
Acknowledgements
O.P., T.K.L., and the Lu lab acknowledges support from the National Institutes of Health (1DP2OD008435 and 1P50GM098792), the National Science Foundation (1124247), the Defense Advanced Research Projects Agency, the Ellison Foundation, the Office of Naval Research (N00014-11-1-0725, N00014-11-1-0687, and N00014-13-1-0424), the Army Research Office (W911NF-11-1-0281), and the United States Presidential Early Career Award for Scientists and Engineers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benenson Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 2012;13:455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- 2.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J. Biotechnol. 2007;130:329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Inniss MC, Silver PA. Building Synthetic Memory. Curr. Biol. 2013;23:R812–R816. doi: 10.1016/j.cub.2013.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial Synthesis of Genetic Networks. Science. 2002;296:1466–1470. doi: 10.1126/science.1067407. [DOI] [PubMed] [Google Scholar]
- 5.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 6.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lienert F, et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinaudo K, et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 10.Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. nnano.2010.135. 2010:1–5. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. This paper showed the creation of a multi-input circuit capable of discriminating between different human cell types for cancer detection based on miRNA profiles.
- 12.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 14.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301:1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 15. Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. The authors construct circuits capable of XOR logic and both half-addition and half-subtraction, in mammalian cells.
- 16. Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. The work demonstrated the construction of a 4-input, 2-layer AND gate in E. coli which is, to date, the largest published synthetic gene circuit. The authors also investigate the potential issue of signalling delays between circuit layers.
- 17.Tabor JJ, et al. A synthetic genetic edge detection program. Cell. 2009;137:1272–1281. doi: 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regot S, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 19.Tamsir A, Tabor JJ, Voigt CA. Robust multicellular computing using genetically encoded NOR gates and chemical “wires”. Nature. 2011;469:212–215. doi: 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic Analog Computation in Living Cells. Nature. 2013 doi: 10.1038/nature12148. The first demonstration of synthetic gene networks capable of complex analog computations. The authors created circuits that performed logarithms, addition, subtraction, division, and power laws.
- 21.Gillespie DT. Deterministic limit of stochastic chemical kinetics. J. Phys. Chem. B. 2009;113:1640–1644. doi: 10.1021/jp806431b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillespie DT. Stochastic simulation of chemical kinetics. Annu. Rev. Physcial Chem. 2007;58:35–55. doi: 10.1146/annurev.physchem.58.032806.104637. [DOI] [PubMed] [Google Scholar]
- 23.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 24.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 25.Wu M, et al. Engineering of regulated stochastic cell fate determination. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10610–10615. doi: 10.1073/pnas.1305423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Atkinson MR, Savageau MA, Myers JT, Ninfa AJ. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell. 2003;113:597–607. doi: 10.1016/s0092-8674(03)00346-5. [DOI] [PubMed] [Google Scholar]
- 28.Ajo-Franklin CM, et al. Rational design of memory in eukaryotic cells. Genes & Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burrill DR, Silver PA. Synthetic circuit identifies subpopulations with sustained memory of DNA damage. Genes & Dev. 2011;25:434–439. doi: 10.1101/gad.1994911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilaboa N, Fenna M, Munson J, Roberts SM, Voellmy R. Novel gene switches for targeted and timed expression of proteins of interest. Mol. Ther. 2005;12:290–298. doi: 10.1016/j.ymthe.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 31.Burrill DR, Inniss MC, Boyle PM, Silver PA. Synthetic memory circuits for tracking human cell fate. Genes & Dev. 2012;26:1486–1497. doi: 10.1101/gad.189035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sera T, Uranga C. Rational design of artificial zinc-finger proteins using a nondegenerate recognition code table. Biochemistry. 2002;41:7074–7081. doi: 10.1021/bi020095c. [DOI] [PubMed] [Google Scholar]
- 33.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hillman RT, Calos MP. Site-specific integration with bacteriophage ΦC31 integrase. Cold Spring Harb. Protoc. 2012;2012 doi: 10.1101/pdb.prot069211. [DOI] [PubMed] [Google Scholar]
- 35.Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One. 2008;3:e2815. doi: 10.1371/journal.pone.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. By balancing and controlling the concentrations of Bxb1 recombinase and RDF (Recombinase Directionality Factor), the authors described controllable bidirectional memory.
- 38.Lou C, et al. Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Mol. Syst. Biol. 2010;6:350. doi: 10.1038/msb.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013 doi: 10.1038/nbt.2510. This paper demonstrated the first implementation of all 16 2-input logic gates integrated with memory using recombinases, as well as digital-to-analog conversion circuits.
- 40. Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying Genetic Logic Gates. Science. 2013 doi: 10.1126/science.1232758. This paper created a number of 2-input logic gates using recombinases and passed logic gate state between cells using M13 phage.
- 41.Lu TK, Khalil AS, Collins JJ. Next-generation synthetic gene networks. Nat. Biotechnol. 2009;27:1139–1150. doi: 10.1038/nbt.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farzadfard F, Perli SD, Lu TK. Tunable and Multifunctional Eukaryotic Transcription Factors Based on CRISPR/Cas. ACS Synth. Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou C, Stanton B, Chen Y-J, Munsky B, Voigt CA. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purcell O, Jain B, Karr JR, Covert MW, Lu TK. Towards a whole-cell modeling approach for synthetic biology. CHAOS. 2013;23:25112. doi: 10.1063/1.4811182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roquet N, Lu TK. Digital and analog gene circuits for biotechnology. Biotechnol. J. 2014 doi: 10.1002/biot.201300258. [DOI] [PMC free article] [PubMed] [Google Scholar]