Abstract
The pro-apoptotic BH3-only BCL-2 family member BIM is a critical determinant of hematopoietic cell development and homeostasis. It has been argued that the striking hematopoietic abnormalities of BIM-deficient mice (accumulation of lymphocytes and granulocytes) may be the result of the loss of the protein throughout the whole animal rather than a consequence intrinsic to the loss of BIM in hematopoietic cells. To address this issue and allow the deletion of BIM in specific cell types in future studies, we have developed a mouse strain with a conditional Bim allele as well as a new Cre transgenic strain, Vav-CreER, in which the tamoxifen-inducible CreER recombinase (fusion protein) is predominantly expressed in the hematopoietic system. We show that acute loss of BIM in the adult mouse rapidly results in the hematopoietic phenotypes previously observed in mice lacking BIM in all tissues. This includes changes in thymocyte subpopulations, increased white blood cell counts and resistance of lymphocytes to BIM-dependent apoptotic stimuli, such as cytokine deprivation. We have validated this novel conditional Bim knockout mouse model using established and newly developed CreER strains (Rosa26-CreER and Vav-CreER) and will make these exciting new tools for studies on cell death and cancer available.
The mitochondrial (also called intrinsic, stress or BCL-2 regulated) apoptotic pathway is regulated by members of the BCL-2 family.1 This protein family can be divided into pro-survival (A1, MCL-1, BCL-2, BCL-XL and BCL-W) and pro-apoptotic members. The latter can be further sub-divided into the BH3-only (BIM, PUMA, BID, BAD, NOXA, HRK, BMF, BIK) and the multi-BH domain members (BAX, BAK and possibly BOK).2 Pro-survival BCL-2 family members protect cells from dying through binding and neutralizing the pro-apoptotic BCL-2 family members. Upon an apoptotic stimulus, the levels of certain BH3-only proteins increase as a result of transcriptional and/or post-transcriptional upregulation. These BH3-only proteins activate the multi-domain members BAX and BAK either directly, or indirectly through neutralizing the pro-survival BCL-2 family members.3,4 Activated BAX and BAK oligomerize and form pores into the outer mitochondrial membrane, leading to the release of apoptogenic factors, such as cytochrome c, provoking the activation of the so-called caspase cascade with subsequent demolition of the cell.3
BIM is a critical initiator of the mitochondrial apoptotic pathway, particularly in hematopoietic cells.5 During B- and T-cell development, BIM activity is required to eliminate autoreactive lymphocytes.6,7 Constitutive loss of BIM (in all cell types) leads to the accumulation of lymphocytes that infiltrate non-hematopoietic organs, such as lungs, kidneys, liver and salivary glands, produce autoantibodies and on a mixed C57BL/6x129SV background this causes severe autoimmune disease resembling systemic lupus erythematosus.5 BIM is also an important factor in peripheral T-cell apoptosis during the shutdown of an immune response.8,9 BIM is a tumor suppressor in mantle cell lymphoma, where the gene is lost,10 as well as Burkitt's lymphoma and renal carcinoma in which the gene is silenced.11,12 Loss of BIM renders cells resistant to several pro-apoptotic stimuli, such as withdrawal of growth factors, treatment with calcium ionophores5 or ER stress.13
This information about BIM's physiological function was derived from the study of a mouse strain in which the Bim gene was constitutively inactivated in all cell types.5 The complete absence of a gene product during the entire life of an animal can lead to phenotypes that are not cell-autonomous and may also lead to compensatory events (for example, upregulation of genes with overlapping function). Therefore techniques have been developed so that a gene of interest can be conditionally inactivated in a tissue-specific and/or temporally controllable manner using the Cre/Lox system.14 A further refinement of this technique consisted in engineering an inducible Cre recombinase by fusing it with a modified hormone-binding domain of the estrogen receptor (CreER).15 In the absence of 4-hydroxytamoxifen (4-OHT), the CreER protein is sequestered in an inactive state in the cytosol. Upon administration of 4-OHT, the ER domain of the CreER fusion protein changes conformation, prompting the translocation of the fusion protein into the nucleus where the recombinase can delete DNA sequences flanked by loxP sites.15
We developed a conditional Bim allele allowing for temporally and spatially controllable deletion of this critical apoptosis initiator. In addition, we developed a novel strain, which expresses the CreER fusion protein under the control of the pan-hematopoietic Vav promoter.16 We show that the deletion of Bim in the adult mouse through activation of the CreER recombinase, using the Vav-CreER or the ubiquitously expressed Rosa-CreER transgene, caused hematopoietic abnormalities that were similar to those found in the constitutive Bim knockout animals. These novel strains (conditional Bim ko and Vav-CreER) will allow further detailed genetic investigations of cell death and tumourigenesis.
Results
BIM expression and phenotype in mice with floxed Bim alleles before and after Cre-mediated recombination
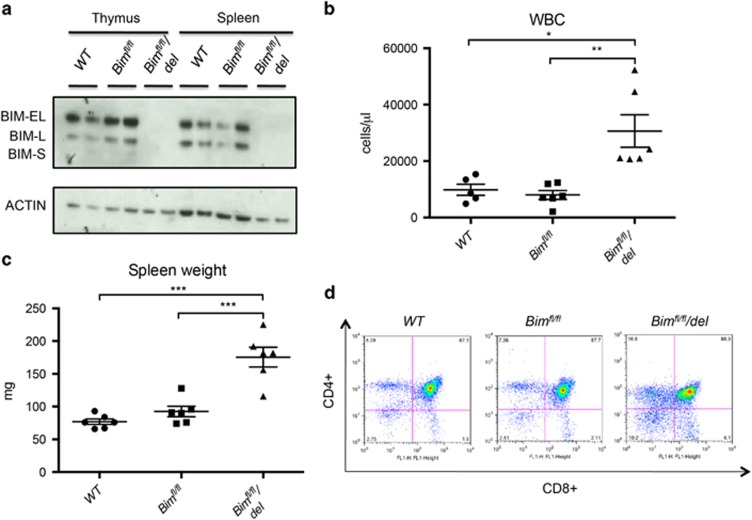
The BH3-only protein BIM is the most critical initiator of apoptosis in hematopoietic cell development and homeostasis.5 Mice constitutively deficient for BIM show many abnormalities, including increased white blood cell (WBC) counts, splenomegaly and defects in thymic T-cell selection.6 As these defects could potentially be affected by the absence of BIM (i) in non-hematopoietic cells or (ii) be dependent on absence of BIM during embryonic development, we decided to generate mice with a conditional Bim allele (Bimfl), to allow specific deletion of Bim in a time- and/or tissue-specific manner. Coding exons 2, 3 and 4 of Bim17 were flanked by loxP sites.18 As expected, BIM protein expression and hematopoietic cell composition, WBC counts, spleen weights and thymic cell subset distribution, were comparable between Bimfl/fl and wt mice (Figure 1). Crossing Bimfl/fl mice with the CMV-Cre deleter strain19 resulted in the complete loss of BIM protein (Bimfl/fl/del; in this strain, Cre is active in the early embryo) (Figure 1a) and the concomitant increase of WBC counts (Figure 1b) and spleen weights (Figure 1c), as well as the altered distribution of thymocyte populations (Figure 1d). These results demonstrate that (i) the loxP sequences do not alter the expression of the BIM protein or its function and (ii) that deletion of the floxed Bim allele recapitulates the phenotype observed in the constitutive Bim knockout mice.
Figure 1.
Influence and deletion of a conditional Bimfl allele in mice. (a) Lysates from splenocytes from wt, floxed Bim (Bimfl/fl) and BIM-deficient mice (Bimfl/fl/del) were immunoblotted for BIM. Probing for β-actin served as a loading control. N=2 mice per genotype were tested, six independent experiments were performed. (b) White blood cell counts and (c) spleen weights from wt, Bimfl/fl and Bimfl/fl/del mice were determined. N=5–6 mice for each genotype. Data represent mean±S.E.M. *P<0.05, **P<0.01, ***P<0.001 (paired t-test). (d) Thymocytes from mice of the indicated genotypes were isolated and single-cell suspensions stained with anti-CD4-PE (clone# YTA3.2.1) and anti-CD8-FITC (clone# 53.6.7.2). Dead cells were excluded from analysis by staining with 10 μg/ml propidium iodide (PI; Sigma-Aldrich)
Induced deletion of Bim in adult mice results in phenotypic alterations similar to those observed in constitutive Bim knockout mice
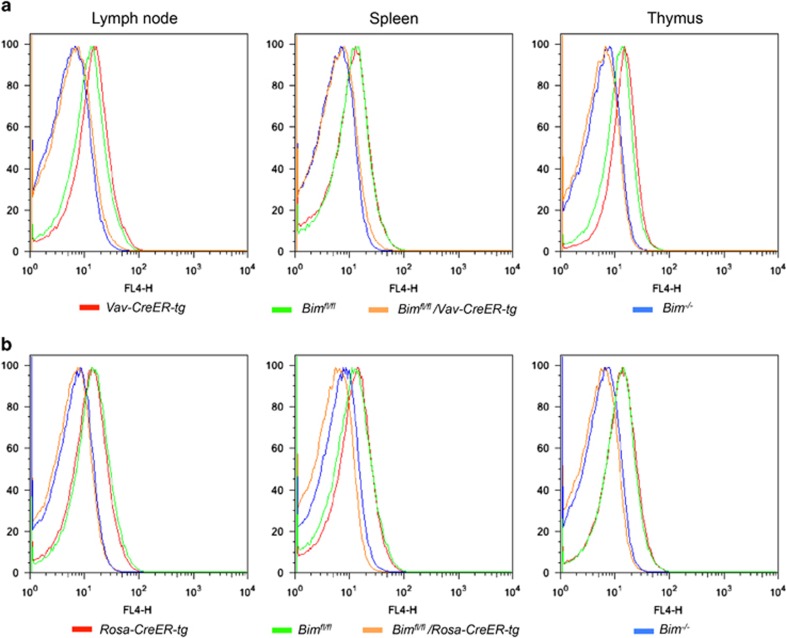
The advantage of a conditional Bim allele is the possibility to delete the gene in a temporally and cell type-controllable manner. In order to delete Bim at a predetermined time specifically in hematopoietic cells of adult mice, we generated a new transgenic mouse model, in which the tamoxifen-inducible CreERT2 recombinase20 is expressed under the control of the pan-hematopoietic Vav promoter (Vav-CreER).16 To induce Bim deletion, 12–20-week-old Bimfl/fl/Vav-CreER-tg mice were administered 4-OHT by oral gavage. Four weeks after the treatment, lymph nodes, spleens and thymi were collected from 4-OHT-treated Bimfl/fl, Vav-CreER, Bimfl/fl/Vav-CreER as well as Bim−/− mice, and BIM protein levels were measured by intracellular FACS analysis (Figure 2a). As expected, the BIM protein levels in cells from 4-OHT-treated Bimfl/fl/Vav-CreER mice were comparable to those seen in cells from Bim−/− mice, whereas the presence of the floxed Bim (Bimfl/fl) alleles or CreER (Vav-CreER) transgene had no impact on BIM protein expression when CreERT2 was not activated. This indicated that both Bimfl alleles had been successfully recombined in Bimfl/fl/Vav-CreER hematopoietic cells of tamoxifen-treated mice, explaining why the BIM protein was no longer present in these cells. To further validate the efficiency of the Vav-CreER strain, we crossed this strain to mice harboring a floxed allele of Mcl-1 (Mcl1fl) and treated Mcl-1fl/+/Vav-CreER-tg animals with three doses of 4-OHT. As the deletion of the floxed Mcl-1 allele leads to the expression of the hCD4 reporter,21,22 we analyzed the blood of Mcl-1fl/+/Vav-CreER-tg two days post treatment by flow cytometry for hCD4 expression (Supplementary Figure 1). This revealed that ~20% of cells had recombined the floxed Mcl-1 allele within 1 day, validating the utility of the Vav-CreER strain.
Figure 2.
Induced Bimfl deletion in adult mice. (a) Bimfl/fl/Vav-CreER-tg and control mice were treated for 5 days and (b) Bimfl/fl/Rosa-CreER-tg and control mice were treated three times within 5 days (1 day break in between dosing) with a daily dose of 4.8 mg Tamoxifen (4-OHT; Sigma) by oral gavage and then left untreated for 1 month or 3 weeks, respectively. Each FACS plot is representative of N>3 repeat experiments
We also crossed the Bimfl/fl mice with the Rosa-CreER transgenic strain, in which the ubiquitously expressed CreER protein causes the deletion of Bim in the entire organism upon 4-OHT treatment.23 Three to four weeks after 4-OHT treatment, BIM protein levels were examined by intracellular FACS analysis. Cells from the lymph nodes, thymus and spleen of Bimfl/fl/Rosa-CreER-tg and Bim−/− mice showed complete absence of the BIM protein, whereas cells from Bimfl/fl or Bim+/+/Rosa-CreER-tg mice expressed similar levels of BIM protein as the corresponding cells from wt mice (Figure 2b). As in the Rosa-CreER-tg mice, the CreER transgene is ubiquitously expressed, we also analyzed the deletion of the BIM protein in non-haematopoietic tissues (liver, kidney) of 4-OHT-treated Bimfl/fl/Rosa-CreER-tg mice. As expected, BIM protein was almost undetectable in these organs, whereas treatment of Rosa-CreER-tg mice with 4-OHT had no impact on BIM protein levels (Supplementary Figure 2).
These results demonstrate that recombination of the floxed Bim locus can be successfully achieved in the adult mouse using two different 4-OHT-inducible Cre strains (Vav-CreER and Rosa-CreER).
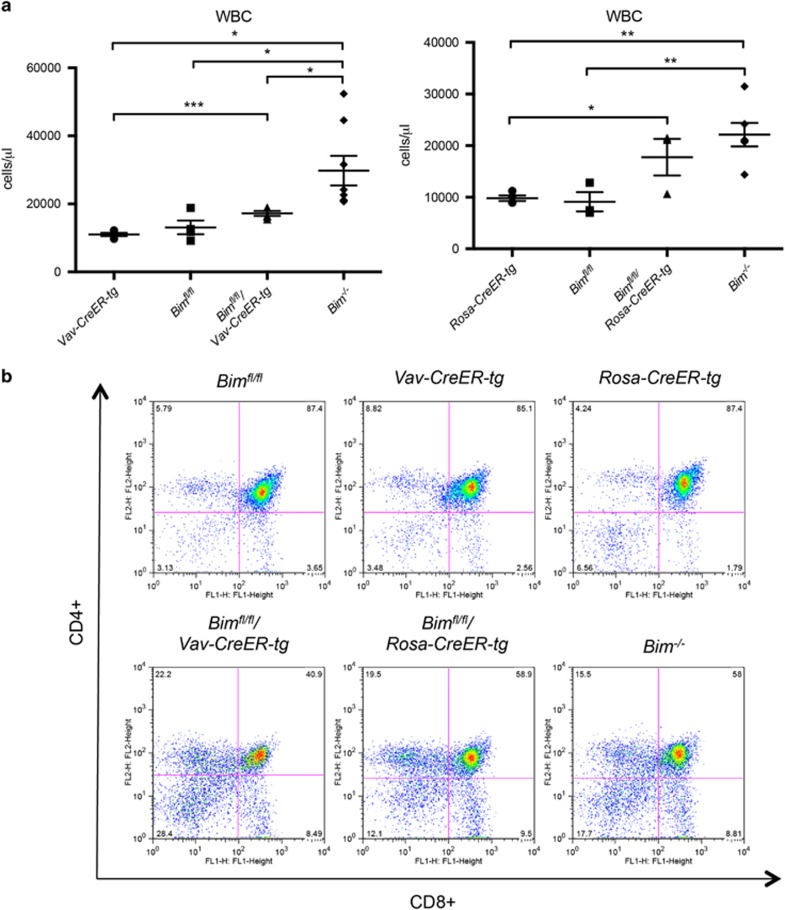
As loss of BIM in the entire mouse leads to increased WBC numbers,5 we analyzed the blood from Bimfl/fl/Vav-CreER-tg and Bimfl/fl/Rosa-CreER-tg mice 3–4 weeks after 4-OHT treatment (Figure 3). As anticipated, induced loss of BIM caused an increase in WBC numbers in both Bimfl/fl/Vav-CreER-tg and Bimfl/fl/Rosa-CreER-tg animals, albeit to different levels (Figure 3a). The larger increase observed in the Bimfl/fl/Rosa-CreER-tg animals probably reflects a faster deletion of Bimfl alleles in these animals. 4-OHT-treated Bimfl/fl/Vav-CreER-tg and Bimfl/fl/Rosa-CreER-tg mice also showed similarly abnormal distribution of thymocyte subpopulations, comparable to what is seen in Bim−/− mice. This is characterised by abnormally high frequencies of double-negative (CD4−CD8−) and single-positive (CD4+CD8−, CD4−CD8+) thymocytes and reduced proportions of double-positive (CD4+CD8+) thymocytes compared with control (wt) mice (Figure 3b). This indicates that, although Bimfl deletion might occur at a slower rate in peripheral lymphoid organs of Bimfl/fl/Vav-CreER-tg mice compared with the Bimfl/fl/Rosa-CreER-tg animals, Bimfl recombination in thymocytes (or their precursors) appears to occur at similar rates in both strains.
Figure 3.
Induced deletion of Bimfl in mature animals leads to increased WBC numbers and abnormalities in the composition of thymocyte subpopulations. Blood from (a) Bimfl/fl/Vav-CreER-tg and control mice as well as from (b) Bimfl/fl/Rosa-CreER-tg and control mice was analyzed with an ADVIA blood analyzer after the animals had been treated with 4-OHT, as described in Figure 2. (c) Thymocytes of mice of the indicated genotypes from a and b were stained with anti-CD4-PE and anti-CD8-FITC and analyzed by flow cytometry. N=4–8 mice for each genotype. Data represent mean±S.E.M. *P<0.05, **P<0.01, ***P<0.001 (paired t-test)
Induced deletion of Bim in the adult mouse protects thymocytes from BIM-dependent apoptotic stimuli
BIM-deficient thymocytes are resistant to a variety of pro-apoptotic stimuli.5 We therefore compared the response of thymocytes from Bim−/−, Bimfl/fl/Vav-CreER-tg, Bimfl/fl/Rosa-CreER-tg, Bimfl/fl, Vav-CreER and Rosa-CreER mice, which had been treated with 4-OHT four weeks prior to organ isolation, to diverse cytotoxic stimuli. Thymocytes of the different genotypes were cultured in medium (medium; mimicking cytokine withdrawal) or treated with BIM-dependent (Ionomycin=Iono) and BIM-independent (phorbol ester=PMA) apoptotic stimuli (Figure 4). Although thymocytes from mice of all genotypes were killed at a similar rate when treated with PMA (kills in a PUMA-dependent manner24), untreated (medium) and Ionomycin-treated thymocytes from 4-OHT-treated Bim−/−, Bimfl/fl/Vav-CreER-tg and Bimfl/fl/Rosa-CreER-tg mice all showed a marked survival advantage when compared with thymocytes from 4-OHT-treated Bimfl/fl, Rosa-CreER-tg and Vav-CreER-tg mice. This demonstrates that constitutive or induced deletion of Bim in thymocytes leads to a similar resistance to these apoptotic stimuli, and that the observed change in thymocyte distribution is a consequence of the loss of BIM and not due to other abnormalities caused by Cre-mediated recombination.
Figure 4.
Induced deletion of Bimfl in adult animals renders thymocytes resistant to BIM-dependent apoptotic stimuli. Thymocytes from 4-OHT-treated Bimfl/fl/Vav-CreER-tg, Bimfl/fl/Rosa-CreER-tg and control mice were cultured (a) in medium only, or were (b) treated with 1 μg/ml Ionomycin (Sigma) or (c) with 2 ng/ml PMA (Sigma). At the indicated time points, the percentages of live cells were determined by staining with AnnexinV/PI (AnnexinV-PI=living cells). N=3–7 mice for each genotype. Data represent mean±S.E.M. *P<0.05, **P<0.01, ***P<0.001 (paired t-test)
Discussion
We report here the development of a new floxed Bim allele, which allows the deletion of this pro-apoptotic BH3-only protein in a cell type-restricted and temporally controllable manner. Using the well-established Rosa-CreER and newly developed Vav-CreER transgenic strains, deletion of Bim in the adult mice results in the same phenotype as that observed in the constitutive Bim knockout mice. It thus appears that the hematopoietic phenotype associated with the constitutive loss of BIM in all cell types reported previously5 is intrinsic to the hematopoietic system rather than a consequence of an unrecognized developmental defect due to the absence of BIM.
Our studies clearly show that the floxed Bim allele is functional and can be efficiently deleted by CreER recombinases. Importantly, these Bimfl/fl mice have recently been used in two other studies,18,25 in which Bim was specifically deleted in the regulatory T-cell lineage (Treg) only. This new mouse strain will thus be a valuable tool to further dissect the role of BIM in various cell types.
Importantly, our new Vav-CreER transgenic strain showed the same deletion efficiency of the floxed Bim alleles 4 weeks after 4-OHT treatment as the very well-characterised and widely used Rosa-CreER strain.23 However, when we tested the loss of BIM protein 5 days after 4-OHT treatment, we observed residual BIM protein in the lymphoid cells from the Bimfl/fl/Vav-CreER mice, whereas it was almost completely absent in the cells from the Bimfl/fl/Rosa-CreER animals at this time point (data not shown). This indicates that deletion of Bimfl alleles occurs more rapidly and probably also more efficiently in the Bimfl/fl/Rosa-CreER strain than in the Bimfl/fl/Vav-CreER strain, possibly because of the lower expression of CreER in the Vav-CreER strain.
However, the higher efficiency of Cre-mediated recombination of floxed target genes in the Rosa-CreER strain is accompanied by a higher toxicity observed in the animals upon treatment with 4-OHT. Accordingly, treatment of Rosa-CreER or Vav-CreER transgenic mice with 4-OHT for 5 consecutive days led to substantial destruction of lymphoid organs in the former, whereas these organs remained unaffected in the latter (Supplementary Figure 3 and data not shown). Both CreER strains are highly efficient at recombining the floxed Bim allele and, depending on the experimental requirements, are valuable tools for induced deletion of floxed alleles in mature or developing animals.
A significant leakage in CreER recombinase activity was observed in the Bimfl/fl/Rosa26-CreER strain, as tail DNA obtained at weaning showed evidence of Cre-mediated deletion of the floxed Bim allele in ~20% of these animals (data not shown). This was never observed in Bimfl/fl/Vav-CreER-tg mice. Note that only mice with no evidence of Bimfl recombination prior to tamoxifen administration were used in this study. Pertinently, we observed no abnormalities in splenic weights and thymocyte sub-population distribution in Bimfl/fl/Vav-CreER-tg and Bimfl/fl/Rosa-CreER-tg animals prior to 4-OHT treatment (data not shown). We therefore conclude that the occasional leakiness of CreER recombinase activity in the Rosa-CreER-tg mice must be limited to embryonic development and that despite this issue (which can be identified by tail DNA analysis), both this inducible CreER mouse strain and the Vav-CreER-tg strain are suitable tools for the inducible deletion of floxed genes.
Materials and Methods
Mice
Experiments with mice were conducted according to the guidelines of The Walter and Eliza Hall Institute Animal Ethics Committee. The generation of the conditional Bimfl, Mcl-1fl and the RosaCreER mice, all on a C57BL/6 background, has been described previously.18,21,26 Vav-CreER transgenic mice were generated by replacing the hCD4 sequence of the Vav hematopoietic vector27 with the CreERT220 sequence using Sfi1/Not1 restriction sites. The Vav-CreER construct was linearized with HindIII before pro-nuclear injection of the DNA into zygotes derived from C57BL/6 mice. Positive offspring were identified by PCR for the genomic integration of the Vav-CreER transgene.
To activate the latent CreER recombinase, mice were given 200 mg/kg tamoxifen (Sigma-Aldrich, Rowville, VIC, Australia) in peanut oil/10% ethanol each day for 5 days by oral gavage.28
Western blotting and WBC analysis
Cell extracts for western blot analysis were prepared in ONYX lysis buffer (20 mM Tris-HCl pH 7.4, 135 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10% glycerol). Antibodies used include: rat anti-BIM (clones 3C5 and CF7, ENZO Life Sciences,29 Waterloo, NSW, Australia), mouse anti-β-actin (Sigma AC-40, Rowville, VIC, Australia). Horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rat IgG antibodies (both from Southern Biotech, Birmingham, AL, USA) served as secondary reagents and the enhanced chemoluminescence (ECL; GE Healthcare, Rydalmere, NSW, Australia) system was used for detection. WBC analysis was performed with an ADVIA blood analyzer (Siemens Healthcare Diagnostics, Tarrytown, NY, USA).
Intracellular immunofluorescent staining of BIM for flow cytometric analysis
Cells (1 × 106) from lymph nodes, spleen and thymus were isolated from mice of the indicated genotypes, fixed and permeabilized by suspension in 100 μl of BD Cytofix/Cytoperm (BD Biosciences, San Jose, CA, USA) solution for 20 min on ice. Cells were then washed twice in Perm/Wash buffer (BD Biosciences) and stained with Alexa-647-conjugated BIM antibody (rat, clone # 3C5,29 ENZO Life Sciences). Analysis was performed in a FACSalibur (BD Biosciences).
Statistical analysis
Statistical comparisons were made using a two-tailed Student's t-test with Prism v.5.0 (GraphPad Software, Inc., La Jolla, CA, USA). P-values <0.05 were considered to indicate a statistically significant difference. n indicates the number of mice analyzed for each genotype.
Acknowledgments
We thank M Robati for technical assistance, Dr LA O'Reilly for sharing reagents and expertise, G Siciliano and S O'Connor for mouse care, J Corbin for automated blood analysis, B Helbert and C Young for genotyping. This work was supported by the Australian NHMRC (Program Grant 461221, Independent Research Institutes Infrastructure Support Scheme Grant 361646, Research Fellowship 1042629 and Project Grant 1049720 (MJH)), the Leukemia and Lymphoma Society (Specialised Center of Research Grant 7015), and infrastructure support from the NHMRC (IRISS) and the Victorian State Government (OIS).
Glossary
- 4-OHT
4-hydroxytamoxifen
- WBC
white blood cells
- PMA
phorbol ester
- Iono
ionomycin
- Treg
regulatory T cells
- Tg
transgenic
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- 1Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2013; 15: 49–63. [DOI] [PubMed] [Google Scholar]
- 2Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 2008; 9: 47–59. [DOI] [PubMed] [Google Scholar]
- 3Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene 2008; 27: S128–S136. [DOI] [PubMed] [Google Scholar]
- 4Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O'Reilly LA et al. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol 2009; 186: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Köntgen F et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 1999; 286: 1735–1738. [DOI] [PubMed] [Google Scholar]
- 6Bouillet P, Purton JF, Godfrey DI, Zhang L-C, Coultas L, Puthalakath H et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature 2002; 415: 922–926. [DOI] [PubMed] [Google Scholar]
- 7Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreative B cells. J Exp Med 2003; 198: 1119–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Pellegrini M, Belz G, Bouillet P, Strasser A. Shut down of an acute T cell immune response to viral infection is mediated by the pro-apoptotic Bcl-2 homology 3-only protein Bim. Proc Natl Acad Sci USA 2003; 100: 14175–14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Hughes PD, Belz GT, Fortner K, Budd RC, Strasser A, Bouillet P. Apoptosis regulators Fas and Bim cooperate in shutdown of chronic immune responses and prevention of autoimmunity. Immunity 2008; 28: 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 2005; 24: 1348–1358. [DOI] [PubMed] [Google Scholar]
- 11Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood 2010; 116: 2531–2542. [DOI] [PubMed] [Google Scholar]
- 12Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S et al. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene 2007; 26: 7038–7048. [DOI] [PubMed] [Google Scholar]
- 13Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell 2007; 129: 1337–1349. [DOI] [PubMed] [Google Scholar]
- 14Schwenk F, Kuhn R, Angrand PO, Rajewsky K, Stewart AF. Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res 1998; 26: 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA 1996; 93: 10887–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Ogilvy S, Elefanty AG, Visvader J, Bath ML, Harris AW, Adams JM. Transcriptional regulation of vav, a gene expressed throughout the hematopoietic compartment. Blood 1998; 91: 419–430. [PubMed] [Google Scholar]
- 17Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P et al. Gene structure alternative splicing, and chromosomal localization of pro-apoptotic Bcl-2 relative Bim. Mamm Genome. 2001; 12: 163–168. [DOI] [PubMed] [Google Scholar]
- 18Raynor J, Sholl A, Plas DR, Bouillet P, Chougnet CA, Hildeman DA. IL-15 fosters age-driven regulatory T cell accrual in the face of declining IL-2 levels. Front Immunol 2013; 4: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995; 23: 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res 1999; 27: 4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Glaser S, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD et al. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Gene Dev 2012; 26: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Kelly GL, Grabow S, Glaser SP, Fitzsimmons L, Aubrey BJ, Okamoto T et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Gene Dev 2014; 28: 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 1999; 21: 70–71. [DOI] [PubMed] [Google Scholar]
- 24Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 2003; 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- 25Pierson W, Cauwe B, Policheni A, Schlenner SM, Franckaert D, Berges J et al. Antiapoptotic Mcl-1 is critical for the survival and niche-filling capacity of Foxp3(+) regulatory T cells. Nat Immunol 2013; 14: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res 2003; 31: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Ogilvy S, Metcalf D, Gibson L, Bath ML, Harris AW, Adams JM. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood 1999; 94: 1855–1863. [PubMed] [Google Scholar]
- 28Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol 2010; 477: 109–123. [DOI] [PubMed] [Google Scholar]
- 29O'Reilly LA, Kruse EA, Puthalakath H, Kelly PN, Kaufmann T, Huang DC et al. MEK/ERK-mediated phosphorylation of Bim is required to ensure survival of T and B lymphocytes during mitogenic stimulation. J Immunol 2009; 183: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.