Abstract
Loss of imprinting (LOI) is an epigenetic alteration involving loss of parental origin-specific expression at normally imprinted genes. A LOI for Igf2, a paracrine growth factor, is important in cancer progression. Epigenetic modifications may be altered by environmental factors. However, is not known whether changes in imprinting occur with aging in prostate and other tissues susceptible to cancer development. We found a LOI for Igf2 occurs specifically in the mouse prostate associated with increased Igf2 expression during aging. In older animals, expression of the chromatin insulator protein CTCF and its binding to the Igf2-H19 imprint control region was reduced. Forced down-regulation of CTCF leads to Igf2 LOI. We further show that Igf2 LOI occurs with aging in histologically normal human prostate tissues and that this epigenetic alteration was more extensive in men with associated cancer. This finding may contribute to a postulated field of cancer susceptibility that occurs with aging. Moreover, Igf2 LOI may serve as a marker for the presence of prostate cancer.
Introduction
Our understanding of how aging predisposes to the frequent development of prostate and other major cancers is limited. Histologic prostate cancer is found in over 60% of men in their 70s at autopsy (1). DNA damage accumulates in aging human prostate tissues that results from multiple exogenous and endogenous stressors (2). Conversely, it has been proposed that the modification of epigenetic factors, defined as heritable changes in information passed during cellular replication that do not involve altered DNA sequence, underlie the gene expression changes that occur with aging (3). Differences in DNA methylation have been shown when identical twins are compared in older age (4). In the colon, and recently in the prostate, DNA hypermethylation with increasing age has been found within various genes (5, 6). Epigenetic modifications, including histone/chromatin modifications and genomic imprinting, seem uniquely sensitive to alterations in the cellular and organismal environment (7, 8). Degradation of these epigenetic patterns may provide insight into the susceptibility of cancer that develops with aging.
One manifestation of epigenetic modification is genomic imprinting, or the allele-specific expression of a gene based on its parental origin. The insulin-like growth factor 2 (Igf2) gene and its closely linked 3′ neighbor, H19, display imprinting and are expressed solely from either the paternal or maternal allele, respectively (9). The Igf2-H19 imprinted cluster shares enhancers, as well as an intergenic imprint control region (ICR) that coordinates gene expression (10). A major element in the control of Igf2 imprinting at the ICR is the presence and binding of CTCF, a well-characterized chromatin insulator that is required for repression of the maternal Igf2 allele (11). CpG methylation at this ICR blocks the binding of CTCF to DNA and allows the downstream enhancer to stimulate Igf2 promoter activity across the inert boundary site. Deletion or hypermethylation of this ICR prevents CTCF binding and results in a reactivation of the maternal allele and the biallelic expression of Igf2 (12, 13). Strict imprinting is generally maintained in normal adult tissues. However, the loss of imprinting (LOI) at Igf2, a potent autocrine and paracrine growth stimulator, seems to provide an important early switch in the progression of neoplasia (14, 15). This has lead to the proposition that an epigenetic loss of allelic silencing plays a role in cancer progression. In the Apc+/Min mouse, animals displaying LOI developed twice as many intestinal tumors as did control littermates (16). Given the plasticity of epigenetic controls, we examined whether imprinting of the Igf2 gene undergoes alteration during aging in the prostate, an organ susceptible to cancer development.
Materials and Methods
Mice and diet
B6 (cast H19-p57) mice, a congenic strain heterozygous for distal chromosome 7 sequences from C57BL/6 and Mus castaneus, were obtained from Dr. Shirley Tilghman (Princeton University, Princeton, NJ). Male mice homozygous for Mus castaneus alleles (H19-p57) were bred with female C57BL/6 and male mice from each litter entered randomly into aging groups. The animals were individually housed in the Veteran’s Administration and University of Wisconsin Shared Aging Rodent Facility and fed on an 84 kcal/wk diet (TD91349 [Teklad]), which is ~ 10% less than the average ad libitum intake, to prevent obesity and maintain health. Ten animals per time point were euthanized at intervals (every 8 mo) beginning at ages 3 mo. Tissues were microdissected, including the coagulating glands, dorsolateral prostate (DLP), and ventral prostate (VP), and placed in RNase-free PBS and snap frozen in liquid nitrogen. Nonprostate tissues were treated identically.
cDNA preparation and quantitative PCR
Total RNA was extracted from frozen tissues using the RNeasy Mini kit (Qiagen) as described by the manufacturer. Total RNA was treated with DNaseI before reverse transcription. cDNA was made from 1 μg of total RNA, and oligo (dT), using Omniscript RT reagents (Qiagen) per the manufacturer’s protocol. Reverse transcription without reverse transcriptase was carried out to detect genomic DNA contamination. Quantitative PCR (QPCR) was performed with SYBR Green (Applied Biosystems) on a MyiQ QPCR machine (Bio-Rad; ref. 17). Both cyclophilin and GAPDH were used as internal controls and neither showed altered expression with aging (data not shown). Primer sequences for Igf2, H19, p57, CTCF, GAPDH, and cyclophilin are available on request.
Fluorescent primer extension assay
Primer extension assays were performed as previously described (18). A single nucleotide polymorphism was identified on Igf2 exon 6 (G/A) and used to identified individual alleles. A 700-bp intron spanning PCR product was generated from cDNA using primer sequences CTCTCAGGCCGTACTTCCGGAC (forward) and GCGCCGAATTAGTTGATTT (reverse). ExoSAP-IT (U.S. Biochemical) was used to remove excess deoxynucleotide triphosphates (dNTP) and primers from the PCR reaction. A FAM-labeled fluorescent primer extension assay (FluPE) primer was generated (5′-ACCATCGGGCAAGGGGATCTCAGC). One hudred nanograms of PCR product were added to 10 nmol/L of FAM-labeled primer, 0.75 U of HotStarTaq Polymerase (Qiagen), 200 μmol/L of each specific dNTP/ddNTP, 5% of DMSO, and 5 mmol/L of magnesium in a 20 μL volume. Reactions were cycled at 95°C for 15 min, then 25 cycles of 95°C 30 s and 62°C 20 s. The combination of the FluPE primer and specific dNTP/ddNTPs (dATP, dTTP, ddCTP, and ddGTP) gives either a 2-bp or 5-bp extension for an imprinted allele or both if imprinting is no longer maintained. The reaction mix was loaded on ABI PRISM 377 DNA Sequencer, which performs electrophoresis, laser detection, and imaging. Quantitation of spectral emissions generated by laser-induced fluorescence of fluorophore-labeled C57BL/6 and castaneus amplicons was performed as described by Berres and colleagues (19). Differences in the relative concentration of C57BL/6 and castaneus alleles in each sample were determined by calculating the ratio of their respective spectral intensities [repressed allele (Ai)/active allele (Aa)]. For Igf2, mixing experiments using purified allelic templates determined background and confirmed the assay to be linear for the concentration of the products analyzed. Fluorescent primer extension was used to differentiate allelic expression with high sensitivity (<5 parts in 100) in the linear range.
DNA methylation analyses
Bisulfite treatment of DNA, cloning, and sequencing was performed as we have previously described (17). Primers used to amplify bisulfite treated DNA within the H19 ICR for cloning included CTCF 3 and CTCF 4 (available upon request). For each region, 10 to 20 colonies were picked and sequenced. Methylation-specific QPCR (MS-qPCR) was performed as described (20). Briefly, 100 ng of bisulfilte-treated DNA were placed in a QPCR reaction with primer pairs recognizing only methylated sequences at CTCF3, CTCF4, and MyoD (control; sequences available upon request). Bisulfite-treated mouse tail DNA was treated with SssI methylase and used to generate a standard curve to quantify the amount of fully methylated sequences in each reaction. The normalized index of methylation was defined as the ratio of the normalized amount of methylated template at the gene of interest to the normalized converted MyoD template (20). Samples were run in triplicate and the mean used for statistical comparison.
Chromatin immunoprecipitation assays
Fresh tissues were cross-linked by mincing in 1% Formaldehyde-PBS solution for 20 min. Tissue samples from 2 mice for each age group were pooled; washed in PBS; resuspended in 150 mmol/L NaCl, 10% glycerol, and 50 mmol/L Tris 8.0 (with 1:200 dilution of Sigma protease inhibitor cocktail); and homogenized using the Tissue Tearor (Biospec Products). Crosslinked chromatin was prepared, sonicated, and immunoprecipitated using 2 μg of anti-CTCF (Upstate), or normal rabbit IgG control antibody. After the reversal of crosslinks and DNA purification, QPCR was performed on samples in triplicate. Values are expressed as percent recovery compared with the input into the immunoprecipitation. Primers used included the following: CTCF 2 (Forward, ACGGCGGCAGTGAAGTCTC; Reverse, CAGTTGCAATCCGTTTCAGGA); CTCF 3 (Forward, CCCAAATGCTGCCAACTTG; Reverse, CTGGGATATTGCTGGGAATGA); CTCF 4 (Forward, GTCTTGCGCCCTTCACGAT; Reverse, AAATCTGCACAGCGTGGAGAG); and H19 (Forward, AGGCCCTGTCTAAGGATTCCA; Reverse, CCAAAGACAGCCTCACCACAA).
Immunofluorescence and quantitation
Frozen prostate tissues from mice were sectioned and immunostained using fluorescence-conjugated secondary antibodies (21). Briefly, slides were fixed in PBS containing 4% paraformaldehyde/0.2% Triton X-100/10 mmol/L NaF/1 mmol/L Na3VO4 at room temperature for 15 min. After washing, slides were blocked in PBS containing 0.1% bovine serum albumin/10% fetal bovine serum/0.2% Triton X-100/10 mmol/L NaF/1 mmol/L Na3VO4 at room temperature for 1 h. Rabbit polyclonal antibodies to IGF2 (sc-5622; Santa Cruz Biotechnology), CTCF (Upstate), or mouse monoclonal antibodies to α-tubulin (Ab-1; Oncogene Research Products) were diluted in blocking buffer at a 1:500 ratio (the linear range of detection) and incubated at 4°C overnight. Negative controls included no primary antibodies. After washing, Alexa 594–conjugated anti-rabbit (Invitrogen) and FITC-conjugated anti-mouse (Becton Dickson) secondary antibodies were diluted 1:2,000 in blocking buffer containing 10 μg/mL Hoechst 33342 (Molecular Probes) as a DNA counter stain and incubated at room temperature for 1 h. Sections were then washed twice in blocking buffer, aspirated, mounted beneath coverslips, and allowed to dry overnight (21).
Stained tissues were analyzed by quantitative fluorescence microscopy as described (21, 22). Using an Olympus BX51 microscope, digital images were acquired using a RT Color digital camera and ProSPOT Advanced software (Diagnostic Instruments, Inc.). Using identical camera settings, images from five different random fields were acquired per section and the integrated density of each whole single-color image was measured using NIH ImageJ. Each IGF2 measurement was normalized to that of α-tubulin, a constitutively expressed, cell-associated control.
siRNA transfection
Human prostate epithelial cells (HPEC) and mouse prostate epithelial cells were harvested and cultured as described (23). Cell lines were seeded to 50% confluence on collagen-coated 6-well plates 24 h before transfection. One hundred to 200 pmol of CTCF SMARTpool (DHARMACON, Inc) siRNAs (mouse or human, respectively) were combined in medium with Lipofectamine 2000 (Invitrogen Life Technologies) transfection reagent following the manufacture’s protocol. The mixture was then added dropwise to the cells in complete DMEM medium and mixed by gentle rocking. Cells were retreated with siRNAs 12 h after the initial transfection. RNA and protein were harvested at 48 h. Experiments were performed in duplicate with similar results.
Statistics
Expression ratios were expressed as mean ± SE and compared using Student’s t tests. These were felt to be appropriate because there were no observed differences in the variability between groups. We examined the influence of age and cancer on expression ratio using an ANOVA. P values of <0.05 were considered as significant. All analyses were performed using SAS statistical software version 9.1, SAS Institute, Inc.
Results
To test whether Igf2 undergoes a LOI during aging in the prostate, we determined the imprinting status of mouse prostate tissues using a sensitive primer extension assay. C57BL/6 mice containing a Mus castaneus Igf2-P57 locus were used to allow differentiation between the paternal and maternal alleles based on a polymorphism (G/A) within Igf2 exon 6. Mice have multiple prostate lobes. The DLP most closely corresponds to the human peripheral prostate, where prostate cancer regionally develops, based on anatomic and RNA signature analyses (24). We observed that DLP tissues from 3-month-old sexually mature mice mostly retain the imprinted status of Igf2 (Fig. 1A). Igf2 and its closely linked 3′ neighbor H19 display imprinting in adult tissues and are expressed solely from either the paternal or maternal allele respectively (9). In contrast, we found older cohorts developed progressively higher average expression from the once inactive Igf2 chromosome. To establish whether the marked LOI for Igf2 observed in the DLP of aging mice is unique to that prostate lobe, and to the prostate in general, we analyzed VP, liver, and kidney in aging C57BL/6 (Cast Igf2-P57) cohorts (Fig. 1B). A relaxation of imprinting was not observed in any of these tissues indicating that this age-associated LOI was lobe-specific for the mouse DLP.
Figure 1.
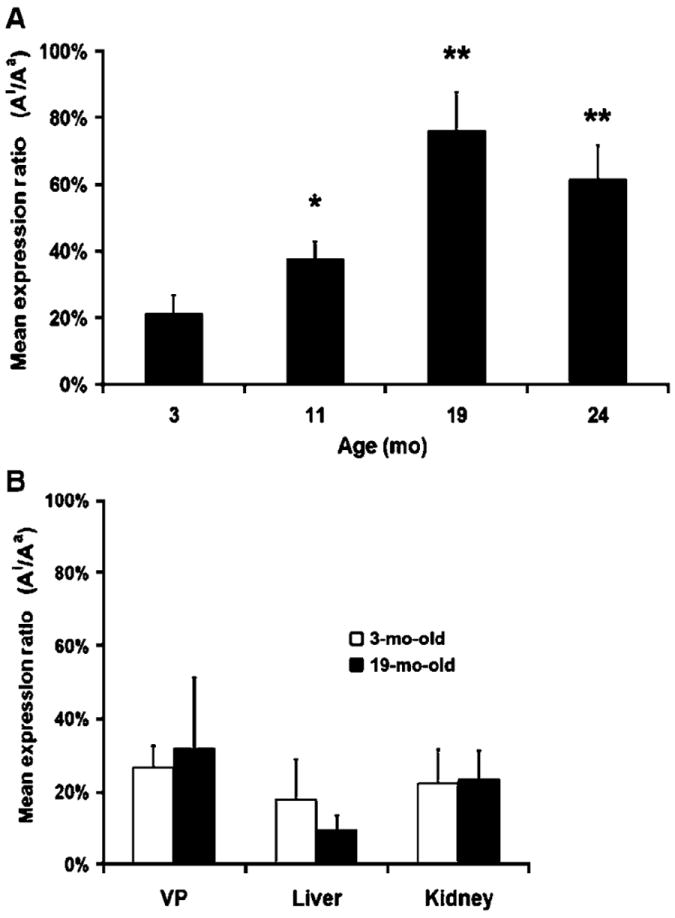
Re-expression of inactive Igf2 allele in aging mouse tissues. A, DLP tissues were microdissected from aging mice (3, 11, 19, and 24 mo) and the maternal and paternal allelic expression was measured using FluPE. The ratio of the inactive allele (Ai) to active allele (Aa) was calculated for each aging cohort (n = 6). All DLP samples in the 11-, 19-, and 24-month cohorts have significantly higher Ai/Aa ratios when compared with the 3-mo-old cohort (**, P < 0.01; *, P < 0.05). B, Igf2 imprinting is maintained with aging in the VP and other mouse tissues. Allelic expression from the liver, kidney, and VP was determined using FluPE. Tissues from 3- and 19-mo-old cohorts (n = 6) were compared and no age-associated relaxation of Igf2 imprinting was seen.
Relaxation of Igf2 imprinting has been linked to increases in Igf2 gene expression (12, 25). Igf2 functions as a potent autocrine and paracrine growth stimulator (15) and is important in the progression to neoplasia (14). We used qPCR initially to detect and quantify mRNA levels in the 3-, 11-, 19-, and 24-month cohorts. We observed age-associated increases in Igf2 mRNA levels in DLP tissues (Fig. 2A). H19 cDNA expression decreased and approached significance (P = 0.07) with aging. Both results are consistent with coordinate regulation from the H19 ICR (10). In the aging cohorts, Igf2 protein expression was evaluated in the DLP using quantitative immunofluorescence. DLP tissues from 24-month-old mice express significantly more Igf2 (30%; P = 0.01) when compared with 3-month-old mice (Fig. 2B). The prostate and other tissues examined did not show macroscopic morphologic changes or tumors (Supplementary Fig. S1A). However, histologic analysis of the DLP revealed both epithelial and stromal hyperplasia within the individual glands, as well as rare glandular atrophy. Inflammation, as assessed by CD45 a marker of lymphocytes (26), was minimally increased in older mouse prostate tissues on this moderate caloric restriction protocol used to engender longevity and health (Supplementary Fig. S1B).
Figure 2.
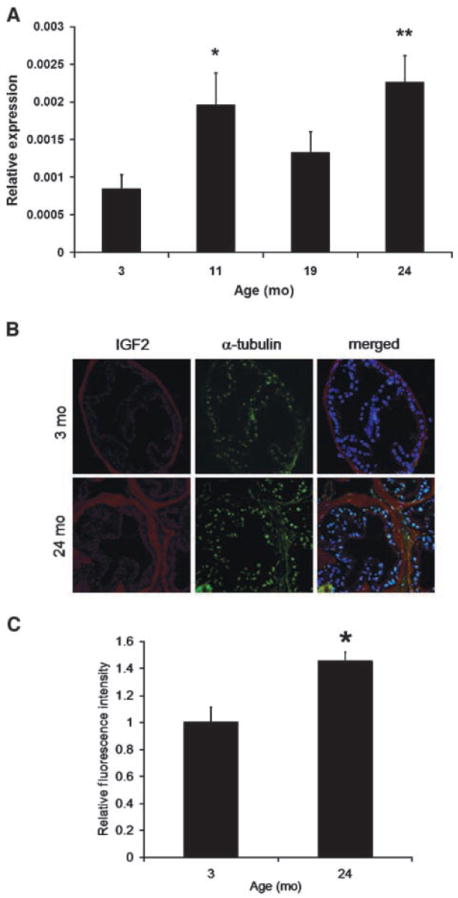
Igf2 expression increases in aging mouse DLP. A, QPCR was used to measure Igf2 expression levels in the mouse DLP of the 3-, 11-, 19-, and 24-mo-old cohorts (n = 6; **, P < 0.01; *, P < 0.05). B and C, mouse DLP sections were analyzed using immunofluorescence for Igf2 and α-tubulin (control). Images from five different random fields were acquired per section (n = 3) and the integrated density of each whole single-color image was measured with NIH ImageJ as described. Igf2 measurements were then normalized to that of α-tubulin. The older 24-mo cohort expresses significantly higher levels of Igf2 protein (P < 0.01) when compared with the 3-mo group.
To understand the mechanism involved in the relaxation of Igf2 imprinting seen in the prostate, we assessed CpG methylation at multiple CTCF binding sites within the H19 ICR (Fig. 3A). Previous studies in mice showed that methylation or deletion of the ICR results in re-expression of the maternal allele and biallelic Igf2 expression (12, 13). Utilizing MS-qPCR after bisulfite treatment of DNA, we found no significant change in methylation at CTCF binding sites between young (3 months) and old (24 months) DLP tissues. These results were additionally confirmed by sequencing after bisulfite treatment. Other regions, such as the H19 promoter, show increased methylation in the older cohort (28% ± 4% versus 38% ± 2%; P = 0.04) consistent with the hypothesis of aging-related regional DNA hypermethylation (6).
Figure 3.
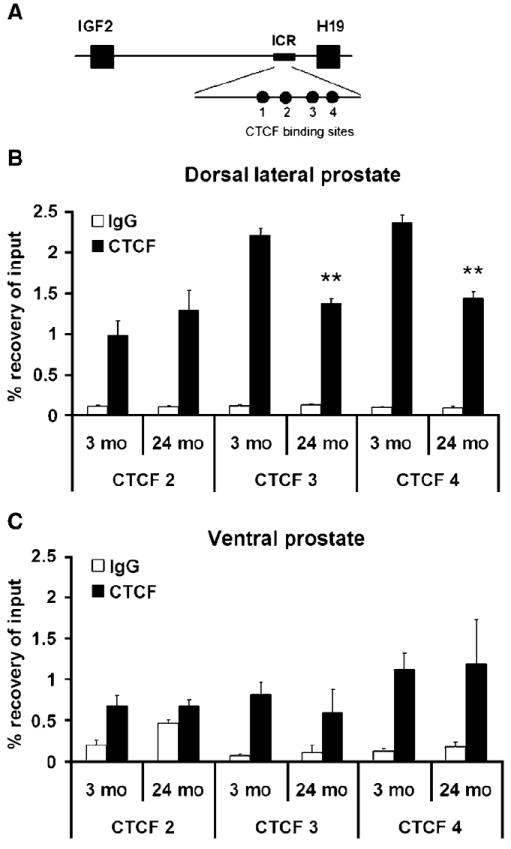
Decrease of CTCF binding in aging mouse DLP. A, schematic of the Igf2-H19 genomic region showing the ICR containing four CTCF binding sites. Methylation analysis using Methylation-sensitive PCR and bisulfite cloning of CTCF binding sites 3 (63% ± 6% versus 52% ± 3%; P = 0.16) and 4 (58% ± 5% versus 47% ± 5%; P = 0.26) did not show any differences when young and old animals were compared (n = 5). CTCF 3 and 4 represent the major target sites for CTCF binding in this region, whereas CTCF 2 does not display nuclease hypersensitivity. B, ChIP assay was performed to examine CTCF protein binding alterations at CTCF binding sites 2, 3, and 4 within the H19 ICR in aging mouse DLP. A significant decrease in binding is noted in older (24 mo) versus younger (3 mo) cohorts at both CTCF 4 and CTCF 3 (**, P < 0.01; n = 6). No alteration in CTCF 2 binding (a site that does not regulate Igf2 promoter activity) is seen. C, no significant change in CTCF binding is seen when the older cohort is compared with younger in the VP, a tissue found to retain imprinting. Binding to IgG was used as a nonspecific control for the experiments.
We next examined the extent to which CTCF protein binding is altered by aging at binding sites 2, 3, and 4 within the H19 ICR in DLP tissues. CTCF acts as an insulator by binding to the H19 ICR and preventing enhancer binding to and expression of the Igf2 promoter (10). Using chromatin immunoprecipitation (ChIP) assays, a significant age-associated decrease in binding was observed at both CTCF 3 (41%; P = 0.01) and CTCF 4 (40%; P = 0.01; Fig. 3B). No alteration in binding was seen at CTCF 2, a site that does not regulate Igf2 promoter activity (27). In VP tissue from the same animals, which does not show an age-related LOI, no alteration in CTCF binding was found (Fig. 3C).
This decrease in CTCF binding to the ICR directed us to inquire whether CTCF expression levels change in mouse DLP tissues with aging and whether they modulate Igf2 imprinting. Expression analyses showed an age-associated decrease of CTCF mRNA as well as protein levels in mouse DLPs (Supplementary Fig. S2A). To determine the effect of CTCF expression on Igf2 imprinting, we knocked down CTCF levels in mouse prostate epithelial cells cultured from the DLP, and HPECs using siRNA. CTCF protein expression was reduced 30% to 50% in transfected cell lines using pooled CTCF siRNAs specific for the human or mouse gene (Supplementary Fig. S2B). After 48 h, no morphologic changes were noted in the transfected cells. QPCR showed a ~30% to 70% silencing of CTCF RNA expression in siRNA-transfected mouse and human cultures when compared with controls (data not shown). Increased expression (Ai/Ae = 8–18%) of the silenced/imprinted Igf2 allele was reproducibly shown after siRNA transfection using FluPE (Supplementary Fig. S2). Hence, a loss of CTCF expression and binding to the H19 ICR resulted in biallelic Igf2 expression. To understand better the implications of this prostate-specific epigenetic change, we next focused on determining whether Igf2 imprinting alterations occur in the peripheral prostate of aging men.
Prostate tissues from men ages 17 to 81 years old with and without associated prostate cancer were analyzed. Given the predilection of human prostate cancer for the peripheral zone, we focused our analysis on this region. Using FluPE, a polymorphism (G to C) within the Igf2 coding sequence was used to quantitate allele-specific expression. Overall, a significant trend toward LOI with aging was seen when all samples were included and statistically analyzed in a linear fashion (P = 0.02; Fig. 4A). We then analyzed only those samples without associated prostate cancer. The imprint status of peripheral prostate tissue from a cohort of older men (55–81 years; mean, 64 years) without cancer (Ai/Aa = 28%) was suggestively more relaxed than from younger (18–40 years; mean, 27 years) patients (Ai/Aa = 24%; P = 0.2). When the allelic expression ratios were compared for older men with associated cancer to the age-matched group without prostate cancer (60% versus 28%), a significant difference (P < 0.001) was seen. Thus, we find a more extensive relaxation in imprinting develops in the prostates of men who have associated cancer.
Figure 4.
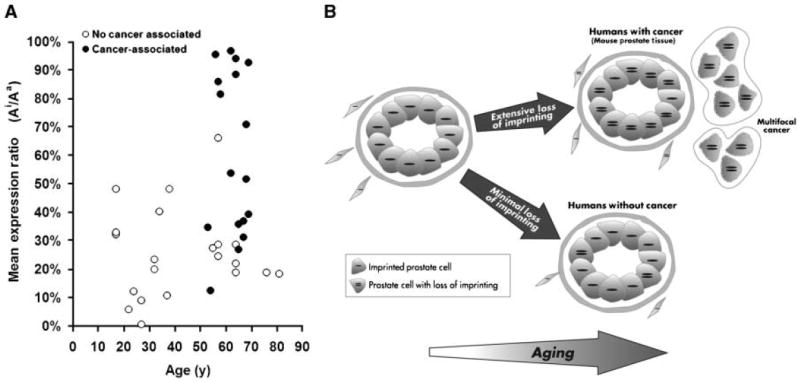
Relaxation of Igf2 imprinting in aging histologically normal human prostate tissues with and without associated cancer. A, prostate tissues from donors of various ages were analyzed using FluPE. The imprint status from younger patients ages <40 y (mean, 27 y) was significantly (P = 0.02) more imprinted when compared with a cohort of tissues from patients older than 55 y (mean, 64 y). Histologically normal peripheral prostate tissues from men with associated prostate cancer (mean, 63 y) showed a greater associated LOI (60%) than an age-matched noncancer associated cohorts (**, P < 0.01). Interestingly, variations in Ai/Aa ratios within groups of younger men even without associated cancer are seen. These were not associated with the presence of inflammation on histology. B, model of epigenetic changes in the prostate with aging. The imprinting of Igf2 (expression of single allele depicted in cell) is maintained in prostate tissues from young mammals. In rodents, who poorly maintain epigenetic patterns, and in humans with associated cancer, a marked loss of Igf2 imprinting (LOI) occurs. This LOI generates a “field effect” throughout the peripheral zone of the prostate that is associated with the development of prostate cancer in multiple areas. Other genetic and epigenetic factors are required for the development of malignancy. Igf2 imprinting patterns are generally maintained and cancer risk is lower in individuals who maintain Igf2 imprinting.
Discussion
Our findings confirm that the Igf2 imprint is not stable in adult mammals, but rather undergoes degradation with aging. Furthermore, in humans, it is associated with a “field effect” that develops in the peripheral prostate containing cancer. Several factors implicated in aging may play a role in this LOI including oxidative stress, diet, hormones, and environmental toxicants (7, 8, 28). There is remarkable variation in the rate of epigenetic degradation within and among organs with aging. Although the extent of this LOI with aging in the mouse prostate is extensive (>60%), we do not show a significant change in the kidney or liver. Minor changes in Igf2 imprinting (<6%), not linked to expression, have been recognized in the heart tissue from aging mice (18). We also find that Igf2 LOI with aging extensively involves the DLP of mice but not the VP. This is of interest, owing to the anatomic homology of the DLP to the peripheral prostate in the human (24, 29), the region where prostate cancer most commonly develops. One observation that has led to the development of the hypothesis that tissue-specific imprinting degrades with aging was our finding that Igf2 LOI is limited to the peripheral zone of human prostates, and not found in the transition zone where benign prostatic hyperplasia occurs (30).
A number of factors are involved in Igf2 imprinting control, including DNA methylation, the covalent modification of histone proteins, and the binding of the insulator protein CTCF (10, 11). One target of this observed loss of epigenetic control is the chromatin-associated protein CTCF, a zinc finger binding protein. We show that a forced decrease in CTCF expression in both human and mouse prostate epithelial cells leads to a re-expression of the paternal Igf2 allele. In the mouse prostate, a decrease in CTCF expression with aging was associated with a loss of CTCF binding at the H19 ICR. This decrease in CTCF expression has also been recognized in vitro with the development of cellular senescence (17). Our finding that substantial changes Igf2 imprinting occur in the absence of significant methylation changes at the H19 ICR, an observation seen in other human genitourinary tissues (31), challenge the central dogma that DNA methylation solely controls Igf2 imprinting in the human (13). Imprinting in Wilms’ tumors may also be maintained in the presence of aberrant methylation at CTCF core sites within the H19 ICR (32). It has been also found that polycomb group proteins expressed from the maternal genome, not DNA methylation, control paternal MEAEA silencing (33). These data emphasize the emerging importance of chromatin factors, including CTCF, in the regulation of imprinting.
Histologic prostate cancer is pervasive in aging men (1). Our finding of a significantly greater relaxation of Igf2 imprinting in histologically normal tissues from men with associated prostate cancer compared with men without the disease suggests a role for Igf2 LOI in the postulated field effect encompassing prostate tissues and influencing them to develop cancer (Fig. 4B). Further support for this field effect is evidenced by the typical finding of 5 separate foci of cancer in found in human prostates removed for malignancy (34). Other gene expression alterations may mark this global tissue defect in the peripheral prostate (35). The finding that Apc+Min mice that biallelically express Igf2 have enhanced intestinal tumor development compared with monoallelic expression suggests LOI is a risk factor for tumor development in the susceptible organ (16). We anticipate this epigenetic alteration involving LOI may find use as a marker for patients with prostate cancer. Furthermore, it is likely that men who are better able to maintain Igf2 imprinting status with aging have a decreased risk of prostate cancer development.
Supplementary Material
Acknowledgments
Grant support: NIH (R01CA97131), the University of Wisconsin George M. O’Brien Urology Research Center (1P50DK065303), the John Livesey endowment, the Department of Defense Prostate Cancer Research Program (DAMD17-02-1-0163), NIH training grant T32 AG000213-16 to the Biology of Aging and Age-related Diseases Training Program (J.R. Dobosy), and T32 CA009681 to the University of Wisconsin McArdle Laboratory Cancer Biology Training Progarm (J.A. Ewald).
We thank Dr. Raj Dhir and Dr. Joel B. Nelson at the University of Pittsburgh for their contribution of samples used in this work, Dr. William Dove for his review and helpful comments, Dr. Terry Oberley for interpretation of the mouse prostate histology, and Dr. Glen Leverson for the statistical support.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
R. Weindruch is a founder of LifeGen Technologies, LLC. The other authors disclosed no potential conflicts of interest.
References
- 1.Sakr WA, Grignon DJ, Crissman JD, et al. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: an autopsy study of 249 cases. In Vivo. 1994;8:439–43. [PubMed] [Google Scholar]
- 2.Malins DC, Johnson PM, Wheeler TM, et al. Age-related radical-induced DNA damage is linked to prostate cancer. Cancer Res. 2001;61:6025–8. [PubMed] [Google Scholar]
- 3.Bandyopadhyay D, Medrano EE. The emerging role of epigenetics in cellular and organismal aging. Exp Gerontol. 2003;38:1299–307. doi: 10.1016/j.exger.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 6.Kwabi-Addo B, Chung W, Shen L, et al. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 7.Thompson SL, Konfortova G, Gregory RI, et al. Environmental effects on genomic imprinting in mammals. Toxicol Lett. 2001;120:143–50. doi: 10.1016/s0378-4274(01)00292-2. [DOI] [PubMed] [Google Scholar]
- 8.McLachlan JA, Burow M, Chiang TC, Li SF. Gene imprinting in developmental toxicology: a possible interface between physiology and pathology. Toxicol Lett. 2001;120:161–4. doi: 10.1016/s0378-4274(01)00295-8. [DOI] [PubMed] [Google Scholar]
- 9.Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C. Parental genomic imprinting of the human IGF2 gene. Nat Genet. 1993;4:98–101. doi: 10.1038/ng0593-98. [DOI] [PubMed] [Google Scholar]
- 10.Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113:81–9. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 11.Kanduri C, Pant V, Loukinov D, et al. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol. 2000;10:853–6. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 12.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. comment. [DOI] [PubMed] [Google Scholar]
- 14.Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–8. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- 15.Rogler CE, Yang D, Rossetti L, et al. Altered body composition and increased frequency of diverse malignancies in insulin-like growth factor-II transgenic mice. J Biol Chem. 1994;269:13779–84. [PubMed] [Google Scholar]
- 16.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–8. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 17.Fu VX, Schwarze SR, Kenowski ML, et al. A loss of insulin-like growth factor-2 imprinting is modulated by CCCTC-binding factor down-regulation at senescence in human epithelial cells. J Biol Chem. 2004;279:52218–26. doi: 10.1074/jbc.M405015200. [DOI] [PubMed] [Google Scholar]
- 18.Bennett-Baker PE, Wilkowski J, Burke DT. Age-associated activation of epigenetically repressed genes in the mouse. Genetics. 2003;165:2055–62. doi: 10.1093/genetics/165.4.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berres MR, Engels WR, Kirsch JAW. A method for genotyping ostensibly dominant markers in AFLP fingerprints. Genetics. 2008 In press. [Google Scholar]
- 20.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–86. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 21.Inman CF, Rees LE, Barker E, et al. Validation of computer-assisted, pixel-based analysis of multiple-colour immunofluorescence histology. J Immunol Methods. 2005;302:156–67. doi: 10.1016/j.jim.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Smith PD, McLean KJ, Murphy MA, et al. A brightness-area-product-based protocol for the quantitative assessment of antigen abundance in fluorescent immunohistochemistry. Brain Res Brain Res Protoc. 2005;15:21–9. doi: 10.1016/j.brainresprot.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Jarrard DF, Sarkar S, Shi Y, et al. p16/pRb pathway alterations are required for bypassing senescence in human prostate epithelial cells. Cancer Res. 1999;59:2957–64. [PubMed] [Google Scholar]
- 24.Berquin IM, Min Y, Wu R, Wu H, Chen YQ. Expression signature of the mouse prostate. J Biol Chem. 2005;280:36442–51. doi: 10.1074/jbc.M504945200. [DOI] [PubMed] [Google Scholar]
- 25.Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 26.Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanduri C, Holmgren C, Pilartz M, et al. The 5′ flank of mouse H19 in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr Biol. 2000;10:449–57. doi: 10.1016/s0960-9822(00)00442-5. [DOI] [PubMed] [Google Scholar]
- 28.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price D. Comparative aspects of development and structure in the prostate. Natl Cancer Inst Monogr. 1963;12:1–27. [PubMed] [Google Scholar]
- 30.Jarrard DF, Bussemakers MJ, Bova GS, Isaacs WB. Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Cancer Res. 1995;1:1471–8. [PubMed] [Google Scholar]
- 31.Byun HM, Wong HL, Birnstein EA, et al. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–8. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 32.Cui H, Niemitz EL, Ravenel JD, et al. Loss of imprinting of insulin-like growth factor-II in Wilms’ tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001;61:4947–50. [PubMed] [Google Scholar]
- 33.Gehring M, Huh JH, Hsieh TF, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epstein JI, Carmichael MJ, Partin AW, Walsh PC. Small high grade adenocarcinoma of the prostate in radical prostatectomy specimens performed for non-palpable disease: pathogenetic and clinical implications. J Urol. 1994;151:1587–92. doi: 10.1016/s0022-5347(17)35309-0. [DOI] [PubMed] [Google Scholar]
- 35.Chandran UR, Dhir R, Ma C, et al. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


