Abstract
Background
De novo transcriptome sequencing is a robust method of predicting miRNA target genes, especially for organisms without reference genomes. Differentially expressed miRNAs had been identified previously in kidney samples collected from susceptible and resistant grass carp (Ctenopharyngodon idella) affected by Aeromonas hydrophila. Target identification for these differentially expressed miRNAs poses a major challenge in this non-model organism.
Results
Two cDNA libraries constructed from mRNAs of susceptible and resistant C. idella were sequenced by Illumina Hiseq 2000 technology. A total of more than 100 million reads were generated and de novo assembled into 199,593 transcripts which were further extensively annotated by comparing their sequences to different protein databases. Biochemical pathways were predicted from these transcript sequences. A BLASTx analysis against a non-redundant protein database revealed that 61,373 unigenes coded for 28,311 annotated proteins. Two cDNA libraries from susceptible and resistant samples showed that 721 unigenes were expressed at significantly different levels; 475 were significantly up-regulated and 246 were significantly down-regulated in the SG samples compared to the RG samples. The computational prediction of miRNA targets from these differentially expressed genes identified 188 unigenes as the targets of 5 conserved and 4 putative novel miRNA families.
Conclusion
This study demonstrates the feasibility of identifying miRNA targets by transcriptome analysis. The transcriptome assembly data represent a substantial increase in the genomic resources available for C. idella and will provide insights into the gene expression profile analysis and the miRNA function annotations in further studies.
Background
Next-generation sequencing (NGS) -based RNA sequencing for transcriptome methods (RNA-seq) allow simultaneous acquisition of sequences for gene discovery as well as identification of transcripts involved in specific biological processes. This is especially suitable for non-model organisms whose genomic sequences are unknown [1], [2]. In addition, the dynamic range, sensitivity and specificity of RNA-seq also make it ideal for quantitatively analyzing various aspects of gene regulation [3]. These techniques do not require prior knowledge of genomic sequence and are much advanced in terms of time, cost, labor, amount of data produced, data coverage, sensitivity, and accuracy compared to traditional sequencing methods [4], [5].
The grass carp (Ctenopharyngodon idella) is one of the most important farmed fish species in China, with a cultural history dating back to the 7th century CE (Tang Dynasty) [6]. According to the FAO, the value of farmed C. idella reached more than 6.46 billion USD for a production of 5.03 billion tons in 2012, thus accounting for the highest production and third highest value of major cultured fish species worldwide at single species level [7]. Despite favorable growth traits, farmed C. idella are rather susceptible to various disease. Outbreaks of disease associated with bacteria such as Aeromonas hydrophila have caused high mortality, resulting in reduced production and considerable economic losses [8].
A. hydrophila is a causative agent of a wide spectrum of diseases in humans and animals [9]. While originally thought to be an opportunistic pathogen in immunocompromized humans, an increasing number of intestinal and extraintestinal disease cases suggest that it is an emerging human pathogen irrespective of the immune status of the host [10]. The pathogenesis, pathogenic mechanism, and virulence factors responsible for selected A. hydrophila infections in different species are not well understood [11]. A. hydrophila is a Gram-negative motile bacillus widely distributed in aquatic environments. It causes motile aeromonad septicemia (MAS), which results in great economic losses in worldwide freshwater fish farming [12]. Thus, more effective measures against A. hydrophila infection in fish are needed. Identification of differentially expressed genes (DGEs) following A. hydrophila infection is important for an improved understanding of fish MAS.
MicroRNAs (miRNAs) are 20–22 nt non-coding RNAs that play important roles in post-transcriptional gene regulation. In animal cells, miRNAs regulate their targets by translational inhibition and mRNA destabilization [13]. MicroRNAs (miRNAs) are key effectors in mediating host-pathogen interactions and constitute a family of small RNA species; they are considered a promising candidate for regulating the interaction between host and pathogen [14], [15]. Therefore, dissecting the biological functions of miRNAs may help us understand the pathogenic mechanism of motile aeromonad septicemia in C. idella. Many studies have identified miRNAs and mRNA transcriptome in fish species, like common carp [16], [17], nile tilapia [18], [19] rainbow trout [20], [21] channel catfish [22], [23] and silver carp [24], [25]. To thoroughly interpret the biological functions of these miRNAs, a first step is to predict their targets. Therefore, establishing a more powerful transcriptome data for target identification is preferred.
Although two parallel C. idella expressed sequence tag analyses have already been conducted using head kidney tissue [26], [27], the data presented here represent the first effort to analyze the transcriptome of C. idella affected by A. hydrophila. Two cDNA libraries from SG and RG C. idella used for our miRNA analysis were constructed and sequenced with Illumina Hiseq 2000. The obtained reads were assembled into transcripts and annotated by BLAST analysis against various databases before screening the results for differentially expressed genes and the prediction of miRNA targets. Our work will provide an approach to identify the target genes of miRNAs and to characterize their functional/regulatory network to increase our understanding of hemorrhagic septicemia outbreaks in C. idella.
Materials and Methods
Ethics statement
All handling of fishes was conducted in accordance with the guidelines on the care and use of animals for scientific purposes set up by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Ocean University, Shanghai, China. The IACUC has specially approved this study within the project “Breeding of Grass Carp” (approval number is SHOU-09-007).
Sample collection
C. idella with an average weight of 50 g were cultured individually at the Wujiang National Farm of Chinese Four Family Carps (Jiangsu Province, China). Animals were raised at 28°C in 400-L aerated tanks for one week before the experiment and fed twice daily (in the morning and late in the afternoon) at a ratio of 5% of total biomass. Two groups (30 animals in each group) were maintained in two aquariums and intraperitoneally injected with A. hydrophila AH10 (Aquatic Pathogen Collection Center of Ministry of Agriculture, China) at a dose of 7.0×106 cells suspended in 100 µl PBS per fish. All fish were observed every 4 h for any mortality and samples were taken until the termination of the experiment at 240 h post-challenge. C. idella died in the first 72 h post-challenge were classified as susceptible group (SG) for their high sensitivity to A. hydrophila, while the animals that survived over 240 h post-challenge were considered as resistant group (RG) [28]. Spleen and kidney samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until further use.
cDNA library construction and sequencing
Experimental protocols for the cDNA sequence were performed according to the manufacturer's technical instructions. The spleen and kidney tissues of randomly-selected three fish from both the susceptible and resistant groups were collected and, labeled as SG and RG, respectively. The RNA from the same tissue of three fishes of SG and RG C. idella was pooled with equal quantity for the construction of SG and RG cDNA libraries. The pooled total RNA was isolated from each spleen and kidney samples with TRIZOL reagent (Invitrogen, Grand Island, NY, USA). RNA integrity was confirmed using a 2100 Bioanalyzer (Agilent Technologies, Inc.) by RNA 6000 nano with a minimum RNA integrity number (RIN) value of 7.0. Poly (A) mRNA was purified from the total RNA using oligo (dT) magnetic beads. Equal amounts of the high-quality mRNA samples were obtained from each group for cDNA library preparation using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and purified using Agencount AMPure XP beads (Beckman Coulter, Krefeld, Germany) according to the manufacturer's recommendations. The concentration of the cDNA library was determined on an Agilent Technologies 2100 Bioanalyzer by Agilent DNA-1000. Libraries were sequenced, at the Novogene Bioinformatics Institute (Beijing, China) on an Illumina HiSeq 2000 instrument (Illumina, San Diego CA, USA) that generated paired-end reads of 101 nucleotides.
Data processing, assembly, and functional annotation
Raw reads generated by Illumina Hiseq 2000 were then cleaned by removing the adaptor containing sequences, any ambiguous base>10% reads and low quality reads (1/2 reads with Q-value≤5) to get clean reads. Then, all clean reads were assembled using the de novo assembly program Trinity [29]: first, short reads were assembled into high-coverage contigs that could not be extended farther in either direction in a k-mer-based approach for fast and efficient transcript assembly. Then, the related contigs were clustered and a de Bruijn graph for each cluster was constructed. Finally, in the context of the corresponding de Bruijn graph and all plausible transcripts, alternatively spliced isoforms and transcripts were derived.
All assembled transcripts were compared with publicly available databases including Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences) [30], KOG/COG (Clusters of Orthologous Groups of proteins) [31], Swiss-Prot (a manually annotated and reviewed protein sequence database) [32], KO (KEGG ortholog database) [33], Pfam (protein family) [34] and GO (Gene Ontology) (http://www.geneontology.org/). Nr, Nt, KOG/COG, Swiss-Prot, and KO used the BLASTx analysis with a cut-off E-value of 10−5, Pfam used Hmmerscan and GO used Blast2GO [35]. The best Blast hits from all Blast results were parsed for a homology-based functional annotation. For the nr annotations, the Blast2GO program was used to obtain GO annotations of unique assembled transcripts to describe biological processes, molecular functions, and cellular components.
Differentially expressed genes between the SG/RG libraries
High-quality reads were mapped to reference sequences (unigenes from the transcriptome data of the cDNA library) using RSEM [36]. Gene expression levels were calculated using the fragments per kilo bases per million mapped reads (FPKM) method [37]. The calculation of unigene expression levels and the identification of unigenes that were differentially expressed between the libraries were performed by DEGseq [38] based on TMM normalized counts. The settings “q.value <0.005” and “|log2.Fold change.normalized|>2” were used as thresholds for judging significant differences in transcript expression. Differentially expressed genes across the samples were further annotated by GO and KEGG pathway analysis
MiRNA target prediction
The SG and RG kidney miRNA-seq analysis were conducted in the same biological samples as mRNA-seq. Small RNA libraries were constructed using a Small RNA Cloning Kit (Takara). RNA was purified by polyacrylamide gel electrophoresis (PAGE) to enrich for the molecules in the range of 17–27 nt, then was ligated with 5' and 3′ adapters. The resulting samples were used as templates for cDNA synthesis followed by PCR amplification. The obtained sequencing libraries were subjected to Solexa sequencing-by-synthesis method. After the run, image analysis, sequencing quality evaluation and data production summarization were performed with Illumina/Solexa pipeline. The sequencing data was pretreated to discard low quality reads, no 3′-adaptor reads, 5′-adaptor contaminants and sequences shorter than 18 nucleotides. After trimming the 3′ adaptor sequence, sequence tags were mapped onto the transcriptome of C. idella using bowtie. Any small RNAs having exact matches to transcriptome of C. idella were used from further analysis. The mapped reads were compared to the miRBase (19.0) to annotate conserved miRNAs. To predict novel miRNAs, the miREvo [39] and mirdeep2 [40] were used.
Computational identification of differentially expressed miRNA targets was performed using the miRanda toolbox [41], using the complementary region between miRNAs and mRNAs and the thermodynamic stability of the miRNA-mRNA duplex. All mRNAs used for target prediction came from the differentially expressed unigenes obtained as described above. The miRanda toolbox employed a dynamic programming algorithm to search the complementary regions between the miRNA and the 3′-UTR of the mRNA, and the scores were based on sequence complementary as well as minimum free energy of RNA duplexes, and were calculated with the Vienna RNA package [42]. All detected targets with scores and energies less than the threshold parameters of S>90 (single-residue pair scores) and ΔG <−17 kcal/mol (minimum free energy) were selected as potential targets.
Real time PCR validation
The sequencing results were validated by real time PCR using One Step PrimeScript miRNA cDNA Synthesis Kit for miRNA (TaKaRa) reversely transcribed, PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa) for mRNA and SYBR Premix Ex Taq II (2x) (Takara) for qPCR according to the manufacturer protocols. Specific primer assays for miR-21 [F: 5′-TAGCTTATCAGACTGGTGTTGGC-3′, R: Uni-miR qPCR Primer (TaRaKa)], JNK1 (F: 5′- TGGTCAGAGGTAGTGTGTTG-3′, R: 5′-AGTTTGTTGTGGTCCGAGTC-3′) and ccr7 (F: 5′-CAAGCCAAGAACTTTGAGAGG-3′, R: 5′-GGCATAAAGGCGAATGTTGTC-3′) were purchased from sangon biotech and real time PCR quantification was carried out in CFX96 Real-Time PCR System (Bio-Rad, CA, USA). To normalize the expression values, miR-22a for miRNA and 18s for mRNA were used as housekeeping control [43], [44]. Expression levels were quantitatively analyzed using the 2−△△CT method. One-way ANOVA tests were performed using SPSS 17.0 to determine significant differences. Each experiment was repeated in triplicates.
Results and Discussion
Antagonistic bacteria, such as A. hydrophila, enhance non-specifically immune-related enzyme activities and disease resistance in C. idella and provide a theoretical basis for disease prevention in aquaculture. However, the molecular mechanisms of this disease are still far from fully understood. The identification and characterization of candidate genes involved in MAS would represent the first step in understanding the genetic basis of this process in C. idella.
De novo assemblies and unigenes annotation
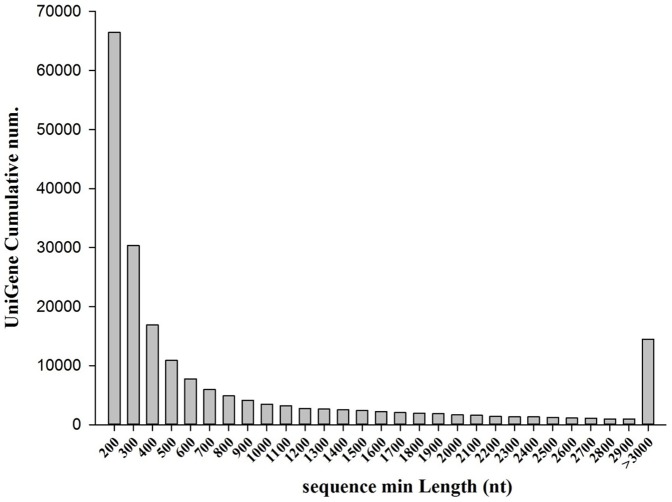
The sequencing generated 135.80 million raw reads. After trimming, 130.88 million clean reads remained, corresponding to 13.22 GB clean bases. The dataset of each sample, SG and RG, was represented by over 60 million clean reads, a read density sufficient for the subsequent quantitative analysis of genes. The raw sequencing reads have been submitted to the NCBI Short Read Archive under the accession number of SRR1124206 and SRR1125014. Then, all clean reads were assembled using a de novo assembly program Trinity [45]. These short reads were further assembled into 199,554 transcripts with an average length of 977 bp (Table 1). The size distribution of these transcripts ranged from 201 to 27,185 bp, of which 27,334 were larger than 2,000 bp (Figure 1). The assembled transcriptome data were deposited in NCBI's Transcriptome Shotgun Assembly (TSA) database under the accession numbers from SUB583458.
Table 1. Summaries of sequencing cDNA library.
| Sample name | SG | RG |
| Total reads | 73,063,654 | 62,737,669 |
| Clean reads | 70,210,307 | 60,668,815 |
| Total mapped to unigenes readcounts | 61,396,052.97 | 51,785,549.97 |
| Reads length (bp) | 101 | |
| GC content (%) | 48.43 | 47.13 |
| Number of unigenes | 199,554 | |
| Total length of unigenes (bp) | 195,075,872 | |
| Mean length of unigenes (bp) | 977 | |
| N50 of unigenes (bp) | 2,117 | |
| Maximal length of unigenes (bp) | 27,185 | |
Figure 1. Length distribution of assembled unigenes in the sequenced cDNA library.
Annotation of predicted proteins
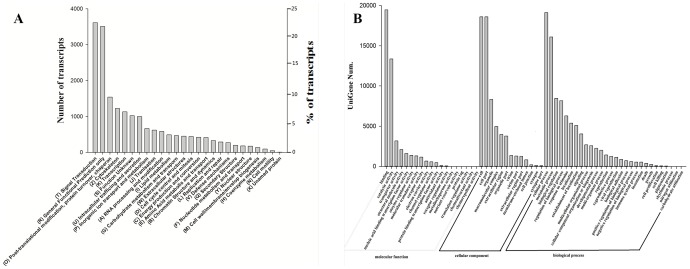
A total of 61,373 distinct sequences (30.75% of the transcripts) matched known genes corresponding to 28,311 of the annotated proteins (Table 2, Table S1). An additional functional annotation of the unigenes of C. idella was performed searching for putative orthologs and paralogs within the KOG database [31]. A total of 16980 unigenes (27.7%) were assigned to 26 eukaryotic orthologous groups (Figure 2A). The category “signal transduction”, which contained 3,611 unigenes (21.27% of 16980 unigenes), was the largest, followed by the categories “general functional prediction only” (3506, 20.65%), “post-translational modification, protein turnover, chaperone” (1539, 9.06%) and “cytoskeleton” (1226, 7.22%).
Table 2. Statistics of the annotation results for the C. idella unigenes.
| All | Nr | Nt | Pfam | KOG | Swiss-Prot | KO | GO | |
| Number of unigenes | 61373 | 28311 | 46653 | 33013 | 16980 | 22674 | 27775 | 34207 |
| % of unigenes | 100 | 46.1 | 76.0 | 53.8 | 27.7 | 36.9 | 45.3 | 55.7 |
Nr: NCBI non-redundant protein sequences, Nt: NCBI non-redundant nucleotide sequences, Pfam: Protein family, KOG: Clusters of Orthologous Groups of proteins, Swiss-Prot: A manually annotated and reviewed protein sequence database, KO: KEGG Ortholog database and GO: Gene Ontology.
Figure 2. Functional annotations of the unigenes of C. idella.
(A) KOG annotation. (B) Level 2 GO term distribution for the biological process, cellular component and molecular function categories.
GO annotation and KEGG pathway analyses
After GO annotation, C. idella transcripts could be assigned to three categories: biological processes, molecular functions and cellular components. Within the various biological processes, cellular processes (19,121 unigenes) metabolic processes (16,091) and biological regulation (8,463) were the most highly represented members (Figure 2B). Important functions, such as cell death (389) and immune system processes (540), were also identified in this category. Similarly, cell (18,586) as well as cell part (18,586) and binding (19,460) were the most represented sub-categories in the cellular component and molecular function categories, respectively.
Searching against the Kyoto Encyclopedia of Genes and Genomes Pathway database (KEGG) [33] revealed that 10,561 unigenes could be matched to 298 KEGG pathways. The most-represented pathways hierarchy 2 were the “infectious diseases” pathway (3210 unigenes) and the “signal transduction” pathway (2316) (Table 3). Some pathways related to immune system were also identified, such as the “Toll-like receptor signaling” pathway (110) [46] and the “chemokine signaling” pathway (224) [47] (Table 4).
Table 3. Top 10 list of the gene number of Pathway Hierarchy 2.
| Pathway Hierarchy 2 | Unigene Number |
| Infectious Diseases | 3210 |
| Signal Transduction | 2316 |
| Cancers | 2235 |
| Nervous System | 1648 |
| Immune System | 1554 |
| Neurodegenerative Diseases | 932 |
| Digestive System | 875 |
| Endocrine System | 861 |
| Cell Communication | 859 |
| Signaling Molecules and Interaction | 763 |
Table 4. Top 10 list of pathways related to immune system.
| Pathway Hierarchy 2 | KEGG Pathway | Unigene Numbers |
| Immune System | Chemokine signaling pathway | 224 |
| Immune System | Leukocyte transendothelial migration | 199 |
| Immune System | T cell receptor signaling pathway | 156 |
| Immune System | Fc gamma R-mediated phagocytosis | 155 |
| Immune System | Natural killer cell mediated cytotoxicity | 129 |
| Immune System | B cell receptor signaling pathway | 110 |
| Immune System | Toll-like receptor signaling pathway | 110 |
| Immune System | Antigen processing and presentation | 86 |
| Immune System | Fc epsilon RI signaling pathway | 84 |
| Immune System | RIG-I-like receptor signaling pathway | 67 |
Digital gene expression library sequencing
Based on the transcriptome sequence data, two DGE libraries were constructed to identify the differentially expressed unigenes between the SG and RG samples. After removing low-quality reads, 70,210,307 and 60,668,815 clean reads were generated from the SG and RG libraries, respectively (Table 1). Among these clean reads, 61,396,052.97 of the SG and 51,785,549.97 of the RG readcounts were mapped to unigenes.
Differential gene expression between the SG and RG libraries
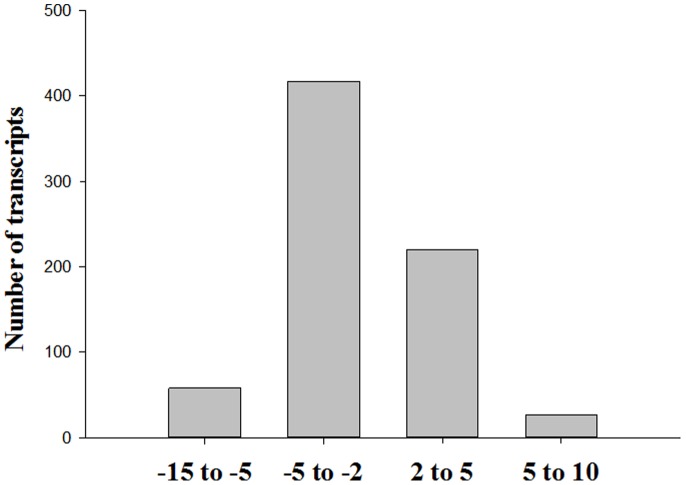
The results suggest that the expression of 721 genes differed significantly between the SG and RG groups of C. idella. Of these genes, 475 were up-regulated and 246 were down-regulated in the SG samples compared to the RG samples (Figure 3 and Table S2). GO enrichment analysis of DEGs indicated that these genes were significantly enriched in oxidation-reduction processes (biological process), integral to membrane (cellular components), and protein binding (molecular function) (Table S3). Pathway enrichment analysis found the DEGs to be mainly enriched in complement and coagulation cascades, Staphylococcus aureus infection and porphyrin and chlorophy II metabolism (Table S4). Notably, several genes involved in the immune and inflammatory response were also identified, such as C-type lectin [48] and matrix metalloproteinase-9 [49].
Figure 3. Number and Fold change distribution of differentially expressed genes between the SG/RG libraries.
MiRNA target prediction
The identification of miRNAs and their targets is important for understanding the physiological functions of miRNAs and the functional roles of differentially expressed miRNAs between healthy and diseased fish. We were thus interested in predicting miRNA target genes involved in the immune response or immune system, according to the KEGG analysis. In a previous study, small RNA deep-sequencing data were aligned with miRBase 18.0 to search for known miRNAs with complete matches, namely, conserved miRNAs (data unpublished). Meanwhile, miRNAs predicted by miRDeep 2.0 that could form stable secondary structures, were identified as novel miRNAs.
We have used a single algorithm, miRanda [50] to predict miRNA targets. As miRNA binding to target 3′ UTR generally results in mRNA destabilization and degradation [51], we chose to narrow down potential targets to those showing differential expression in the opposite direction as the mRNA. This approach increases the strength to discover true target genes and functions affected by miRNA dysregulation. In total, 188 of the target genes predicted by miRanda were differentially expressed in the opposite direction in the target tissue. The identification of 188 unigenes (Table S5) as the predicted target genes of 5 conserved and 4 putative novel miRNA families (Table 5). The identified target genes involved in biological processes, molecular functions, and cellular components were defined using GO annotations. GO analysis demonstrated that these targets were involved in a broad range of physiological processes, including gene expression, transcription regulation, immune system processes, and responses to stress or stimuli (Table S6).
Table 5. Differentially expressed miRNAs in C. idella kidney between SG and RG.
| miRNA | Sequence (5′-3′) | SG (TMP) | RG (TMP) | log2 (Fold_change) normalized | p-value |
| let-7i | UGAGGUAGUAGUUUGUGCUGUU | 3419.67473 | 1475.401006 | 1.212751983 | 4.62E-174 |
| miR-142a-3p | UGUAGUGUUUCCUACUUUAUGGA | 4770.81118 | 9798.344345 | −1.038303405 | 0 |
| miR-21 | UAGCUUAUCAGACUGGUGUUGGC | 3431.54242 | 1640.502276 | 1.064719594 | 4.12E-142 |
| miR-217 | UACUGCAUCAGGAACUGAUUGG | 240.9140969 | 2983.647091 | −3.630486183 | 0 |
| miR-223 | UGUCAGUUUGUCAAAUACCCC | 1460.912578 | 6800.683353 | −2.218809871 | 0 |
| novel_115 | UGAAGGCCGAAGUGGAGA | 3.560306851 | 17.95531055 | −2.334337113 | 0.001253505 |
| novel_131 | UGCCCGCAUUCUCCACCA | 7.713998177 | 40.28996513 | −2.384869847 | 9.85E-07 |
| novel_154 | CCCAGCCAUAUUUGUUUGAAC | 16.02138083 | 0 | 4.768565529 | 4.44E-05 |
| novel_3 | UGUUUCUGGCUCUGAUAUUUGCU | 32.04276166 | 71.82124218 | −1.164412111 | 8.07E-05 |
Searching against the KEGG indicated that 188 unigenes mapped to 48 KEGG pathways. 26 pathways were related to immune and diseases in all pathways (Table 6). These included the categories “Toll-like receptor signaling pathway”, “Fc epsilon RI signaling pathway”, “Chemokine signaling pathway”, “Fc gamma R-mediated phagocytosis”, “Antigen processing and presentation”, “Natural killer cell mediated cytotoxicity”, “T cell receptor signaling pathway” and “Complement and coagulation cascades” related immune functions. Toll-like receptor signaling pathway induce the expression of a variety of host defense genes. These include chemokine signaling pathway and other effectors necessary to arm the host cell against the invading pathogen [52]. Cytokine-cytokine receptor interaction play a pivotal role in the generation of immunological responses during bacterial infection [53]. This implicated functions that are likely regulated by miRNAs and suggests regulation of different pathways during immune activation in susceptible and resistant C. idella.
Table 6. 26 pathways were related to immune and diseases in all pathways.
| KEGG Pathway | Pathway Hierarchy 2 | Target gene of different expression miRNA |
| Toll-like receptor signaling pathway | Immune system | JNK, TLR5, TBK1 |
| Hepatitis C | Infectious diseases: Viral | JNK, TBK1, LDLR |
| Salmonella infection | Infectious diseases: Bacterial | JNK, TLR5 |
| Fc epsilon RI signaling pathway | Immune system | JNK, FYN |
| Tuberculosis | Infectious diseases: Bacterial | JNK, PLK3, CTSS |
| Influenza A | Infectious diseases: Viral | JNK, DNAJB1, TBK1 |
| Measles | Infectious diseases: Viral | FYN, TBK1 |
| MAPK signaling pathway | Signal transduction | JNK, MKP |
| Toxoplasmosis | Infectious diseases: Parasitic | JNK, LDLR |
| Chemokine signaling pathway | Immune system | CCR7, ADCY7, DOCK2 |
| HTLV-I infection | Infectious diseases: Viral | JNK, ADCY7, SLC2A1 |
| Herpes simplex infection | Infectious diseases: Viral | JNK, TBK1 |
| Fc gamma R-mediated phagocytosis | Immune system | DOCK2 |
| Prion diseases | Neurodegenerative diseases | FYN |
| Viral myocarditis | Cardiovascular diseases | FYN |
| Pathways in cancer | Cancers: Overview | JNK, SLC2A1 |
| Transcriptional misregulation in cancer | Cancers: Overview | CCR7 |
| Type II diabetes mellitus | Endocrine and metabolic diseases | JNK |
| Cytokine-cytokine receptor interaction | Signaling molecules and interaction | CCR7, TNFSF12 |
| Antigen processing and presentation | Immune system | CTSS |
| Pertussis | Infectious diseases: Bacterial | JNK |
| Natural killer cell mediated cytotoxicity | Immune system | FYN |
| T cell receptor signaling pathway | Immune system | FYN |
| Chagas disease | Infectious diseases: Parasitic | JNK |
| Staphylococcus aureus infection | Infectious diseases: Bacterial | CFB |
| Complement and coagulation cascades | Immune system | CFB |
List of gene abbreviations: JNK: c-Jun N-terminal kinase, TLR5: toll-like receptor 5, TBK1: TANK-binding kinase 1, LDLR: low-density lipoprotein receptor, FYN: tyrosine-protein kinase Fyn, CTSS: cathepsin S, PLK3: polo-like kinase 3, DNAJB1: DnaJ homolog subfamily B member 1, MKP: dual specificity MAP kinase phosphatase, DOCK2: dedicator of cytokinesis protein 2, ADCY7: adenylate cyclase 7, SLC2A1: MFS transporter, SP family, solute carrier family 2 (facilitated glucose transporter), member 1, CCR7: C-C chemokine receptor type 7, TNFSF12: tumor necrosis factor ligand superfamily member 12, CFB: component factor B.
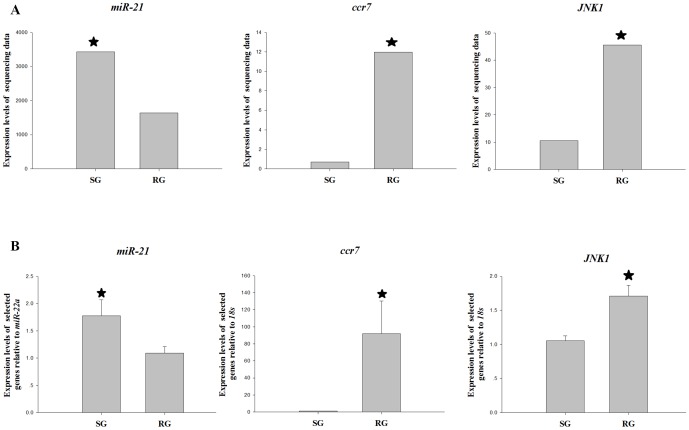
Of particular interest to our study is the fact that several of the most highly expressed miRNAs in SG have been shown to have a role in immunity. let-7i and associated TLR4 expression are involved in cholangiocyte immune responses against C. parvum infection [54]. miR-21 targets multiple genes associated with the immunologically localized disease [55]. miR-21 which is up-regulated in SG is predicted to target 28 differentially expressed genes (Table S7) in C. idella. Frequently represented in the top immune functions were protein kinase JNK1 (JNK1) and chemokine (C-C motif) receptor 7 (ccr7) (Table 6). JNK1 and ccr7 showed clearly down-regulated expression profiles in the SG samples compared to the RG samples (Figure 4A http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0073506-pone.0073506.s003 ). The decreased expression profile of these targets in the susceptible samples supported our previous finding that the expression of miR-21 was significantly up-regulated, with TMP of 3,432 and 1,642 in the SG and RG groups, respectively (Table 5). We validated the miR-21, JNK1 and ccr7 which had expression change in the sequencing data by performing real time PCR in the same samples used for the sequencing (Figure 4B).
Figure 4. The expression analysis of selected genes from the expression profile by relative quantitative real-time PCR.
A Transcriptome sequencing data, B Real-time PCR data. Increases and decreases in relative levels of transcripts with respect to the control 18s for mRNA and miR-22a for miRNA are shown. The settings “q.value <0.005” and “|log2.Fold change.normalized|>2” were used as thresholds for judging significant differences in transcript expression. One-way ANOVA tests were performed using SPSS 17.0 to determine significant differences for Real-time PCR data. Statistical significance of the relative expression ratio is indicated *.
JNK1 is involved in apoptosis, neurodegeneration, cell differentiation and proliferation, inflammatory conditions and cytokine production mediated by AP-1 (activation protein 1), such as Regulated upon activation normal T cell expressed and presumably secreted (RANTES), Interleukin 8 (IL-8), and Granulocyte-macrophage colony-stimulating factor (GM-CSF) [56]. It has been reported that JNK plays an important role in the innate immune response to microbial challenge [57], [58]. Most importantly, JNK1 serves as a negative regulatory factor for MAP kinase phosphatase 5 (MKP5) that plays an essential role in innate immune responses [59]. The chemokine receptor CCR7 acts as an important organizer of the primary immune response [60]. A previous study demonstrated a discrete CCR7 requirement in the activation of different T cell subsets during bacterial infection [61]. CCR7 is differentially regulated by macrophages in exposure to bacteria, as it is triggered by exposure to both Gram-negative and Gram-positive bacteria [62].
The discovery of microRNAs dramatically changed our perspective on eukaryotic gene expression regulation [63]. MicroRNAs play important gene-regulatory roles in animals and plants by pairing to the mRNAs of protein-coding genes to direct their posttranscriptional repression [64], [65]. The identification of miRNA target genes is an important step in understanding their role in gene regulatory networks. Most miRNA-associated computational methods comprise the prediction of miRNA genes and their targets, and an increasing number of computational algorithms and web-based resources are being developed to fulfill the needs of scientists performing miRNA research, like miRanda [42], TargetScan [66], RNAhybrid [67] and PicTar [68]. However, animal miRNA targets are difficult to predict since miRNA: mRNA duplexes often contain several mismatches, gaps, and G:U base pairs in many positions, thus limiting the maximum length of contiguous sequences of matched nucleotides [69]. The predicted interactions using these computational methods are inconsistent and the expected false positive rates are still high. Recently, several authors suggested integrating expression profiles from both miRNA and mRNA with in silico target predictions to reduce the number of false positives and increase the number of biologically relevant targets [70]. These methods have been shown to be effective in identifying the most prominent interactions from the databases of putative targets [71]. To minimize false positive rates in our study, the RNA-seq and miRNA-seq analysis were conducted in the same biological samples. Likewise, to reduce the number of putative target genes, the miRNA targets were predicted from differentially expressed genes. However, some false positive predictions proved inevitable. Further studies will focus on the experimental validation of the differentially expressed mRNA and miRNAs identified in this study. They are likely to be central regulators of the innate immune response to A. hydrophila and thus represent potential therapeutic targets or novel biomarkers of infection and inflammation.
Conclusions
In this study, we used high-throughput sequencing data to characterize the transcriptome of C. idella, a species for which little genomic data are available. Further, DGE tags were mapped to the assembled transcriptome for further gene expression analysis. A large number of candidate genes involved in MAS were identified. This represents a fully characterized transcriptome, and provides a valuable resource for genetic and genomic studies in C. idella. Additionally, DGE profiling provides new leads for functionally studies of genes involved in MAS.
Finally, comparison with our previous miRNA profiling, this study strongly indicates that miRNA is a critical factor in determining mRNA abundance and regulation during MAS. Our on-going effort using experimental approach such as knock-down or over-express candidate miRNAs and mRNAs in vitro is expected to provide new evidence in understanding these regulatory mechanisms of MAS in C. idella.
Supporting Information
Summary of unigene annotation against Nr, Nt, Pfam, KOG, Swiss-Prot, KO and GO database.
(XLSX)
721 genes differed significantly between the SG and RG groups of C. idella.
(XLSX)
GO enrichment analysis of 721 genes differed significantly.
(XLSX)
Pathway enrichment analysis of 721 genes differed significantly.
(XLSX)
188 genes as the predicted target genes of 9 different expression miRNA between SG and RG groups of C. idella.
(XLSX)
GO enrichment analysis of 188 genes as the predicted target genes of different expression miRNA.
(XLSX)
28 target gene information of miR-21 .
(XLSX)
Acknowledgments
We thank Zhiwei Wang and Liang Zhang (Novogene Bioinformatics Technology Co. Ltd) for their help in sequencing and data analysis.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The raw sequencing reads have been submitted to the NCBI Short Read Archive under the accession number of SRR1124206 and SRR1125014. The assembled transcriptome data were deposited in NCBI's Transcriptome Shotgun Assembly (TSA) database under the accession numbers from SUB583458.
Funding Statement
This work was supported by grants from National Key Technology R&D Program of China (2012BAD26B02), the China's Agricultural Research System (CARS-46-04), the Agricultural Seed Development Program of Shanghai City (2012NY10), Shanghai Ocean University Doctoral Research Foundation, Shanghai Universities First-class Disciplines Project of Fisheries and the Innovation Plan of Shanghai Graduate Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang Y, Pan Y, Liu Z, Zhu X, Zhai L, et al. (2013) De novo transcriptome sequencing of radish (Raphanus sativus L.) and analysis of major genes involved in glucosinolate metabolism. BMC Genomics 14: 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Graveley BR (2008) Molecular biology: power sequencing. Nature 453: 1197–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, et al. (2010) Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bhardwaj J, Chauhan R, Swarnkar MK, Chahota RK, Singh AK, et al. (2013) Comprehensive transcriptomic study on horse gram (Macrotyloma uniflorum): De novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genomics 14: 647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renkui C (1991) Development History of Freshwater Culture in China. China Press of Science & Technology, Beijing, China.
- 7.FAO (2014) FAO Yearbook of Fishery and Aquaculture Statistics Summary tables. Rome: FAO.
- 8.Huang Q, Tang S, Zhang J (1983) Ichthyopathology. Shanghai, China: Shanghai Press of Science & Technology.
- 9. Igbinosa I, Igumbor E, Aghdasi F, Tom M, Okoh A (2012) Emerging Aeromonas species infections and their significance in public health. The Scientific World Journal 2012: 625023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Figueras MJ (2005) Clinical relevance of Aeromonas sM503. Reviews in Medical Microbiology 16: 145–153. [Google Scholar]
- 11. Rahman M, Colque-Navarro P, Kühn I, Huys G, Swings J, et al. (2002) Identification and characterization of pathogenic Aeromonas veronii biovar sobria associated with epizootic ulcerative syndrome in fish in Bangladesh. Applied and environmental microbiology 68: 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu X-Y, Shen Y-B, Fu J-J, Liu F, Guo S-Z, et al. (2012) Matrix metalloproteinase 2 of grass carp Ctenopharyngodon idella (CiMMP2) is involved in the immune response against bacterial infection. Fish & shellfish immunology 33: 251–257. [DOI] [PubMed] [Google Scholar]
- 13.Bushati N, Cohen SM (2007) MicroRNA functions. Annual Review of Cell and Developmental Biology. pp.175–205. [DOI] [PubMed]
- 14. Zhang P, Li C, Zhu L, Su X, Li Y, et al. (2013) De novo assembly of the sea cucumber Apostichopus japonicus hemocytes transcriptome to identify miRNA targets associated with skin ulceration syndrome. PLoS One 8: e73506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 16. Zhu YP, Xue W, Wang JT, Wan YM, Wang SL, et al. (2012) Identification of common carp (Cyprinus carpio) microRNAs and microRNA-related SNPs. BMC Genomics 13: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ji P, Liu G, Xu J, Wang X, Li J, et al. (2012) Characterization of common carp transcriptome: sequencing, de novo assembly, annotation and comparative genomics. PloS one 7: e35152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan B, Guo J-T, Zhao L-H, Zhao J-L (2012) microRNA expression signature in skeletal muscle of Nile tilapia. Aquaculture 364–365: 240–246. [Google Scholar]
- 19. Tao W, Yuan J, Zhou L, Sun L, Sun Y, et al. (2013) Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS One 8: e63604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mennigen JA, Panserat S, Larquier M, Plagnes-Juan E, Medale F, et al. (2012) Postprandial regulation of hepatic microRNAs predicted to target the insulin pathway in rainbow trout. PLoS One 7: e38604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salem M, Rexroad CE 3rd, Wang J, Thorgaard GH, Yao J (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11: 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mu X, Pridgeon JW, Klesius PH (2011) Transcriptional profiles of multiple genes in the anterior kidney of channel catfish vaccinated with an attenuated Aeromonas hydrophila. Fish Shellfish Immunol 31: 1162–1172. [DOI] [PubMed] [Google Scholar]
- 23. Barozai MY (2012) The MicroRNAs and their targets in the channel catfish (Ictalurus punctatus). Mol Biol Rep 39: 8867–8872. [DOI] [PubMed] [Google Scholar]
- 24. Fu B, He S (2012) Transcriptome analysis of silver carp (Hypophthalmichthys molitrix) by paired-end RNA sequencing. DNA Res 19: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chi W, Tong C, Gan X, He S (2011) Characterization and comparative profiling of MiRNA transcriptomes in bighead carp and silver carp. PLoS One 6: e23549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen J, Li C, Huang R, Du F, Liao L, et al. (2012) Transcriptome analysis of head kidney in grass carp and discovery of immune-related genes. BMC Vet Res 8: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu F, Wang D, Fu J, Sun G, Shen Y, et al. (2010) Identification of immune-relevant genes by expressed sequence tag analysis of head kidney from grass carp (Ctenopharyngodon idella). Comp Biochem Physiol Part D Genomics Proteomics 5: 116–123. [DOI] [PubMed] [Google Scholar]
- 28. Heng JF, Su JG, Huang T, Dong J, Chen LJ (2011) The polymorphism and haplotype of TLR3 gene in grass carp (Ctenopharyngodon idella) and their associations with susceptibility/resistance to grass carp reovirus. Fish & Shellfish Immunology 30: 45–50. [DOI] [PubMed] [Google Scholar]
- 29. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, et al. (2004) A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kanehisa M (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, et al. (2002) The Pfam protein families database. Nucleic Acids Res 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic acids research 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 38. Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138. [DOI] [PubMed] [Google Scholar]
- 39. Wen M, Shen Y, Shi S, Tang T (2012) miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics 13: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. John B, Enright AJ, Aravin A, Tuschl T, Sander C, et al. (2004) Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Enright AJ, John B, Gaul U, Tuschl T, Sander C, et al. (2003) MicroRNA targets in Drosophila. Genome Biol 5: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu XY, Shen YB, Fu JJ, Lu LQ, Li JL (2014) Determination of reference microRNAs for relative quantification in grass carp (Ctenopharyngodon idella). Fish & Shellfish Immunology 36: 374–382. [DOI] [PubMed] [Google Scholar]
- 44. Su J, Zhang R, Dong J, Yang C (2011) Evaluation of internal control genes for qRT-PCR normalization in tissues and cell culture for antiviral studies of grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol 30: 830–835. [DOI] [PubMed] [Google Scholar]
- 45. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29: 644–U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Akira S, Takeda K (2004) Toll-like receptor signalling. Nature Reviews Immunology 4: 499–511. [DOI] [PubMed] [Google Scholar]
- 47. Rot A, von Andrian UH (2004) Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol 22: 891–928. [DOI] [PubMed] [Google Scholar]
- 48. Liu F, Li J, Fu J, Shen Y, Xu X (2011) Two novel homologs of simple C-type lectin in grass carp (Ctenopharyngodon idellus): potential role in immune response to bacteria. Fish Shellfish Immunol 31: 765–773. [DOI] [PubMed] [Google Scholar]
- 49. Xu XY, Shen YB, Fu JJ, Liu F, Guo SZ, et al. (2013) Characterization of MMP-9 gene from grass carp (Ctenopharyngodon idella): An Aeromonas hydrophila-inducible factor in grass carp immune system. Fish Shellfish Immunol 35: 801–807. [DOI] [PubMed] [Google Scholar]
- 50. Enright AJ, John B, Gaul U, Tuschl T, Sander C, et al. (2004) MicroRNA targets in Drosophila. Genome Biology 5: R1–R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Janeway CA, Medzhitov R (2002) Innate immune recognition. Annual Review of Immunology 20: 197–216. [DOI] [PubMed] [Google Scholar]
- 53. Plata-Salamán C, Ilyin S, Gayle D, Flynn MC (1998) Gram-negative and gram-positive bacterial products induce differential cytokine profiles in the brain: Analysis using an integrarive molecular-behavioral in vivo model. International journal of molecular medicine 1: 387–398. [DOI] [PubMed] [Google Scholar]
- 54. Chen XM, Splinter PL, O'Hara SP, LaRusso NF (2007) A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J Biol Chem 282: 28929–28938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, et al. (2012) MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat Med 18: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oltmanns U, Issa R, Sukkar MB, John M, Chung KF (2003) Role of c-jun N-terminal kinase in the induced release of GM-CSF, RANTES and IL-8 from human airway smooth muscle cells. Br J Pharmacol 139: 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boutros M, Agaisse H, Perrimon N (2002) Sequential Activation of Signaling Pathways during Innate Immune Responses in Drosophila. Developmental Cell 3: 711–722. [DOI] [PubMed] [Google Scholar]
- 58. Lee J, Mira-Arbibe L, Ulevitch RJ (2000) TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J Leukoc Biol 68: 909–915. [PubMed] [Google Scholar]
- 59. Zhang Y, Dong C (2005) MAP kinases in immune responses. Cell Mol Immunol 2: 20–27. [PubMed] [Google Scholar]
- 60. Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, et al. (1999) CCR7 Coordinates the Primary Immune Response by Establishing Functional Microenvironments in Secondary Lymphoid Organs. Cell 99: 23–33. [DOI] [PubMed] [Google Scholar]
- 61. Kursar M, Hopken UE, Koch M, Kohler A, Lipp M, et al. (2005) Differential requirements for the chemokine receptor CCR7 in T cell activation during Listeria monocytogenes infection. J Exp Med 201: 1447–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, et al. (2002) Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci U S A 99: 1503–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mendes ND, Freitas AT, Sagot MF (2009) Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res 37: 2419–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics 11: 597–610. [DOI] [PubMed] [Google Scholar]
- 65. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787–798. [DOI] [PubMed] [Google Scholar]
- 67. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, et al. (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495–500. [DOI] [PubMed] [Google Scholar]
- 69. Stark A, Brennecke J, Russell RB, Cohen SM (2003) Identification of Drosophila microRNA targets. PLoS biology 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, et al. (2013) Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res 41: 2817–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Muniategui A, Pey J, Planes FJ, Rubio A (2013) Joint analysis of miRNA and mRNA expression data. Brief Bioinform 14: 263–278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of unigene annotation against Nr, Nt, Pfam, KOG, Swiss-Prot, KO and GO database.
(XLSX)
721 genes differed significantly between the SG and RG groups of C. idella.
(XLSX)
GO enrichment analysis of 721 genes differed significantly.
(XLSX)
Pathway enrichment analysis of 721 genes differed significantly.
(XLSX)
188 genes as the predicted target genes of 9 different expression miRNA between SG and RG groups of C. idella.
(XLSX)
GO enrichment analysis of 188 genes as the predicted target genes of different expression miRNA.
(XLSX)
28 target gene information of miR-21 .
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The raw sequencing reads have been submitted to the NCBI Short Read Archive under the accession number of SRR1124206 and SRR1125014. The assembled transcriptome data were deposited in NCBI's Transcriptome Shotgun Assembly (TSA) database under the accession numbers from SUB583458.